Significance
To probe the effect of heme ruffling on electron transport, we studied three cytochromes that display wide variation in the heme ruffling distortion. Ruffling is characterized by a low-frequency heme mode in the region 45–60 cm−1 and by a photoreduction cross-section that displays strong variation as a function of the magnitude of the distortion. Given the similarity in the distance between the heme and the nearest aromatic amino acid for all three proteins, the order-of-magnitude changes in photoreduction rate demonstrate that the ruffling coordinate can serve as a control mechanism for electron transport in heme proteins. Major differences in heme ruffling are noted for cytochrome c when bound to the mitochondrial membrane compared with its solution structure.
Abstract
Cytochrome (cyt) c is an important electron transfer protein. The ruffling deformation of its heme cofactor has been suggested to relate to its electron transfer rate. However, there is no direct experimental evidence demonstrating this correlation. In this work, we studied Pseudomonas aeruginosa cytochrome c551 and its F7A mutant. These two proteins, although similar in their X-ray crystal structure, display a significant difference in their heme out-of-plane deformations, mainly along the ruffling coordinate. Resonance Raman and vibrational coherence measurements also indicate significant differences in ruffling-sensitive modes, particularly the low-frequency γa mode found between ∼50–60 cm−1. This supports previous assignments of γa as having a large ruffling content. Measurement of the photoreduction kinetics finds an order of magnitude decrease of the photoreduction cross-section in the F7A mutant, which has nearly twice the ruffling deformation as the WT. Additional measurements on cytochrome c demonstrate that heme ruffling is correlated exponentially with the electron transfer rates and suggest that ruffling could play an important role in redox control. A major relaxation of heme ruffling in cytochrome c, upon binding to the mitochondrial membrane, is discussed in this context.
Cytochrome (cyt) c is an important electron transfer protein that is involved in a variety of biological functions such as photosynthesis, respiration, and apoptosis (1). The heme group (Fe-protoporphyrin IX) is the functional center of cyt c. The heme iron is axially coordinated to His18 (proximal ligand) and Met80 (distal ligand) in its native solution state. The porphyrin ring is also covalently anchored to the protein by two thioether linkages with Cys-14 and Cys-17, which form a Cys-X-X-Cys-His (CXXCH) pentapeptide unit that is a unique feature shared by nearly all c-type hemes (1) [“XX” refers to other amino acids, e.g., Val and Ala, as in Pseudomonas aeruginosa (Pa) cyt c551].
The heme in cyt c has a geometry that is dominated by a large ruffling distortion, induced by both the protein fold and by the CXXCH motif (2, 3). Systematic analysis of X-ray crystal structures of heme proteins has shown that the proteins belonging to the same functional class share similar out-of-plane (OOP) heme distortions (4–6). These protein-induced OOP distortions are energetically unfavorable for the heme, and their evolutionary conservation implies that they have biological significance. Among them, doming and ruffling have been reasonably well characterized and correlated with protein functions. Doming is typically observed in oxygen storage or transport proteins such as hemoglobin (7, 8) and myoglobin (9). Moreover, the coupling of heme doming to the protein conformational substates has been shown to be functionally significant in a variety of heme protein systems (10–12). However, heme ruffling, which is the primary topic of this paper, is the dominant OOP deformation found in c-type cytochromes (4–6, 13) and nitrophorins (14–16), which are involved in electron and NO transport, respectively.
As seen in Fig. 1, ruffling involves a pyrrole-ring twisting about the Fe–N bond. The ruffling distortion tilts the pz orbitals of the porphyrin nitrogens away from the heme normal and increases overlap of the porphyrin a2u and iron dxy orbitals. It has been shown (17) via NMR experiments and density functional theory computation that, in the absence of a strong π-acceptor axial ligand (18), a ruffling deformation increases the Fe 3dπ-based electron density on the iron center, which makes the heme meso-carbon electron donation to the iron 3dxy orbital less energetically favorable (17). Ruffling destabilizes all three occupied Fe 3d-based molecular orbitals and decreases the positive and negative spin density on the β-pyrrole and meso-carbon, respectively (17). Consequently, the electron transfer rate to the ferric heme is expected to decrease as a function of the ruffling deformation (17). In addition, when ruffling is considered in isolation, it decreases the reduction potential of ferric cyt c (19–22).
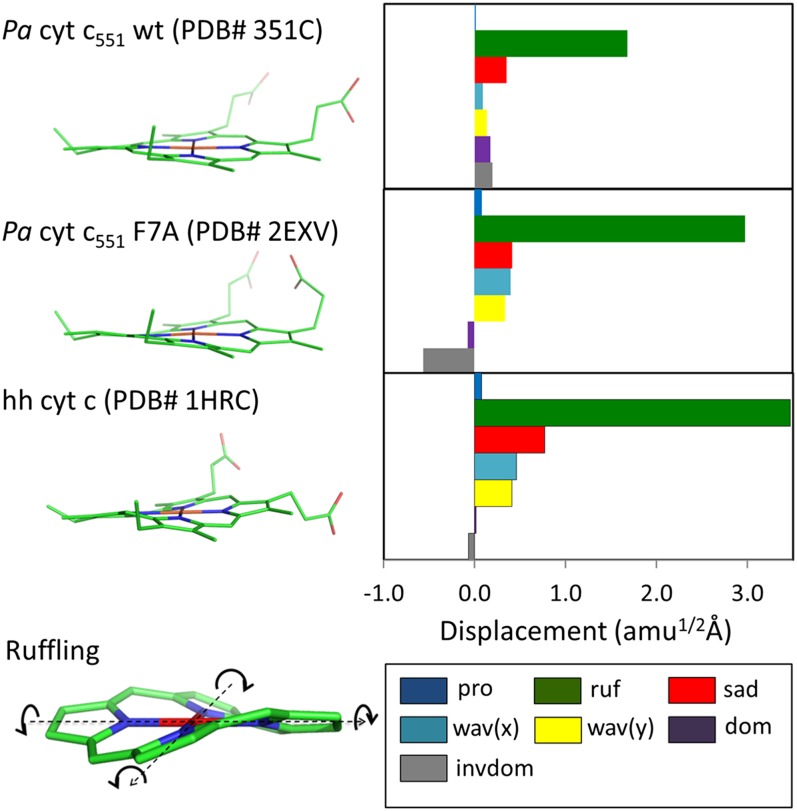
Fig. 1.
Crystal structure and NSD analysis of hemes in ferric Pa cyt c551 and its F7A mutant are compared with hh cyt c. The minus sign of displacement is defined only for doming and inverse doming to indicate the direction of Fe displacement (+, proximal; −, distal). The ruffling mode is shown at the lower left part of the figure and the arrows indicate the rotation of pyrrole rings with respect to Fe–N axis (dotted black lines).
The CXXCH pentapeptide in cyt c may be critical to the ruffled structure and the function of cyt c (2, 3, 23). The CXXCH unit is thought to affect the heme reduction potential (1), and it can influence heme deformation through the covalent bonding of the thioether groups and by hydrogen bonding within the pentapeptide (2, 3). Furthermore, the CXXCH pentapeptide may have a biologically important role related to its proximity to the electron transfer partner binding site, as in the yeast cyt c peroxidase/cyt c complex (24). The local vibrational modes of heme in the 250–400 cm−1 region have been shown to strongly mix with the vibrational modes of the CXXCH motif (23). This suggests that the heme–CXXCH vibrational dynamic couplings can play a role in electron transfer by coupling the vibrations of the heme directly to vibrations of the CXXCH unit at the protein–protein interface. This coupling could help to transduce thermal energy or alter the reorganization energy and the barrier for electron tunneling (23).
Despite the great deal of work that has been done to investigate electron transfer and heme deformation in cyt c, no experiment has directly demonstrated a quantitative correlation between heme deformation and the electron transfer rate. Generally, the functionally important heme modes, such as doming and ruffling, are delocalized and involve many nuclei and lie in the low-frequency region below 200 cm−1. Infrared and resonance Raman spectroscopy cannot reliably detect heme modes below ∼150 cm−1 in the aqueous phase, owing to the strong absorbance, Rayleigh scattering, and quasi-elastic scattering of water (25). In contrast, impulsive stimulated Raman driven vibrational coherence, or vibrational coherence spectroscopy (VCS), makes it possible to extract vibrational modulations of the third-order polarization of the heme at very low frequency, which provides access to this relatively unexplored region.
We have used this technique previously to investigate the low-frequency modes of a variety of heme proteins, using Soret band excitation (26–29). Unlike the higher frequency modes (>200 cm−1), the low-frequency modes (which have weaker force constants) are more easily distorted from equilibrium by the protein surroundings. These modes are activated in VCS when the protein induces symmetry-breaking nonplanar heme distortions (29). In addition, these modes take on a special functional significance because of their thermal accessibility. The low-frequency coherence spectra offer a unique window into how the surrounding protein environment can alter these important thermally active heme modes.
In this work, we studied Pa cyt c551 and its F7A mutant using absorption spectroscopy, resonance Raman spectroscopy, and VCS. Pa cyt c551 and its F7A mutant have very similar crystal structures, but the mutant has a more ruffled heme geometry than the WT. The investigations of this very similar pair of proteins revealed a clear difference between their resonance Raman and VCS spectra, reflecting the different degree of heme ruffling deformation. These observations support the previous assignment that γa (45∼60 cm−1) is a mode with major ruffling content in the c-type heme of cyt c. We also investigated the photoreduction kinetics of the two cyt c551 proteins as well as horse heart cyt c (hh cyt c hereafter). The photoreduction cross-section determined for WT cyt c551 is an order of magnitude larger than for the more ruffled F7A mutant and approximately two orders of magnitude larger than hh cyt c. Although the details of photoreduction in heme proteins are not fully understood (30–32), these measurements provide direct quantitative evidence that correlate dramatic increases in the photoinduced electron transfer rate with only approximately a factor of two decrease in the ruffling distortion.
Results and Discussion
Normal Coordinate Structural Decomposition Analysis and Resonance Raman Spectra.
The WT Pa cyt c551 and its F7A mutant share a very similar crystal structure (33), with a ∼0.6 Å deviation of the mean square position of the Cα atoms (33). However, a significant structural difference is found in the porphyrin OOP deformation along the ruffling coordinate. Using the normal coordinate structural decomposition (NSD) method (4–6), the nonplanar heme distortions can be quantitatively characterized. NSD describes OOP distortions by using the lowest frequency OOP mode of each symmetry type. In our NSD analysis (29) we use the optimized planar reduced porphine (including the iron atom) with D4h symmetry as the reference structure. The displacement along each normal coordinate is calculated in the mass-weighted coordinate space using the scalar product (X − X0) · Qα, where X and X0 are the mass-weighted atomic coordinates of the X-ray input and reference structures. The unit vectors, Qα, are taken from the mass-weighted normal modes, α, of the reference structure.
As shown in Fig. 1, WT cyt c551 displays a prominent ruffling distortion of 1.6 amu1/2Å, and its distortions along the other normal coordinates are less than 0.5 amu1/2Å. The mutation at position 7 leads to a 1.4 amu1/2Å increase in the ruffling distortion compared with the WT, whereas the distortions along the other normal coordinates remain less than 0.5 amu1/2Å. Although there is a small variation in the doming coordinates, the difference along the ruffling coordinate accounts for ∼90% of the overall difference between WT and the F7A mutant, making these samples an ideal model system in which to study the effect of ruffling on electron transfer in the c-type heme. The NSD structure of hh cyt c can also be seen in Fig.1, where it displays even more ruffling than cyt c551, but without any doming contribution.
Despite the 1.4-Å change in ruffling distortion, the optical absorption spectra display only a very small difference between the two c551 proteins and both spectra are very similar to that of hh cyt c. The peak of the Soret absorption band is at 408 nm for the cyt c551, compared with 409 nm for native hh cyt c. The FWHM of the Soret band for the two c551 proteins is the same as hh cyt c, as can be seen from the absorption spectra shown in SI Appendix, Figs. S2.1 and S2.2. Upon mutation, the removal of the Phe-7 residue produces a larger cavity near Cys12, which hosts three water molecules (33). This, along with a 0.3-Å decrease in distance between the residue 7 backbone CO and the Cys-12 HN, affects the CXXCH pentapeptide H-bonding and leads to changes in the ruffling of the heme (2, 3).
The low-frequency resonance Raman spectra of oxidized WT Pa cyt c551 and its F7A mutant are compared with hh cyt c in SI Appendix, Fig. S2.3. As discussed previously (23), the low-frequency spectra reflect coupling between the heme vibrations, the axial ligands, and the CXXCH motif. Thus, the change of the axial Met ligand orientation from R-Met in hh cyt c to S-Met in Pa cyt c551 along with the slightly different CXXCH configuration are likely responsible for subtle changes of the low-frequency modes. The difference between the spectra of cyt c551 WT and the F7A mutant include the weaker γ21 modes in the less ruffled WT. (The higher frequency resonance Raman spectra are shown in SI Appendix, Figs. S2.4 and S2.5).
The γ21 mode near 568 cm−1 is thought to be a ruffling distortion marker band for c-type hemes because it shows intensity changes roughly proportional to the amount of the ruffling deformation (SI Appendix, Fig. S2.6). This relationship is consistent with previous cyt c unfolding studies (3, 34, 35) and with studies (SI Appendix, Fig. S2.7) using live mitochondria (36).
Low-Frequency VCS Spectra.
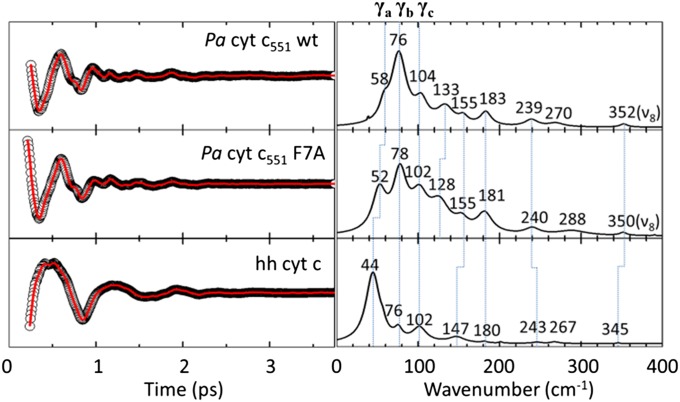
The low-frequency open-band VCS spectra of WT Pa cyt c551 and its F7A mutant are compared with hh cyt c in Fig. 2. The degenerate pump and probe pulses are centered at 412 nm. The linear predictive singular value decomposition (LPSVD) spectra derived from the time domain data agree very well with frequency domain resonance Raman spectra in the region where overlap is possible (200∼400 cm−1). Comparisons between resonance Raman spectra and the VCS spectra are given in SI Appendix, Fig. S2.8. The modes γb, γc, 155 cm−1, 183 cm−1, γ24 (240 cm−1), and ν8 are in excellent agreement among the three proteins. However, the mode at 133 cm−1 for WT and 128 cm−1 for F7A are not seen in hh cyt c, which might be due to the axial Met orientation difference between Pa cyt c551 and hh cyt c. However, the general similarity of the low-frequency spectra of Pa cyt c551 and hh cyt c is consistent with the higher-frequency resonance Raman spectra and implies that their porphyrin configurations are very similar.
Fig. 2.
Open-band VCS spectra of ferric Pa cyt c551 WT and its F7A mutant at 412 nm. LPSVD fits (Left) and LPSVD power spectra (Right) are shown.
In a previous study of cyt c unfolding, the mode labeled γa (Fig. 2) was suggested to have significant ruffling content (3). Because the WT Pa cyt c551 and its F7A mutant have very similar Soret bands, and all other conditions in the VCS experiment were fixed, we can normalize the LPSVD spectra and compare the relative amplitude of the low-frequency modes by using the peak of the coherence coupling signal (3). Using this approach, we find the amplitude ratios γa(WT)/γa(F7A) = 0.2 and γb(WT)/γb(F7A) = 0.9. The small amplitude change of γb is consistent with a prior study where γb was found to be insensitive to changes associated with relaxation of the ruffling distortion owing to unfolding (3). As a result, γb was assigned to a mode with very little or no ruffling content. The ratio of γa(WT)/γa(F7A) = 0.2 is roughly equal to the square of the ratio of the ruffling distortion in the two proteins (∼0.25). This result supports the assignment of γa as primarily a ruffling mode and is consistent with the explanation for the Raman activity of “soft” low-frequency modes proposed by Kubo et al. (29), where the amplitude is proportional to the square of its protein-induced distortion from the planar reference state.
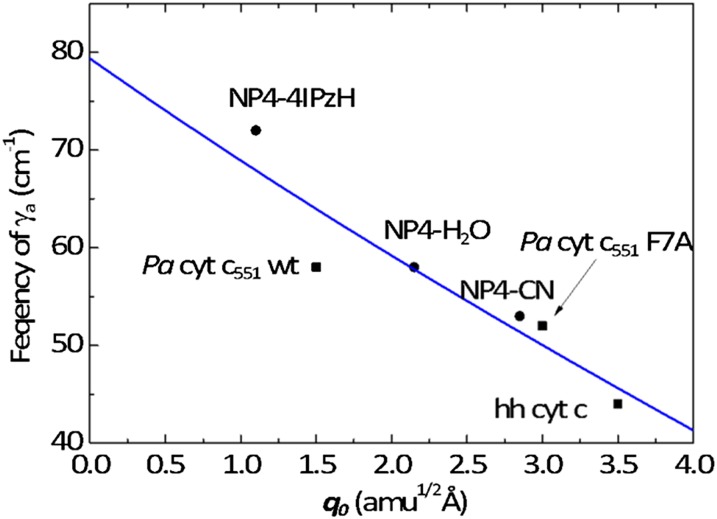
Inspection of Fig. 2 reveals an inverse correlation between the frequency of γa and the magnitude of the porphyrin ruffling distortion. In WT cyt c551, with a moderately ruffled heme, γa is located at 58 cm−1, which downshifts to 52 cm−1 in the more ruffled F7A mutant. In hh cyt c, with its strongly ruffled heme, this mode is located at an even lower frequency (44 cm−1). A similar inverse correlation was observed for the ruffling mode in nitrophorin (NP4) as different axial ligands were added (29). The data from both works are plotted in Fig. 3 and are fitted with a straight line.
Fig. 3.
Frequency of γa is plotted as a function of ruffling deformation. Data points are from this work and NP4 work (29). Cyt c and Pa cyt c551 data are in black squares, and NP4 data are in black circles. The data are fit by the solid curve using Eq. S3.3, as discussed in SI Appendix, section S3. The fit results in values of a = 0.09 (amu1/2Å)−1 and a2De/μr = 3.15 × 103 (rad/s)2.
This inverse correlation demonstrates an OOP distortion-induced mode frequency downshift, which probably reflects the anharmonic nature of the potential energy surface along the low-frequency ruffling coordinate (27, 37). Generally, under an external force from the surroundings, the potential along a given coordinate, q, can be described as 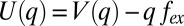 , where fex is the applied external force and V(q) is the ground state potential in its absence. The equilibrium position, q0, is determined by setting
, where fex is the applied external force and V(q) is the ground state potential in its absence. The equilibrium position, q0, is determined by setting 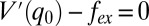 . For small oscillations near q0, the frequency is
. For small oscillations near q0, the frequency is 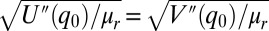 , where
, where 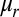 is the reduced mass of the ruffling coordinate. If V(q) is a harmonic potential, there will be no change of frequency, because V″(q0) is constant. However, the large distortion along the ruffling coordinate in cyt c551 and hh cyt c may exceed the limit for the harmonic approximation so that the observed frequency probes the anharmonicity of the potential surface (27). We discuss such a possibility in SI Appendix, section S3, where we provide a simple example demonstrating how relatively small anharmonic corrections can lead to observable shifts in a low-frequency heme mode such as ruffling.
is the reduced mass of the ruffling coordinate. If V(q) is a harmonic potential, there will be no change of frequency, because V″(q0) is constant. However, the large distortion along the ruffling coordinate in cyt c551 and hh cyt c may exceed the limit for the harmonic approximation so that the observed frequency probes the anharmonicity of the potential surface (27). We discuss such a possibility in SI Appendix, section S3, where we provide a simple example demonstrating how relatively small anharmonic corrections can lead to observable shifts in a low-frequency heme mode such as ruffling.
Photoreduction Kinetics.
The difference in the ruffling distortion shown in Fig. 1 makes WT Pa cyt c551 and its F7A mutant an ideal system to investigate the relationship between the ruffling distortion and electron transfer. The structural similarity between the two proteins limits the factors that can alter the electron transfer rate. As noted above, the aromatic Phe7 side chain of the WT is replaced in the mutant by three waters over 10 Å away from the β-pyrrole carbon and the solvent-exposed edge of the heme (17). In addition, the relative orientation of the plane of Phe7 is nearly perpendicular to the heme (SI Appendix, Fig. S5.1). This means that Phe7 is an unlikely electron donor in the photoreduction process (38–40). However, the relative position of the heme and the other aromatic side chains such as Trp56 and Tyr27 stays nearly unchanged upon mutation. These residues are closer to the heme plane and much more likely candidates to be the primary electron donor. Because of this we suggest that the mutation of Phe7 to Ala influences the photoreduction of heme primarily by modulation of its ruffling distortion.
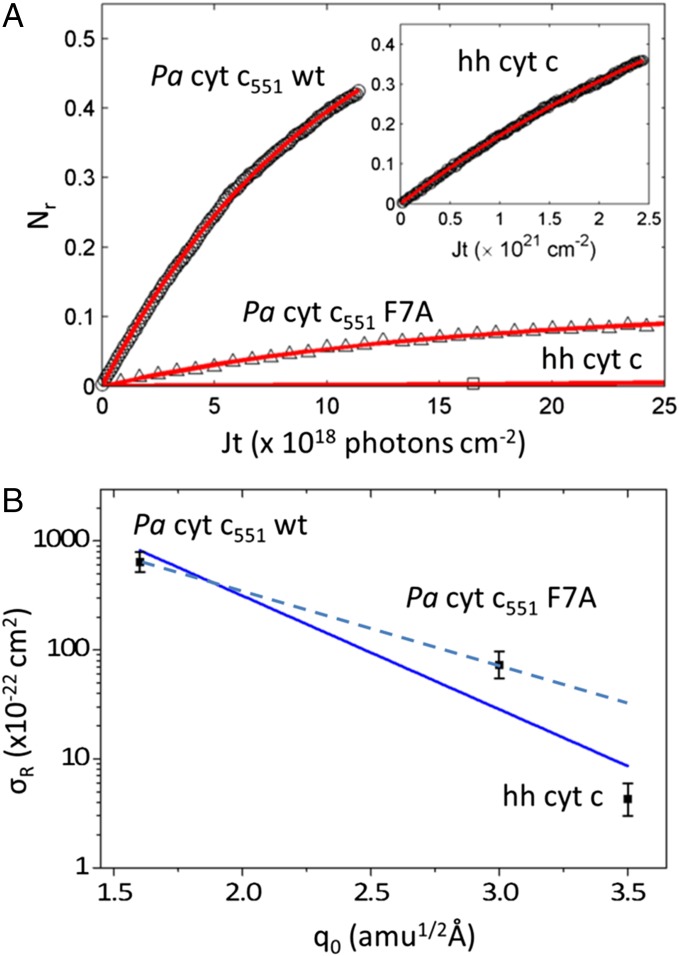
We carried out experiments that measure the photoreduction kinetics of Pa cyt c551 using 413.1-nm excitation. Some results are plotted in Fig. 4A. The intensity of the sharp α-band absorption of the ferrous form near 550 nm (SI Appendix, Figs. S2.1 and S2.2) was recorded as a function of time. We also measured the hh cyt c photoreduction kinetics to compare with a previous result (30). Photoreduction of hh cyt c is a much slower process than that observed for Pa cyt c551, as seen in Fig. 4A, Inset.
Fig. 4.
(A) Kinetic measurements of photoreduction. Nr(t) increases with irradiation time and is fitted with Nr = Nr(∞)(1− exp(−σJt)) and Nr(∞) = σr / σ with σ = σr + σo. The data for Pa cyt c551 WT, and F7A, and cyt c are shown as circles, triangles, and squares, respectively. The fitted curves are shown in red. Parameters used in the fits are summarized in Table 1. The photon flux was J = 7.3 × 1015 photons/s-cm2 and 4.5 × 1016 photons/s-cm2 for the Pa cyt c551 samples and for hh cyt c, respectively. Division by J yields the data as a function of time. (B) The photoreduction cross-sections are plotted as a function of ruffling distortion and fit using an exponential function  with both hh cyt c and Pa cyt c551 samples (solid) or only Pa cyt c551 (dashed).
with both hh cyt c and Pa cyt c551 samples (solid) or only Pa cyt c551 (dashed).
The photoreduction of Pa cyt c551 can be described by a four-state model with vanishing excited state population (30). We can determine the photoreduction cross-section for cyt c551 and hh cyt c using the photoreduction and photooxidation kinetic analysis provided previously (30) where 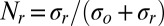 and
and 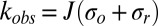 . The quantity J is the average photon flux, and kobs is the observed rate of population change. The quantities σo and σr are the oxidation and reduction cross-sections, respectively, and Nr is the normalized photoreduced population in the photostationary state. From the fits to the data, we can determine the cross-sections for photoreduction and photooxidation, which are listed in Table 1. In addition to cyt c551, the photoreduction experiments were carried out on hh cyt c as a control and the rates agree within error with the values found by Gu et al. (30), who used a somewhat different methodology. SI Appendix, section S1 discusses control experiments to determine the rate of reduction in the absence of photons, which can be ignored as in previous work (30).
. The quantity J is the average photon flux, and kobs is the observed rate of population change. The quantities σo and σr are the oxidation and reduction cross-sections, respectively, and Nr is the normalized photoreduced population in the photostationary state. From the fits to the data, we can determine the cross-sections for photoreduction and photooxidation, which are listed in Table 1. In addition to cyt c551, the photoreduction experiments were carried out on hh cyt c as a control and the rates agree within error with the values found by Gu et al. (30), who used a somewhat different methodology. SI Appendix, section S1 discusses control experiments to determine the rate of reduction in the absence of photons, which can be ignored as in previous work (30).
Table 1.
Summary of photoreduction (σr) and photooxidation (σo) cross-section determined from Fig. 4A
| Cross-section | Pa cyt c551 WT | Pa cyt c551 F7A | hh cyt c |
| σr (cm2) | 6.4 × 10−20 | 7.3 × 10−21 | 4.3 × 10−22 |
| σo (cm2) | 3.6 × 10−20 | 5.8 × 10−20 | 0.88 × 10−22 |
The photooxidation cross-sections are expected to be less sensitive to the ruffling distortion because photooxidation probably involves electron loss from the ferrous π* excited state. The ferrous iron is low-spin and already has fully filled iron t2g orbitals, so that there is no localized or delocalized hole state (associated with an unpaired iron d electron) available to modulate the tunnel distance.
The results demonstrate that the photoreduction cross-section of Pa cyt c551 is nearly two orders of magnitude larger than that of hh cyt c. Although the heme of hh cyt c exceeds the ruffling of WT Pa cyt c551 by over a factor of two, it is only ∼17% more ruffled than the F7A mutant (SI Appendix, Fig. S2.6). This is suggestive of a nonlinear correlation, where ruffling modulates the rate exponentially. There are several possibilities that may account for this observation (41). One involves a modulation of the tunneling matrix element associated with the overlap of the donor and the (iron dπ) acceptor orbitals. Another source could be exponential effects associated with the “Marcus barrier” (42), 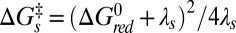 , which might also respond to the ruffling perturbation.
, which might also respond to the ruffling perturbation.
Although hh cyt c has a similar tertiary structure to Pa cyt c551 (1), we must consider the possibility that differences in the arrangement of the primary sequence of amino acids could be responsible for its significantly reduced photoreduction cross-section compared with cyt c551. This is discussed in more depth in SI Appendix, section S5. Even though the aromatic amino acids are arranged differently in hh cyt c compared with c551, the closest aromatic amino acid in hh cyt c (Tyr67), based on edge-to-edge distance, is located in a near parallel geometry 3.3 Å from the closest heme carbon atom. In Pa cyt c551 the closest amino acid is Tyr27, which also has a near parallel geometry and has a 3.5 Å edge-to-edge distance to the heme. These very similar distances suggest that the order of magnitude difference in the photoreduction cross-sections depends on an additional factor, such as heme ruffling. Even when multiple donors are considered (SI Appendix, section S5), we find that the effects of summing the parallel channels are essentially identical for the cyt c551 samples and very similar for hh cyt c.
As mentioned earlier, the F7 residue of Pa cyt c551 is perpendicular to the heme and 9.9 Å away from the heme iron and 7.6 Å from the closest heme carbon atom, making it highly unlikely to be the primary electron donor in the WT protein. The F7 distance from the heme in Pa cyt c551 is larger than the heme–aromatic residue distances in hh cyt c and it has a perpendicular orientation (38–40). Thus, if F7 were the primary electron donor, we would expect the photoreduction cross-section of Pa cyt c551 to be smaller than hh cyt c, instead of two orders of magnitude larger. Once we eliminate the F7 residue as the primary electron donor in Pa cyt c551, it seems that the different experimental photoreduction rates for WT and the F7A mutant are reflecting the difference in the ruffling distortion between these two nearly identical proteins. Given the similarity in donor–acceptor distances for cyt c and Pa cyt c551, it also seems that the even more ruffled heme structure in hh cyt c is implicated in the additional order of magnitude decrease of its photoreduction cross-section compared with F7A Pa cyt c551. These results offer a direct confirmation of the correlation between the heme ruffling distortion and the electron transfer rate for heme reduction.
Exponential Effect of Ruffling on Rates.
In Fig. 4B we plot the photoreduction cross-section (which, for a given laser flux, is directly proportional to the photoreduction rate) as a function of the ruffling distortion, 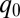 . The two orders of magnitude change can be fit using an exponential function,
. The two orders of magnitude change can be fit using an exponential function, 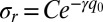 , where the quantity
, where the quantity 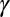 is meant to represent all factors that act to exponentially reduce the rate as the ruffling is increased. One factor to consider is the electron tunneling matrix element (43),
is meant to represent all factors that act to exponentially reduce the rate as the ruffling is increased. One factor to consider is the electron tunneling matrix element (43), 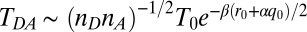 , which is written here to expose the possibility that an extra tunnel distance
, which is written here to expose the possibility that an extra tunnel distance 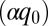 , beyond the distance
, beyond the distance 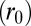 between the donor edge and the heme periphery, can result from hole localization on the iron atom when the heme becomes more ruffled (see SI Appendix, section S5 and Fig. S5.2). The normalization factor,
between the donor edge and the heme periphery, can result from hole localization on the iron atom when the heme becomes more ruffled (see SI Appendix, section S5 and Fig. S5.2). The normalization factor, 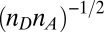 , indicates the number of atoms on the donor
, indicates the number of atoms on the donor 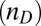 and acceptor
and acceptor 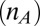 over which the electron wave function is delocalized (43). Another important factor that can depend upon ruffling is the reorganization energy,
over which the electron wave function is delocalized (43). Another important factor that can depend upon ruffling is the reorganization energy, 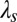 , which is expected to increase with ruffling owing to the increased hole localization on the iron atom. This factor will act to increase the semiclassical Marcus barrier,
, which is expected to increase with ruffling owing to the increased hole localization on the iron atom. This factor will act to increase the semiclassical Marcus barrier, 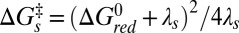 , assuming the photoreduction takes place in the “normal” region of electron transfer kinetics (42). Similarly,
, assuming the photoreduction takes place in the “normal” region of electron transfer kinetics (42). Similarly, 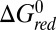 may increase (becoming less negative) to some degree upon heme ruffling, which is consistent with the cyt c551 reduction potentials (19). Moreover, treatment of inelastic processes (41) that lead to excitation of the ruffling vibration during the reaction will also effectively increase
may increase (becoming less negative) to some degree upon heme ruffling, which is consistent with the cyt c551 reduction potentials (19). Moreover, treatment of inelastic processes (41) that lead to excitation of the ruffling vibration during the reaction will also effectively increase 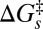 . The ruffling mode can be viewed as effectively modulating the donor–acceptor distance, so its excitation by the thermal bath will also affect the temperature dependence of the rate (41). For the purpose of the estimates given below, we assume that the spectral density and thermal distribution of the initial vibrational states is very similar for these nearly identical samples.
. The ruffling mode can be viewed as effectively modulating the donor–acceptor distance, so its excitation by the thermal bath will also affect the temperature dependence of the rate (41). For the purpose of the estimates given below, we assume that the spectral density and thermal distribution of the initial vibrational states is very similar for these nearly identical samples.
Because of the various ruffling-induced effects that can potentially lead to an exponential decrease in the photoreduction rate, we can only estimate a rough upper limit for 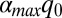 , the maximum tunnel distance change due to ruffling. We do this to determine whether the tunnel distance change is on the order of, or less than, the scale of the heme dimension. We note that the factor
, the maximum tunnel distance change due to ruffling. We do this to determine whether the tunnel distance change is on the order of, or less than, the scale of the heme dimension. We note that the factor 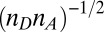 will increase the rate upon ruffling-induced hole localization, so to some degree it will partially cancel the exponential effect of an increasing Marcus barrier. Because we want an order of magnitude estimate for the tunnel distance change, we fix all other factors and take β ∼2.0 Å−1, which is a reasonable value for through space aromatic donor–acceptor systems (39, 44, 45). Ionic donors, where orientation issues are usually neglected, and covalently connected donor–acceptors, give somewhat smaller values for β ∼1.0–1.5 Å−1 (46). However, because this is an order of magnitude estimate, even if we decrease the value of β from ∼2.0 Å−1 to ∼1.5 Å−1, it will only increase the upper limits of the tunnel distance change by a factor of 1.33.
will increase the rate upon ruffling-induced hole localization, so to some degree it will partially cancel the exponential effect of an increasing Marcus barrier. Because we want an order of magnitude estimate for the tunnel distance change, we fix all other factors and take β ∼2.0 Å−1, which is a reasonable value for through space aromatic donor–acceptor systems (39, 44, 45). Ionic donors, where orientation issues are usually neglected, and covalently connected donor–acceptors, give somewhat smaller values for β ∼1.0–1.5 Å−1 (46). However, because this is an order of magnitude estimate, even if we decrease the value of β from ∼2.0 Å−1 to ∼1.5 Å−1, it will only increase the upper limits of the tunnel distance change by a factor of 1.33.
Upon fitting all of the data in Fig. 4B (solid line), we find 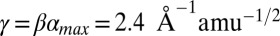 and, for β = [1.5–2.0] Å−1, we find
and, for β = [1.5–2.0] Å−1, we find 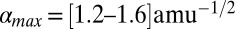 , which (using the values of q0 in Fig. 4) leads to
, which (using the values of q0 in Fig. 4) leads to 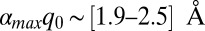 , [3.6–4.8] Å, and [4.2–5.6] Å for Pa cyt c551, its F7A mutant, and hh cyt c, respectively. Because these upper-limit distance changes are on the order of the distance between the iron atom and the β-carbon atoms on the edge of the heme, they are not inconsistent with the idea that the d-orbital hole state of the ferric heme can be delocalized or localized, depending upon the magnitude of the ruffling distortion, q0. If the data in Fig. 4B are analyzed by excluding the hh cyt c rate (dashed line), because of its differing amino acid structure and possible difference in
, [3.6–4.8] Å, and [4.2–5.6] Å for Pa cyt c551, its F7A mutant, and hh cyt c, respectively. Because these upper-limit distance changes are on the order of the distance between the iron atom and the β-carbon atoms on the edge of the heme, they are not inconsistent with the idea that the d-orbital hole state of the ferric heme can be delocalized or localized, depending upon the magnitude of the ruffling distortion, q0. If the data in Fig. 4B are analyzed by excluding the hh cyt c rate (dashed line), because of its differing amino acid structure and possible difference in 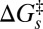 , we find
, we find 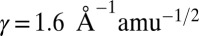 and
and 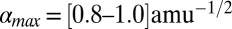 , which reduces the magnitude of the tunnel distance variation for the Pa cyt c551 samples to 1.2–1.6 Å and 2.3–3.0 Å, well within the heme radius.
, which reduces the magnitude of the tunnel distance variation for the Pa cyt c551 samples to 1.2–1.6 Å and 2.3–3.0 Å, well within the heme radius.
The fact that ruffling can increase 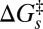 , which also reduces the rate exponentially, means that the exponential decay of the tunnelling matrix element may vary less dramatically than if it were the only factor involved in modulating the rate. These order of magnitude estimates simply demonstrate that the observed changes in the photoreduction rate are consistent with hole localization on a length scale that is equal to, or smaller than, the heme dimension. They are not meant to be taken as a precise measure of tunnel distance changes due to ruffling.
, which also reduces the rate exponentially, means that the exponential decay of the tunnelling matrix element may vary less dramatically than if it were the only factor involved in modulating the rate. These order of magnitude estimates simply demonstrate that the observed changes in the photoreduction rate are consistent with hole localization on a length scale that is equal to, or smaller than, the heme dimension. They are not meant to be taken as a precise measure of tunnel distance changes due to ruffling.
Photoreduction Process.
Although transient vibrational heating of the heme acceptor states may play a role (30, 47), as discussed in SI Appendix, section S6, it is more likely that the actual electron transfer takes place while the heme is in an excited electronic state when there is a short-lived “hole” in the porphyrin π-electron system with a lifetime, τeg ∼100 fs (48, 49). If electron transfer from a nearby aromatic donor amino acid takes place with a characteristic heme reduction time that is on the order of τr ∼1 ns, it results in a quantum yield of 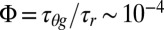 . Given that the heme Soret band absorption cross-section is ∼10−16 cm2, this leads to an overall photoreduction cross-section that is close to the measured value of ∼10−20 cm2.
. Given that the heme Soret band absorption cross-section is ∼10−16 cm2, this leads to an overall photoreduction cross-section that is close to the measured value of ∼10−20 cm2.
The heme excited state can undergo nonradiative relaxation via an intermediate state with one electron transferred from the porphyrin π* to the iron centered orbitals (50, 51). In the intermediate state, the half-filled π orbital will be delocalized on the porphyrin periphery, allowing for an increased electron transfer rate from the environmental electron donor to the heme. In the electronic ground state, the ruffling deformation tends to localize the ferric Fe dπ hole state on the iron atom (17) and analogous effects are expected for the short-lived π-orbital hole created by photoexcitation. For example, one can imagine that ruffling increases τr (by localizing the hole state at the iron and reducing the donor–acceptor overlap at the heme periphery as well as by increasing the Marcus barrier). This leads to a reduction in the photoreduction cross-section, which is proportional to 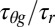 as more ruffling is introduced to the heme. This interpretation is consistent with the suggestion by Liptak et al. (17) that a more ruffled ground-state heme will decrease the delocalization of Fe 3dπ-based molecular orbitals onto the β-pyrrole carbons. This will effectively decrease the overlap integral between the “hole” in the dπ orbital and the electron donor orbital, which lowers the electron transfer rate. It is noteworthy that we draw the same conclusion even if the photoreduction process is presumed to involve photoexcitation of the donor residue, where the overlap of the electronically excited donor orbital with the heme ground-state d-electron hole orbital is the pertinent quantity to consider.
as more ruffling is introduced to the heme. This interpretation is consistent with the suggestion by Liptak et al. (17) that a more ruffled ground-state heme will decrease the delocalization of Fe 3dπ-based molecular orbitals onto the β-pyrrole carbons. This will effectively decrease the overlap integral between the “hole” in the dπ orbital and the electron donor orbital, which lowers the electron transfer rate. It is noteworthy that we draw the same conclusion even if the photoreduction process is presumed to involve photoexcitation of the donor residue, where the overlap of the electronically excited donor orbital with the heme ground-state d-electron hole orbital is the pertinent quantity to consider.
Summary.
The photoreduction kinetics of WT Pa cyt c551 and its F7A mutant show a strong correlation between heme ruffling and the photoreduction cross-section. The photoreduction rate is a reflection of the electron transfer process from a given donor to the heme (30) and therefore directly probes the effect of protein-induced ruffling perturbations on the heme structure. When the even more ruffled heme of hh cyt c is also considered, the results are consistent with an electron transfer rate that depends exponentially on the magnitude of heme ruffling. The maximum values for the change in tunnel distance, obtained by holding the other ruffling-dependent parameters fixed, are found to be on the length scale of the heme, which is consistent with the hypothesis that heme ruffling affects hole delocalization (17).
We also note that when the protein distorts the heme along the ruffling coordinate, which is a normal coordinate of the planar Fe-porphine core, the “ruffling mode” will mix differentially with other normal coordinates. In addition to introducing anharmonicity (37), this mixing can tune the mode–mode coupling resonances and allow interactions between the electron transfer partners that might affect the electron transfer rate (23). Moreover, thermal excitations of the ruffling mode, which effectively modulates the donor–acceptor distance, are expected to contribute to the temperature dependence of the rate (41).
The observations presented here have obvious biological relevance for protein control of the electron transfer process. Because the ruffling deformation of the heme cofactor is believed to arise primarily from constraints associated with the CXXCH pentapeptide (2, 3), which lies close to the protein–protein binding site (24), the binding event can perturb the CXXCH structure and can lead to changes in the heme ruffling deformation that helps to control the electron transfer process. With regard to this latter point, it is shown (3, 36) in SI Appendix, Fig. S2.7 that when cyt c is bound to the living mitochondrial membrane the strong γ21 mode observed in solution vanishes. This demonstrates that the heme relaxes to a much more planar state in the mitochondrial membrane and suggests that the ruffling distortion may act as a “switch,” making it much easier for cyt c to accept electrons when it is bound in the mitochondrial membrane than when it is floating freely in the cytoplasm.
Materials and Methods
Pa cyt c551 WT and F7A mutant was prepared as previously reported (33, 52). The resonance Raman system (29) and the femtosecond VCS system used in this work have been described in detail elsewhere (26–29). A crossed-beam transient absorption detection scheme was used to measure the photoreduction kinetics of Pa cyt c551 and hh cyt c. A detailed description of the setups and experimental procedure are provided in SI Appendix, section S1.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Award CHE-1243948 (to P.M.C.) and National Institutes of Health Grant R01-GM63170 (to K.L.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322274111/-/DCSupplemental.
References
- 1.Bowman SEJ, Bren KL. The chemistry and biochemistry of heme c: Functional bases for covalent attachment. Nat Prod Rep. 2008;25(6):1118–1130. doi: 10.1039/b717196j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma JG, Vanderkooi JM, Zhang J, Jia SL, Shelnutt JA. Resonance Raman investigation of nickel microperoxidase-11. Biochemistry. 1999;38(9):2787–2795. doi: 10.1021/bi982332a. [DOI] [PubMed] [Google Scholar]
- 3.Sun Y, Karunakaran V, Champion PM. Investigations of the low-frequency spectral density of cytochrome c upon equilibrium unfolding. J Phys Chem B. 2013;117(33):9615–9625. doi: 10.1021/jp404881k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jentzen W, Ma JG, Shelnutt JA. Conservation of the conformation of the porphyrin macrocycle in hemoproteins. Biophys J. 1998;74(2 Pt 1):753–763. doi: 10.1016/S0006-3495(98)74000-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jentzen W, Song XZ, Shelnutt JA. Structural characterization of synthetic and protein-bound porphyrins in terms of the lowest-frequency normal coordinates of the macrocycle. J Phys Chem B. 1997;101(9):1684–1699. [Google Scholar]
- 6.Shelnutt JA, et al. Nonplanar porphyrins and their significance in proteins. Chem Soc Rev. 1998;27(1):31–41. [Google Scholar]
- 7.Perutz MF. Regulation of oxygen affinity of hemoglobin: Influence of structure of the globin on the heme iron. Annu Rev Biochem. 1979;48:327–386. doi: 10.1146/annurev.bi.48.070179.001551. [DOI] [PubMed] [Google Scholar]
- 8.Perutz MF, Wilkinson AJ, Paoli M, Dodson GG. The stereochemical mechanism of the cooperative effects in hemoglobin revisited. Annu Rev Biophys Biomol Struct. 1998;27:1–34. doi: 10.1146/annurev.biophys.27.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Evans SV, Brayer GD. High-resolution study of the three-dimensional structure of horse heart metmyoglobin. J Mol Biol. 1990;213(4):885–897. doi: 10.1016/S0022-2836(05)80270-0. [DOI] [PubMed] [Google Scholar]
- 10.Srajer V, Reinisch L, Champion PM. Protein fluctuations, distributed coupling, and the binding of ligands to heme-proteins. J Am Chem Soc. 1988;110(20):6656–6670. [Google Scholar]
- 11.Benabbas A, Karunakaran V, Youn H, Poulos TL, Champion PM. Effect of DNA binding on geminate CO recombination kinetics in CO-sensing transcription factor CooA. J Biol Chem. 2012;287(26):21729–21740. doi: 10.1074/jbc.M112.345090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benabbas A, et al. Ultrafast dynamics of diatomic ligand binding to nitrophorin 4. J Am Chem Soc. 2010;132(8):2811–2820. doi: 10.1021/ja910005b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hobbs JD, Shelnutt JA. Conserved nonplanar heme distortions in cytochromes c. J Protein Chem. 1995;14(1):19–25. doi: 10.1007/BF01902840. [DOI] [PubMed] [Google Scholar]
- 14.Maes EM, Walker FA, Montfort WR, Czernuszewicz RS. Resonance Raman spectroscopic study of nitrophorin 1, a nitric oxide-binding heme protein from Rhodnius prolixus, and its nitrosyl and cyano adducts. J Am Chem Soc. 2001;123(47):11664–11672. doi: 10.1021/ja0031927. [DOI] [PubMed] [Google Scholar]
- 15.Walker FA. Nitric oxide interaction with insect nitrophorins and thoughts on the electron configuration of the FeNO6 complex. J Inorg Biochem. 2005;99(1):216–236. doi: 10.1016/j.jinorgbio.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Roberts SA, et al. Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4. Biochemistry. 2001;40(38):11327–11337. doi: 10.1021/bi0109257. [DOI] [PubMed] [Google Scholar]
- 17.Liptak MD, Wen X, Bren KL. NMR and DFT investigation of heme ruffling: Functional implications for cytochrome c. J Am Chem Soc. 2010;132(28):9753–9763. doi: 10.1021/ja102098p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura M. Electronic structures of highly deformed iron(III) porphyrin complexes. Coord Chem Rev. 2006;250(17-18):2271–2294. [Google Scholar]
- 19.Takayama SJ, et al. Electron transfer from cytochrome c to cupredoxins. J Biol Inorg Chem. 2009;14(6):821–828. doi: 10.1007/s00775-009-0494-8. [DOI] [PubMed] [Google Scholar]
- 20.Ma JG, et al. The structural origin of nonplanar heme distortions in tetraheme ferricytochromes c3. Biochemistry. 1998;37(36):12431–12442. doi: 10.1021/bi981189i. [DOI] [PubMed] [Google Scholar]
- 21.Matsuura Y, Takano T, Dickerson RE. Structure of cytochrome c551 from Pseudomonas aeruginosa refined at 1.6 A resolution and comparison of the two redox forms. J Mol Biol. 1982;156(2):389–409. doi: 10.1016/0022-2836(82)90335-7. [DOI] [PubMed] [Google Scholar]
- 22.Gianni S, et al. Demonstration of long-range interactions in a PDZ domain by NMR, kinetics, and protein engineering. Structure. 2006;14(12):1801–1809. doi: 10.1016/j.str.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Galinato MG, et al. Heme-protein vibrational couplings in cytochrome c provide a dynamic link that connects the heme-iron and the protein surface. Proc Natl Acad Sci USA. 2012;109(23):8896–8900. doi: 10.1073/pnas.1200345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelletier H, Kraut J. Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c. Science. 1992;258(5089):1748–1755. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- 25.Reinisch L, Schomacker KT, Champion PM. Anomalous resonance enhanced quasielastic light scattering of cytochrome-c. J Chem Phys. 1987;87(1):150–158. [Google Scholar]
- 26.Gruia F, et al. Coherence spectroscopy investigations of the low-frequency vibrations of heme: effects of protein-specific perturbations. J Am Chem Soc. 2008;130(15):5231–5244. doi: 10.1021/ja7104027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosca F, et al. Investigations of anharmonic low-frequency oscillations in heme proteins. J Phys Chem A. 2002;106(14):3540–3552. [Google Scholar]
- 28.Gruia F, Kubo M, Ye X, Champion PM. Investigations of vibrational coherence in the low-frequency region of ferric heme proteins. Biophys J. 2008;94(6):2252–2268. doi: 10.1529/biophysj.107.122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubo M, et al. Low-frequency mode activity of heme: Femtosecond coherence spectroscopy of iron porphine halides and nitrophorin. J Am Chem Soc. 2008;130(30):9800–9811. doi: 10.1021/ja800916d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu YG, Li PS, Sage JT, Champion PM. Photoreduction of heme-proteins - spectroscopic studies and cross-section measurements. J Am Chem Soc. 1993;115(12):4993–5004. [Google Scholar]
- 31.Ogura T, Yoshikawa S, Kitagawa T. Resonance Raman study on photoreduction of cytochrome c oxidase: Distinction of cytochromes a and a3 in the intermediate oxidation states. Biochemistry. 1985;24(26):7746–7752. doi: 10.1021/bi00347a037. [DOI] [PubMed] [Google Scholar]
- 32.Ozaki Y, Iriyama K, Ogoshi H, Kitagawa T. Ligand-aided photoreduction of iron-porphyrin complexes probed by resonance Raman spectroscopy. J Am Chem Soc. 1987;109(19):5583–5586. [Google Scholar]
- 33.Borgia A, Bonivento D, Travaglini-Allocatelli C, Di Matteo A, Brunori M. Unveiling a hidden folding intermediate in c-type cytochromes by protein engineering. J Biol Chem. 2006;281(14):9331–9336. doi: 10.1074/jbc.M512127200. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi S, et al. Folding of cytochrome c initiated by submillisecond mixing. Nat Struct Biol. 1997;4(1):44–50. doi: 10.1038/nsb0197-44. [DOI] [PubMed] [Google Scholar]
- 35.Jordan T, Eads JC, Spiro TG. Secondary and tertiary structure of the A-state of cytochrome c from resonance Raman spectroscopy. Protein Sci. 1995;4(4):716–728. doi: 10.1002/pro.5560040411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berezhna S, Wohlrab H, Champion PM. Resonance Raman investigations of cytochrome c conformational change upon interaction with the membranes of intact and Ca2+-exposed mitochondria. Biochemistry. 2003;42(20):6149–6158. doi: 10.1021/bi027387y. [DOI] [PubMed] [Google Scholar]
- 37.Tsai HH, Simpson MC. Isolated impact of ruffling on the vibrational spectrum of Ni porphyrins. Diagnosing out-of-plane distortions. J Phys Chem A. 2004;108(7):1224–1232. [Google Scholar]
- 38.Wasielewski MR. Photoinduced electron-transfer in supramolecular systems for artificial photosynthesis. Chem Rev. 1992;92(3):435–461. [Google Scholar]
- 39.Cave RJ, Siders P, Marcus RA. Mutual orientation effects on electron transfer between porphyrins. J Phys Chem. 1986;90(7):1436–1444. [Google Scholar]
- 40. Tusell JR (2011) Computation of tryptophan fluorescence quenching by amide and histidine. PhD thesis (Montana State University, Bozeman, MT)
- 41.Skourtis SS, Waldeck DH, Beratan DN. Fluctuations in biological and bioinspired electron-transfer reactions. Annu Rev Phys Chem. 2010;61:461–485. doi: 10.1146/annurev.physchem.012809.103436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcus RA, Sutin N. Electron transfers in chemistry and biology. Biochim Biophys Acta. 1985;811:265–322. [Google Scholar]
- 43.Hopfield JJ. Electron transfer between biological molecules by thermally activated tunneling. Proc Natl Acad Sci USA. 1974;71(9):3640–3644. doi: 10.1073/pnas.71.9.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis FD, et al. Donor-bridge-acceptor energetics determine the distance dependence of electron tunneling in DNA. Proc Natl Acad Sci USA. 2002;99(20):12536–12541. doi: 10.1073/pnas.192432899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith DMA, Rosso KM, Dupuis M, Valiev M, Straatsma TP. Electronic coupling between heme electron-transfer centers and its decay with distance depends strongly on relative orientation. J Phys Chem B. 2006;110(31):15582–15588. doi: 10.1021/jp057068r. [DOI] [PubMed] [Google Scholar]
- 46.Gray HB, Winkler JR. Electron flow through proteins. Chem Phys Lett. 2009;483(1-3):1–9. doi: 10.1016/j.cplett.2009.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Löwenich D, Kleinermanns K, Karunakaran V, Kovalenko SA. Transient and stationary spectroscopy of cytochrome c: Ultrafast internal conversion controls photoreduction. Photochem Photobiol. 2008;84(1):193–201. doi: 10.1111/j.1751-1097.2007.00219.x. [DOI] [PubMed] [Google Scholar]
- 48.Champion PM, Lange R. On the quantitation of light-emission from cytochrome-c in the low quantum yield limit. J Chem Phys. 1980;73(12):5947–5957. [Google Scholar]
- 49.Champion PM, Perreault GJ. Observation and quantitation of light-emission from cytochrome c using Soret band laser excitation. J Chem Phys. 1981;75(1):490–491. [Google Scholar]
- 50.Bräm O, Consani C, Cannizzo A, Chergui M. Femtosecond UV studies of the electronic relaxation processes in Cytochrome c. J Phys Chem B. 2011;115(46):13723–13730. doi: 10.1021/jp207615u. [DOI] [PubMed] [Google Scholar]
- 51.Negrerie M, Cianetti S, Vos MH, Martin JL, Kruglik SG. Ultrafast heme dynamics in ferrous versus ferric cytochrome c studied by time-resolved resonance Raman and transient absorption spectroscopy. J Phys Chem B. 2006;110(25):12766–12781. doi: 10.1021/jp0559377. [DOI] [PubMed] [Google Scholar]
- 52.Russell BS, Zhong L, Bigotti MG, Cutruzzolà F, Bren KL. Backbone dynamics and hydrogen exchange of Pseudomonas aeruginosa ferricytochrome c(551) J Biol Inorg Chem. 2003;8(1-2):156–166. doi: 10.1007/s00775-002-0401-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.