Significance
New antifungal compounds are needed due to an increasing incidence of invasive fungal infections and resistance to many currently used drugs. Here we show that cyanobacteria are a rich source of antifungal compounds such as glycosylated lipopeptides, called hassallidins, which are commonly produced by filamentous nitrogen-fixing cyanobacteria. A diverse group of hassallidins and their complex nonribosomal biosynthesis were characterized in detail. Hassallidins and their previously unidentified biosynthetic enzymes offer new material for drug development. In addition, these compounds may have an ecological role in protecting cyanobacteria from parasitic fungi.
Keywords: nonribosomal peptide synthesis, natural product discovery, genome mining, secondary metabolites, bioactive peptide
Abstract
Cyanobacteria produce a wide variety of cyclic peptides, including the widespread hepatotoxins microcystins and nodularins. Another class of peptides, cyclic glycosylated lipopeptides called hassallidins, show antifungal activity. Previously, two hassallidins (A and B) were reported from an epilithic cyanobacterium Hassallia sp. and found to be active against opportunistic human pathogenic fungi. Bioinformatic analysis of the Anabaena sp. 90 genome identified a 59-kb cryptic inactive nonribosomal peptide synthetase gene cluster proposed to be responsible for hassallidin biosynthesis. Here we describe the hassallidin biosynthetic pathway from Anabaena sp. SYKE748A, as well as the large chemical variation and common occurrence of hassallidins in filamentous cyanobacteria. Analysis demonstrated that 20 strains of the genus Anabaena carry hassallidin synthetase genes and produce a multitude of hassallidin variants that exhibit activity against Candida albicans. The compounds discovered here were distinct from previously reported hassallidins A and B. The IC50 of hassallidin D was 0.29–1.0 µM against Candida strains. A large variation in amino acids, sugars, their degree of acetylation, and fatty acid side chain length was detected. In addition, hassallidins were detected in other cyanobacteria including Aphanizomenon, Cylindrospermopsis raciborskii, Nostoc, and Tolypothrix. These compounds may protect some of the most important bloom-forming and globally distributed cyanobacteria against attacks by parasitic fungi.
Cyanobacteria are known for their propensity to form toxic blooms that have caused the deaths of wild and domestic animals all over the world (1). However, they are also a rich source of natural products that exhibit antimicrobial, anticancer, and immunosuppressive activities that can be exploited in drug development (2, 3). These secondary metabolites have versatile and often highly complex chemical structures with diverse biosynthetic origins. The majority of bioactive compounds reported from cyanobacteria are cyclic or linear peptides that can be heavily modified, including derivatization such as epimerization, glycosylation, acylation, formylation, methylation, halogenation, or sulphation (4). Many of these peptides are made nonribosomally with peptide cores consisting of an array of proteinogenic or nonproteinogenic amino acids as well as components of polyketide origin. They are assembled on large enzyme complexes by a thiotemplate mechanism in which the nonribosomal peptide synthetases (NRPSs) act simultaneously as template and biosynthetic machinery (5, 6). NRPSs are organized into modules, each of which is responsible for one amino acid activation and peptide bond formation (5, 7–9). The selectivity of NRPS adenylation domains combined with NRPS catalytic domain organization that generally follows the colinearity principle (7, 10) offers a means to predict the amino acid building blocks and thus the putative peptide structure.
Microbial genomes are replete with NRPS gene clusters (11, 12). However, the majority of these gene clusters are cryptic and their products are unknown (4, 12). The mining of microbial genomes provides a useful approach for natural product discovery. The initial genome annotation of the toxic bloom-forming cyanobacterium Anabaena sp. 90 identified gene clusters for anabaenopeptins, anabaenopeptilides, and microcystins (13–15). Surprisingly, a fourth cryptic inactive gene cluster was found and proposed to be responsible for the biosynthesis of hassallidins (16). In the present study, we report the detailed characterization of the gene cluster from the genome of Anabaena sp. SYKE748A and show that it is responsible for the production of numerous novel glycosylated lipopeptides. These compounds resemble antifungal hassallidins A and B, which were isolated from an epilithic cyanobacterium Hassallia sp. (17, 18). A large number of new hassallidin variants was detected, showing that the ability to produce these compounds is widespread in heterocyst-forming cyanobacteria.
Results
The Hassallidin Biosynthetic Gene Cluster.
The assembly and annotation of the Anabaena sp. 90 genome revealed a cryptic nonribosomal peptide gene cluster, coding for a previously unknown natural product that was proposed to be hassallidin (16). To identify the hassallidin gene cluster from an active producer of hassallidins, we obtained a draft genome of Anabaena sp. SYKE748A. The identified hassallidin gene cluster was located on three contigs and had a sequence similarity greater than 99.8% to that of Anabaena sp. 90. The hassallidin synthetase (has) gene clusters from Anabaena sp. 90 and SYKE748A are bidirectional and span 59 kb of genomic DNA (Fig. 1A). They consist of 26 ORFs, designated as hasA–hasZ, that are flanked by two transposase genes (SI Appendix, Table S1). The cluster encodes four multidomain NRPS proteins, HasN, HasO, HasV, and HasY, with sizes of 345 kDa, 412 kDa, 244 kDa, and 323 kDa, respectively. The NRPSs contain nine modules, which are responsible for the activation and incorporation of nine amino acids into the growing polypeptide chain (Fig. 1B). Each of the NRPS modules bears a condensation (C), adenylation (A), and thiolation (T) domain. In addition, modules HasN1, HasN2, and HasO2 each contain an epimerization (E) domain suggesting that the end-product would contain three d-amino acids. The HasY1 module encodes an N-methyltransferase (NMT) domain, indicating the presence of an N-methylated amino acid in the end-product. The HasY2 module bears a C-terminal thioesterase domain that catalyzes the cyclization and release of the peptide backbone by forming an ester bond between amino acids incorporated by modules HasV2 and HasY2. Predictions of the amino acid backbone of the peptide were made using the substrate specificity-conferring nature of the adenylation domains (SI Appendix, Table S2). The substrate specificity codes of the first eight modules have an exact or nearly exact match to those of adenylation domains with known substrate specificities (SI Appendix, Table S2). The last HasY2 module shows lower identity to known adenylation domains.
Fig. 1.
Biosynthetic pathway of hassallidin. (A) Organization of the has gene cluster in Anabaena sp. SYKE748A showing the location of the deletion in the has gene cluster of Anabaena sp. 90. (B) Proposed biosynthetic pathway of hassallidins and predicted domain structure of HasV1–2, HasN1–2, HasO1–3, and HasY1–2. Stereochemistries of Thr-2, 3, and 4 are putative. A, adenylation domain; C, condensation domain; Dhfa, C14 to C18 dihydroxy fatty acid; E, epimerisation domain; M, methylation domain; T, thiolation domain; Te, thioesterase domain.
Sequence comparison with database proteins suggests the involvement of four glycosyltransferases (HasD, HasQ, HasT, and HasX), which are predicted to catalyze the addition of sugars to hassallidin (Fig. 1 and SI Appendix, Table S1). The putative acyltransferase HasR in the gene cluster could have a role in acetylation reactions (SI Appendix, Table S1). Moreover, the predicted gene products of hasG, hasH, and hasL show similarity to enzymes involved in fatty acid synthesis. HasG exhibits conserved domains typical of the acyl-protein synthetase superfamily, including synthetase and ligase, whereas the deduced function for HasH is acyl carrier protein, proposing a role in transfer of the fatty acid of hassallidin. The putative tailoring enzyme HasL shows high similarity to 3-oxoacyl-(acyl-carrier-protein) reductase, also suggesting a role in lipid side chain biosynthesis (SI Appendix, Table S1).
Structural Elucidation of Hassallidins.
The genome assembly of Anabaena sp. 90 revealed that the putative hassallidin gene cluster contained a 526 bp deletion in the condensation domain of hasV (Fig. 1A) (16). This deletion would render the gene cluster inactive. No hassallidins were detected from Anabaena sp. 90. We screened 99 Anabaena strains (SI Appendix, Table S3) using PCR primers specific for hasV (SI Appendix, Table S4) to identify other strains that might produce hassallidins. The cell extracts from Anabaena cultures were analyzed by liquid chromatography/mass spectrometry (LC/MS). A group of ions between m/z 1,220 and 1,311 was deduced to represent different aglyconic lipopeptide variants from the known hassallidin A and B with an m/z of 1,220 (Fig. 2 and SI Appendix, Fig. S1). A series of ions with increased m/z values of 132, 162, and 203 and their sodium adducts were found together with ions representing aglyconic lipopeptide structures (SI Appendix, Fig. S2 A and B). A mass of 132 Da is equal to a pentose, and 162 Da is equivalent to a hexose residue. In addition, series of ions were detected with aglyconic lipopeptide plus 162, 204, 246, 288, and 330 mass units (SI Appendix, Fig. S2). The difference between these fragments of 42 Da equates with an acetyl group, indicating that the hexose sugar is decorated with 0–4 acetyl groups. Moreover, the mass difference of 203 Da between ions indicates an acetylated hexosamine residue (161 + 42). MS2 analysis showed that these monosaccharides together with the aglyconic lipopeptide form the native hassallidin (SI Appendix, Fig. S2 C and D). MS2 of the aglyconic lipopeptide variants indicate that the chain length of dihydroxy fatty acid1 and amino acid10 were the origins of variation (SI Appendix, Table S5). Fragmentation of the protonated aglyconic lipopeptide was conducted, revealing Thr, Tyr, dehydrobutyrine (Dhb), Gln, Gly, and N-MeThr residues that are in agreement with the hassallidin A and B peptide aglyconic lipopeptide structures (Fig. 3 and SI Appendix, Fig. S1 and Table S5). The acid hydrolysis of Anabaena hassallidin C and D for the determination of amino acid chirality confirmed the presence of l-Thr, d-Thr, d-allo-Thr, d-Tyr, d-Gln, Gly, N-MeThr, l-Gln (hassallidin C), and l-Tyr (hassallidin D). This amino acid content is fully compatible with the prediction given by the hassallidin gene cluster (Fig. 1 and SI Appendix, Table S2), with two exceptions. In position 10, there is either l-Gln or l-Tyr (Tyr from NMR data) instead of Lys. Second, the formation of d-Thr from l-Thr requires epimerization of both of its chiral centers, which is not explainable by the enzyme activities encoded in the hassallidin NRPS. We defined the stereochemistries of l-Thr2 and d-allo-Thr4 in Fig. 1 based on the location of epimerase in the HasN1 module, but still the stereochemistries of the three threonines in positions 2, 3, and 4 remain putative.
Fig. 2.
Total ion current and extracted ion chromatograms (TICCs and EICs) showing all of the hassallidins and their aglyconic core variants produced by a single strain of Anabaena sp. SYKE748A. The arrows indicate the retention times of different hassallidin variants, which are grouped into eight categories: A to H. Numbers 0–4 above the arrows represent the acetylation degree of the sugar (acetyl) hexose in hassallidin variants. The multiplier of the EIC signal is in parentheses.
Fig. 3.
The structure of hassallidins A–D. Dhfa, dihydroxy fatty acid, dihydroxy tetradecanoic acid in hassallidin A and B, and dihydroxy hexadecanoic acid (Dhh) in hassallidin C and D. Dhb, dehydrobutyric acid.
NMR analysis of Anabaena sp. SYKE748A hassallidin D confirmed the structures and sequence of subunits 1–10 and identified the mono- and disaccharides and their positions, mannose bound to 3-OH of N-MeThr9 and GlcNAc-(1→3)-pentopyranose bound to 3-OH of 2,3-dihydroxy-hexadecanoic acid1 (Dhh1) (Fig. 3 and SI Appendix, NMR Results, Figs. S3 and S4, and Table S6).
Anabaena Genus as a Producer of Diverse Structural Variants of Hassallidins.
Anabaena strains carrying the hassallidin gene cluster produced hassallidins with various modifications both within and between the strains. Anabaena sp. SYKE748A produces at least 40 different structural variants of hassallidin (Fig. 2). Structurally, all of the newly discovered hassallidins produced by Anabaena contain a peptide ring with eight amino acids, a side chain with one amino acid, 2,3-dihydroxy fatty acid, and two to three sugar moieties (Fig. 3 and Table 1). The amino acid backbone was otherwise invariable and comparable to hassallidins A and B (17, 18), with the exception of the amino acid in position 10 being either glutamine (hassallidin C) or tyrosine (hassallidin D) (Fig. 3). The length of the 2,3-dihydroxy fatty acid in position 1 varied, containing either 14, 15, 16, or 18 carbon atoms. Hassallidins from Anabaena possessed a previously unidentified pattern of glycosylation. The sugar variants of hassallidins discovered were disaccharide N-acetylhexosamine–pentose (M1–M2; 203 Da–132 Da), mono or dipentose (M1 and M2), deoxyhexose (M1 and/or M2; 146 Da), and acetylhexose possessing different degrees of acetylation with 0–4 acetyl groups attached (M1 and M3; 162, 204, 246, 288, or 330 Da) (Fig. 3 and Table 1). The main variants produced by Anabaena sp. SYKE748A and many other Anabaena strains were hassallidin C and hassallidin D (Fig. 3).
Table 1.
The structural variation of hassallidins in strains of cyanobacteria from Anabaena, Nostoc, Aphanizomenon, Tolypothrix, and Cylindrospermopsis genera
 |
A–M, 1,220, 1,234, 1,248, 1,276, 1,255, 1,269, 1,283, 1,311, 1,249, 1,232, 1,241, 1,282, and 1,298 m/z. Variation in aglyconic lipopeptides (A–M) can be explained by the hydrocarbon chain length (Cn, 14–18; U, unknown) of the dihydroxy fatty acid in position 1 and by the amino acid in position 10 (Aa10; Q, glutamine; X, unknown amino acid; Y, tyrosine). Monosaccharides M1, M2, and M3 of hassallidins are N-acetylhexosamine (purple triangle), pentose (green inverted triangle), deoxyhexose (blue diamond), and hexose (red circle), which contains 0–4 acetyl (Ac) groups. Monosaccharide positions have been deduced based on the identified positions in hassallidins A, B, and D. Black squares indicate presence. Strain numbers are as in Table 2. Detailed variation is shown in SI Appendix, Table S7.
We screened 99 Anabaena strains for the presence of hasV (SI Appendix, Table S3), and we amplified two other NRPS genes, hasO and hasN, from the Anabaena strains that contained hasV. We were able to identify the hassallidin genes from 23 Anabaena strains and to show that 20 of these strains also produce hassallidins (Table 2). No hassallidin production was detected in the remaining three strains despite the presence of the hassallidin genes. Among these three inactive strains is the present-day culture of Anabaena sp. 90 that, due to a 526-bp deletion in hasV, no longer produces hassallidin (16). Using PCR primers specific for hasV, we found that the deletion occurred between 2003 and 2006; Anabaena sp. 90 was isolated in 1986 (SI Appendix, Fig. S5). Using 10-y-old freeze-dried material, we were able to show that Anabaena sp. 90 produced hassallidins before the deletion event. In addition, we found that a culture of Anabaena sp. 90M3, an apdA mutant of Anabaena sp. 90 constructed in 1999 (13), produces hassallidins (SI Appendix, Fig. S5).
Table 2.
Detected (LC/MS) hassallidin production of cyanobacterial strains and results of PCR amplification of has NRPS genes indicating the presence of the has gene cluster
| No. | Strain code | Origin, location | Year | has NRPS genes | Hassallidins LC/MS |
| Anabaena sp. | |||||
| 1 | 90, y 2009 | Lake Vesijärvi, Finland | 1986 | ■ | □ |
| 2 | 90, y 1998 (freeze dried) | Lake Vesijärvi, Finland | 1986 | ■ | ■ |
| 3 | 90 M3 (apdA mutant) | Lake Vesijärvi, Finland | 1986 | ■ | ■ |
| 4 | 299A | Lake Vesijärvi, Finland | 1992 | ■ | □ |
| 5 | 299B | Lake Vesijärvi, Finland | 1992 | ■ | ■ |
| 6 | 258 | Lake Tuusulanjärvi, Finland | 1990 | ■ | ■ |
| 7 | SYKE748A | Lake Tuusulanjärvi, Finland | 1999 | ■ | ■ |
| 8 | SYKE763A | Lake Tuusulanjärvi, Finland | 1999 | ■ | ■ |
| 9 | 0TU33S16 | Lake Tuusulanjärvi, Finland | 2000 | ■ | ■ |
| 10 | 0TU43S8 | Lake Tuusulanjärvi, Finland | 2000 | ■ | ■ |
| 11 | 1TU33S8 | Lake Tuusulanjärvi, Finland | 2001 | ■ | ■ |
| 12 | 1TU35S12 | Lake Tuusulanjärvi, Finland | 2001 | ■ | ■ |
| 13 | 1TU44S9 | Lake Tuusulanjärvi, Finland | 2001 | ■ | ■ |
| 14 | 1TU44S16 | Lake Tuusulanjärvi, Finland | 2001 | ■ | ■ |
| 15 | SYKE971/6 | Lake Kotojärvi, Finland | 1999 | ■ | ■ |
| 16 | PH256 | Lake Knud, Denmark | 1994 | ■ | □ |
| 17 | NIVA-CYA 269/2 | Lake Frøylandsvatnet, Norway | 1990 | ■ | ■ |
| 18 | NIVA-CYA 269/6 | Lake Frøylandsvatnet, Norway | 1990 | ■ | ■ |
| 19 | XPORK5C | Porkkala Cape, the Baltic Sea coast, Finland | 1999 | ■ | ■ |
| 20 | XSPORK7B | Porkkala Cape, the Baltic Sea coast, Finland | 1999 | ■ | ■ |
| 21 | XSPORK36B | Porkkala Cape, the Baltic Sea coast, Finland | 1999 | ■ | ■ |
| 22 | XSPORK14D | Porkkala Cape, the Baltic Sea coast, Finland | 1999 | ■ | ■ |
| 23 | BECID19 | The Gulf of Finland, Vuosaari, Finland | 2001 | ■ | ■ |
| Cylindrospermopsis raciborscii | |||||
| 24 | ATC-9502 | Lake Balaton, Hungary | 1994 | ■ | ■ |
| 25 | CS-505 | Freshwater, Solomon Dam, Australia | 1996 | ■ | ■ |
| Aphanizomenon gracile | |||||
| 26 | Heaney/Camb 1986 140 1/1 | Freshwater, Lough Neagh, Ireland | 1986 | ■ | ■ |
| Nostoc sp. | |||||
| 27 | 159 | Lake Haukkajärvi, Finland | 1986 | ■ | ■ |
| 28 | 113.5 | Lichen associated | ? | ■ | ■ |
| Tolypothrix sp. | |||||
| 29 | PCC 9009 | Watkins Glen State Park, New York, United States | ? | ■ | ■ |
| 30 | PCC 7504 | Aquarium, Stockholm, Sweden | 1972 | ■ | □ |
| 31 | PCC 7101 | Borneo, soil | 1950 | ■ | □ |
Filled square, positive result; unfilled square, negative result. See also SI Appendix, Table S5.
Diversity of Cyanobacterial Hassallidin Producers.
Bioinformatic analysis of the Anabaena sp. SYKE748A hassallidin gene clusters strongly suggests that Cylindrospermopsis raciborskii CS-505 also encodes a cryptic hassallidin gene cluster, as it showed significant resemblance with 65–72% identity between HasN, HasO, HasV, and HasY and 55–84% identity between some of the additional ORFs (SI Appendix, Table S1). To test this possibility, we analyzed the cell extract from two C. raciborskii strains by LC/MS and discovered that C. raciborskii strains CS-505 and ATC 9502 both produce hassallidins (Tables 1 and 2 and SI Appendix, Table S7). The hassallidins produced by C. raciborskii strains contained a C18 dihydroxy fatty acid side chain and two sugars, a pentose (M2) and a hexose (M3). The amino acid in position 10 in strain CS-505 was threonine according to the MS2 analysis of the aglyconic lipopeptide unit (SI Appendix, Table S7) fitting to the predicted amino acid based on substrate specificity (SI Appendix, Table S2). This prompted us to screen other heterocyst-forming cyanobacterial genera for the ability to produce hassallidins. By screening 104 cyanobacterial strains (SI Appendix, Table S3) from different heterocyst-forming genera other than Anabaena for hasN and analyzing the cell extracts by LC/MS, we identified has genes and hassallidins from an Aphanizomenon strain, two Nostoc strains, and a Tolypothrix strain but only has genes from two other Tolypothrix strains (Table 2). However, there was less structural variation in hassallidins identified from strains of Cylindrospermopsis, Nostoc, Aphanizomenon, and Tolypothrix than Anabaena (Table 1 and SI Appendix, Table S7).
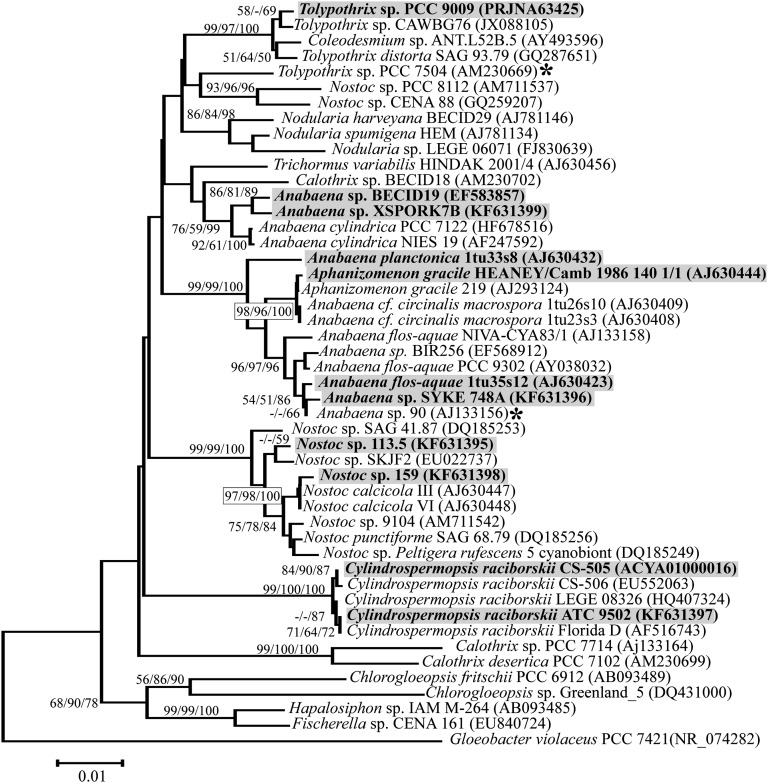
To find the phylogenetic relationship between hassallidin-producing strains, we constructed a 16S rRNA-based phylogenetic tree (Fig. 4). The producing strains were evenly distributed throughout the Nostocales order (subsection IV), comprising five separate clusters.
Fig. 4.
Evolutionary history of cyanobacteria-producing hassallidin based on 16S rRNA genes. Phylogenetic tree was inferred using MEGA 5, indicating in the nodes of the bootstrap values above 50% for maximum parsimony, maximum likelihood, and neighbor-joining bootstrap replicates. Strains detected to produce hassallidins are highlighted. An asterisk indicates two strains that have the hasN gene but no hassallidins were detected by LC/MS due to deletion in the hassallidin gene cluster in Anabaena sp. 90 and possible mutation in Tolypothrix sp. PCC 7504.
Antifungal Activity.
The minimum inhibitory concentration (MIC) of hassallidin D was ≤2.8 µg/mL (1.5 µM) against all Candida strains tested (Fig. 5). The half maximum inhibitory concentration IC50 was 0.55 (0.29), 1.53 (0.82), and 1.86 (1.00) µg/mL (µM) for Candida albicans HAMBI 261 (ATCC 11006), C. albicans HAMBI 484 (ATCC 10231), and Candida krusei HAMBI 486 (ATCC 6258), respectively (Fig. 5). Interestingly, the MIC of the linear form of hassallidin D was found to be 36 µg/mL (20 µM) against C. albicans HAMBI 261 (ATCC 11006). The disk diffusion bioassay using 10 µg of hassallidin D shows inhibition of Cryptococcus albidus HAMBI 264 (ATCC 10666) and Filobasidiella neoformans (ATCC 10226), respectively, with inhibition zones of 17 and 11 mm. No inhibition of Aspergillus strains by hassallidin D was observed. Antifungal activity of cell extracts containing 10 µg of hassallidin D was tested against C. albicans HAMBI 261 (ATCC 11006) using the disk diffusion method. The cell extracts of Anabaena strains 90M3, SYKE748A, XSPORK7B, XSPORK36B, Nostoc sp. 159, and C. raciborskii ATC 9502 inhibited the growth of C. albicans HAMBI 261 (ATCC 11006) (SI Appendix, Fig. S6). Nostoc sp. 159 extract had the largest inhibition zone of all of the strains tested (SI Appendix, Table S8). However, Anabaena sp. SYKE748A proved to produce the highest amount of hassallidins. Anabaena sp. 90, Tolypothrix sp. PCC 7415 and PCC 7504 strains, C. raciborskii CS-505, and Aphanizomenon gracile Heaney/Camb 1986 140 1/1 did not exhibit antifungal activity against C. albicans HAMBI 261 (ATCC 11006) with the tested amount of cell extract. The cell extracts of Anabaena sp. SYKE748A and XSPORK36B strains showed weak activity against Aspergillus flavus HAMBI 829, whereas extracts from other tested strains did not inhibit this fungus (SI Appendix, Fig. S7).
Fig. 5.
MIC and half maximum inhibitory concentration (IC50) of hassallidin D against C. albicans HAMBI 261 (ATCC 11006), C. albicans HAMBI 484 (ATCC 10231), and C. krusei HAMBI 486 (ATCC 6258).
Discussion
A multitude of previously unidentified hassallidin variants are produced among heterocystous cyanobacteria. They exhibit antifungal activity against important opportunistic pathogenic fungi. Particularly Anabaena strains proved to be a good source of hassallidins, with multiple different variants produced simultaneously (Fig. 2 and Table 1). The newly discovered hassallidin variants possessed unusually diverse structures distinct from the previously found hassallidins A and B (17, 18), due to the co-occurrence of variable lengths of the dihydroxy fatty acid chain (constituting 14, 15, 16, or 18 carbons) and complex glycosylation patterns with three variable monosaccharide moieties comprised of five optional monosaccharides: N-acetylhexosamine (M1), pentose (M1 and M2), hexose (M1 and M3), deoxyhexose (M1 or M2), or acetylhexose possessing different degrees of acetylation (M3) (Table 1). The dihydroxy fatty acid side chain of hassallidins A and B contains 14 carbons decorated with mannose (17), and hassallidin B also has an additional rhamnose (18). The peptide backbone of the newly discovered hassallidins is identical to hassallidins A and B, except that the amino acid in position 10 was either glutamine (hassallidin C), tyrosine (hassallidin D), or in one case threonine (Fig. 3 and Table 1). Dihydroxy fatty acid side chain compounds such as cormycin A with antifugal bioactivity has been previously reported from Pseudomonas corrugata, but the family of those compounds does not contain any sugars (19). In a majority of glycosylated natural products, the sugar moieties belong to the 6-deoxyhexose family (20). The most exceptional feature of hassallidin produced by Anabaena is the presence of an unusual acetylhexose with an acetylation degree ranging from 0 to 4. Acetylated sugar structures have been previously found only in a few natural products (21–23) and never in natural products produced by cyanobacteria to date, and overall they are rare among secondary metabolites produced by microbes.
All tested Anabaena strains inhibited the growth of C. albicans, with the exception of Anabaena sp. 90 wild type, which, due to a deletion, no longer produces hassallidins (16). A few natural products produced by cyanobacteria have been previously described to exhibit antifungal activity—for example, cryptophycin (24, 25), tanikolide (26), and nostofungicidine (27). These compounds and hassallidins share a lactone structure and a long hydrocarbon side chain, 5-hydroxy-5-hydroxymethylpalmitic acid in tanikolide, 3-amino-6-hydroxystearic acid in nostofungicidine, and 2,3-dihydroxymyristic, 2,3-dihydroxypalmitic or 2,3-dihydroxystearic acid in hassallidins. Both could be essential elements for the antifungal activity. The MIC value for hassallidin D isolated from Anabaena sp. SYKE748A against C. albicans and C. krusei was measured to be ≤2.8 µg/mL (1.5 µM). Previously, MIC values of 4.8 µg/mL for hassallidins A and B against C. albicans have been reported (28). Interestingly, the linear form of hassallidin D has an MIC of ≤36 µg/mL (20 µM), showing the importance of ring structure for bioactivity. Nevertheless, this result shows that hassallidin is also slightly active in its linear form. Many other bioactive compounds lose their bioactivity when the ring structure is opened (e.g., refs. 29, 30). The structural differences between the hassallidin variants produced might influence their bioactivity. For example, the sugar moieties of many glycosylated compounds play a significant or even fundamental role in their bioactivity (31). The number of sugars has also been reported to influence the activity. The antitumor activity of landomycin increases with the length of the deoxysugar chain, although its mode of action is yet not fully understood (32). On the other hand, the additional sugar group of hassallidin B was not reported to have an influence on its antifungal activity, although no aglycon form was examined (28). The function of the acetyl groups of acetylhexose in hassallidin is unknown, but for antitumor compound chromomycin A3, the acetylation of sugars was essential for its biological activity (31). Previous study indicates that hassallidin A affects the integrity of the plasma membrane (33). This mechanism of action differs from the inhibition of the cell wall synthesis by echinocandins, as caspofungin (33). The incidence of invasive fungal infections in humans has been increasing, and the need for new selective antifungals is growing due to the toxicity, low efficacy, and drug resistance of many currently used drugs (34).
The structural characterization, genetic organization, and predicted substrate specificities of adenylation domains attest to the involvement of the gene cluster from the draft genome of cyanobacterium Anabaena sp. SYKE748A in hassallidin biosynthesis (Fig. 1). We verified this by the discovery of the natural mutation during the genome sequencing process and loss of hassallidin production by the Anabaena sp. 90 (Fig. 1 and Table 2) (16). The complex structure of hassallidins indicates that in addition to the four key NRPS proteins, several putative tailoring enzymes are required to complete the biosynthesis. Four proteins—HasD, HasQ, HasT, and HasX—exhibit conserved motifs toward glycosyltransferase with fairly high similarities (SI Appendix, Table S1) and are proposed to be involved in the glycosylation of hassallidins. In most known cases, the number of glycosyltransferases corresponds to the number of sugars attached (35). Some glycosyltransferases are described to work iteratively, catalyzing the attachment of more than one sugar to the aglycon (36). However, the has gene cluster seems to encode one additional glycosyltransferase, whose function remains unclear. In addition, a predicted acyltransferase, HasR, was found to be encoded in the has gene cluster. Acyl/acetyltransferases have been shown to be responsible for the acetylation of the sugar moieties in an antifungal glycolipid flocculosin (23). An antitumor polyketide chromomycin A3 contains two O-acetylated sugars, but only one acyltransferase is found in the biosynthetic gene cluster, which is proposed to catalyze both sugar O-acetylation reactions (21). Thus, it is possible that HasR could be a candidate for the transfer of one or more acetyl groups required to decorate the hassallidin sugars. The acylation of hassallidins most likely includes the action of the additional C domain in the first HasV1 module. The atypical N-terminal C domain has been reported in lipopeptide biosynthetic gene clusters (37), and thus we postulate that it is catalyzing the N-acylation of the first threonine in the peptide chain and represents the first step in the assembly of hassallidin. Based on the sequence similarities, lipidation might involve the action of proteins HasG, HasH, and HasL to work in concert with the first C domain. How Dhb is enzymatically formed from threonine is generally unknown. However, this modification has been detected in cyanobacteria in nodularin and microcystin biosyntheses (38, 39).
C. raciborskii has previously been reported to produce only two different types of toxins: tricyclic alkaloidic hepatotoxin cylindrospermopsin and neurotoxic tetrahydropurine saxitoxin (40, 41). The genome of C. raciborskii CS-505 contains a cryptic NRPS gene cluster in addition to the NRPS/PKS hybrid gene cluster responsible for the production of cylindrospermopsin (41, 42). The significant similarity between the amino acid sequences of the has gene cluster and the genes in the genome of C. raciborskii CS-505 (SI Appendix, Table S1) led us to discover that C. raciborskii produces hassallidins. This has gene cluster (SI Appendix, Table S1) carries four NRPS genes that code for nine modules, which is in accordance with the number of amino acids in hassallidin. Cylindrospermopsin-producing C. raciborskii has been reported to be the cause of the outbreak of hepatoenteritis in 1979, in Palm Island, Australia, where 148 people were poisoned due to a contamination of a drinking water reservoir (43–45). Interestingly, when tested in mice, pure cylindrospermopsin damaged only the liver, but the cylindrospermopsin-containing cell extract caused injuries also to the kidneys, adrenal glands, lungs, and intestines. It has been suggested that the cell lysates might have contained an unknown compound in addition to cylindrospermopsin (45). Considering that hassallidins have also shown cytotoxicity against eukaryotic human acute T-cell leukemia with IC50 0.2 µg/mL (28), they may have contributed to the additional symptoms.
In addition to Anabaena and Cylindrospermopsis, we discovered that hassallidin production ability is even further dispersed among cyanobacteria. Some of the strains from genera Nostoc, Aphanizomenon, and Tolypothrix also produced hassallidins (Table 2). However, no hassallidin production of two other strains of Tolypothrix (PCC 7101 and PCC 7504) containing hasN was detected. The phylogenetic tree showed that the hassallidin-producing strains fell into five separate clusters (Fig. 4). Moreover, the strains producing hassallidins are widely dispersed to various different habitats, including freshwater, brackish water, and lichen association, in distant geographic areas (Table 2 and SI Appendix, Table S3). Little is known about the importance of secondary metabolites for the producer strains, but they might be a way to combat rival organisms or parasites. It is known that some genera of cyanobacteria are susceptible to infections caused by parasitic chytrid fungi (46, 47). It has been shown that oligopeptides produced by Planktothrix cyanobacteria disturb the infection caused by the fungi Rhizophydiales (46) and the wild type harboring an entire oligopeptide repertoire is more resistant to fungal infection than the oligopeptide-minus mutants (47). It has also been reported that the presence of Anabaena spiroides in the water reduces the number of Hyphomycetes fungi (48). Due to their antifungal activity, it is possible that hassallidins could be a means of defense against fungal infections or play a role as an allelochemical. Although hassallidins show promising antifungal activity, little is known about their effect on other organisms.
Materials and Methods
Cyanobacterial Cultures.
Strains of Anabaena, Nostoc, Cylindrospermopsis, Tolypothrix, and Aphanizomenon were grown in Z8 medium either with or without nitrogen (49). Strains were cultivated in 40 mL culture at 17–25 °C under continuous illumination of 3–15 µmol⋅m−2⋅s1. Anabaena sp. SYKE748A was mass cultivated in 5 × 2.7 L batches with aeration of filter-sterilized pressurized air. After cultivation, the clear medium was decanted away, and the rest of the culture was freeze dried.
Sequencing of the Hassallidin Gene Cluster.
DNA extraction from the Anabaena sp. SYKE748A strain was made using the phenol-chloroform method (15, 16). Ultra high-coverage sequence data were produced by Illumina MiSeq. The genome was assembled using Newbler (version 2.8), after adapter removal and short read filtering. The hassallidin gene cluster was located on three contigs, and the gaps were closed by PCR and Sanger sequencing to produce the final intact 59-kb gene cluster.
Identification of the Hassallidin Gene Cluster.
The gene cluster from strain SYKE748A was annotated according to that in Anabaena sp. 90 (16), due to their extremely high sequence similarities (99.8%). The putative roles of the has gene cluster encoded proteins were assigned by sequence similarity using the National Center for Biotechnology Information (NCBI) BLASTp and InterProScan Sequence searches (48). An in silico approach was used for prediction of domain organization (50). The substrate specificity conferring code of adenylation domains in NRPSs (51) was performed using NRPSpredictor software (52, 53). Identification of the hassallidin gene cluster in C. raciborskii was based on the genome of strain CS-505 (41) (NCBI database accession no. NZ_ACYA01000025, www.ncbi.nlm.nih.gov/nuccore/NZ_ACYA01000025).
DNA Isolation, PCR Amplification, and Sequencing.
Genomic DNA from the cyanobacterial strains was extracted using either DNeasy Plant Mini Kit (Qiagen Gmbh) or E.Z.N.A SP Plant DNA Mini Kit (Omega Bio-Tek Inc.). PCR amplifications of DNA from Anabaena were performed in iCycler (Bio-Rad) using primers hasV-fw and hasV-rev, hasN-fw and hasN-rev, and hasO-fw and hasO-rev, designed to amplify parts from hasV, hasN, and hasO (SI Appendix, Table S4). PCR conditions are described in SI Appendix. The PCR products were sequenced by Macrogen, Inc.
Primers AF and AR (SI Appendix, Table S4) were used in PCR to screen cyanobacteria from other genera for the gene hasN. PCR conditions are described in SI Appendix. PCR products were cloned into the pCR2.1-TOPO cloning vector (TOPO-TA Cloning-Kit, Invitrogen) and transformed to TOP10 Escherichia coli cells (Invitrogen). Colonies were tested by PCR, and plasmids containing inserts of the right size were purified using QiaprepSpin Miniprep Kit (Qiagen). The sequencing was performed by ABI PRISM BigDye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems) and analyzed on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems). Internal primers (1194Rc, 16S545F, 16S979F, 16S1092R, and 359F; SI Appendix, Table S4) were used to sequence 16S rRNA gene fragments. The contigs were assembled using Phred/Phrap/Consed (Philip Green), considering bases with quality ≥20. The 16S rRNA phylogenetic tree was constructed in Molecular Evolutionary Genetic Analysis (MEGA) 5 (54) using maximum likelihood (K2+G+I model), neighbor-joining (MCL+G model), and maximum parsimony (closest neighbor interchange on random trees) methods. The 16S rRNA partial gene sequences data have been submitted to the GenBank database.
Extraction, Chemical Analyses, Purification, and Amino Acid Analysis of Hassallidin D.
Extraction and chemical analyses were performed as described in ref. 55 with the following exceptions: Luna C18 (2) column (100 × 4.6 mm) was eluted with a 30 min gradient from 10% to 100% of acetonitrile in 0.1% aqueous formic acid. The LC/MS parameters were optimized with hassallidin ion m/z 1,862 with a negative mode of polarity as follows: capillary voltage, 3,650 V; capillary offset value, 250 V; skimmer potential, 58.0 V; trap drive value, 112.0. Commercial hassallidins A and B (Alexis Biochemicals, Ezno Life Science Inc.) were used as references.
For bioactivity tests, biomass of Anabaena sp. SYKE748A (2.7 g) was divided into two 50 mL plastic tubes. We added 2 × 35 mL of acetonitrile/dimethyl sulfoxide (1:1) solution, and suspensions were treated for 30 s at 18,000 rpm with a Silent Crusher homogenizer at an ambient temperature. Suspensions were centrifugated at 10,000 × g for 5 min, and supernatants were pooled. Acetonitrile was evaporated with a rotary evaporator at 20 °C, and the residue was diluted 20 times with water. The solution was divided into four and then passed through four primed (first methanol, then 5% (vol/vol) aqueous methanol) solid phase extraction cartridges (Strata C18-E, 5 g/20 mL, 55 μm, 70 Å; Phenomenex Inc.). Cartridges were eluted with 50 mL of 90% (vol/vol) aqueous acetonitrile, which was then removed with a rotary evaporator from the effluent. The residue was freeze dried and dissolved to HPLC eluent. Solution was injected into a semipreparative Luna C8 (2) column (10 × 150 mm, 5 μm, 100 Å; Phenomenex Inc.) in 1–2 mL batches. The column was eluted isocratically with acetonitrile/0.1% ammoniumacetate (40/60) at an ambient temperature. Acetonitrile was removed with a nitrogen stream from the collected hassallidin fractions, and the residue was freeze dried. Purification yielded 2.4 mg of native hassallidin D for testing the antifungal activity. Hassallidin D purification for the NMR and bioassay (linear hassallidin D) is described in SI Appendix. Amino acid analysis of hassallidin C and D was made using the Marfey method as previously described in ref. 56.
NMR Spectroscopy.
All NMR spectra were collected using a Varian Unity INOVA 600 MHz NMR spectrometer at 40 °C in DMSO-d6. Detailed analysis is presented in SI Appendix.
Antifungal Assay.
Microdilution assay was used to investigate the hassallidin D MIC and the half maximum inhibitory concentration (IC50) of C. albicans and C. krusei. A stock solution of hassallidin D with a concentration of 2.24 mg/mL in DMSO was used for a serial dilution assay. Twofold dilutions from the stock solution in DMSO were prepared, and this serial dilution ranged from 2.24 to 0.0044 mg/mL. The final concentration in the well was from 22.4 to 0.04 µg/mL. Two microliters of each dilution containing hassallidin D in DMSO was used per well containing 98 µL of RPMI-1640 medium, in triplicates (R6504, with l-glutamine and without NaHCO3; Sigma). Two microliters of DMSO was used as the negative control. Potato dextrose agar medium plates containing C. albicans HAMBI 261 (ATCC 11006), C. albicans HAMBI 484 (ATCC 10231), and C. krusei HAMBI 486 (ATCC 6258) grown at 35 °C for 24 h were used to prepare the inoculum. The test inoculum was prepared as described in the guidelines of the Clinical and Laboratory Standards Institute document M27-A3 (57). We added 100 µL of the test inoculum per well, allowing a final concentration of 0.5 × 103 to 2.5 × 103 cells/mL. The MIC value was the lowest concentration in which no growth of the organism was observed. The IC50 was calculated by linear interpolation of the concentrations using the following formula: IC50 = (50% – LowI%)/(HighI% – LowI%) × (HighC – LowC) + LowC, in which C is concentration and I% is inhibition percentage calculated with the formula I% = 100 – [(ODsample – ODmedium)/(ODcontrol – ODmedium)] × 100. LowI% is the inhibition percentage correspondent to the value directly below the MIC, and HighI% is the one correspondent to the MIC. In addition, hassallidin D was found to linearize during the storage, and this form was used in a microdilution assay. The assay was performed as described in ref. 28. Twofold dilutions with a hassallidin concentration between 4.6 and 38 µg/mL were dissolved in 50% (vol/vol) aqueous methanol. Negative controls containing 50% methanol and medium alone and positive controls containing 10 µg of nystatin were tested. The MIC was considered the minimum concentration of hassallidin that strongly inhibited the growth of C. albicans HAMBI 261 (ATCC 11006). The bioactivity assay performed using crude extract of different tested strains are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Lyudmila Saari for maintaining and handling the cyanobacteria strains and the anonymous reviewers and the editor for their constructive feedback and help. This work was supported by Grants 118637 and 258827 from the Academy of Finland (to K.S.). T.K.S. was partially funded by the Helsinki Graduate Program in Biotechnology and Molecular Biology, the São Paulo Research Foundation (2009/13455-0), the Centre for International Mobility (TM-09-6506), and the Finnish Cultural Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. KJ502174, KF631395, KF631396, KF631397, KF631398, and KF631399.).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320913111/-/DCSupplemental.
References
- 1.Sivonen K. In: The Encyclopedia of Microbiology. 3rd Ed. Schaechter M, editor. Oxford: Academic; 2009. pp. 290–307. [Google Scholar]
- 2.Burja AM, Banaigs B, Abou-Mansour E, Burgess JG, Wright PC. Marine cyanobacteria—Profilic source of natural products. Tetrahedron. 2001;57(46):9347–9377. [Google Scholar]
- 3.Sivonen K, Börner T. In: The Cyanobacteria: Molecular Biology, Genetics and Evolution. Herrero A, Flores E, editors. Norfolk, UK: Academic; 2008. pp. 159–198. [Google Scholar]
- 4.Welker M, von Döhren H. Cyanobacterial peptides—Nature’s own combinatorial biosynthesis. FEMS Microbiol Rev. 2006;30(4):530–563. doi: 10.1111/j.1574-6976.2006.00022.x. [DOI] [PubMed] [Google Scholar]
- 5.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides1. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 6.Sieber SA, Marahiel MA. Molecular mechanisms underlying nonribosomal peptide synthesis: Approaches to new antibiotics. Chem Rev. 2005;105(2):715–738. doi: 10.1021/cr0301191. [DOI] [PubMed] [Google Scholar]
- 7.Marahiel MA, Stachelhaus T, Mootz HD. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97(7):2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 8.Schwarzer D, Finking R, Marahiel MA. Nonribosomal peptides: From genes to products. Nat Prod Rep. 2003;20(3):275–287. doi: 10.1039/b111145k. [DOI] [PubMed] [Google Scholar]
- 9.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: Logic, machinery, and mechanisms. Chem Rev. 2006;106(8):3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 10.Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7(3):211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 11.Walsh CT, Fischbach MA. Natural products version 2.0: Connecting genes to molecules. J Am Chem Soc. 2010;132(8):2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shih PM, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA. 2013;110(3):1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouhiainen L, et al. Genes encoding synthetases of cyclic depsipeptides, anabaenopeptilides, in Anabaena strain 90. Mol Microbiol. 2000;37(1):156–167. doi: 10.1046/j.1365-2958.2000.01982.x. [DOI] [PubMed] [Google Scholar]
- 14.Rouhiainen L, et al. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl Environ Microbiol. 2004;70(2):686–92. doi: 10.1128/AEM.70.2.686-692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouhiainen L, Jokela J, Fewer DP, Urmann M, Sivonen K. Two alternative starter modules for the non-ribosomal biosynthesis of specific anabaenopeptin variants in Anabaena (Cyanobacteria) Chem Biol. 2010;17(3):265–273. doi: 10.1016/j.chembiol.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, et al. Genome-derived insights into the biology of the hepatotoxic bloom-forming cyanobacterium Anabaena sp. strain 90. BMC Genomics. 2012;13:613. doi: 10.1186/1471-2164-13-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuhof T, et al. Hassallidin A, a glycosylated lipopeptide with antifungal activity from the cyanobacterium Hassallia sp. J Nat Prod. 2005;68(5):695–700. doi: 10.1021/np049671r. [DOI] [PubMed] [Google Scholar]
- 18.Neuhof T, Schmieder P, Seibold M, Preussel K, von Döhren H. Hassallidin B—Second antifungal member of the Hassallidin family. Bioorg Med Chem Lett. 2006;16(16):4220–4222. doi: 10.1016/j.bmcl.2006.05.094. [DOI] [PubMed] [Google Scholar]
- 19.Scaloni A, et al. Structure, conformation and biological activity of a novel lipodepsipeptide from Pseudomonas corrugata: Cormycin A. Biochem J. 2004;384(Pt 1):25–36. doi: 10.1042/BJ20040422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salas JA, Méndez C. Engineering the glycosylation of natural products in actinomycetes. Trends Microbiol. 2007;15(5):219–232. doi: 10.1016/j.tim.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Menéndez N, et al. Biosynthesis of the antitumor chromomycin A3 in Streptomyces griseus: Analysis of the gene cluster and rational design of novel chromomycin analogs. Chem Biol. 2004;11(1):21–32. doi: 10.1016/j.chembiol.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Teichmann B, Linne U, Hewald S, Marahiel MA, Bölker M. A biosynthetic gene cluster for a secreted cellobiose lipid with antifungal activity from Ustilago maydis. Mol Microbiol. 2007;66(2):525–533. doi: 10.1111/j.1365-2958.2007.05941.x. [DOI] [PubMed] [Google Scholar]
- 23.Teichmann B, et al. Identification of a biosynthesis gene cluster for flocculosin a cellobiose lipid produced by the biocontrol agent Pseudozyma flocculosa. Mol Microbiol. 2011;79(6):1483–1495. doi: 10.1111/j.1365-2958.2010.07533.x. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz RE, et al. Pharmaceuticals from cultured algae. J Ind Microbiol. 1990;5(2–3):113–124. [Google Scholar]
- 25.Edelman MJ, et al. Phase 2 study of cryptophycin 52 (LY355703) in patients previously treated with platinum based chemotherapy for advanced non-small cell lung cancer. Lung Cancer. 2003;39(2):197–199. doi: 10.1016/s0169-5002(02)00511-1. [DOI] [PubMed] [Google Scholar]
- 26.Singh IP, Milligan KE, Gerwick WH. Tanikolide, a toxic and antifungal lactone from the marine cyanobacterium Lyngbya majuscula. J Nat Prod. 1999;62(9):1333–1335. doi: 10.1021/np990162c. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi A, Kajiyama S-I. Secondary metabolites of cyanobacteria Nostoc sp. Chin J Oceanology Limnol. 1998;16(Suppl 1):109–117. [Google Scholar]
- 28.Neuhof T, et al. 2006. Lipopeptides having pharmaceutical activity. Patent analysis, 1698638 EP.
- 29.Choi BW, et al. Isolation of linear peptides related to the hepatotoxins nodularin and microcystins. Tetrahedron Lett. 1993;34(49):7881–7884. [Google Scholar]
- 30.Herfindal L, et al. Nostocyclopeptide-M1: A potent, nontoxic inhibitor of the hepatocyte drug transporters OATP1B3 and OATP1B1. Mol Pharm. 2011;8(2):360–367. doi: 10.1021/mp1002224. [DOI] [PubMed] [Google Scholar]
- 31.Menéndez N, et al. Tailoring modification of deoxysugars during biosynthesis of the antitumour drug chromomycin A by Streptomyces griseus ssp. griseus. Mol Microbiol. 2004;53(3):903–915. doi: 10.1111/j.1365-2958.2004.04166.x. [DOI] [PubMed] [Google Scholar]
- 32.Ostash B, et al. Generation of new landomycins by combinatorial biosynthetic manipulation of the LndGT4 gene of the landomycin E cluster in S. globisporus. Chem Biol. 2004;11(4):547–555. doi: 10.1016/j.chembiol.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Neuhof T, et al. Comparison of susceptibility and transcription profile of the new antifungal hassallidin A with caspofungin. Biochem Biophys Res Commun. 2006;349(2):740–749. doi: 10.1016/j.bbrc.2006.08.110. [DOI] [PubMed] [Google Scholar]
- 34.Vicente MF, Basilio A, Cabello A, Peláez F. Microbial natural products as a source of antifungals. Clin Microbiol Infect. 2003;9(1):15–32. doi: 10.1046/j.1469-0691.2003.00489.x. [DOI] [PubMed] [Google Scholar]
- 35.Luzhetskyy A, Bechthold A. Features and applications of bacterial glycosyltransferases: Current state and prospects. Appl Microbiol Biotechnol. 2008;80(6):945–952. doi: 10.1007/s00253-008-1672-2. [DOI] [PubMed] [Google Scholar]
- 36.Luzhetskyy A, et al. Iteratively acting glycosyltransferases involved in the hexasaccharide biosynthesis of landomycin A. Chem Biol. 2005;12(7):725–729. doi: 10.1016/j.chembiol.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol Biol. 2007;7:78. doi: 10.1186/1471-2148-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffitt MC, Neilan BA. Characterization of the nodularin synthetase gene cluster and proposed theory of the evolution of cyanobacterial hepatotoxins. Appl Environ Microbiol. 2004;70(11):6353–6362. doi: 10.1128/AEM.70.11.6353-6362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sano T, Beattie KA, Codd GA, Kaya K. Two (Z)-dehydrobutyrine-containing microcystins from a hepatotoxic bloom of Oscillatoria agardhii from Soulseat Loch, Scotland. J Nat Prod. 1998;61(6):851–853. doi: 10.1021/np980047m. [DOI] [PubMed] [Google Scholar]
- 40.Lagos N, et al. The first evidence of paralytic shellfish toxins in the fresh water cyanobacterium Cylindrospermopsis raciborskii, isolated from Brazil. Toxicon. 1999;37(10):1359–1373. doi: 10.1016/s0041-0101(99)00080-x. [DOI] [PubMed] [Google Scholar]
- 41.Stucken K, et al. The smallest known genomes of multicellular and toxic cyanobacteria: Comparison, minimal gene sets for linked traits and the evolutionary implications. PLoS ONE. 2010;5(2):e9235. doi: 10.1371/journal.pone.0009235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mihali TK, Kellmann R, Muenchhoff J, Barrow KD, Neilan BA. Characterization of the gene cluster responsible for cylindrospermopsin biosynthesis. Appl Environ Microbiol. 2008;74(3):716–722. doi: 10.1128/AEM.01988-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawkins PR, Runnegar MT, Jackson AR, Falconer IR. Severe hepatotoxicity caused by the tropical cyanobacterium (blue-green alga) Cylindrospermopsis raciborskii (Woloszynska) Seenaya and Subba Raju isolated from a domestic water supply reservoir. Appl Environ Microbiol. 1985;50(5):1292–1295. doi: 10.1128/aem.50.5.1292-1295.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtani I, Moore RE, Runnegar MTC. Cylindrospermopsin: A potent hepatotoxin from the blue-green alga Cylindrospermopsis raciborskii. J Am Chem Soc. 1992;114(20):7941–7942. [Google Scholar]
- 45.Griffiths DJ, Saker ML. The Palm Island mystery disease 20 years on: A review of research on the cyanotoxin cylindrospermopsin. Environ Toxicol. 2003;18(2):78–93. doi: 10.1002/tox.10103. [DOI] [PubMed] [Google Scholar]
- 46.Sønstebø JH, Rohrlack T. Possible implications of chytrid parasitism for population subdivision in freshwater cyanobacteria of the genus Planktothrix. Appl Environ Microbiol. 2011;77(4):1344–1351. doi: 10.1128/AEM.02153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohrlack T, Christiansen G, Kurmayer R. Putative antiparasite defensive system involving ribosomal and nonribosomal oligopeptides in cyanobacteria of the genus Planktothrix. Appl Environ Microbiol. 2013;79(8):2642–2647. doi: 10.1128/AEM.03499-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Czeczuga B, Orlowska M. Investigations on the joint occurrence of Anabaena spiroides Klebahn and hyphomycetes in various types of water bodies. Acta Hydrochim Hydrobiol. 2000;28(3):162–165. [Google Scholar]
- 49.Kótai J. 1972. Instructions for Preparation of Modified Nutrient Solution Z8 for Algae, Blindern B–11/69 (Norwegian Institute for Water Research, Oslo)
- 50.Zdobnov EM, Apweiler R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 51.Bachmann BO, Ravel J. Chapter 8. Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. Methods Enzymol. 2009;458:181–217. doi: 10.1016/S0076-6879(09)04808-3. [DOI] [PubMed] [Google Scholar]
- 52.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6(8):493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 53.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33(18):5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leikoski N, et al. Analysis of an inactive cyanobactin biosynthetic gene cluster leads to discovery of new natural products from strains of the genus Microcystis. PLoS ONE. 2012;7(8):e43002. doi: 10.1371/journal.pone.0043002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leikoski N, et al. Genome mining expands the chemical diversity of the cyanobactin family to include highly modified linear peptides. Chem Biol. 2013;20(8):1033–1043. doi: 10.1016/j.chembiol.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 57.Clinical and Laboratory Standards Institute . Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard. 3rd Ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2008. pp. 1–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.