Significance
Filopodia act as organelles for sensing and exploring the environment, as well as producing traction forces during cellular locomotion. Myosin-10 is a molecular motor crucial for intrafilopodial trafficking and filopodia formation. To decipher how myosin-10 generates force and movement, we used electron microscopy and a combination of ensemble biochemical and single molecule mechanical techniques to help elucidate its structure and mechano-chemical coupling. Our results clarify current controversies about myosin-10 structure and function by revealing that it generates an unexpectedly large biphasic power stroke and moves processively along actin, but detaches rapidly at relatively low force. These adaptations may be advantageous features for a myosin motor that carries bulky cargo within the narrow confines of the filopodium.
Keywords: actomyosin, stable single alpha-helix, optical trapping, myosin X, myosin-5a
Abstract
Myosin-10 is an actin-based molecular motor that participates in essential intracellular processes such as filopodia formation/extension, phagocytosis, cell migration, and mitotic spindle maintenance. To study this motor protein’s mechano-chemical properties, we used a recombinant, truncated form of myosin-10 consisting of the first 936 amino acids, followed by a GCN4 leucine zipper motif, to force dimerization. Negative-stain electron microscopy reveals that the majority of molecules are dimeric with a head-to-head contour distance of ∼50 nm. In vitro motility assays show that myosin-10 moves actin filaments smoothly with a velocity of ∼310 nm/s. Steady-state and transient kinetic analysis of the ATPase cycle shows that the ADP release rate (∼13 s−1) is similar to the maximum ATPase activity (∼12–14 s−1) and therefore contributes to rate limitation of the enzymatic cycle. Single molecule optical tweezers experiments show that under intermediate load (∼0.5 pN), myosin-10 interacts intermittently with actin and produces a power stroke of ∼17 nm, composed of an initial 15-nm and subsequent 2-nm movement. At low optical trap loads, we observed staircase-like processive movements of myosin-10 interacting with the actin filament, consisting of up to six ∼35-nm steps per binding interaction. We discuss the implications of this load-dependent processivity of myosin-10 as a filopodial transport motor.
Myosins are a superfamily of actin-based molecular motors comprising ∼35 distinct classes (1, 2), responsible for a wide variety of cellular functions. They convert the chemical energy derived from ATP hydrolysis into directed motion along actin and are responsible for a wide variety of intracellular motilities and for muscle contraction. Mammalian myosin-10 was originally found to localize to the tips of filopodia (3, 4) and is known to be important for filopodial formation and extension (4–6), phagocytosis (7), cell migration (8), and mitotic spindle length maintenance and anchoring (9). It is an ∼240-kDa protein consisting of an N-terminal, consensus motor region (which binds actin, hydrolyses ATP, and produces force and movement), a light chain binding domain (LCBD) with three IQ motifs (10), and a stable single alpha-helix (SAH) followed by a tail region containing, an antiparallel coiled-coil (APCC) region, a proline, glutamate, serine, and threonine rich (PEST) domain, three pleckstrin homology (PH) domains, a myosin tail homology 4 (MyTH4) domain, and a band 4.1, ezrin, radixin, moesin (FERM) domain (Fig. 1A). Recently, Umeki et al. (11) expressed native full-length myosin-10 in an Sf9/baculovirus expression system, and electron micrographs of this protein indicate that most molecules are single-headed monomers and that the tail region can fold back onto the head, switching off the ATPase and actin-binding activity. ATPase activity was found to be regulated both by Ca2+ and phospholipids. Knight et al. (12) also found that a heavy meromyosin (HMM)-like fragment terminating at residue 953, which would include the SAH domain and the APCC region, was mostly monomeric, with only about 10% of the molecules clearly dimerized.
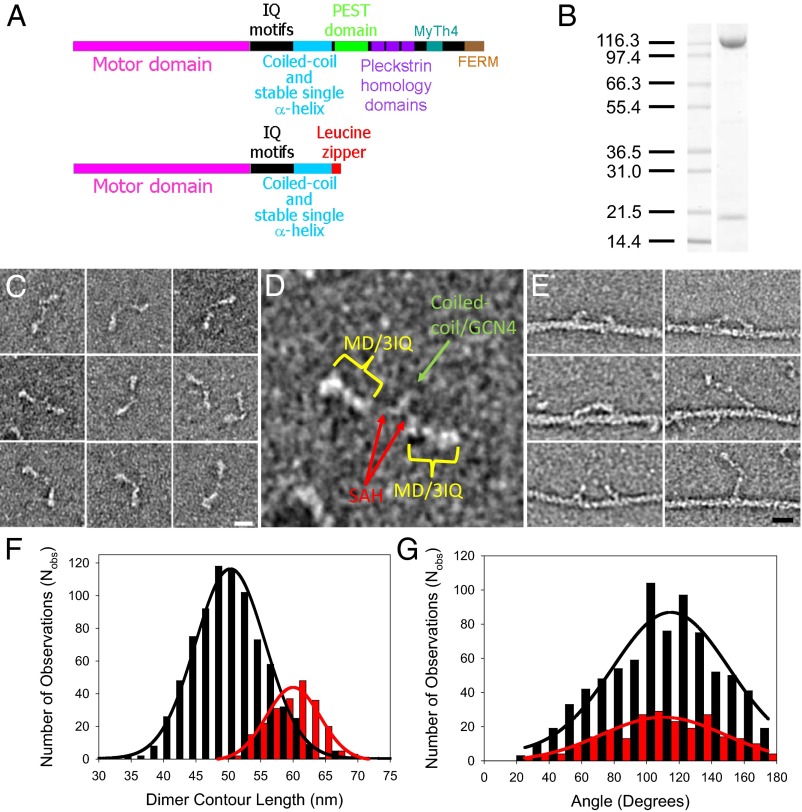
Fig. 1.
Myosin-10 heavy meromyosin-like (M10HMM) construct design. (A) Illustration of the full-length myosin-10 (Upper) and the M10HMM construct (Lower) structural organization. (B) Image of a typical SDS-polyacrylamide gel electrophoretogram (4–20%) of M10HMM after elution from an anti-FLAG resin. Approximately 0.4–0.8 mg of protein was purified per preparation. (C) Collage of M10HMM dimers negatively stained with 1% uranyl acetate (Each window = 92 × 92 nm). From electron micrographs it was determined that 87.2% of heavy chains were dimerized (Nobservations = 1,089). (Scale bar for all panels, 20 nm.) (D) An example M10HMM image from a panel from C with identification of regions, including motor domain (MD)/3 IQ motifs, the proximal coiled-coil region, and regions that most likely represents the stable SAH domains. Window size = 92 × 92 nm. (E) Images of M10HMM bound to actin in the presence of 1 μM ATP. Both single-headed binding and double-headed binding are seen. Examples can be seen in which the two heads of M10HMM span the actin pseudorepeat. (Scale bar for all panels, 20 nm.) (F) Histogram showing the contour length of M10HMM dimers (black bins), measured from the tip of one motor domain to the tip of the other [black line = Gaussian fit; peak = 51 ± 5.5 nm (SD); Nobs = 795; R2 = 0.99]. As a comparison, the contour length of myosin-5a-HMM (M5aHMM) (red bins) are shown [red line = Gaussian fit; peak = 60 ± 3.8 nm (SD); Nobs= 221; R2 = 0.96]. (G) Histogram showing the distribution of head-head angular variation of M10HMM (black bins; Nobs = 782) and M5aHMM (red bins; Nobs = 230) dimers. Gaussian fits showed peaks at 115 ± 2.7° (SEM) for M10HMM (black line; R2 = 0.89) and 110 ± 3.1° (SEM) (red line; R2 = 0.86) for M5aHMM.
Solution kinetic studies of truncated recombinant myosin-10, which resembles the soluble subfragment-1 produced by proteolytic cleavage of muscle myosin (i.e., S1-like), have shown that release of product, ADP, is slow relative to the overall ATPase cycle time, making it a high duty cycle ratio motor, spending a significant fraction of its ATPase cycle tightly bound to actin (13, 14). Consistent with these kinetic studies, single molecule fluorescence imaging studies using artificially dimerized GFP-tagged myosin-10 (15–19) constructs that resemble HMM (i.e., HMM-like) shows that it moves processively along filamentous actin (F-actin), presumably using its two heads to take one step after another. Using surface-immobilized F-actin, Nagy et al. (15) showed that myosin-10 is more processive (i.e., walks further before detaching) on fascin-bundled F-actins compared with single F-actins, implying that the two heads might move along by binding alternately on neighboring F-actins, described as a “waddling” movement. Sun et al. (18) found that processive motion along a single F-actin is increased if it is held suspended in midsolution, allowing the myosin molecule to take a left-handed helical path around the filament as it moves along, representing a “whirling” movement around the F-actin. Each ATP-driven step taken by such truncated chimeras of myosin-10 has been reported to be in the range of 18–34 nm long (16–19). Furthermore, live cell fluorescence imaging studies (20, 21) using a full-length GFP-tagged myosin-10 construct showed that single molecules move in a highly directed manner within the filopodia. The fact that myosin-10 moves processively along actin filaments is perhaps surprising because, although the motor has the correct kinetic properties, the lever arm region, initially thought of to be composed of only the three IQ binding motifs (3), might be expected to produce a power stroke that falls short of the F-actin helix pseudorepeat of 36 nm (22). However, recent structural, biochemical, and molecular modeling studies (12, 23, 24) indicate that myosin-10 has a 70-amino acid residue-long (amino acids 813–882) SAH domain immediately following its three IQ motifs, which might serve to extend the lever arm. A structural study (25) using a peptide corresponding to amino acid residues 883–934 showed this later region can form a relatively low affinity (Kd = 0.6 μM) APCC. The authors postulated that the APCC region might mediate dimerization of myosin-10 heavy chains within the filopodia, where the local concentration is highest. Rotary-shadowed electron micrographs of a truncated myosin-10 genetic construct that encoded the first 953 amino acids (12) showed a minority of dimeric (two-headed) molecules (∼10%), consistent with the notion that myosin-10 has a weak propensity to dimerize at low protein concentration. Although a picture is emerging about how myosin-10 might be regulated and how it may switch between monomeric and dimeric states (26), little is known about its mechanical properties or how it generates force and movement. To study the mechano-chemistry of dimeric myosin-10, we generated a recombinant protein with a leucine zipper motif appended after amino acid residue 936 to augment dimerization. This protein construct gave rise to a soluble (HMM-like) protein that formed a majority of monodispersed, two-headed (dimeric) molecules at submicromolar protein concentrations. This protein allowed us to perform bulk biochemical and single molecule mechanical experiments to probe the kinetic and mechanical properties of the two-headed myosin-10. Using optical tweezers, we found that the myosin-10 construct produced a power stroke of ∼17 nm, which occurred in two distinct phases (15 + 2-nm displacements). The actin-attached lifetimes measured at different ATP concentrations showed Michaelis–Menten rate dependence with a limiting detachment rate of ∼13 s−1, similar to both the ATP turnover and ADP release rates measured in bulk biochemical studies. Together, these data imply that both mechanical and biochemical cycles are most likely limited by the rate of ADP release. Finally, under conditions of low load, myosin-10 moved processively along a single actin filament, taking ∼36-nm steps in the single molecule optical tweezers assays.
Results
Production and Characterization of an HMM-Like Myosin-10 Recombinant Protein.
We expressed and purified a truncated bovine myosin-10 heavy chain construct (amino acids 1–936) consisting of the N-terminal motor domain, the LCBD consisting of three IQ motifs, the SAH domain and the APCC domain fused to a GCN4 leucine-zipper and FLAG epitope tag (Fig. 1A). This construct is referred to as M10HMM throughout this article. Protein was coexpressed with calmodulin using the Sf9/baculovirus expression system yielding 0.4–0.8 mg of protein from ∼1 × 109 cells (Fig. 1B) (the genetic construct we used and its likely structure are described in more detail in SI Text).
Electron micrographs (EMs) showed (Fig. 1 C–E and Fig. S1B) that most (∼87%) of the molecules were dimeric, and the material was monodispersed and did not aggregate into higher oligomers. The contour length measured from the tip of one motor domain to the tip of the other motor domain was 51 ± 5.5 nm (SD) (Fig. 1F), which is only slightly shorter than the contour length measured for a negatively stained myosin-5a-6IQ-HMM (M5aHMM) construct, which was 60 ± 3.8 nm (SD) (Fig. 1F and Fig. S1A). This measurement is consistent with the SAH domain increasing the lever length as has been previously noted in rotary shadowed EM images of a truncated myosin-10 construct (amino acids 1–953) (12) and the GCN4 leucine zipper motif forcing dimerization. EM images of M10HMM show (Fig. 1 C and D and Fig. S1B) the two myosin heads with associated light chains, connected by an often indistinct region to a section that dimerizes the heavy chains via their C termini. At this level of resolution, we cannot distinguish whether the dimerization is via an APCC, a parallel coiled-coil, or a combination of these structures (Fig. S2). We measured the angular disposition of the two heads relative to one another and found a broad distribution of angles, centered around 120°, similar to measurements of M5aHMM (Fig. 1G). We did not find a significant number of heads pointing 180° away from each other. Negatively stained EMs of M10HMM bound to actin, in the presence of 1 µM ATP (Fig. 1E), show M10HMM is capable of binding to F-actin via both heads, spanning the 36-nm helical pseudorepeat of the actin filament, similar to images of acto-myosin-5a (27, 28). Single-headed M10HMM binding to F-actin was also occasionally observed (Fig. 1E, right bottom two panels).
In Vitro F-Actin Gliding Assays.
To confirm that the purified M10HMM was mechanically active, in vitro motility assays (29) were performed at 23 °C and at 37 °C. The fluorescently labeled actin filaments showed smooth gliding motion at both temperatures, and the translocation speeds were 310 ± 70 nm/s (SD) at 23 °C and 1,500 ± 120 nm/s (SD) at 37 °C (Fig. 2 A and B and Movies S1 and S2). The speeds are similar to those measured for EGFP labeled myosin-10 molecules tracked within filopodia by live cell fluorescence imaging, reported as 580 and 840 nm/s at 25 °C and 37 °C, respectively (20).
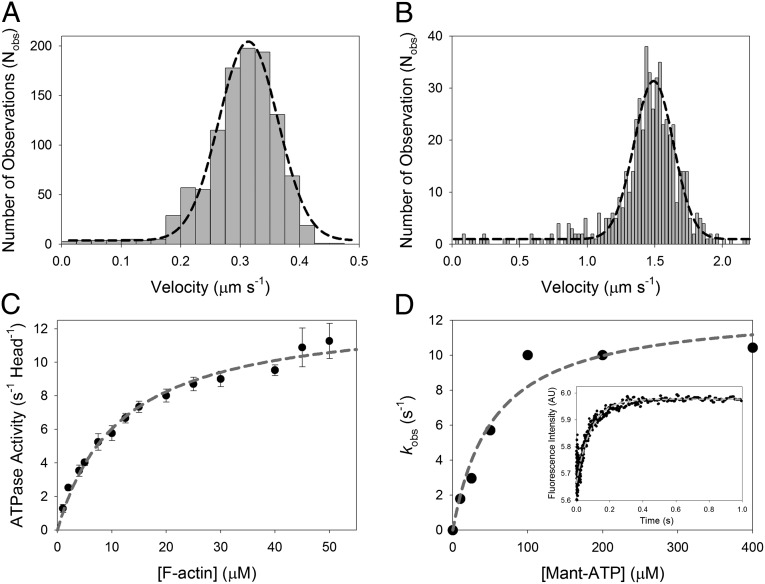
Fig. 2.
Ensemble mechanics and solution kinetics data of acto-M10HMM. (A) In vitro actin gliding assay using M10HMM construct at 23 °C. Average velocity = 310 ± 70 nm/s (SD) (Nfilaments tracked = 1,076). (B) In vitro actin gliding assay using M10HMM at 37 °C. Average velocity = 1,500 ± 120 nm/s (SD) (Nfilaments tracked = 541). (C) Plot of the actin-activated steady-state ATPase of M10HMM. In the presence of actin, the steady-state activity of M10HMM reached a maximal rate of 13.0 ± 0.47 s−1 (SD) (Vmax). Half-maximal activation was achieved at 12 ± 1.2 μM (SD) (KATPase) (Npreparations = 5). In the absence of actin, the basal steady-state activity of the construct was 0.04 ± 0.01 s−1 (SD). Data were collected at 25 °C. (D) To determine the ADP release rate from the acto-M10HMM complex, we used various mant-ATP concentrations to chase the dissociation of ADP from the M10HMM-ADP complex. A premixture of 1 μM M10HMM with 12 μM filamentous actin was incubated with 30 μM ADP. The acto-M10HMM-ADP complex was chased with different concentrations of mant-ATP. Fitting the kobs at different mant-ATP concentrations with a hyperbola was used to determine the maximal kobs, ∼12.7 s−1. (Inset) Transient record at 200 μM mant-ATP (postmixture concentration) with a kobs = 11.6 s−1. Data were collected at 25 °C.
Solution Kinetic Measurements.
In the absence of actin, the steady-state ATPase activity of M10HMM was 0.04 ± 0.01 s−1⋅head−1. In the presence of actin, the steady-state activity increased ∼300-fold, exhibiting a Vmax of 13.0 ± 0.5 s−1⋅head−1 (Fig. 2C) and half-maximal activation (Kactin) at 12 ± 1.2 μM F-actin. Using stopped-flow, 2′-(or-3′)-O-(N-methylanthraniloyl) adenosine 5′-triphosphate (mant-ATP) chase, transient kinetic experiments, we found the release of ADP could be fitted by a single exponential process and that the maximal rate of ADP release, kobs, was ∼12.7 s−1 (Fig. 2D, Inset), which is similar to the Vmax of the ATPase.
Single Molecule Detachment Rates Exhibit Michaelis–Menten Kinetics.
Optical tweezers experiments were performed using the well-established three-bead assay (30–32) that allows both the kinetic parameters (i.e., detachment rate) and mechanical parameters (i.e., average power stroke size) associated with the actomyosin-10 ATPase cycle to be measured (Fig. 3A and Fig. S3A). Initial experiments conducted under our standard conditions using an optical trap stiffness of ∼0.015–0.045 pN/nm showed mostly unitary acto-M10HMM interactions or events, reminiscent of the nonprocessive behavior of myosins such as muscle myosin-2 and nonmuscle myosin-1 (33–36). The experiments were then repeated in the presence of sinusoidal oscillations (200 Hz; ∼200 nm peak-to-peak) that were applied to one of the optical tweezers, with the aim being twofold: (i) to improve our ability to discriminate between detached and attached states and (ii) to determine the stiffness of a single actomyosin-10 attachment (32, 34, 37, 38).
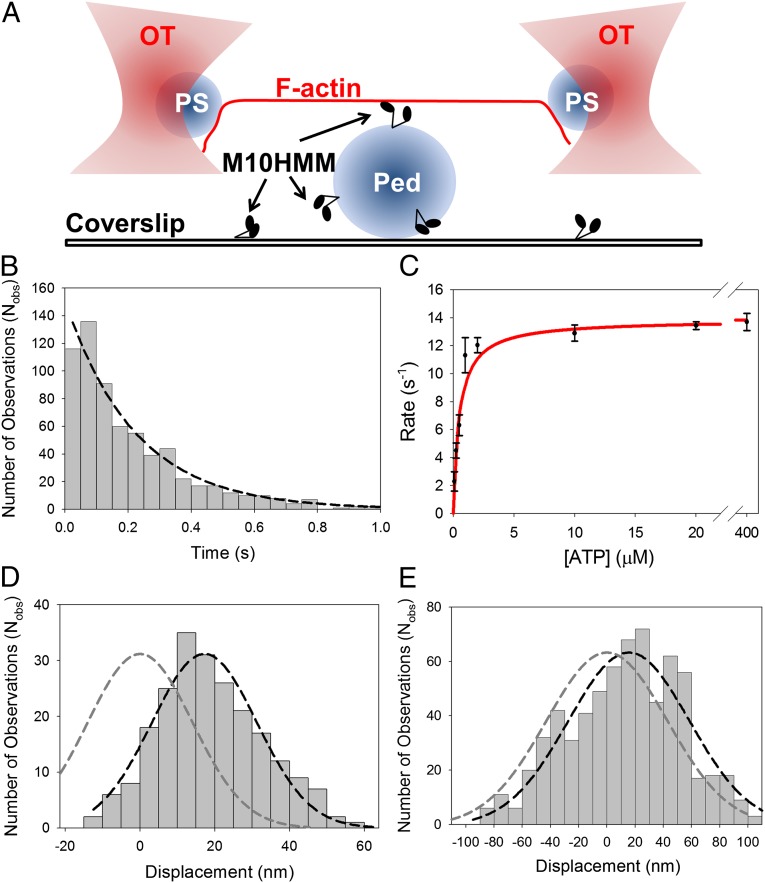
Fig. 3.
Single molecule optical tweezers assay. (A) Illustration of the three-bead assay. Two 1-μm polystyrene (PS) beads, coated with NeutrAvidin, are trapped in independent optical tweezers (OT), and a single actin filament (F-actin), containing 10% biotinylated globular-actin, is attached to form a dumbbell structure using a biotin-NeutrAvidin linkage at either end. The dumbbell is then brought to close proximity to a 2.4-μm glass pedestal (Ped) attached to the coverslip, sparsely coated with M10HMM (∼0.005–0.02 μg/mL). The bead positions are measured with nanometer precision using split photodiodes. In some experiments the optical tweezer was oscillated back and forth; this increases the signal-to-noise ratio and also allows the acto-myosin stiffness to be measured (SI Text). (B) An example of attached lifetime histogram for (Nobs= 667) acto-M10HMM events (measured at 250 nM ATP). The detachment rate was determined by fitting a single-exponential to the histogram [4.5 ± 0.5 s−1 (SD); R2 = 0.97; Table S1]. (C) ATP dependence of detachment rate, determined from event lifetime histograms measured at different ATP concentrations, exhibited Michaelis–Menten type kinetics with Vmax = 14.0 ± 0.5 s−1 (SD); KM = 0.44 ± 0.08 μM (SD) (R2 = 0.97). (D) Displacement histogram for acto-M10HMM (measured at 250 nM ATP) in the absence of oscillations. Power stroke determined from the Gaussian fit (black dashed line) was ∼17 nm [17.2 ± 0.8 nm (SEM); R2 = 0.95] (Nmyosins = 6; Nobs = 220) shifted from 0 nm (dark gray dashed line). (E) Example displacement histogram for acto-M10HMM (at 250 nM ATP) in the presence of oscillations. Power stroke was determined as the shift in the Gaussian peak (black dashed line) from 0 nm (dark gray dashed line). In this example, the average power-stroke is ∼16 nm [16 ± 3.1 nm (SEM); R2 = 0.9] (Nmyosins = 8; Nobs = 667).
Using data from the three-bead assay, acto-M10HMM attachment events were identified from sudden changes in thermal motion of the optically trapped beads caused by increased system stiffness as myosin binds to actin (30–32). For each event, the bead displacement from its mean rest position was measured to determine the average power stroke amplitude (see below), and event duration was measured to estimate the average attached lifetimes and hence detachment rate as a function of ATP concentration (100 nM to 400 μM ATP). Attached lifetime histograms were fitted to a single exponential process (39, 40) (Fig. 3B and SI Text), and the rate constants were plotted against the ATP concentration (Fig. 3C and Table S1) and fitted to the Michaelis–Menten equation. This fit gave a Vmax of 14.0 ± 0.5 s−1 (SD) and a KM of 0.44 ± 0.08 μM (SD).
At low ATP concentrations, detachment rates were linearly related to the ATP concentration, consistent with, but not necessarily proof of, the acto-myosin complex being dissociated by binding of a single ATP molecule. At high ATP concentrations, the detachment rate was limited by a slower step on the ATPase pathway. At low ATP, the second-order rate of ATP binding was determined as 9.7 μM−1⋅s−1. The maximal rate (Vmax) of ∼14 s−1 is similar to the rate of ADP release measured in stopped-flow experiments. Detachment rates, determined from lifetime duration histograms, were unaffected by the application of mechanical oscillations (described above).
Power Stroke Determined by Optical Tweezers.
The average power stroke size was extracted from the three-bead assay data by fitting a Gaussian function to a histogram of event amplitudes. In the absence of applied oscillations, M10HMM generated an average power-stroke of 17.2 ± 0.8 nm (SEM) (Fig. 3D), and in the presence of oscillations, a similar value was obtained: 16 ± 3.1 nm (SEM) (Fig. 3E). Thus, the applied optical tweezer oscillations made during the measurements did not greatly affect the power stroke measurement.
Interestingly, we occasionally observed staircase-like movements consisting of up to three steps per binding interaction (start of the record shown in Fig. S3B), a behavior indicative of a processive molecular motor, such as myosin-5a. However, such observations were rare (<2% of the total number of acto-M10HMM interactions), and they were excluded from the data set used to compile our power stroke histograms.
Ensemble Averaging Indicates a Biphasic Power Stroke.
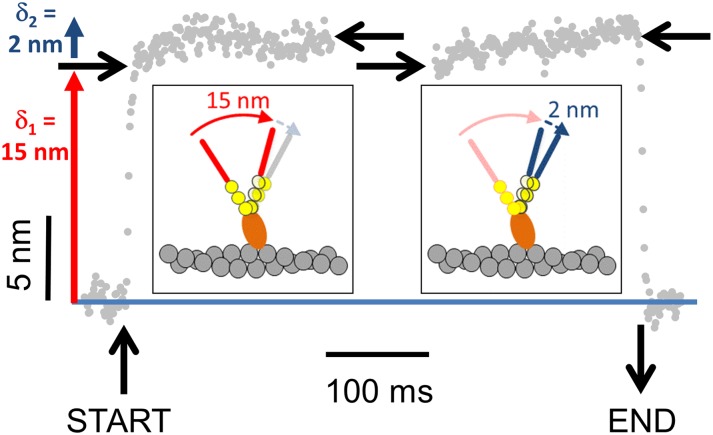
Previous studies have shown that the power strokes of some myosins are composed of more than one discrete motion of the lever arm (33, 35, 36, 38, 41–44). To test whether myosin-10 exhibits this behavior, individual binding events (recorded at 250 nM ATP) were identified in the raw data traces and synchronized (n = 220 interactions) to their starting and ending point based on changes in signal variance. Data were then ensemble averaged so that random (thermal) noise was cancelled out (reduced by a factor N0.5), whereas consistent movements produced by the power stroke were reinforced. The ensemble averaged displacement data reveal that the power stroke occurs in two phases: the initial phase generates ∼15-nm displacement, and the subsequent phase generates an additional ∼2 nm (Fig. 4).
Fig. 4.
Ensemble average showing the time course of the M10HMM power stroke. Binding events were synchronized to their start-and end times (n = 220, from six myosin molecules) by changes in signal variance. The position values were then averaged so that random thermal noise cancelled out, whereas movement due to the M10HMM power stroke was reinforced. The power stroke of shows two phases. First, a 15-nm step (δ1) then a second 2-nm step (δ2). Vertical black lines show the start and end of the synchronized interaction. Horizontal black arrows show the initial and final positions of start and end times for the synchronized plots. The cartoon (Inset) illustrates how a two-step power stroke might occur (for simplicity, only one myosin-10 head/lever is shown).
M10HMM Stiffness Measured Using Optical Tweezers.
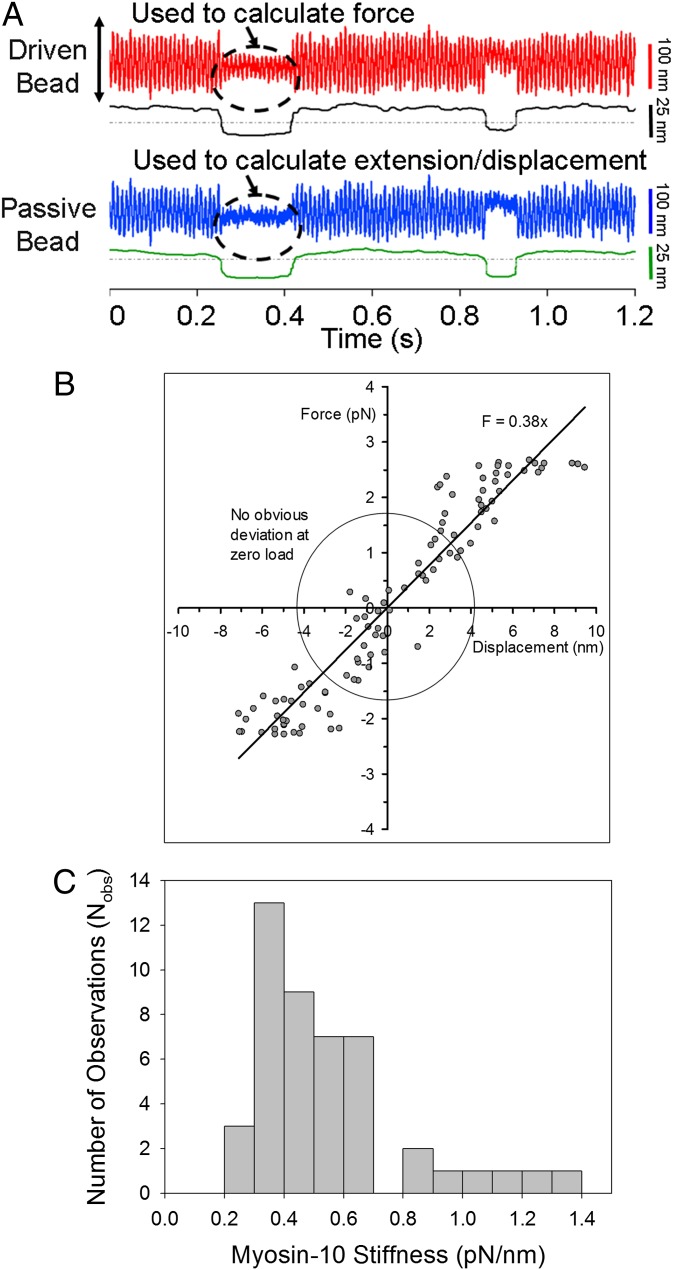
We next measured the force-extension property of the acto-M10HMM complex during individual attachment events by driving one of the optically trapped beads back and forth with a large-amplitude triangular waveform while simultaneously measuring the bead positions at either end of the actin filament (32, 38) (Fig. 5A). The difference in position between the bead held in the driven tweezer and the tweezer centroid allowed the applied force to be calculated, whereas the position of the bead at the other end of the actin filament allowed the distortion of the acto-M10HMM complex to be determined. Following a mathematical correction for actin bead connection compliance (end compliance) (32), force-extension diagrams for the acto-M10HMM complex during an attachment event could then be plotted (Fig. 5B and SI Text), and the gradient gives the acto-M10HMM stiffness. Individual estimates were histogrammed (Fig. 5C), and the median stiffness value was 0.45 pN/nm (n = 59 molecules).
Fig. 5.
Single molecule acto-M10HMM stiffness. (A) Raw data for stiffness measurement of M10HMM using the three-bead assay. In each trace (driven and passive bead traces), top signal shows the raw data (red and blue) and the bottom signal (black) is variance. (B) Example force-displacement diagram of acto-M10HMM. The slope determines the stiffness of a single M10HMM bound to F-actin, which is ∼0.38 pN/nm, for this example. Within the circle (force ∼ ±1.5 pN and displacement ∼ ±4 nm), there seems to be no obvious nonlinearity behavior. (C) Distribution of acto-M10HMM stiffnesses spans from 0.2 to 1.5 pN/nm, with a median ∼0.45 pN/nm (Nmolecules = 59).
Myosin-10 Is a Processive Motor Under Low Load.
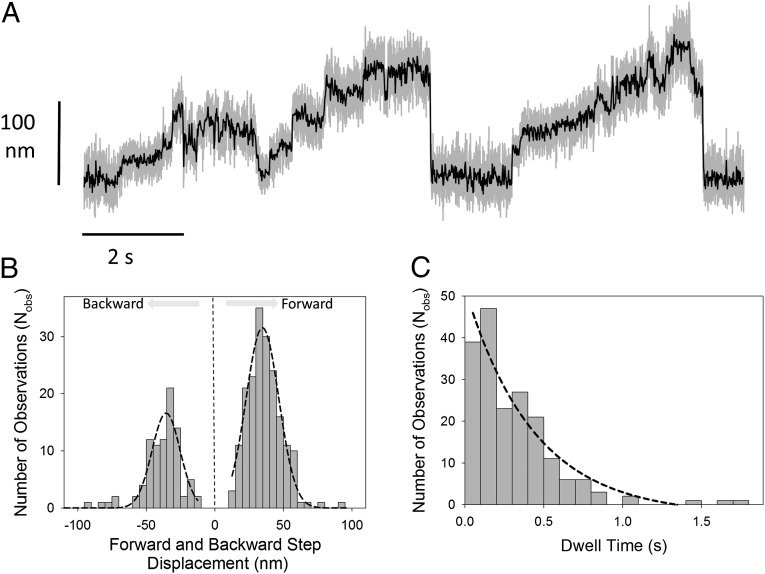
To test whether the processive staircase-like movements observed in the initial optical trapping experiments depended on external load, we decreased the combined optical tweezers stiffness from ∼0.03 to ∼0.005 pN/nm. This lower optical tweezer stiffness means that, for a given bead movement, the force applied by the optical tweezers would be approximately sixfold lower (Fig. S4). We found that, using the same protein preparation under otherwise identical experimental conditions, the interactions were now predominantly staircase-like (Fig. 6A). The step size and dwell time distributions (Fig. 6 B and C) were determined using a t-test step-finder algorithm (45) (SI Text and Fig. S5). The average forward step size was approximately +35 nm [34.7 ± 0.6 nm (SEM)], and the average dwell time was ∼0.44 s (recorded using 250 nM ATP). Although the staircase motions observed with any given actin filament had a clear directionality, closer inspection revealed occasional backward steps that had an average amplitude of approximately −36 nm [−36 ± 1.1 nm (SEM)], close to F-actin pseudorepeat distance (46). The maximum processive run length was approximately six steps per processive interaction, indicating that the maximum force that could be generated before detachment occurred was (6 × 36 nm × 0.005 pN/nm) = ∼1 pN.
Fig. 6.
Processive behavior at low optical tweezers stiffness. (A) Using a very compliant (i.e., low stiffness) optical trap (∼0.005–0.008 pN/nm), we could observe staircase-like movements generated by a single M10HMM molecule (here recorded at 100 nM ATP). Gray lines are raw data sampled at 20 kHz; black lines are following a fourth-order Butterworth, 200 Hz low-pass filter. (B) Individual step amplitudes, identified by a t test step-finder algorithm (SI Text) were histogrammed, and Gaussian fitting showed that the forward step size had an averaged ∼35 nm [34.7 ± 0.6 nm (SEM); R2 = 0.97; Nobs = 198] and the backward steps averaged ∼36 nm [36 ± 1.1 nm (SEM); R2 = 0.86; Nobs = 91]. (C) Exponential fitting to the forward stepping dwell time histogram gave a step rate of ∼2.3 s−1 [2.3 ± 0.5 s−1 (SEM); R2 = 0.93; Nobs = 198].
Discussion
The biochemical, mechanical, and structural studies of myosin-10 have been enigmatic. When its amino acid sequence was first published, conventional wisdom assumed that it would be dimerized by a predicted coiled-coil domain, but the first expression studies showed that a HMM-like fragment was largely monomeric (12). The same proved true when the full-length molecule was expressed (11). The proximal portion of the putative coiled-coil forming domain was later predicted to be a stable SAH domain, and this explained both the largely single-headed nature of the native molecule and also the unexpectedly long neck length observed in rotary-shadowed EMs of a truncated (amino acids 1–953) myosin-10 construct (12). A recent structural study has shown that the region from amino acids 883–934 forms a weakly associating (Kd = 0.6 μM) APCC (25), consistent with the earlier finding of a minority (∼10%) population of dimeric (two-headed) molecules in rotary-shadowed EMs using the amino acid 1–953 construct (12). In the present study, we wanted to determine the mechanical and biochemical properties of the dimeric form of the molecule because it seems most likely that this is the functional form of the motor within the filopodium in vivo (26). To do this, we appended a GCN4 leucine zipper motif shortly after the APCC region. The nanomolar affinity of the GCN4 motif (47) ensured effective dimerization of the molecule into a two-headed species (Fig. S1B) (15, 18, 48, 49). Our negatively stained EMs confirm the long neck length of M10HMM and allow direct comparison with images of myosin-5a (Fig. S1A). Baboolal et al. (23) showed that a SAH domain can serve to extend the lever arm. They created a chimeric molecule in which a SAH domain from Dictyostelium MyoM (∼16.8 nm long) was inserted in place of the last four IQ motifs of myosin-5a. EMs showed that this chimeric molecule had head-to-head separation similar to the native myosin-5a (which has six IQ motifs). It also had similar mechanical properties to those of myosin-5a, giving a power stroke of ∼21.5 nm in optical tweezers experiments and taking strides of ∼70 nm (i.e., center of mass translocation of ∼35 nm) in single molecule tracking assays (50–52).
Another potentially novel feature of myosin-10 has been the recent finding that a 52-residue fragment of the molecule (amino acids 883–934) forms an APCC dimer (25). However, it is worth noting that earlier work using a 93 amino acid peptide from the coiled-coil forming region of scallop muscle myosin-2 also gave rise to an APCC structure (53), but this was known to form a parallel coiled-coil in the context of the full-length molecule (54). At present, the resolution of our negatively stained EMs, obtained using recombinant M10HMM with the leucine zipper sequence after amino acid 936, does not allow us to discern the tail structure directly. We know that the majority of molecules dimerize into two-headed motors, and our structural models [based on the published structures Protein Data Bank (PDB) ID code 2LW9 (25) and 2ZTA (55), as shown in Fig. S2 A and B] indicate that the APCC motif can still form, but it is not clear if the region is instead triggered to form a parallel coiled-coil or some other conformation altogether. The average head-to-head contour distance measured from our negatively stained EM images was 51 ± 5.5 nm, whereas the predicted length of two myosin-10 motor domains each with three IQ motifs (3.4 nm long each) is predicted to be ∼2 × [8 nm + (3 × 3.4 nm)] = 36.4 nm. This discrepancy can be explained if we assume that part or all of the 70 amino acid residues 813–882 function as a stable single alpha helix (70 × 0.14) = 9.8 nm and that the two heavy chains are then linked via the APCC region (amino acids 883–910; ∼4 nm long), giving an expected head-to-head distance of 36.4 + (9.8 × 2) + 4 nm = 60 nm. However, contour distances measured from negatively stained micrographs are somewhat error prone because of variation in stain thickness between samples. An alternative configuration in which the APCC region is forced into a parallel coiled-coil by the adjoining GCN4 would give an expected head-to-head distance of 56 nm, which also lies within the error of our measurements.
Our optical trapping studies are consistent with the stable SAH domain forming part of the lever arm. We found the M10HMM power stroke was ∼17 nm, and it has been shown previously that for myosins with two IQ motifs, the power stroke ranges from 5 to 11 nm (30–32, 41, 51, 56–58), therefore averaging to ∼8 nm, and for a six IQ myosin, ∼20–25 nm (38, 59, 60). Therefore, assuming that the power stroke is linearly related to the number of IQ motifs and that the conformational change driving the power stroke is similar to that of myosin-5a and myosin-2, then the expectation for a three IQ motor would be a power stroke of ∼12.4 nm (61). Our myosin-10 power stroke measurement is therefore 25–40% larger than that predicted for a myosin with three IQs, and we propose this may be explained if the SAH domain acts to extend the lever arm length. An alternative explanation of our data might be that the lever arm rotation angle is larger than 60°, as reported for some members of the myosin-1 family (62).
On the basis of the above arguments and observations, we hypothesize that the lever arm of myosin-10 is composed of three structurally distinct elements: (i) the converter region; (ii) the three IQ motifs with associated light chains; and (iii) the stable SAH domain. A possible outcome of this composite mechanical system is that the force-extension property might show nonlinear behavior. However, the force-extension diagrams of the acto-M10HMM complex showed no obvious nonlinearities within the range of forces (∼±3 pN) and extensions (∼±10 nm) used in this study. The least-squares linear fits indicate an acto-M10HMM stiffness of 0.3–0.5 pN/nm with a median value of 0.45 pN/nm (SI Text). If the stiffness of the acto-myosin structure is dominated by cantilever bending of the lever arm, then stiffness should decrease with lever arm length. The stiffness we measure for acto-M10HMM is consistent with this idea because it lies between previous values measured for the shorter stiffer two IQ myosin-2 lever arm (∼0.7–1.7 pN/nm) (31, 32, 41, 63, 64) and the longer and more compliant six IQ myosin-5a lever arm (∼0.2 pN/nm) (38). In addition, single-headed M10HMM binding to F-actin observed via EM (Fig. 1G, right bottom two panels) illustrates that the lever arm of M10HMM is reasonably rigid, showing a structure remarkably similar to images shown in Walker et al. (27) of single-headed binding of myosin-5a to F-actin, despite the very different lever arm configuration. The acto-M10HMM cross-bridge therefore has similar mechanical properties to the other previously studied myosin cross-bridges.
Ensemble averaging of acto-M10HMM interactions measured using optical tweezers shows the power stroke occurs in two discrete phases, as found for several other myosin types (33, 35, 36, 38, 41). A large amplitude (15 nm) initial movement occurred within the first 5 ms of myosin binding to actin, presumably while ADP is still bound at the catalytic site. This rapid movement was then followed, over the next 50 ms or so, by an additional movement of 2 nm. Although the time course of the second movement is similar to the slow rate (13 s−1) of ADP release measured in solution, it is not clear from our data whether the mechanical movement corresponds directly to the product release step. The second phase of the event is then terminated when ATP binds, causing myosin to detach from actin, and this process would have an expected lifetime of ∼200 ms at 250 nM ATP (Fig. 3B). Because the amplitude of the second phase is small (2 nm) relative to the residual noise, we could not produce reliable kinetic fits to the data. However, the approximate time courses of the movements observed agree with the above interpretations.
The duty cycle ratio of a myosin is defined as the fraction of time it spends bound strongly to F-actin during its ATPase cycle. Skeletal muscle myosin-2 is a good example of a low duty ratio motor because it spends only about 5% of its time bound to actin. This duty ratio is an ideal functional adaptation in the context of an array of myosins in a muscle sarcomere thick filament because individual motors can then act independently and asynchronously to translocate the actin thin filaments at high speed. Conversely, motors such as myosin-5a that act alone or in small numbers to transport cellular cargos generally have high duty ratios (65, 66) so that they can move processively on F-actin as single molecules, taking multiple steps before dissociating. Two previous studies have measured the duty ratio of single-headed myosin-10 using transient kinetic techniques. Kovacs et al. (14) generated a single-headed recombinant form of myosin-10 by truncating the molecule at residue 810 and obtained a duty cycle ratio of 16%, whereas Homma and Ikebe (13) used a construct truncated at residue 773 and obtained a value of 60%. In the present study, we find that our predominantly two-headed M10HMM construct has an actin-activated ATP turnover rate, Vmax = 14 s−1, which is similar to the cross-bridge detachment rate measured using optical tweezers (Vmax ∼ 13 s−1), and both of these values are close to the rate of ADP dissociation from the acto⋅M10HMM⋅ADP complex measured by transient kinetic techniques (kobs ∼13 s−1). These data suggest ADP release is rate-limiting, so acto⋅M10HMM⋅ADP will be the predominant steady-state intermediate, making this a high duty ratio motor. Regarding the difference between the current results with HMM and the earlier results with subfragment-1 (S1), it is worth noting that a recent paper has produced evidence of smooth muscle myosin-2 S1 has a much lower steady-state actin-activated ATPase activity than HMM (67).
We found that release of ADP from acto⋅M10HMM, measured in transient kinetic experiments, fitted well to a single exponential process, implying that both M10HMM catalytic sites release ADP at similar rates, and there is little kinetic gating of ADP release from either head due to internal strain (Fig. 2D, Inset). This kinetic behavior is in marked contrast to experiments using acto⋅myosin-5a HMM, which require two exponential processes to fit the data, consistent with the internal strain caused by binding of the two heads along the same actin filament, causing one head to release ADP faster than the other (52, 68, 69). Notably, the chimeric myosin-5a/SAH HMM construct used by Baboolal et al. (23) also released ADP in a monoexponential manner. Because both the myosin-5a/SAH–containing HMM (23) and M10HMM (15, 17–19) constructs exhibit processive motion at low load, we conclude that strong kinetic gating of ADP release is not a prerequisite for processivity (23).
The hallmark of processive myosins studied using the optical tweezers/three-bead assay is their ability to generate staircase-like translocations of F-actin, often over many hundreds of nanometers (37, 70), whereas nonprocessive motors exhibit isolated intermittent binding events (39). We found little evidence of processive movements when probing M10HMM in the optical trap under our usual protocol, which uses a trap stiffness of ∼0.03 pN/nm. However, on lowering the trap stiffness, short processive runs could then be seen, showing both forward and backward steps of ∼35 nm, similar to the center of mass translocation value obtained via single molecule TIRF imaging assays [∼34 (18) and ∼31 nm (19)], but significantly longer than values obtained by Nagy et al. (15) and Ricca and Rock (17), who found steps of 18 nm on F-actin bundles and a mixture of 10- to 20-nm steps on single surface-immobilized F-actin filaments. Nagy and Rock (16) also carried out optical tweezers experiments using F-actin bundles and found a load-dependent step size that was 18 nm at higher loads and 12 nm at lower loads. Our ∼35-nm step size is supported by the EM images of M10HMM on F-actin (Fig. 1G), showing that the heads are separated by ∼36 nm. Furthermore, M10HMM made frequent reverse-directed or backward steps of ∼36 nm. The frequent backward stepping and broad distribution of step sizes is reminiscent of observations made with myosin-6 (48) and similar to results obtained by Nagy and Rock (15).
One possible explanation of increased processivity under lower load may be that the azimuthal geometrical constraint exerted onto the actin filament due to the elongated (ellipsoidal) optical tweezer profile along the z axis would be lower at lower trap stiffness. The increased rotational freedom may, in turn, increase the ability of two M10HMM heads to bind actin simultaneously because the required azimuthal distortion of the two heads would be lower if the actin filament could rotate after the first head binds.
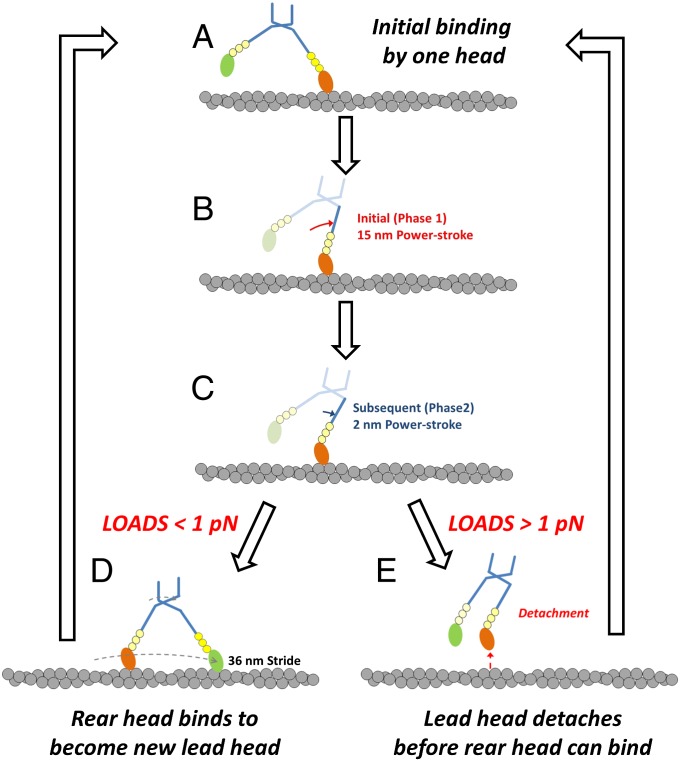
The maximum force generated by M10HMM during a processive run (6 steps × 36 nm × 0.005 pN/nm ∼ 1 pN) is substantially less than the stall force measured for myosin-5a, which is ∼3 pN (35). Such a low maximum force might also explain why processive interactions were rarely observed at high optical tweezer stiffness because just a single processive step would then produce a force close to the maximum (i.e., 1 step × 36 nm × 0.03 pN/nm ∼ 1 pN). Termination of processive movement along actin by complete detachment of the motor at relatively low loads (Fig. 7) may be an adaptive mechanism that allows myosin-10 to quickly detach/switch to another F-actin and change its path when the cargo it is carrying becomes snagged. This behavior might be especially important for intrafilopodial trafficking where the environment is crowded at the molecular length scale and a highly processive motor, which stalls (and pauses) at high load, would essentially remain stuck and make slower net progress than a motor that detached and perhaps moved sideways to navigate around an obstruction.
Fig. 7.
Proposed model of the M10HMM translocation along a single actin filament incorporating mechanical data from study. Initial binding of M10HMM to F-actin occurs via a leading head (A), is followed by an initial rapid phase of the power stroke giving 15 nm of movement (phase 1; B), and then a subsequent 2 nm (phase 2; C). Then, if the external load on the molecule is <1 pN, the trailing M10HMM head may swing forward and bind at the next available actin site (36 nm) in front and therefore becomes the new leading head (D), allowing processive stepping movement to proceed. However, if the load exceeds 1 pN, the leading head detaches before the trailing head has a chance to bind so that the M10HMM molecule then detaches from F-actin, which terminates the processive movement (E).
In this article, we addressed a number of physical aspects of myosin-10 force generation and motion; however, the most pressing question related to the in vivo function of myosin-10 that has not been addressed in our current study is whether myosin-10 forms a dimer or higher oligomer inside cells. Myosin-10 has only a weak tendency to do so when expressed without the artificial dimerization mechanisms in Sf9 cells (12), but expression of full-length myosin-10 constructs in cells does result in the accumulation of the molecule at the tips of filopodia, which suggests that it oligomerizes by some yet to be determined mechanism inside the cell. Some of the discrepancies observed in the in vitro experiments among the different groups may arise from the different means to dimerize the molecule in different studies.
In conclusion, despite having an unusual architecture, the lever arm of myosin-10 still functions in a conventional manner. Using structural and single molecule techniques, here we show directly that the power stroke can be accounted for by assuming that the SAH domain is an integral part of the lever arm and that it is sufficiently stiff over the expected ranges of forces. Its lever arm allows the two heads to bind simultaneously to F-actin, separated by ∼36 nm. The duty ratio is sufficiently high as to allow a dimeric myosin-10 to move processively along actin, taking ∼36-nm steps. Myosin-10, therefore, is an adaptable molecular motor that can either (1) move processively or (2) produce intermittent forces (Fig. 7), depending on the prevailing loads encountered by the motor in vivo.
Materials and Methods
Protein Expression, Purification, and Reagents.
A truncated bovine myosin-10 construct was created that encoded the first 936 amino acids of the myosin heavy chain fused, in the same reading frame, to a GCN4 leucine zipper motif followed by a FLAG epitope tag. Recombinant baculovirus was created and amplified using standard methods, and protein was coexpressed with Xenopus calmodulin. Protein purification was performed as described previously (71) (SI Text). Myosin-5a HMM (M5HMM) was prepared as described previously (59).
Negative Stain EM.
M10HMM (or M5aHMM for control experiments) was diluted to 50 nM in EM buffer (EMB), which consisted of 10 mM MOPS (pH 7.0), 2 mM MgCl2, 0.1 mM EGTA, and 50 mM KCl. Samples were applied to a UV light pretreated, carbon-coated copper grid and stained with 1% uranyl acetate. For actin binding experiments, M10HMM was diluted to 100 nM in EMB supplemented with 2 μM ATP, and actin was diluted to 1 μM in EMB. Equal volumes of myosin and actin solutions were then mixed to give final concentrations of 50 nM M10HMM, 0.5 μM actin, and 1 μM ATP. Protein was applied to grids and stained rapidly (<10 s) to avoid ATP depletion. Micrographs were recorded at a magnification of ×40,000 on a JEOL 1200EX II microscope using the AMT XR-60 CCD camera. As a size calibration standard, catalase crystals were used. Head-to-head contour lengths were measured using ImageJ.
Kinetic Measurement.
Steady-state ATPase measurements were measured using an NADH-coupled assay as previously described (72–74). Stopped flow experiments were performed using a SF-2001 stopped-flow apparatus (KinTek Corp.) at 25 °C. All experiments were performed in a buffer containing 20 mM MOPS (pH 7.0), 5 mM MgCl2, 0.05 mM EGTA, and 50 mM KCl supplemented with different concentrations of mant-ATP (Results). Data analyses were performed using SigmaPlot 11.0 (Systat Software), OriginPro 7.5 (Microcal Corp.), and KinTek SF-2004 data analysis software.
In Vitro Actin Gliding Assay.
All assays were performed using an Olympus IX51 inverted fluorescence microscope system, and methods are described by Homsher et al. (75). The incubation buffer contained 25 mM imidazole (pH 7.4), 25 mM KCl, 4 mM MgCl2, 1 mM EGTA, and 2 mM ATP. Temperature was controlled (at 23 °C or 37 °C) using an objective heater with a thermocouple sensor for calibration. Data analysis was performed as shown previously (75) to track the leading ends of the labeled F-actin.
Single Molecule Optical Tweezers Experiments.
A dual beam optical tweezers setup was used to measure the force and movement produced by individual myosin-10 molecules as they interacted with a single actin filament. The methods have been described previously (32), and additional details are provided in SI Text, including data acquisition and analysis.
Supplementary Material
Acknowledgments
We thank Keir C. Neuman and Earl Homsher for critical reading of the manuscript, as well as for helpful discussions; David P. Corey (Harvard University) for the cDNA clone of the Bos taurus myosin-10 heavy chain; the Electron Microscopy Core Facility at the National Heart, Lung and Blood Institute (NHLBI), National Institutes of Health (NIH), for the use of the equipment; the members of the Laboratory of Molecular Physiology, NHLBI, NIH; and the Division of Physical Biochemistry, Medical Research Council National Institute for Medical Research, for technical assistance. Y.Y. was supported by National Natural Science Foundation of China Grant 31270819. This work was funded by the intramural funds from NHLBI Grant HL004243 12 (to J.R.S.) and Medical Research Council Grant U1175.70592 (to J.E.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320122111/-/DCSupplemental.
References
- 1.Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8(9):R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436(7054):1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 3.Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci. 2000;113(Pt 19):3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- 4.Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4(3):246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 5.Zhu XJ, et al. Myosin X regulates netrin receptors and functions in axonal path-finding. Nat Cell Biol. 2007;9(2):184–192. doi: 10.1038/ncb1535. [DOI] [PubMed] [Google Scholar]
- 6.Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103(33):12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox D, et al. Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat Cell Biol. 2002;4(7):469–477. doi: 10.1038/ncb805. [DOI] [PubMed] [Google Scholar]
- 8.Nie S, Kee Y, Bronner-Fraser M. Myosin-X is critical for migratory ability of Xenopus cranial neural crest cells. Dev Biol. 2009;335(1):132–142. doi: 10.1016/j.ydbio.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woolner S, O’Brien LL, Wiese C, Bement WM. Myosin-10 and actin filaments are essential for mitotic spindle function. J Cell Biol. 2008;182(1):77–88. doi: 10.1083/jcb.200804062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sousa AD, Berg JS, Robertson BW, Meeker RB, Cheney RE. Myo10 in brain: Developmental regulation, identification of a headless isoform and dynamics in neurons. J Cell Sci. 2006;119(Pt 1):184–194. doi: 10.1242/jcs.02726. [DOI] [PubMed] [Google Scholar]
- 11.Umeki N, et al. Phospholipid-dependent regulation of the motor activity of myosin X. Nat Struct Mol Biol. 2011;18(7):783–788. doi: 10.1038/nsmb.2065. [DOI] [PubMed] [Google Scholar]
- 12.Knight PJ, et al. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J Biol Chem. 2005;280(41):34702–34708. doi: 10.1074/jbc.M504887200. [DOI] [PubMed] [Google Scholar]
- 13.Homma K, Ikebe M. Myosin X is a high duty ratio motor. J Biol Chem. 2005;280(32):29381–29391. doi: 10.1074/jbc.M504779200. [DOI] [PubMed] [Google Scholar]
- 14.Kovács M, Wang F, Sellers JR. Mechanism of action of myosin X, a membrane-associated molecular motor. J Biol Chem. 2005;280(15):15071–15083. doi: 10.1074/jbc.M500616200. [DOI] [PubMed] [Google Scholar]
- 15.Nagy S, et al. A myosin motor that selects bundled actin for motility. Proc Natl Acad Sci USA. 2008;105(28):9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy S, Rock RS. Structured post-IQ domain governs selectivity of myosin X for fascin-actin bundles. J Biol Chem. 2010;285(34):26608–26617. doi: 10.1074/jbc.M110.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricca BL, Rock RS. The stepping pattern of myosin X is adapted for processive motility on bundled actin. Biophys J. 2010;99(6):1818–1826. doi: 10.1016/j.bpj.2010.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, et al. Single-molecule stepping and structural dynamics of myosin X. Nat Struct Mol Biol. 2010;17(4):485–491. doi: 10.1038/nsmb.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao J, Huck D, Gunther LK, Sellers JR, Sakamoto T. Actin structure-dependent stepping of myosin 5a and 10 during processive movement. PLoS ONE. 2013;8(9):e74936. doi: 10.1371/journal.pone.0074936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerber ML, et al. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr Biol. 2009;19(11):967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe TM, Tokuo H, Gonda K, Higuchi H, Ikebe M. Myosin-X induces filopodia by multiple elongation mechanism. J Biol Chem. 2010;285(25):19605–19614. doi: 10.1074/jbc.M109.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard J. Molecular motors: Structural adaptations to cellular functions. Nature. 1997;389(6651):561–567. doi: 10.1038/39247. [DOI] [PubMed] [Google Scholar]
- 23.Baboolal TG, et al. The SAH domain extends the functional length of the myosin lever. Proc Natl Acad Sci USA. 2009;106(52):22193–22198. doi: 10.1073/pnas.0909851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peckham M. Coiled coils and SAH domains in cytoskeletal molecular motors. Biochem Soc Trans. 2011;39(5):1142–1148. doi: 10.1042/BST0391142. [DOI] [PubMed] [Google Scholar]
- 25.Lu Q, Ye F, Wei Z, Wen Z, Zhang M. Antiparallel coiled-coil-mediated dimerization of myosin X. Proc Natl Acad Sci USA. 2012;109(43):17388–17393. doi: 10.1073/pnas.1208642109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerber ML, Cheney RE. Myosin-X: A MyTH-FERM myosin at the tips of filopodia. J Cell Sci. 2011;124(Pt 22):3733–3741. doi: 10.1242/jcs.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker ML, et al. Two-headed binding of a processive myosin to F-actin. Nature. 2000;405(6788):804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- 28.Burgess S, et al. The prepower stroke conformation of myosin V. J Cell Biol. 2002;159(6):983–991. doi: 10.1083/jcb.200208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kron SJ, Spudich JA. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc Natl Acad Sci USA. 1986;83(17):6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molloy JE, Burns JE, Kendrick-Jones J, Tregear RT, White DC. Movement and force produced by a single myosin head. Nature. 1995;378(6553):209–212. doi: 10.1038/378209a0. [DOI] [PubMed] [Google Scholar]
- 31.Takagi Y, Homsher EE, Goldman YE, Shuman H. Force generation in single conventional actomyosin complexes under high dynamic load. Biophys J. 2006;90(4):1295–1307. doi: 10.1529/biophysj.105.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veigel C, Bartoo ML, White DC, Sparrow JC, Molloy JE. The stiffness of rabbit skeletal actomyosin cross-bridges determined with an optical tweezers transducer. Biophys J. 1998;75(3):1424–1438. doi: 10.1016/S0006-3495(98)74061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veigel C, et al. The motor protein myosin-I produces its working stroke in two steps. Nature. 1999;398(6727):530–533. doi: 10.1038/19104. [DOI] [PubMed] [Google Scholar]
- 34.Batters C, et al. Myo1c is designed for the adaptation response in the inner ear. EMBO J. 2004;23(7):1433–1440. doi: 10.1038/sj.emboj.7600169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laakso JM, Lewis JH, Shuman H, Ostap EM. Myosin I can act as a molecular force sensor. Science. 2008;321(5885):133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greenberg MJ, Lin T, Goldman YE, Shuman H, Ostap EM. Myosin IC generates power over a range of loads via a new tension-sensing mechanism. Proc Natl Acad Sci USA. 2012;109(37):E2433–E2440. doi: 10.1073/pnas.1207811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta AD, et al. Myosin-V is a processive actin-based motor. Nature. 1999;400(6744):590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- 38.Veigel C, Wang F, Bartoo ML, Sellers JR, Molloy JE. The gated gait of the processive molecular motor, myosin V. Nat Cell Biol. 2002;4(1):59–65. doi: 10.1038/ncb732. [DOI] [PubMed] [Google Scholar]
- 39.Batters C, Veigel C. Using optical tweezers to study the fine details of myosin ATPase mechanochemical cycle. Methods Mol Biol. 2011;778:97–109. doi: 10.1007/978-1-61779-261-8_7. [DOI] [PubMed] [Google Scholar]
- 40.Nagy A, et al. Kinetic characterization of nonmuscle myosin IIb at the single molecule level. J Biol Chem. 2013;288(1):709–722. doi: 10.1074/jbc.M112.424671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capitanio M, et al. Two independent mechanical events in the interaction cycle of skeletal muscle myosin with actin. Proc Natl Acad Sci USA. 2006;103(1):87–92. doi: 10.1073/pnas.0506830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laakso JM, Lewis JH, Shuman H, Ostap EM. Control of myosin-I force sensing by alternative splicing. Proc Natl Acad Sci USA. 2010;107(2):698–702. doi: 10.1073/pnas.0911426107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lister I, et al. A monomeric myosin VI with a large working stroke. EMBO J. 2004;23(8):1729–1738. doi: 10.1038/sj.emboj.7600180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veigel C, Molloy JE, Schmitz S, Kendrick-Jones J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat Cell Biol. 2003;5(11):980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 45.Carter NJ, Cross RA. Mechanics of the kinesin step. Nature. 2005;435(7040):308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 46.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347(6288):44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 47.Zitzewitz JA, Bilsel O, Luo J, Jones BE, Matthews CR. Probing the folding mechanism of a leucine zipper peptide by stopped-flow circular dichroism spectroscopy. Biochemistry. 1995;34(39):12812–12819. doi: 10.1021/bi00039a042. [DOI] [PubMed] [Google Scholar]
- 48.Rock RS, et al. Myosin VI is a processive motor with a large step size. Proc Natl Acad Sci USA. 2001;98(24):13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y, et al. Dimerized Drosophila myosin VIIa: A processive motor. Proc Natl Acad Sci USA. 2006;103(15):5746–5751. doi: 10.1073/pnas.0509935103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300(5628):2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 51.Sakamoto T, Yildez A, Selvin PR, Sellers JR. Step-size is determined by neck length in myosin V. Biochemistry. 2005;44(49):16203–16210. doi: 10.1021/bi0512086. [DOI] [PubMed] [Google Scholar]
- 52.Sakamoto T, Webb MR, Forgacs E, White HD, Sellers JR. Direct observation of the mechanochemical coupling in myosin Va during processive movement. Nature. 2008;455(7209):128–132. doi: 10.1038/nature07188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Málnási-Csizmadia A, Shimony E, Hegyi G, Szent-Györgyi AG, Nyitray L. Dimerization of the head-rod junction of scallop myosin. Biochem Biophys Res Commun. 1998;252(3):595–601. doi: 10.1006/bbrc.1998.9603. [DOI] [PubMed] [Google Scholar]
- 54.Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: A general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19(8):3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 56.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: Piconewton forces and nanometre steps. Nature. 1994;368(6467):113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 57.Guilford WH, et al. Smooth muscle and skeletal muscle myosins produce similar unitary forces and displacements in the laser trap. Biophys J. 1997;72(3):1006–1021. doi: 10.1016/S0006-3495(97)78753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purcell TJ, Morris C, Spudich JA, Sweeney HL. Role of the lever arm in the processive stepping of myosin V. Proc Natl Acad Sci USA. 2002;99(22):14159–14164. doi: 10.1073/pnas.182539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakamoto T, et al. Neck length and processivity of myosin V. J Biol Chem. 2003;278(31):29201–29207. doi: 10.1074/jbc.M303662200. [DOI] [PubMed] [Google Scholar]
- 60.Moore JR, Krementsova EB, Trybus KM, Warshaw DM. Myosin V exhibits a high duty cycle and large unitary displacement. J Cell Biol. 2001;155(4):625–635. doi: 10.1083/jcb.200103128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warshaw DM, et al. The light chain binding domain of expressed smooth muscle heavy meromyosin acts as a mechanical lever. J Biol Chem. 2000;275(47):37167–37172. doi: 10.1074/jbc.M006438200. [DOI] [PubMed] [Google Scholar]
- 62.Köhler D, Ruff C, Meyhöfer E, Bähler M. Different degrees of lever arm rotation control myosin step size. J Cell Biol. 2003;161(2):237–241. doi: 10.1083/jcb.200212039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewalle A, Steffen W, Stevenson O, Ouyang Z, Sleep J. Single-molecule measurement of the stiffness of the rigor myosin head. Biophys J. 2008;94(6):2160–2169. doi: 10.1529/biophysj.107.119396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehta AD, Finer JT, Spudich JA. Detection of single-molecule interactions using correlated thermal diffusion. Proc Natl Acad Sci USA. 1997;94(15):7927–7931. doi: 10.1073/pnas.94.15.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16(1):61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 66.Hammer JA, 3rd, Sellers JR. Walking to work: Roles for class V myosins as cargo transporters. Nat Rev Mol Cell Biol. 2012;13(1):13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- 67.Ma RN, et al. Cooperation between the two heads of smooth muscle myosin is essential for full activation of the motor function by phosphorylation. Biochemistry. 2013;52(36):6240–6248. doi: 10.1021/bi400554s. [DOI] [PubMed] [Google Scholar]
- 68.Forgacs E, et al. Kinetics of ADP dissociation from the trail and lead heads of actomyosin V following the power stroke. J Biol Chem. 2008;283(2):766–773. doi: 10.1074/jbc.M704313200. [DOI] [PubMed] [Google Scholar]
- 69.Rosenfeld SS, Sweeney HL. A model of myosin V processivity. J Biol Chem. 2004;279(38):40100–40111. doi: 10.1074/jbc.M402583200. [DOI] [PubMed] [Google Scholar]
- 70.Rief M, et al. Myosin-V stepping kinetics: A molecular model for processivity. Proc Natl Acad Sci USA. 2000;97(17):9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang F, Harvey EV, Conti MA, Wei D, Sellers JR. A conserved negatively charged amino acid modulates function in human nonmuscle myosin IIA. Biochemistry. 2000;39(18):5555–5560. doi: 10.1021/bi000133x. [DOI] [PubMed] [Google Scholar]
- 72.Trentham DR, Bardsley RG, Eccleston JF, Weeds AG. Elementary processes of the magnesium ion-dependent adenosine triphosphatase activity of heavy meromyosin. A transient kinetic approach to the study of kinases and adenosine triphosphatases and a colorimetric inorganic phosphate assay in situ. Biochem J. 1972;126(3):635–644. doi: 10.1042/bj1260635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De La Cruz EM, Sweeney HL, Ostap EM. ADP inhibition of myosin V ATPase activity. Biophys J. 2000;79(3):1524–1529. doi: 10.1016/S0006-3495(00)76403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang F, et al. Kinetic mechanism of non-muscle myosin IIB: Functional adaptations for tension generation and maintenance. J Biol Chem. 2003;278(30):27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 75.Homsher E, Wang F, Sellers JR. Factors affecting movement of F-actin filaments propelled by skeletal muscle heavy meromyosin. Am J Physiol. 1992;262(3 Pt 1):C714–C723. doi: 10.1152/ajpcell.1992.262.3.C714. [DOI] [PubMed] [Google Scholar]