Significance
Because bacteria colonize hostile environments they have evolved immune evasion mechanisms, including the expression of enzymes that specifically modify host immune system proteins. Streptococcus pyogenes secretes an enzyme called EndoS that removes carbohydrates specifically from human antibodies, impairing their ability to activate immune defenses. Because of its high substrate specificity, EndoS is also being developed as a treatment for autoimmune diseases and is a key enzyme used in the production of antibodies bearing customized carbohydrates. We have determined the three-dimensional structure of EndoS and present a molecular model depicting how EndoS engages antibodies with high specificity. Our data provide a roadmap for engineering EndoS variants with unique activities for clinical and biotechnological applications.
Abstract
To evade host immune mechanisms, many bacteria secrete immunomodulatory enzymes. Streptococcus pyogenes, one of the most common human pathogens, secretes a large endoglycosidase, EndoS, which removes carbohydrates in a highly specific manner from IgG antibodies. This modification renders antibodies incapable of eliciting host effector functions through either complement or Fc γ receptors, providing the bacteria with a survival advantage. On account of this antibody-specific modifying activity, EndoS is being developed as a promising injectable therapeutic for autoimmune diseases that rely on autoantibodies. Additionally, EndoS is a key enzyme used in the chemoenzymatic synthesis of homogenously glycosylated antibodies with tailored Fc γ receptor-mediated effector functions. Despite the tremendous utility of this enzyme, the molecular basis of EndoS specificity for, and processing of, IgG antibodies has remained poorly understood. Here, we report the X-ray crystal structure of EndoS and provide a model of its encounter complex with its substrate, the IgG1 Fc domain. We show that EndoS is composed of five distinct protein domains, including glycosidase, leucine-rich repeat, hybrid Ig, carbohydrate binding module, and three-helix bundle domains, arranged in a distinctive V-shaped conformation. Our data suggest that the substrate enters the concave interior of the enzyme structure, is held in place by the carbohydrate binding module, and that concerted conformational changes in both enzyme and substrate are required for subsequent antibody deglycosylation. The EndoS structure presented here provides a framework from which novel endoglycosidases could be engineered for additional clinical and biotechnological applications.
Successful infection and colonization by microbes depends on their abilities to evade host immunity. One of the primary routes by which microorganisms escape host immune responses is through the production of enzymes that modify the immune system. Streptococcus pyogenes, a Gram-positive bacterium that is one of the most common human pathogens and the cause of group A streptococcal infections, expresses numerous extracellular enzymes that modulate immune mechanisms, including those that proteolyze antibodies and complement factors, detoxify oxygen free radicals, inhibit T-cell proliferation, and remodel glycans on host proteins (1). This last activity is typically carried out by endo-β-N-acetylglucosaminidases (endoglycosidases), which release N-linked oligosaccharides from glycoproteins by cleaving the β (1–4) glycosidic bond between two N-acetyl glucosamine (GlcNAc) residues of the N,N′-diacetylchitobiose core. X-ray crystal structures of numerous bacterial endoglycosidases (2–7) have shown that these enzymes adopt a common (β/α)8 barrel conformation—a cyclic eightfold repeat comprised of β-strand/loop/α helix motifs in which the parallel β-strands form a central barrel with active site residues located within the open barrel structure. Diversity in the loops connecting the β-strands and α-helices define their specificities for both glycan and protein components of a given substrate.
S. pyogenes secretes a 108-kDa endoglycosidase, EndoS, that specifically hydrolyzes core glycans on human IgG antibodies (8). EndoS has enzymatic activity on natively folded IgG, but not on denatured IgG (9). This activity contributes to increased survival of S. pyogenes in human blood ex vivo, on account of reduced IgG binding to Fc γ receptors and impaired complement pathway activation (10). Injection of EndoS into mice results in the efficient removal of IgG-associated carbohydrate, with a murine IgG subclass specificity of IgG1 = IgG2b > IgG2a (11). EndoS releases the glycan linked to residue Asn297 of the human Fc region CH2 domain, which affects the local structure of IgG (12, 13) and its ability to bind complement factor C1q (14) and Fc γ receptors (15). These binding events regulate two key effector functions induced by IgG antibodies.
The same properties of EndoS that benefit the bacteria can be leveraged for the treatment of autoimmune diseases. When used as an in vivo modulator of IgG glycosylation and effector function activity, EndoS has successfully treated numerous autoimmune conditions in animal models (11, 16–22). Because EndoS is specific for IgG bearing complex-type versus high-mannose carbohydrates, it can also potentially be used to enhance the in vivo efficacy of IgG monoclonal antibodies, when produced recombinantly with high-mannose glycans, by reducing competition for Fc γ receptor binding from serum antibodies (23).
Endoglycosidases, including EndoS, have also been used extensively for in vitro glycan remodeling to modulate the properties of glycoproteins (24). Natural glycoproteins exist as mixtures of glycoforms, of which only one or a few typically exhibit maximal activity. However, individual glycoforms are difficult to purify (25) and recombinant expression of glycoproteins yields heterogeneous glycoforms, even when glycotransferase mutant cell lines are used (26, 27). To circumvent these limitations on glycoprotein homogeneity, recombinant protein expression combined with chemoenzymatic glycan remodeling (28) has been developed. IgG monoclonal antibodies are used extensively as therapeutics and their activities, as mediated by effector functions, depend on the chemistry of their core glycans. EndoS deglycosylates antibody glycoforms that are refractory to processing by other endoglycosidases (29) and glycosynthase mutants of EndoS efficiently transfer predefined N-glycans to intact IgG (30). Together, these catalytic properties of EndoS enzymes allow for customization of IgG glycoforms that can enhance the therapeutic capacities of monoclonal antibodies.
Here, we report the X-ray crystal structure of EndoS and a model of its encounter complex with IgG1 Fc. These findings reveal the structural determinants of EndoS specificity for IgG antibodies, provide a molecular mechanism for its enzymatic activity, and suggest ways in which novel endoglycosidases could be engineered for clinical and biotechnological applications.
Results
Oligomerization and Hydrolytic Activity of EndoS.
Using analytical size exclusion chromatography, we determined the oligomerization state of EndoSWT(37–995), missing the N-terminal signal peptide (residues 1–36) but containing a putative coiled coil (residues 37–97), as ∼20:80 dimer:monomer, whereas EndoSWT(98–995) is entirely monomeric in solution (SI Appendix, Fig. S1A). We examined the Fc glycan hydrolysis activity of these EndoS proteins by SDS/PAGE analysis using the IgG1 monoclonal antibody Rituximab as a substrate. For EndoSWT(37–995), both dimer and monomer fractions exhibited approximately the same hydrolytic activity, whereas the relative activity of EndoSWT(98–995) was reduced at least 50-fold (SI Appendix, Fig. S1B). Thus, whereas the truncated enzyme can process glycosylated IgG1 completely, the putative coiled coil increases the rate of hydrolysis.
Overall Structure of EndoS.
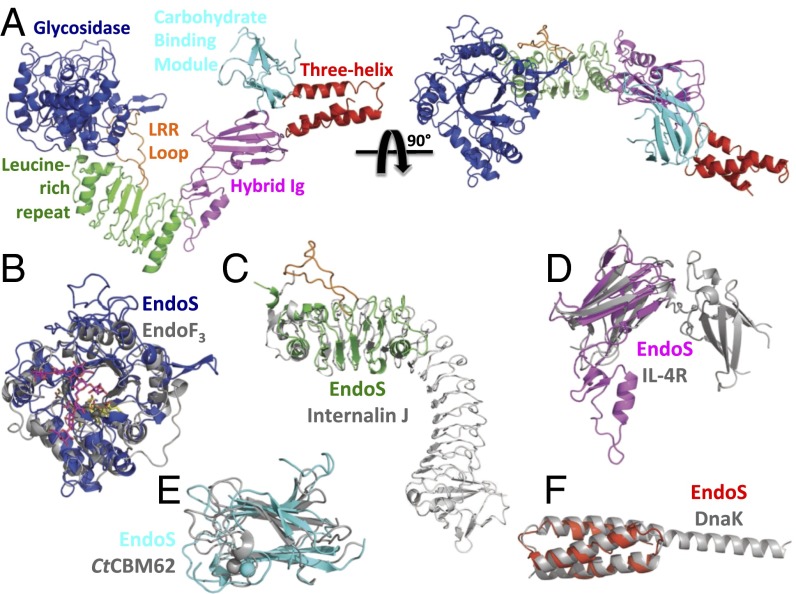
We crystallized SeMet-EndoSD233Q(98–995), a glycosynthase mutant, and determined its structure at 3.2-Å resolution by multiwavelength anomalous dispersion (MAD). We also crystallized native EndoSD233Q(98–995) and EndoSWT(98–995) and solved their structures by molecular replacement at 1.9- and 2.6-Å resolution, respectively, using the partially refined MAD-phased EndoSD233Q(98–995) structure as a search model (SI Appendix, Table S1). With no significant differences in the two structures, we refer to the higher resolution structure throughout the manuscript. The overall morphology of EndoS is that of a letter “V” with a small extension from one end (Fig. 1A). The V shape of the protein measures ∼130 Å across and ∼83 Å high, with a tapered cleft measuring ∼42 Å across its opening. EndoS is composed of five distinct protein domains including, from N to C terminus: (i) an endoglycosidase enzymatic domain, residues 98–445; (ii) a leucine-rich repeat (LRR) domain, residues 446–631; (iii) a hybrid Ig domain, residues 632–764; (iv) a carbohydrate-binding module (CBM), residues 765–923; and (v) a three-helix bundle (3H) domain, residues 924–995 (SI Appendix, Fig. S2). The EndoS structure is not fully globular but, instead, has individual domains arranged akin to beads on a string. Using small angle X-ray scattering (SAXS), we confirmed that the observed V shape of the crystallized EndoS was maintained in solution (SI Appendix, Fig. S3). The SAXS radial distribution, or p(r), function is bimodal, consistent with the overall V shape of the crystal structure. The maximal diameter is 149 ± 7 Å with two peaks centered ∼40 Å apart, values highly similar to the dimensions of crystallized EndoS. Using CRYSOL (31), we observed a nearly perfect fit of the SAXS data to the solution scattering profile calculated from the crystal structure. We also generated ab initio models of the protein envelope, all of which exhibited an open shape, and we fit the EndoS crystal structure into the top-ranked model with a correlation coefficient of 0.87.
Fig. 1.
EndoS structure, domain organization, and comparison with known protein structures. (A) Structure of EndoSD233Q(98–995) viewed in two orientations, inclusive of the glycosidase (blue), leucine-rich repeat (green), hybrid Ig (magenta), carbohydrate binding module (cyan), and three-helix bundle (red) domains. The loop extending from the middle of the leucine-rich repeat is in orange. Superposition of the EndoS glycosidase domain to that of EndoF3 (B), leucine-rich repeat domain to InternalinJ (C), hybrid Ig domain to the IL-4 receptor (D), carbohydrate binding module to CtCBM62 (E), and three-helix bundle domain to DnaK (F).
The Glycosidase Domain Exhibits a Common Fold with Unique Structural Features.
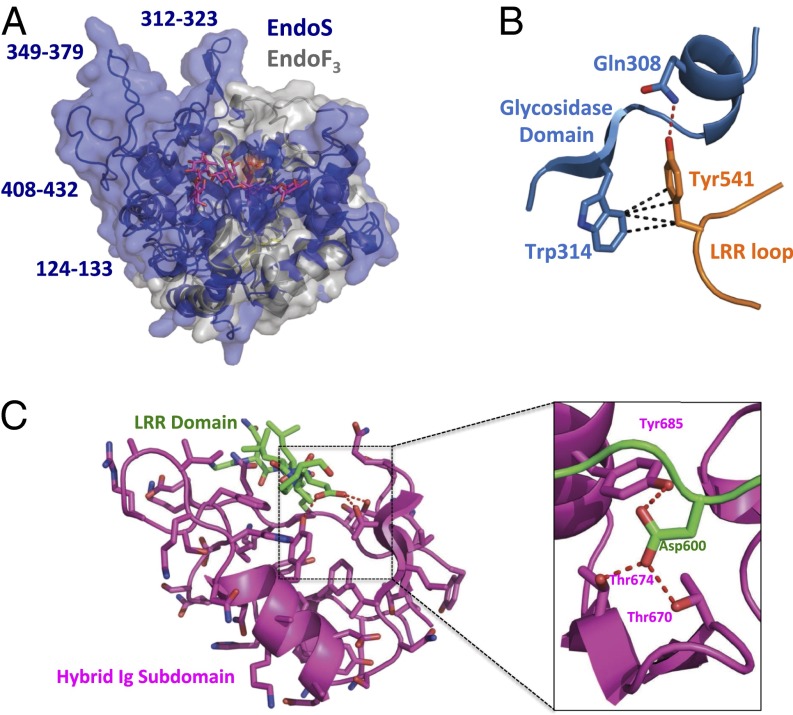
The EndoS glycosidase domain adopts the (β/α)8 barrel conformation typical of bacterial endoglycosidases. Structurally, it is most similar (32) to the analogous domain of EndoF3 (PDB ID code 1E0M; Dali Server Z score = 21.9; Fig. 1B). EndoS cleaves specifically biantennary complex oligosaccharides (29), whereas EndoF3 is specific for both biantennary and triantennary complex oligosaccharides (6); neither have activity on high mannose oligosaccharides. The basic structure of the (β/α)8 barrel fold and the positions of the active site residues are nearly identical in these two enzymes (SI Appendix, Fig. S4A). The loops surrounding the active site barrel opening, however, differ substantially between EndoS and EndoF3, resulting in distinct molecular surfaces that could accommodate divergent glycoprotein substrates (Fig. 2A). In EndoS, tryptophan residues are important for both substrate specificity and enzymatic activity (33), several of which are exposed on the surface surrounding the active site barrel opening (SI Appendix, Fig. S4B), including: Trp314 and Trp358, both of which line the groove that likely accommodates the protein portion of the substrate, and Trp121, Trp153, and Trp161, which line the two grooves in which the oligosaccharide chains in the biantennary complex carbohydrate are likely positioned during hydrolysis.
Fig. 2.
Structural features of the EndoS glycosidase, leucine-rich repeat, and hybrid Ig domains. (A) Superposition of the EndoS glycosidase domain (blue) with that of EndoF3 (gray) with surfaces rendered. The glycan structure as resolved in the EndoF3–glycan complex structure is in magenta. EndoS glycosidase domain loops that diverge most in structure from those in EndoF3 are labeled. (B) Intramolecular contacts between residues in the EndoS glycosidase domain (blue) and the apex of the loop extending from the leucine-rich repeat domain (orange). Van der Waals interactions are shown as black dashed lines; hydrogen bond as a dashed red line. (C) Intramolecular engagement of residues from the leucine-rich repeat (green) and the inserted hybrid Ig subdomain (magenta). (C, Left) Interaction showing the entire hybrid Ig subdomain. (C, Right) Close-up view of contacts made by leucine-rich repeat residue Asp600 (green) with hybrid Ig subdomain residues. Hydrogen bonds are shown as dashed red lines.
The LRR Domain Extends Additional Domains Away from the Catalytic Site.
The LRR domain of EndoS extends from the bottom of the glycosidase domain, opposite the (β/α)8 barrel opening. This domain forms the base of the V-shape structure, keeping the remaining C-terminal domains from folding back onto the glycosidase domain. The LRR domain contains three typical LRR motifs followed by two modified LRR motifs, all of which are capped by several short α-helices. It is structurally most similar to internalins, such as InternalinJ (PDB ID code 3BZ5; Z score = 9.5; Fig. 1C). Despite its limited number of repeat motifs, the EndoS LRR adopts the typical curvature of longer LRR domains.
A Unique LRR Domain Loop Packs Against the Glycosidase Domain.
Within the third LRR motif, a long loop (residues 528–554) extends away from the LRR domain and packs against the side of the glycosidase domain. Residues 534–541 bury 1,112 Å2 of surface area between this loop and the glycosidase domain, approximately one-half of the total 2,173 Å2 of buried surface area between the glycosidase and LRR domains. Known LRR motifs do not typically include such loop insertions. Tyr541, at the tip of the LRR loop, forms a hydrogen bond with Gln308 and several van der Waals interactions with Trp314 of the glycosidase domain (Fig. 2B). This latter residue, as described above, forms the edge of the surface groove on the top of the glycosidase domain that leads to the (β/α)8 barrel opening and the active site. The LRR domain loop effectively extends this groove from the active site and, thus, could potentially play a role in glycoprotein substrate specificity and/or the stability of the protein.
EndoS Exhibits a Topologically Unique Hybrid Ig Domain.
Extending from the LRR domain at the base of the V-shaped EndoS structure is a hybrid Ig domain. This domain is composed of two subdomains that are topologically entwined—the smaller of the two subdomains is inserted within the loop that connects the second and third β-strands of the larger subdomain, which is a typical Ig domain structurally similar to the interleukin-4 receptor (PDB ID code 1IAR; Z score = 5.2; Fig. 1D) and other Ig proteins, including antibodies. The smaller subdomain, conversely, is structurally unique compared with all previously determined protein structures. It acts as a molecular spacer between the LRR domain and the Ig subdomain, the interface between which consists primarily of a loop from the former (residues 598–603) positioned into a deep cleft on the surface of the latter (Fig. 2C), burying 685 Å2 of the 1,562 Å2 of surface area between these two domains. Although most of this interface is comprised of van der Waals interactions, the two terminal oxygen atoms of Asp600 make hydrogen bonds to side-chain oxygen atoms from Thr670, Thr674, and Tyr685 (Fig. 2C).
EndoS Contains a Putative Carbohydrate-Binding Module.
C-terminal to the hybrid Ig domain is a domain that exhibits a high degree of structural homology to noncatalytic CBMs from Clostridium thermocellum, including CtCBM62 (PDB ID code 2YFU; Z score = 7.4; Fig. 1E), which binds galactose-containing polysaccharides. Complex carbohydrates, such as those attached to IgG1 Fc and processed by EndoS, typically contain galactose molecules (12). Like CtCBM62, the EndoS CBM coordinates a single Ca2+ ion by using main-chain oxygen atoms of Lys786, Gly790, Gln791, and Pro915, and a side-chain oxygen atom from Glu916. Neither the oligomerization state nor the hydrolytic activity of EndoS changed in the presence of EDTA. The sole substrate for EndoS is the glycosylated Fc region of IgG (8), an obligate homodimer with Asn-linked glycans attached to each of the two monomer subunits. In the EndoS structure, the two domains, glycosidase and CBM, that most likely bind IgG glycans are located at opposite ends of the V-shaped molecule (Fig. 1A).
The C-Terminal Domain Is a Three-Helix Bundle Pointing Away from All Other Domains.
The final EndoS protein domain, 3H, is a three-helix bundle motif most structurally similar to the substrate binding domain of the Hsp70 chaperone DnaK (PDB ID code 4JNE; Z score = 8.6; Fig. 1E). The 3H domain extends away from the opening of the V-shaped structure (Fig. 1A). However, this domain is not packed extensively against its neighboring EndoS domains, the CBM and hybrid Ig domains, exhibiting 841 and 365 Å2 of buried surface area with each domain, respectively, suggesting that it could potentially rotate and translate in a rigid-body fashion when substrate is bound.
EndoS Requires Multiple Domains for Substrate Specificity.
We expressed catalytically inactive versions of the enzyme, EndoSE235Q(98–995), which includes all five domains, EndoS(446–995), which contains only domains C-terminal to the glycosidase domain, and EndoSE235Q(98–764), which excludes the CBM and 3H domains, and measured their binding affinities to immobilized IgG1 Fc by surface plasmon resonance (SPR) analysis (SI Appendix, Fig. S5). EndoSE235Q(98-995) bound Fc with an affinity (KD) of 22 μM, ∼50-fold lower than the oligomerization-dependent avidity of EndoSD235Q(37–995) (34), essentially equivalent to its relative reduction in hydrolytic activity (SI Appendix, Fig. S1B). Neither EndoS(446–995) nor EndoSE235Q(98–764) exhibited detectable binding to IgG1 Fc. Thus, there exist at least two distinct domains important for the EndoS specificity for IgG1 Fc: (i) between residues 98 and 446, corresponding to the glycosidase domain alone; and (ii) and between residues 766 and 995, inclusive of the CBM and 3H domains.
A Model of the EndoS/IgG1 Fc Encounter Complex.
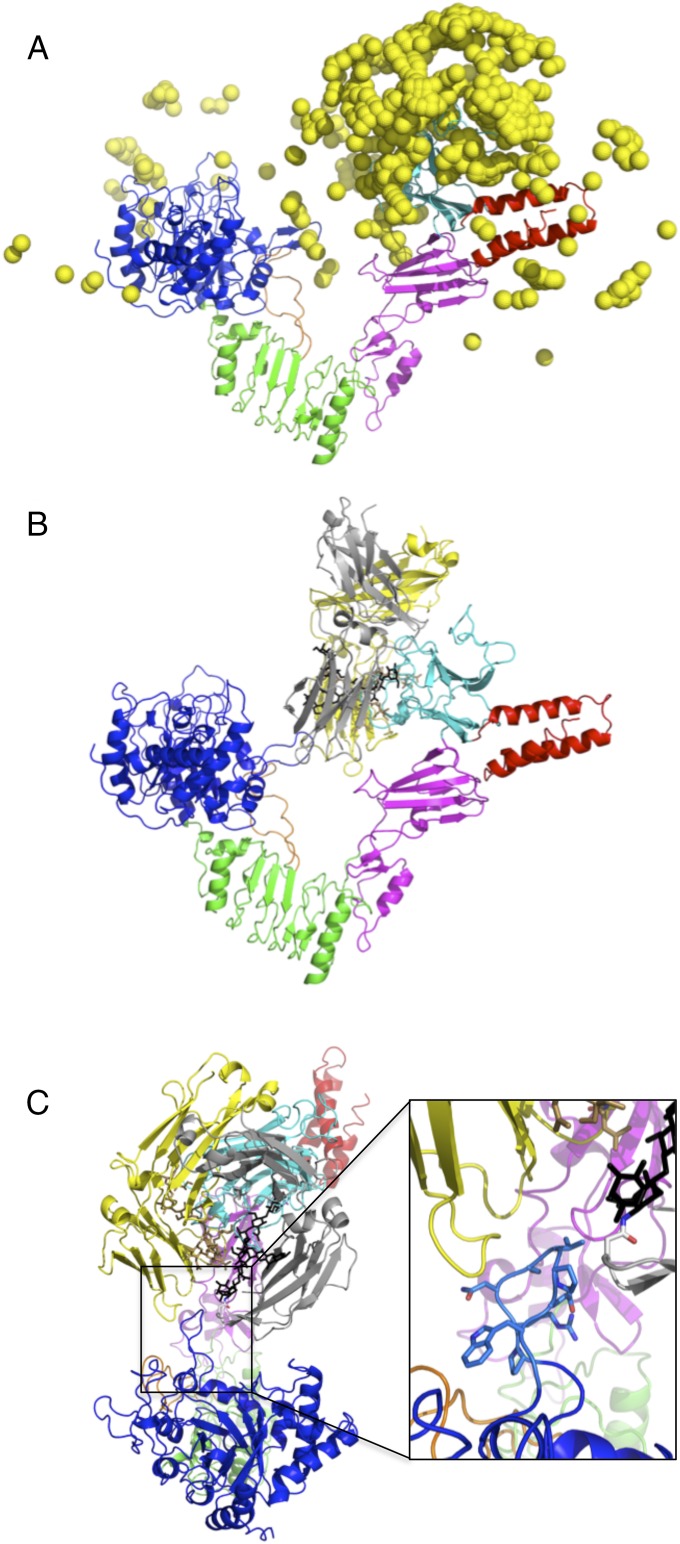
We modeled the EndoS/IgG1 Fc encounter complex by docking an IgG1 Fc homodimer structure (PDB ID code 4BYH) onto the EndoSD233Q(98–995) crystal structure, modified by adding any missing loop residues using ZDOCK 3.0.2 (35). Postprocessing of the docking output was performed in two stages. First, we analyzed the top 2,000 ZDOCK predictions for proximity of the two Asn297IgG1 Fc residues to EndoS glycosidase and CBM domain residues likely involved in glycan binding based on homology to other glycosidase and CBM structures (e.g., residues Leu213EndoS, Gln233EndoS, Glu235EndoS, and Asp827EndoS, Arg830EndoS, Asn836EndoS, respectively). One of the top-ranked ZDOCK models (ZD3, ranked number 3; SI Appendix, Fig. S6A) featured the two Asn297IgG1 Fc residues with relatively small distances to both of these sets of EndoS CBM residues (<10 Å) and glycosidase domain residues (<20 Å). Overall, the vast majority of the initial-stage docking predictions featured Asn297IgG1 Fc residues positioned near the EndoS CBM (Fig. 3A). Second, we performed structural refinement of this model, generating an additional 1,000 refined models. Comparing the refined models with the original model from ZDOCK, we observed an energy funnel-like landscape (SI Appendix, Fig. S6B). In this final model of the EndoS/IgG1 Fc encounter complex (Fig. 3B), the IgG1 Fc is bound predominantly through residues from its CH2 domains and primarily to the EndoS CBM in an orientation in which the plane of the V shape of EndoS is nearly perpendicular to the plane that runs through the two monomer subunits of the Fc homodimer. The EndoS glycosidase domain loop comprised of residues 312–323, unique among all known endoglycosidase structures, bisects the two monomer subunits of the Fc homodimer (Fig. 3C). Positioned as such, there is direct access to the Asn297-linked glycans for the EndoS active site and adequate space between the Fc monomer N-termini and the LRR domain for the remainder of the IgG1 antibody, the Fab domains, to exit. Computational alanine scanning mutagenesis identified CBM domain residues Trp803EndoS, Arg908EndoS, and Glu833EndoS as hot spots for binding.
Fig. 3.
Model of the EndoS/IgG1 Fc encounter complex. (A) Distribution of location of the IgG1 Fc on the EndoS structure from the top 500 initial models of the encounter complex. Colors of EndoS domains are as in previous figures. Each yellow ball represents the center of mass of a single IgG1 Fc dimer. (B) Side view of the final refined EndoS/IgG1 Fc encounter complex model. Monomers of the Fc homodimer are in gray (with corresponding glycan in black) and in yellow (with corresponding glycan in tan). (C) Top view of the encounter complex model with close-up of the EndoS glycosidase loop bisecting the Fc monomers in shown in the box on the right.
Validating the Encounter Complex Model.
Our model of the EndoS/IgG1 Fc encounter complex indicated several EndoS loops that likely interact with the glycoprotein substrate, including the aforementioned glycosidase domain loop (residues 312–323) and loops from the hybrid Ig (residues 742–750) and CBM (residues 793–797) domains (SI Appendix, Fig. S6C). To determine the energetic contributions to binding for each of these loops, and the LRR domain loop (residues 528–554), we produced versions of EndoSE235Q with each loop removed and measured their binding affinities to IgG1 Fc by SPR, all of which exhibited reduced affinities for Fc relative to EndoSE235Q (SI Appendix, Fig. S5). We observed the largest reduction in binding affinity, KD = 489 μM (∼20-fold), for EndoSE235Q/Δ314–323. EndoSE235Q/Δ742–750 bound Fc with a KD of 156 μM (∼10-fold reduction). EndoSE235Q/Δ528–554 and EndoSE235Q/Δ793–797 bound Fc with KD values of 24 and 30 μM, respectively (approximately equivalent to EndoSE235Q). For EndoSE235Q/Δ528–554, we obtained markedly lower protein expression and observed significantly reduced Rmax values for SPR binding isotherms, suggesting that the protein is unstable without this loop. Taken together, these data indicate that loops from the glycosidase and hybrid Ig domains are major determinants of Fc specificity, whereas loops from the LRR domain, which makes no contacts but may be important for the stability of the protein, and the CBM, which makes predominantly glycan interactions, in the encounter complex model are nearly dispensable for Fc binding. We also tested mutants identified by computational alanine scanning mutagenesis as hot spots for binding, including EndoSE235Q/W803A, EndoSE235Q/R908A, and EndoSE235Q/E833A (SI Appendix, Fig. S5), all located in the CBM. EndoSE235Q/W803A and EndoSE235Q/E833A each exhibited no detectable binding to IgG1 Fc at this concentration. EndoSE235QR908A exhibited threefold weaker binding than did EndoSE235Q. The binding capacities of these EndoS mutants largely correlated with their hydrolytic activities (SI Appendix, Fig. S1C). Each mutant that exhibited substantially reduced or no detectable binding also exhibited reduced or no hydrolytic activity, except EndoSE235Q/E833A. In particular, the glycosidase domain loop (residues 314–323) and a point mutation in the CBM domain, EndoSE235Q/E803A, were absolutely required for hydrolytic activity.
Conformation Changes Required for Enzymatic Activity.
According to our modeled EndoS/IgG1 Fc encounter complex, which is transient by definition, there is a clear requirement for conformational changes to carry out the enzymatic reaction. In the encounter complex model, the active site residues are located some 15 Å from the bond between the first and second GlcNAc moieties at which enzymatic cleavage occurs. Thus, rotation and translation of the glycosidase domain relative to the likely more fixed position of the IgG1 Fc bound to the CBM must occur for catalysis to take place. Using HingeProt (36), we found several EndoS regions predicted to act as hinges, including: (i) the residues linking the glycosidase and LRR domains; (ii) the N-terminal portion of the LRR domain; and (iii) the juncture between the two subdomains of the hybrid Ig domain. Concerted movements of EndoS domains around these hinge points would result in a narrowing of its V-shape opening, moving the glycosidase domain closer in space to the CBM (SI Appendix, Fig. S7A). Regardless of the structure of the encounter complex and ensuing conformational changes in EndoS, further conformational changes must take place in the IgG substrate structure to allow the glycan to be properly positioned in the active site for cleavage. Superpositions of IgG1 Fc with the EndoF3–glycan complex by aligning their respective glycans, and of EndoS with the EndoF3–glycan complex by aligning their respective glycosidase domains, indicates that substantial steric clashes would prohibit such a complex (SI Appendix, Fig. S7B). Barring remarkable structural deformation of the EndoS glycosidase domain, protein and/or glycan conformation changes in the antibody would be required for enzymatic activity.
Human IgG Subclass Specificity.
Because EndoS is known to have distinct specificities for individual murine IgG subclasses (11), we determined whether there existed any comparable human IgG subclass specificity. We found that both EndoS(37-995) and EndoS(98-995) exhibited slightly decreased binding to IgG2, IgG3, and IgG4, and reduced hydrolytic activity with these substrates (SI Appendix, Fig. S1C), compared with IgG1. We also tested the hydrolytic activity of our entire panel of EndoS(98-995) mutants and found that, nearly universally, mutants capable of hydrolyzing IgG1 were also capable of hydrolyzing IgG2, IgG3, and IgG4 (SI Appendix, Fig. S1C), and those that could not hydrolyze IgG1 could also not hydrolyze any other IgG subclass. However, each of the hydrolytic EndoS mutants exhibited higher activity on IgG1 relative to IgG2, IgG3, and IgG4. These data indicate that EndoS has a human IgG subclass specificity of IgG1 > IgG2 = IgG3 = IgG4.
Discussion
EndoS is an enzyme secreted by S. pyogenes that removes carbohydrates highly specifically from human IgG antibodies. Because antibodies are central players in many human immune responses and bridge the innate and adaptive arms of immunity, the analysis and manipulation of the enzymatic activity of EndoS impacts diverse fields in biomedicine. Clinically, EndoS contributes to the abilities of S. pyogenes to evade the human immune response (8, 10, 34); the development of specific inhibitors of EndoS activity could improve clinical outcomes of patients suffering a range of inflammatory conditions. Therapeutically, EndoS is already showing great promise in animal models as a treatment for diverse autoimmune diseases that rely on autoantibodies (11, 16–22); fine-tuning the specificity and activity of EndoS will be an important aspect of its further development as a protein therapeutic for use in humans. Biotechnologically, EndoS is a unique glycoprotein-modifying enzyme with the capacity to both remove glycans from and, as a glycosynthase variant, attach glycans to antibodies (29, 30); expanding the repertoire of homogeneous glycosylated antibodies that can be produced with newly designed EndoS variants will be critical for realizing the full potential of engineered antibodies. Our X-ray crystal structure of EndoS provides a platform for future clinical, therapeutic, and biotechnological progress.
EndoS is comprised of five distinct protein domains that adopt a striking V-shaped structure in which the four domains C-terminal to the glycosidase domain extend from the face opposite that used for glycan binding and catalytic activity. Instead of forming a typical globular protein, the additional domains extend away from the glycosidase domain, resulting in the CBM forming one point of the V shape opposite the point formed by the glycosidase domain. The relative positions of these two domains in EndoS known to engage carbohydrate in other proteins suggests that glycosylated antibodies enter the V shape and are held within the concave surface of the enzyme during processing.
Indeed, our molecular modeling predicts that an encounter complex is formed between EndoS and human IgG1 Fc in which the substrate enters the V and is trapped by the CBM, with EndoS making contacts mostly to residues from the IgG1 CH2 domain, in which the N-linked glycan resides. A notable feature of this complex is that the tip of the glycosidase domain loop (residues 312–323) that extends away from the active site barrel opening bisects the two monomer subunits of the IgG1 Fc homodimer. When we measured the binding affinity of the glycosynthase mutant with this loop removed, we observed an ∼20-fold weaker affinity relative to the same protein with this loop intact and complete abolition of hydrolytic activity. Additional components of the EndoS structure implicated by the model in substrate binding and validated by mutagenesis include a loop from the hybrid Ig domain (residues 742–750) and several hot spot residues from the CBM.
Evident from both our structure of the enzyme in the absence of substrate and our model of the EndoS/IgG1 Fc encounter complex, enzymatic activity would require a number of conformational changes, likely in both the enzyme and the substrate. Accordingly, we found, by computational analysis, several hinge points between EndoS domains that predict concerted movements that effectively squeeze the ends of the V shape together, bringing the glycosidase and CBM domains in closer proximity to one another. In the encounter complex model, in which IgG1 Fc is bound predominantly by the CBM, this movement would result in the substrate approaching the active site. Additional conformational changes likely to accompany transition from the encounter complex to a processing–competent complex structure include movement of the glycosidase loop (residues 312–323) that bisects the two IgG monomers and rotation and further translation of the glycosidase domain toward the bound substrate. Regardless of these movements, conformational changes in the substrate are certainly also required for enzymatic activity. Assuming that the N-linked glycan of IgG must eventually occupy a position similar to that observed for the EndoF3–glycan complex (6), the IgG1 Fc homodimer would have to separate and/or the glycans would have to move outside of the space between the CH2 domains that they normally occupy. Whether such concerted enzyme and substrate structural changes would allow the processing of both glycans subsequent to the formation of a single encounter complex or necessitate a unique encounter complex event for the processing of each of the two glycans remains unclear at this point.
EndoS exhibits high specificity for glycosylated IgG antibodies and has no, or only marginal, hydrolytic activity toward N-glycans in the context of other glycoproteins or glycopeptides. Our studies indicate that several structural features scattered throughout the EndoS sequence are important for this high substrate specificity. First, the glycosidase loop (residues 312–323) extending from the barrel opening that engages IgG1 in our encounter complex model has a significant effect on substrate binding and activity. The nature of this loop suggests that simply swapping out glycosidase domains from other endoglycosidases to engineer EndoS variants that maintain IgG specificity but exhibit diverse glycan specificities may not be achievable without incorporating a similar loop in the engineered enzyme. Second, EndoS domains beyond the glycosidase domain are important for substrate specificity. The importance of these domains has previously been implicated by the deactivation of EndoS by SpeB, which releases the glycosidase domain from the remainder of the enzyme (33). That these noncatalytic domains are important for substrate specificity suggests that modifying them might produce EndoS variants with unique substrate specificities, such as those that exhibit distinct human IgG subclass specificity or altered antibody isotype binding.
In summary, we present the X-ray crystal structure of EndoS and a model of the encounter complex formed with IgG1 Fc. Our studies provide a molecular basis for substrate specificity and enzymatic activity and suggest ways in which EndoS variants could be engineered for novel therapeutic and biotechnological purposes.
Materials and Methods
For details, see SI Appendix, SI Materials and Methods.
Protein Production, Oligomerization State, and Hydrolytic Activity Analysis.
All EndoS proteins were expressed in Escherichia coli and purified as described (30, 37). IgG1 Fc was obtained from papain digestion and chemoenzymatic glycoengineering of Rituximab as described (30). IgG2, IgG3, and IgG4 were purchased (Sigma-Aldrich). Oligomerization state was assessed by size exclusion chromatography in comparison with molecular weight standards. Hydrolytic activity was calculated by measuring densitometric changes in high molecular weight glycosylated IgG antibodies over time by using SDS/PAGE.
Protein Structure Analysis.
Crystallization of native EndoS by liquid-liquid diffusion using Crystal Formers (Microlytic) has been described in detail (37). SeMet-EndoSD233Q(98–995) crystals were obtained by microseeding with native crystals in the previously determined crystallization condition. All crystals were flash cooled at 100 K in mother liquor containing 20% ethylene glycol before synchrotron X-ray diffraction data collection. The structure of SeMet-EndoSD233Q(98–995) was solved by MAD at 3.2 Å, whereas those of EndoSD233Q(98–995) and EndoSWT(98–995) were solved at 1.9-Å and 2.6-Å resolution, respectively, by molecular replacement using the SeMet-EndoSD233Q(98–995) structure as a model. SAXS data were measured for EndoS(98–995) at concentrations of 1, 2, 5, and 10 mg/mL.
Binding Analysis.
All SPR experiments were performed by using a Biacore T100 instrument (GE Healthcare) with IgG1 Fc or whole IgG2, IgG3, or IgG4 antibodies immobilized to two flow cell surfaces with one (i.e., negative control) deglycosylated by EndoS pretreatment. Concentration series of all EndoS proteins were injected and affinity constants were calculated by using a general steady-state equilibrium model.
Molecular Modeling.
We used ZDOCK 3.0.2 (35) to dock the glycosylated IgG1 Fc (PDB ID code 4BYH) to the EndoSD233Q(98–995) structure. We performed computational interface alanine scanning of the refined model to determine putative energetic hot spots by using the “interface” protocol of Rosetta 2.0.2 (38).
Supplementary Material
Acknowledgments
We thank the beamline scientists at 23-ID-D and 23-ID-B, Advanced Photon Source (APS) and at 11-1, Stanford Synchrotron Radiation Lightsource (SSRL). These studies were supported by National Institutes of Health Grants R01AI090866 (to E.J.S.) and R01 GM080374 (to L.-X.W.). Use of APS is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. Portions of this research were carried out at SSRL, a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4NUY and 4NUZ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322908111/-/DCSupplemental.
References
- 1.Collin M, Olsén A. Extracellular enzymes with immunomodulating activities: Variations on a theme in Streptococcus pyogenes. Infect Immun. 2003;71(6):2983–2992. doi: 10.1128/IAI.71.6.2983-2992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ling Z, et al. The X-ray crystal structure of an Arthrobacter protophormiae endo-beta-N-acetylglucosaminidase reveals a (beta/alpha)(8) catalytic domain, two ancillary domains and active site residues key for transglycosylation activity. J Mol Biol. 2009;389(1):1–9. doi: 10.1016/j.jmb.2009.03.050. [DOI] [PubMed] [Google Scholar]
- 3.Yin J, et al. Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A. PLoS ONE. 2009;4(3):e4658. doi: 10.1371/journal.pone.0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbott DW, Macauley MS, Vocadlo DJ, Boraston AB. Streptococcus pneumoniae endohexosaminidase D, structural and mechanistic insight into substrate-assisted catalysis in family 85 glycoside hydrolases. J Biol Chem. 2009;284(17):11676–11689. doi: 10.1074/jbc.M809663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Roey P, Rao V, Plummer TH, Jr, Tarentino AL. Crystal structure of endo-beta-N-acetylglucosaminidase F1, an alpha/beta-barrel enzyme adapted for a complex substrate. Biochemistry. 1994;33(47):13989–13996. doi: 10.1021/bi00251a005. [DOI] [PubMed] [Google Scholar]
- 6.Waddling CA, Plummer TH, Jr, Tarentino AL, Van Roey P. Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3) Biochemistry. 2000;39(27):7878–7885. doi: 10.1021/bi0001731. [DOI] [PubMed] [Google Scholar]
- 7.Rao V, Guan C, Van Roey P. Crystal structure of endo-beta-N-acetylglucosaminidase H at 1.9 A resolution: Active-site geometry and substrate recognition. Structure. 1995;3(5):449–457. doi: 10.1016/s0969-2126(01)00178-2. [DOI] [PubMed] [Google Scholar]
- 8.Collin M, Olsén A. EndoS, a novel secreted protein from Streptococcus pyogenes with endoglycosidase activity on human IgG. EMBO J. 2001;20(12):3046–3055. doi: 10.1093/emboj/20.12.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collin M, Olsén A. Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect Immun. 2001;69(11):7187–7189. doi: 10.1128/IAI.69.11.7187-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collin M, et al. EndoS and SpeB from Streptococcus pyogenes inhibit immunoglobulin-mediated opsonophagocytosis. Infect Immun. 2002;70(12):6646–6651. doi: 10.1128/IAI.70.12.6646-6651.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albert H, Collin M, Dudziak D, Ravetch JV, Nimmerjahn F. In vivo enzymatic modulation of IgG glycosylation inhibits autoimmune disease in an IgG subclass-dependent manner. Proc Natl Acad Sci USA. 2008;105(39):15005–15009. doi: 10.1073/pnas.0808248105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 13.Lund J, Takahashi N, Pound JD, Goodall M, Jefferis R. Multiple interactions of IgG with its core oligosaccharide can modulate recognition by complement and human Fc gamma receptor I and influence the synthesis of its oligosaccharide chains. J Immunol. 1996;157(11):4963–4969. [PubMed] [Google Scholar]
- 14.Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Biol. 2003;325(5):979–989. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 15.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8(1):34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 16.Nandakumar KS, et al. Endoglycosidase treatment abrogates IgG arthritogenicity: Importance of IgG glycosylation in arthritis. Eur J Immunol. 2007;37(10):2973–2982. doi: 10.1002/eji.200737581. [DOI] [PubMed] [Google Scholar]
- 17.Collin M, Shannon O, Björck L. IgG glycan hydrolysis by a bacterial enzyme as a therapy against autoimmune conditions. Proc Natl Acad Sci USA. 2008;105(11):4265–4270. doi: 10.1073/pnas.0711271105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Timmeren MM, et al. IgG glycan hydrolysis attenuates ANCA-mediated glomerulonephritis. J Am Soc Nephrol. 2010;21(7):1103–1114. doi: 10.1681/ASN.2009090984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allhorn M, et al. The IgG-specific endoglycosidase EndoS inhibits both cellular and complement-mediated autoimmune hemolysis. Blood. 2010;115(24):5080–5088. doi: 10.1182/blood-2009-08-239020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tradtrantip L, Ratelade J, Zhang H, Verkman AS. Enzymatic deglycosylation converts pathogenic neuromyelitis optica anti-aquaporin-4 immunoglobulin G into therapeutic antibody. Ann Neurol. 2013;73(1):77–85. doi: 10.1002/ana.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirose M, et al. Enzymatic autoantibody glycan hydrolysis alleviates autoimmunity against type VII collagen. J Autoimmun. 2012;39(4):304–314. doi: 10.1016/j.jaut.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Lood C, et al. IgG glycan hydrolysis by endoglycosidase S diminishes the proinflammatory properties of immune complexes from patients with systemic lupus erythematosus: A possible new treatment? Arthritis Rheum. 2012;64(8):2698–2706. doi: 10.1002/art.34454. [DOI] [PubMed] [Google Scholar]
- 23.Baruah K, et al. Selective deactivation of serum IgG: A general strategy for the enhancement of monoclonal antibody receptor interactions. J Mol Biol. 2012;420(1-2):1–7. doi: 10.1016/j.jmb.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L-X. Chemoenzymatic synthesis of glycopeptides and glycoproteins through endoglycosidase-catalyzed transglycosylation. Carbohydr Res. 2008;343(10-11):1509–1522. doi: 10.1016/j.carres.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudd PM, et al. Glycoforms modify the dynamic stability and functional activity of an enzyme. Biochemistry. 1994;33(1):17–22. doi: 10.1021/bi00167a003. [DOI] [PubMed] [Google Scholar]
- 26.Umaña P, Jean-Mairet J, Moudry R, Amstutz H, Bailey JE. Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat Biotechnol. 1999;17(2):176–180. doi: 10.1038/6179. [DOI] [PubMed] [Google Scholar]
- 27.Yamane-Ohnuki N, et al. Establishment of FUT8 knockout Chinese hamster ovary cells: An ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol Bioeng. 2004;87(5):614–622. doi: 10.1002/bit.20151. [DOI] [PubMed] [Google Scholar]
- 28.Wang LX, Lomino JV. Emerging technologies for making glycan-defined glycoproteins. ACS Chem Biol. 2012;7(1):110–122. doi: 10.1021/cb200429n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodfellow JJ, et al. An endoglycosidase with alternative glycan specificity allows broadened glycoprotein remodelling. J Am Chem Soc. 2012;134(19):8030–8033. doi: 10.1021/ja301334b. [DOI] [PubMed] [Google Scholar]
- 30.Huang W, Giddens J, Fan SQ, Toonstra C, Wang LX. Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J Am Chem Soc. 2012;134(29):12308–12318. doi: 10.1021/ja3051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Svergun D, et al. CRYSOL - Evaluation of the solution scattering from macromolecules with known atomic srtructure and fitting to experimental data. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 32.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allhorn M, Olsén A, Collin M. EndoS from Streptococcus pyogenes is hydrolyzed by the cysteine proteinase SpeB and requires glutamic acid 235 and tryptophans for IgG glycan-hydrolyzing activity. BMC Microbiol. 2008;8:3. doi: 10.1186/1471-2180-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allhorn M, Olin AI, Nimmerjahn F, Collin M. Human IgG/Fc gamma R interactions are modulated by streptococcal IgG glycan hydrolysis. PLoS ONE. 2008;3(1):e1413. doi: 10.1371/journal.pone.0001413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE. 2011;6(9):e24657. doi: 10.1371/journal.pone.0024657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emekli U, Schneidman-Duhovny D, Wolfson HJ, Nussinov R, Haliloglu T. HingeProt: Automated prediction of hinges in protein structures. Proteins. 2008;70(4):1219–1227. doi: 10.1002/prot.21613. [DOI] [PubMed] [Google Scholar]
- 37.Trastoy B, Lomino JV, Wang L-X, Sundberg EJ. Liquid-liquid diffusion crystallization improves the X-ray diffraction of EndoS, an endo-β-N-acetylglucosaminidase from Streptococcus pyogenes with activity on human IgG. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69(Pt 12):1405–1410. doi: 10.1107/S1744309113030650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc Natl Acad Sci USA. 2002;99(22):14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.