Significance
Pathogenic bacteria inject effector proteins into the host to suppress its defenses. However, bacteria produce the effector proteins and injection machinery only upon recognition of a potential host. Here we identified an Arabidopsis mutant, mapk phosphatase 1 (mkp1), with decreased levels of chemical signals recognized by the bacterium, thus making the plant more resistant by suppressing the ability of the pathogen, Pseudomonas syringae, to express and inject effector proteins. Reapplying these chemical signals not only eliminated resistance in the mkp1 mutant but also suppressed resistance in wild-type plants with a preinduced immune response. These results demonstrate an important layer in determining the biological outcome during host–pathogen interactions and may provide new targets for enhancing resistance against bacterial pathogens.
Abstract
Genes encoding the virulence-promoting type III secretion system (T3SS) in phytopathogenic bacteria are induced at the start of infection, indicating that recognition of signals from the host plant initiates this response. However, the precise nature of these signals and whether their concentrations can be altered to affect the biological outcome of host–pathogen interactions remain speculative. Here we use a metabolomic comparison of resistant and susceptible genotypes to identify plant-derived metabolites that induce T3SS genes in Pseudomonas syringae pv tomato DC3000 and report that mapk phosphatase 1 (mkp1), an Arabidopsis mutant that is more resistant to bacterial infection, produces decreased levels of these bioactive compounds. Consistent with these observations, T3SS effector expression and delivery by DC3000 was impaired when infecting the mkp1 mutant. The addition of bioactive metabolites fully restored T3SS effector delivery and suppressed the enhanced resistance in the mkp1 mutant. Pretreatment of plants with pathogen-associated molecular patterns (PAMPs) to induce PAMP-triggered immunity (PTI) also restricts T3SS effector delivery and enhances resistance by unknown mechanisms, and the addition of the bioactive metabolites similarly suppressed both aspects of PTI. Together, these results demonstrate that DC3000 perceives multiple signals derived from plants to initiate its T3SS and that the level of these host-derived signals impacts bacterial pathogenesis.
Plants evoke resistance against invading bacteria using plasma membrane-localized pattern recognition receptors (PRRs) to detect the presence of pathogen-associated molecular patterns (PAMPs) in the extracellular space (1). Activation of PRRs by PAMPs results in numerous defense responses that limit bacterial growth (1). However, the actual mechanisms by which plants suppress virulence and restrict bacterial growth remain unclear. Pseudomonas syringae is a model bacterial pathogen that infects a wide range of economically important crops as well as the laboratory model plant Arabidopsis (2). P. syringae uses several different virulence strategies to suppress host defenses, including a type III secretion system (T3SS) that secretes up to 30 effector proteins into plant cells (3, 4). Many effectors function to suppress PRR-induced signaling, thereby allowing the bacteria to avoid detection and proliferate (4). Mutants of P. syringae lacking a functional T3SS are not fully virulent, demonstrating that this system is essential for a successful infection (5, 6). Moreover, recent studies have revealed that PAMP-triggered immunity (PTI) leads to a restriction in the delivery of type III effectors into host cells, suggesting that plants possess an unknown mechanism(s) to block type III secretion (7, 8).
Despite the critical role of the T3SS in P. syringae virulence, T3SS structural components and effectors are not constitutively present but are produced at the onset of infection (9, 10). Early attempts to identify plant signals perceived by P. syringae revealed that synthetic medium mimicking the plant apoplast, namely a minimal nutrient medium with acidic pH and including a sugar such as fructose, is capable of inducing T3SS-associated genes (9–12). However, in some instances expression of the T3SS was higher in planta than in synthetic medium, indicating that additional plant-derived factors likely were required for full induction (10, 12). These results imply the presence of plant-derived signal(s) that induce the T3SS, and various signals have been proposed to be capable of inducing the T3SS in different plant pathogenic bacteria based largely on in vitro experiments (10, 12–17). However, whether any of these signals affect the biological outcome of the host–pathogen interaction remains speculative because of the lack of genetic mutants altering the abundance of these chemical signals in the host.
In the present work, we identify host chemical signals that DC3000 uses to switch to its virulence program and demonstrate that this recognition event plays an important role in a successful infection. The identification of an Arabidopsis mutant, MAPK phosphatase 1 (mkp1), in which the delivery of the P. syringae pv tomato DC3000 effector is suppressed, provided an important genetic model for investigating the basis for T3SS induction. Using a metabolomics comparison of mutant and WT plant exudates, we identified several plant-derived metabolites that are present at lower levels in mkp1 and induce the T3SS in DC3000. The biological significance of these compounds was demonstrated by showing that reintroducing these T3SS-inducing metabolites can overcome both the suppression of effector delivery and the enhanced resistance in mkp1 plants. Furthermore, the addition of these metabolites also can overcome enhanced resistance induced in plants pretreated with PAMPs. Together, these results demonstrate that DC3000 perceives multiple signals derived from plants to initiate its T3SS and that the levels of these host-derived signals contribute to susceptibility or resistance.
Results
Bacterial Effector Delivery Is Reduced in the mkp1 Mutant.
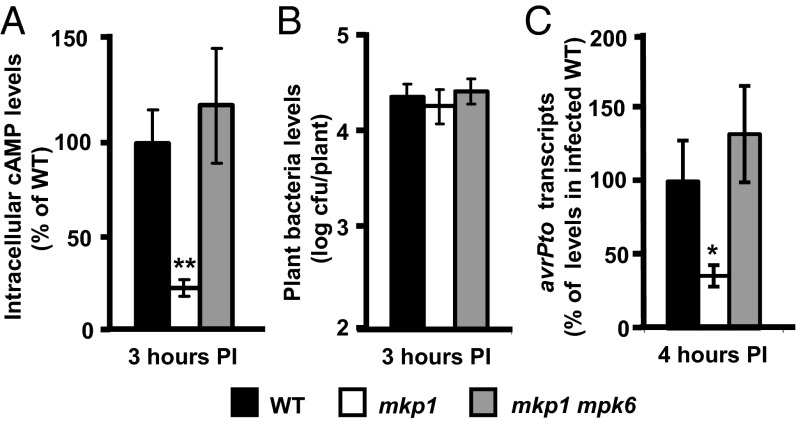
We recently identified MKP1 as a negative regulator of multiple PAMP-induced defense responses in Arabidopsis (18). Consistent with enhanced PAMP-induced responses, loss-of-function mkp1 mutants also were more resistant to infection by the normally virulent strain P. syringae pv tomato DC3000, and this enhanced resistance required the function of a specific MAPK, MPK6 (18). In light of recent reports linking PTI with the restriction of T3SS function (7, 8), we hypothesized that DC3000 may have decreased levels of type III effector delivery in the mkp1 mutant. WT and mkp1 plants were infected with DC3000 expressing the type III effector AvrPto fused to an adenylate cyclase (CyaA) reporter enzyme that produces cAMP only when delivered into eukaryotic cells (19, 20) (Additional details of experimental methods are given in SI Materials and Methods). Three hours postinfection, cAMP significantly increased in infected WT plants, whereas 80% less cAMP was measured in infected mkp1 plants (Fig. 1A). As a control, an increase in cAMP was not detected in WT plants infected with a T3SS-deficient DC3000 hrcC− strain also expressing AvrPto-CyaA (Fig. S1), demonstrating that a functional T3SS is required for cAMP production. In an mkp1 mpk6 double mutant in which the enhanced mkp1 resistance against DC3000 is suppressed (18), cAMP levels were restored to those observed in WT plants (Fig. 1A). No significant difference in the number of bacteria in infected WT and mkp1 plants was observed within this short treatment time (Fig. 1B), indicating that the difference in effector delivery is not a result of differences in bacterial numbers.
Fig. 1.
Delivery and production of the type III effector AvrPto by DC3000 is restricted in mkp1 plants in an MPK6-dependent manner. (A) WT, mkp1, and mkp1 mpk6 plants were infected with 1 × 108 cfu/mL DC3000 expressing AvrPto fused to an adenylate cyclase reporter (AvrPto-CyaA). cAMP levels were measured in plants 3 h postinfection (PI). Graphed data are means ± SE of percent cAMP relative to cAMP in WT plants. Data shown were pooled from four independent experiments, each with six samples per genotype; n = 24. (B) Serial-dilution plating of bacteria isolated from infected plants 3 h postinfection. Graphed data are means ± SE; n = 6. Each sample comprises three infected plants. (C) qRT-PCR analysis of avrPto transcripts in plants infected with 5 × 108 cfu/mL DC3000 for 4 h. avrPto transcript levels were normalized to the levels of bacterial RpoD and 16S rRNA transcripts detected in each sample. Each sample was RNA extracted from three infected plants. Six samples were analyzed for each genotype with technical replication of each. Graphed data are means ± SE of percent avrPto transcripts relative to mean avrPto transcript levels in infected WT plants; n = 12. Asterisks in A and C denote significant differences based on Student's t-test: *P < 0.05, **P < 0.01.
Decreased effector delivery in mkp1 could be caused by increased physical barriers in the plant that prevent efficient T3SS function or by decreased deployment of the T3SS itself. Callose deposits are believed to reinforce the cell wall to prevent access of pathogens to the host cell or cellular contents (21). However, we did not detect any preformed callose or enhanced callose deposition within the short (4-h) infection with DC3000 in mkp1 plants (Fig. S2). Another possibility was that DC3000 failed to induce T3SS gene expression. WT, mkp1, and mkp1 mpk6 plants were infected with DC3000 for 4 h, and the levels of avrPto expression in the infected seedlings were measured by quantitative RT-PCR (qRT-PCR). avrPto transcripts were detected in DC3000-infected WT seedlings (Fig. 1C). Similar to the pattern of effector delivery, avrPto transcripts were reduced to 35% of WT levels in mkp1 plants and were restored to WT levels in mkp1 mpk6 plants (Fig. 1C). Together, these results demonstrate that the loss of MKP1 leads to an MPK6-mediated reduction in both the expression and delivery of a type III effector.
Water-Soluble Plant Signals Stimulate Induction of the Bacterial T3SS.
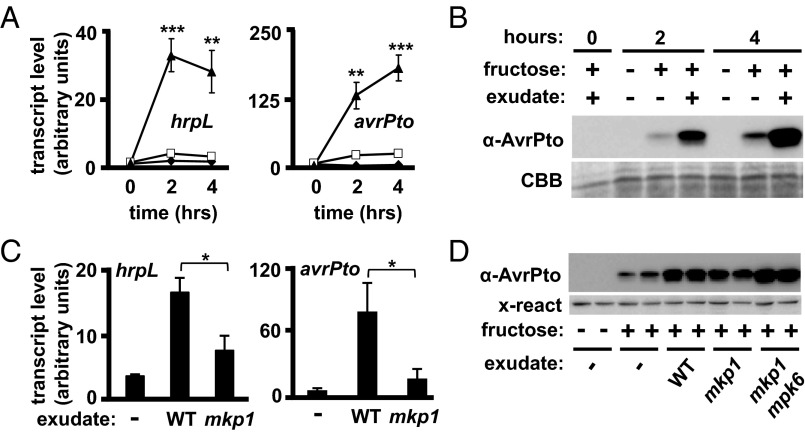
Decreased effector expression and delivery may be caused by altered abundance of plant-derived signals that affect expression of the T3SS. To investigate the existence of these putative signals, water-soluble preparations (herein referred to as “exudate”) from intact Arabidopsis were bioassayed for induction of T3SS expression in DC3000. Addition of exudate to a minimal T3SS-inducing medium consistently resulted in increased AvrPto protein levels (Fig. S3A) as well as the accumulation of both hrpL, an alternative sigma factor that regulates expression of the T3SS (12), and avrPto transcripts (Fig. S3B) after 24 h of treatment. Serial-dilution plating of bacteria after treatment revealed no significant differences in bacteria numbers among all treatment conditions (Fig. S3C), demonstrating that the enhanced responses were not a result of increased bacterial growth. Exudates also enhanced T3SS gene expression within a shorter timeframe relevant to T3SS deployment during infection, because avrPto and hrpL transcript levels were greatly increased within 2 h after treatment with exudate (Fig. 2A), and immunoblot analyses confirmed these results for AvrPto at the protein level (Fig. 2B).
Fig. 2.
Soluble signals in Arabidopsis exudates strongly enhance the expression of T3SS-associated genes in DC3000 and are genetically regulated by MKP1 and MPK6. (A) Arabidopsis exudate was mixed with DC3000 in minimal medium with or without 50 mM fructose. qRT-PCR analysis of hrpL and avrPto transcripts was performed using samples isolated at the indicated times. Graphed data are means ± SE; n = 4. ◆, −fructose/−exudate; □, +fructose/−exudate; ▲, +fructose/+exudate. **P < 0.01, ***P < 0.001 based on a t test comparison of +fructose/−exudate and +fructose/+exudate treatments. (B) Immunoblot of AvrPto from DC3000 treated as described in A. CBB, Coomassie Brilliant Blue staining. (C) DC3000 was incubated with WT or mkp1 exudate, and hrpL (Left) and avrPto (Right) transcript levels were measured by qRT-PCR 2 and 24 h posttreatment, respectively. Graphed data are means ± SE; n = 4. *P < 0.05. (D) Immunoblot of AvrPto in DC3000 24 h posttreatment with WT, mkp1, or mkp1 mpk6 exudate. A cross-reacting (x-react) band shows equal loading.
Type III-Inducing Signals Are Reduced in the mkp1 Mutant.
Comparison of exudates from WT and mkp1 plants showed reduced T3SS-inducing activity in exudates from mkp1 as determined by the accumulation of avrPto and hrpL transcripts (Fig. 2C). In addition, the reduced AvrPto-inducing bioactivity in exudates from mkp1 was restored to WT levels in exudates from mkp1 mpk6 plants (Fig. 2D), indicating that the T3SS-inducing bioactivity in exudates followed the same genotypic pattern observed for the cAMP accumulation assays (Fig. 1A). No significant differences in the size of mkp1 plants could explain the differences in exudate bioactivity (Fig. S4 A and B). Together, these data indicate that an unknown signal(s) produced by Arabidopsis strongly induces the expression of T3SS-associated genes. Moreover, the abundance of the signal(s) can be influenced by the genotype of the plant.
The Abundance of Multiple Type III-Inducing Metabolites Is Genetically Regulated.
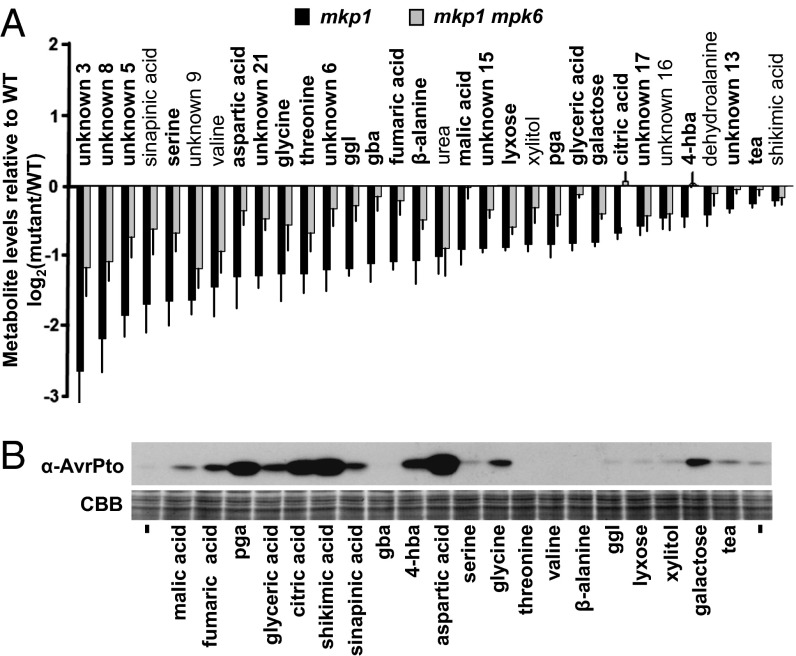
Using differential extraction of the exudates, we found that all bioactivity was recovered in the aqueous rather than in the organic phase (Fig. S4C). Using aqueous-phase extraction as an enrichment step, we prepared exudate samples from WT, mkp1, and mkp1 mpk6 plants for metabolomic analyses by GC-MS. From all metabolites identified (Dataset S1), we selected for subsequent analysis those that followed the pattern of bioactivity, i.e., significantly lower abundance in exudates from mkp1 plants than in WT plants (Fig. 3A). Interestingly, the relative abundance of most of these metabolites was restored at least partially toward WT levels in exudates from mkp1 mpk6 plants (Fig. 3A). Each metabolite identified in Fig. 3A was tested at 200 µM for the ability to induce the accumulation of AvrPto protein in the presence of minimal medium containing fructose (Fig. 3B). Five of these metabolites (pyroglutamic, citric, shikimic, 4-hydroxybenzoic, and aspartic acids) strongly induced AvrPto expression and remained extremely potent inducers of T3SS at 100 µM after only 4 h of treatment (Fig. S5A). Although all five highly bioactive compounds possess carboxyl groups, the observed effects cannot be the result of a change in pH, because buffers were present in at least 25-fold molar excess. All five bioactive compounds required the presence of fructose (Fig. S5A) or other sugars (Fig. S5B) to enhance AvrPto accumulation. Previously, citric and aspartic acids, when used at 10-mM concentrations, were reported to inhibit rather than promote type III expression in P. syringae pv. glycinea (11). To address this apparent discrepancy, we performed a dose–response experiment (Fig. S5C) which revealed that citric acid promotes AvrPto accumulation in DC3000 at lower, more physiological concentrations (22, 23) but becomes inhibitory at higher (>1 mM) concentrations (Fig. S5C). Interestingly, this biphasic effect was not observed for all T3SS-inducing metabolites, because aspartic acid does not become inhibitory even at 5 mM (Fig. S5D). These results indicate that multiple metabolites can influence T3SS expression and that concentration is an important consideration when evaluating each metabolite.
Fig. 3.
Abundance of virulence-inducing metabolites in Arabidopsis exudates is genetically regulated by MKP1 and MPK6. (A) Identification of metabolites present in WT, mkp1, and mkp1 mpk6 exudates by GC-MS. Three biological replicates were analyzed with technical replicates for each. All metabolites shown were significantly decreased in mkp1 versus WT exudates by pairwise t tests; P < 0.05. Metabolites in bold were significantly different in mkp1 and mkp1 mpk6 exudates; P < 0.05. Graphed data are means + SE of log2-transformed peak area values for mutant/WT; n = 6. (B) DC3000 was incubated with 200 μM of individual metabolites in minimal medium plus 50 mM fructose. (Upper) Immunoblot of AvrPto in bacteria 4 h posttreatment. (Lower) Coomassie Brilliant Blue (CBB) staining to confirm equal loading. gba, 4-guanidinobutyric acid; ggl, glucosylglycerol; pga, pyroglutamic acid; 4-hba, 4-hydroxybenzoic acid; tea, triethanolamine.
Exogenous Supplementation of Type III-Inducing Metabolites Is Sufficient to Overcome Enhanced Resistance.
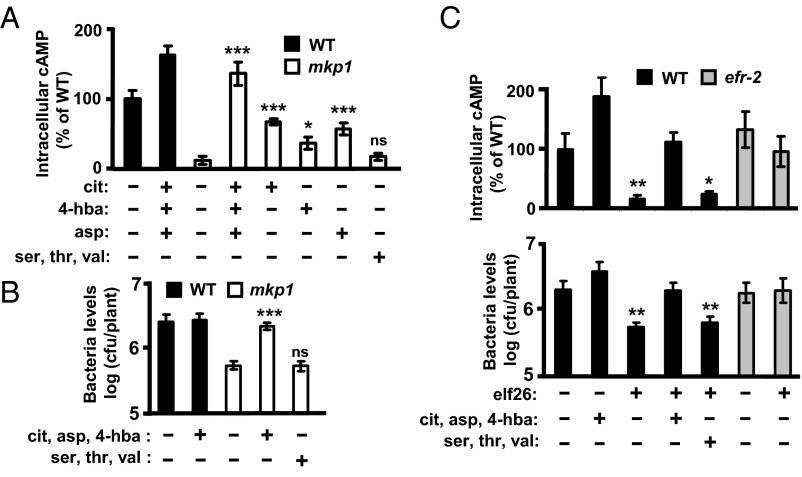
Based on the bioactivity of these metabolites as well as their accumulation patterns in mkp1 exudates, we hypothesized that the decreased levels of one or more of these metabolites may play a role in the decreased effector delivery from DC3000 into mkp1 cells. Adding 50 µM of a T3SS-inducing metabolite (citric, 4-hydroxybenzoic, or aspartic acid) to the cAMP accumulation assay resulted in significantly increased cAMP levels, and therefore effector delivery, in the mkp1 mutant (Fig. 4A). Moreover, mixing all three metabolites in the assay increased cAMP accumulation in the mkp1 mutant back to normal WT levels. As expected of the combined action of both endogenous and exogenous signals, addition of metabolites also increased cAMP levels in WT plants (Fig. 4A). In contrast, a similar mixture of 50 µM each of three other carboxylate-containing compounds (serine, threonine, and valine) had no effect on cAMP accumulation in mkp1 plants (Fig. 4A), demonstrating that not all the metabolites decreased in mkp1 plants (Fig. 3A) can alter cAMP accumulation. In addition to type III effector delivery, the bioactive three-metabolite mixture was sufficient to restore fully both the expression of avrPto transcripts (Fig. S6) and bacterial growth (Fig. 4B) in DC3000-infected mkp1 plants to the levels measured in infected WT plants. Similar to the result from the cAMP assay, the addition of the non–T3SS-inducing amino acid mixture had no effect on bacterial growth in mkp1 plants (Fig. 4B).
Fig. 4.
T3SS-inducing metabolites suppress both mkp1- and elf26-mediated inhibition of type III effector delivery. (A) WT and mkp1 plants were infected with DC3000 expressing AvrPto-CyaA as in Fig. 1 in the presence or absence of 50 μM each of citric acid (cit), aspartic acid (asp), and/or 4-hydroxybenzoic acid (4-hba) or with a non–T3SS-inducing mixture of 50 μM each of serine (ser), threonine (thr), and valine (val). Graphed data are means ± SE of cAMP levels 4 h postinfection; n = 8. (B) WT and mkp1 plants were infected with DC3000 in the presence or absence of the indicated metabolites at concentrations of 100 μM each. Graphed data are means ± SE of bacteria 24 h postinfection; n = 12. (C) WT and efr-2 plants were treated with 1 μM elf26 or a mock control for 24 h. (Upper) WT or elf26-treated plants were infected as described in Fig. 1 in the presence or absence of 50 μM of each of the indicated metabolites. cAMP levels were measured 4 h postinfection. Graphed data are means ± SE of percent cAMP relative to cAMP in infected WT plants; n = 8. (Lower) WT or elf26-treated plants were infected with DC3000 for 30 h in the presence or absence of 100 μM of each of the indicated metabolites. Graphed data are means ± SE of bacteria; n = 12. Asterisks denote t test comparison with mock-treated mkp1 in A and B, and with mock-treated WT in C; *P < 0.05, **P < 0.01, *** P < 0.001. ns, not significant.
We also tested the effect of T3SS-inducing metabolites on type III effector delivery and bacterial growth in soil-grown mkp1 plants infected with DC3000 (Fig. S7A). Similar to the results with agar–grown plants, the addition of a mixture of 10 µM or 20 µM each of citric, aspartic, and 4-hydroxybenzoic acids restored DC3000 delivery of AvrPto-CyaA in mkp1 plants in a dose-dependent manner (Fig. S7B). Furthermore, the addition of the bioactive metabolites suppressed the enhanced resistance to DC3000 in soil-grown mkp1 plants infected by either dip (Fig. S7C) and or inoculation (Fig. S7D). Together, these results demonstrate that the presence of T3SS-inducing metabolites, the expression and delivery of T3SS effectors to the plant, and the ability of the potential pathogenic bacteria to grow in the plant are tightly correlated.
The restriction of type III effector delivery and enhanced resistance in the mkp1 mutant is similar to effects described when PTI is induced in plants (7, 8), raising the possibility that the T3SS-inducing metabolites also may be able to overcome either of these PTI-mediated effects. Consistent with previous reports (8), pretreatment of plants with a PAMP, elf26 (the conserved 26 amino acid eliciting peptide from bacterial elongation factor-Tu), significantly decreased the delivery of type III effector based on reduced cAMP accumulation (Fig. 4C, Upper, lanes 1 and 3, and this decrease was dependent on EF-Tu receptor (24), the receptor for elf26 (Fig. 4C, lanes 6 and 7). The addition of T3SS-inducing metabolites (Fig. 4C, lane 4), but not the inactive metabolites (Fig. 4C, lane 5), restored cAMP levels to those in untreated WT plants. The metabolites do not interfere with PAMP responses, because both transcript accumulation (Fig. S8A) and MAPK activation (Fig. S8B) in response to elf26 were not affected by the T3SS-inducing metabolite mixture. The growth of DC3000 (Fig. 4C, Lower) followed the pattern described for the cAMP accumulation assay. Therefore, these results are consistent with a model in which control of T3SS delivery by metabolite signals present in host plants also plays a role in PTI.
Discussion
When a potential pathogen and host encounter each other, a race ensues to deploy their respective virulence and defense mechanisms. Genetic studies have firmly established the paradigm that PAMP recognition by PRRs is an important component of host nonself recognition leading to initial defense responses (1) and that pathogens may alter the molecular nature of these PAMPs in an attempt to avoid detection (25–27). However, the role of nonself recognition by the pathogen (i.e., pathogen detection of host) in contributing to the deployment of virulence factors is less understood. The T3SS is a critical virulence determinant of many phytopathogenic bacteria. Although a number of molecules have been proposed to induce expression of genes encoding the T3SS, these results are based largely on in vitro assays (10, 12–17). Importantly, plant mutants with altered levels of virulence-inducing signaling molecules have been lacking, making it difficult to ascertain if the host is capable of manipulating these signals or if this manipulation would affect the outcome of a host–pathogen interaction. In characterizing the enhanced resistance response(s) in mkp1 plants, we identified such a mutant, thus providing strong evidence that T3SS-inducing chemical signals play a biologically relevant role in the Arabidopsis-DC3000 pathosystem.
Currently, the rapid delivery of effectors into plant cells is considered a critical step for pathogens to disarm and/or evade both extracellular (PRR-mediated) and intracellular [also called “effector-triggered immunity” (ETI)] plant defenses, and our results support a model in which the speed and/or magnitude of effector delivery by P. syringae is a major determinant of pathogenesis during a compatible interaction. However, given the central role of type III effectors in both virulent and avirulent interactions, the abundance of T3SS-inducing metabolites present in plant tissues at the onset of infection also may influence the outcome of any P. syringae–plant interaction. For example, decreased effector delivery may result in an attenuation of ETI. In this regard, it will be interesting to test whether the presence/absence or relative abundance of specific bioactive metabolites is a contributing factor in processes such as nonhost resistance that determine the host range of P. syringae.
This study demonstrates that DC3000 can perceive multiple signals derived from plants to induce its T3SS rapidly. Although the bioactive metabolites are all organic acids, they are not otherwise highly related in structure. Moreover, they cannot be converted easily to a common chemical intermediate. An interesting question for future research is whether these disparate compounds are perceived by DC3000 through a shared mechanism (e.g., a single receptor that recognizes all metabolites) or through multiple distinct perception mechanisms (e.g., specific receptors for each metabolite) that subsequently converge on T3SS induction. The use of multiple host signals by DC3000 to regulate T3SS deployment and the possibility that multiple receptors may mediate these responses may explain why this layer of signaling has not been found previously in genetic studies: The decreased abundance of a single T3SS-inducing metabolite from the host or a single perception system from DC3000 potentially might be masked by the remaining pathways. These redundant perception strategies are reminiscent of the plant’s ability to recognize multiple PAMP signals, indicating the general importance of successful nonself recognition for both organisms during infection.
Intriguingly, DC3000 did not induce the T3SS in response to plant exudates (Fig. S3) or purified metabolites (Fig. S5A) alone. Instead, the plant exudate or purified metabolites acted as powerful synergistic factors when in the presence of fructose or other sugars (Fig. 2 and Figs. S3 and S5). Similar synergistic activity between simple sugars and plant-derived compounds also enhances vir gene expression in Agrobacterium through the combined action of a two-component system receptor, VirA, together with a periplasmic sugar-binding protein, ChvE (28). Simple sugars such as fructose also strongly enhanced the expression of syringomycin toxin-producing genes in P. syringae pv. syringae induced by plant-derived phenolic glucosides (28, 29). Integrating the perception of multiple nonself signals in phytopathogenic bacteria would appear to be strong fail-safe mechanism to prevent a waste of energy that would occur if virulence genes were activated prematurely, and our results indicate that a similar mechanism may be involved in DC3000.
Several of the bioactive metabolites identified in this work have been reported to regulate aspects of DC3000 virulence other than the T3SS. Citrate, along with succinate and malate, was reported to be a strong chemoattractant for DC3000 (30). In addition, both citrate and shikimate induced the expression of genes responsible for biosynthesis of the phytotoxin coronatine (31). That multiple virulence responses have been associated with perception of citrate and shikimate suggests that these host-derived metabolites can coordinately regulate multiple aspects of DC3000 virulence. However, different requirements for sugars or metabolite signals for induction of these distinct bacterial responses indicate that regulation of virulence-associated responses in DC3000 is likely to be complex. For instance, in contrast to T3SS gene expression, genes involved in the biosynthesis of coronatine were not highly induced in fructose-containing minimal medium (32). In addition, both shikimate and citrate enhanced chemotaxis and coronatine production in the absence of a simple sugar (30, 31). Together, these observations indicate that DC3000 may integrate perception of these metabolites differently to induce different aspects of its virulence.
The lower levels of T3SS-inducing metabolites in the mkp1 mutant correlate with decreased bacterial effector expression and delivery as well as with enhanced resistance. Moreover, exogenous application of these metabolites alone suppressed these defense-related phenotypes. These results provide genetic evidence that the control of host chemical signals defines a possible mechanism involved in resistance. A decrease in levels of T3SS-inducing signals would render the potential pathogen similarly innocuous as the T3SS-deficient hrcC− mutant. If this strategy is correct, the question arises as to why plants produce these extracellular signals if they may have detrimental effects. One explanation is that secreting these metabolites is a normal component of nutrient transport required for the proper growth and development of the plant. Another possibility may involve considering the plant in its ecological environment rather than only in regards to host–pathogen interactions. Recent studies indicate that plants exist in a complex balance with mutualistic microbes and that these interactions may provide an advantage to the plant in the field (33, 34). Therefore, plants may provide nutrients to these colonizers, and DC3000 may have developed recognition strategies to activate the T3SS based on the presence of metabolites that in other respects benefit the plants. An important consideration of our results is that the bioactive metabolites we identified also may serve as nutritional sources for bacteria during infection. For instance, both citric and aspartic acid are relatively abundant in plant tissue and can be catabolized by DC3000 in vitro (22, 23, 35). Therefore, although our data clearly indicate that these metabolites act as signaling cues during early stages of infection (i.e., before changes in bacterial growth occur), the decreased abundance of these metabolites in mkp1 plants may influence bacterial growth during later stages of infection by depriving the bacteria of essential plant nutrients.
Previous studies demonstrated that PTI results in restricted effector delivery, but it was not clear if this restriction arose from the formation of mechanical barriers that block delivery (7, 8). That the addition of T3SS-inducing metabolites can restore effector delivery in PAMP-treated plants argues against mechanical barriers, because the bacteria clearly are capable of transmitting effectors in the presence of the proper stimulating factors. Thus, these results indicate that a component of PTI may involve interference with bacterial T3SS induction. However, this suppression could be achieved by host restriction of T3SS-inducing molecules, by host production of chemicals that interfere with the T3SS, or by a combination of the two.
The genetically altered abundance of T3SS-inducing signals in the mkp1 mutant raises the possibility of novel strategies to introduce resistance in the field. Alternatively, identification of the bacterial receptors for these compounds could provide new targets for antimicrobial treatments that render potential pathogens less infectious. Finally, the investigation of whether other potential pathogens use similar or unique nonself-recognition strategies will be a fertile area of investigation in the future.
Materials and Methods
Preparation of Plant Exudates.
Two-week-old Arabidopsis (ecotype Wassilewskija, Ws) plants grown on Murashige and Skoog agar medium, as well as the loss-of-function mutant mkp1(Ws) and mkp1 mpk6(Ws) plants described previously (18), were placed in water. After incubation for 4 h to overnight, the water (the “exudate”) was removed, filtered (0.22 µM), and stored at −20 °C.
GC-MS Analysis of Plant Exudates.
Water-soluble metabolites in plant exudates underwent a two-stage chemical derivatization as previously described (36) and were analyzed by GC-MS using an Agilent GC 7890A coupled with a MSD 5975C mass spectrometer. The data were processed using Metabolite Detector (37), and metabolites were identified by matching to the Agilent Fiehn Metabolomics Retention Time Locked (RTL) Library (38–40).
Treatments of Bacteria with Plant Exudates and Metabolites.
Five hundred microliters of plant exudate or H2O was mixed with 500 µL of a modified hrp-inducing medium (11) supplemented or not supplemented with 100 mM fructose. For metabolite treatments, 500 µL of a 2×-concentrated stock was substituted for the plant exudate. Then 100 µL of an OD600 = 2.0 solution of Pto DC3000 bacteria was added, and the mixture was incubated at 20 °C for the indicated times. Bacteria were pelleted by centrifugation, flash frozen in liquid nitrogen, and stored at −80 °C until use.
Measurements of Protein and RNA Levels in Bacteria.
RNA and protein were extracted from treated bacteria using TRI reagent (Sigma-Aldrich) and the manufacturer’s protocol. qRT-PCR was performed with the primers and protocol described in SI Materials and Methods. Immunoblotting with anti-AvrPto (41) was performed as described previously (18).
In Planta Bacterial Growth and AvrPto-CyaA Delivery Assays.
Two-week-old plants were incubated for the times indicated in 5 mM Mes-KOH (pH 5.7)–buffered solutions of DC3000 (OD600 = 0.001) or DC3000 expressing pCPP3221 (20) (OD600 = 0.1) supplemented or not supplemented with metabolites. Bacteria levels in infected plants were measured by serial-dilution plating of plant extracts. cAMP levels in infected plants were determined using the Direct cAMP ELISA kit (Enzo Life Sciences).
avrPto Expression in DC3000-Infected Plants.
Two-week-old plants were incubated with 5 mM of Mes-KOH (pH 5.7)–buffered solution of DC3000 (OD600 = 0.5) for 4 h, and avrPto transcript levels in total RNA extracted from the infected plants were measured by SYBR Green-based qRT-PCR. avrPto transcript levels were normalized to the levels of bacterial RpoD and 16S RNA transcripts detected in each sample. Primer sequences and additional protocol details are provided in SI Materials and Methods.
elf26 Treatment of Plants.
An N-terminal acetylated elf26 peptide as described in ref. 42 was synthesized by Genscript Corp. and maintained as 10-mM stocks in DMSO at −20 °C. Polystyrene Petri dishes (100 × 15 mm) containing 15-d-old plants growing on Murashige and Skoog agar at a density of 40 plants per plate were flooded with 20 mL of a sterile 1-μM solution of elf26 or a DMSO only (mock control). After 24 h, plants were removed from the agar plates, rinsed with sterile water, and used for AvrPto-CyaA cAMP assays or DC3000 growth assays as described above.
Supplementary Material
Acknowledgments
We thank Dr. Greg Martin for comments and for providing the DC3000 hrcC− strain, avrPto-CyaA plasmid, and anti-AvrPto antibody. This work was supported by National Science Foundation Grant IOS 1051286 (to S.C.P.). A portion of the research was performed at the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL). PNNL is a multiprogram national laboratory operated by Battelle for the Department of Energy (DOE) under Contract DE-AC05-76RLO 1830. Portions of this research were enabled by capabilities developed by the PNNL Pan-omics Program under support from the DOE Office of Biological and Environmental Research Genome Sciences Program.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403248111/-/DCSupplemental.
References
- 1.Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Curr Opin Microbiol. 2011;14(1):54–61. doi: 10.1016/j.mib.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien HE, Thakur S, Guttman DS. Evolution of plant pathogenesis in Pseudomonas syringae: A genomics perspective. Annu Rev Phytopathol. 2011;49:269–289. doi: 10.1146/annurev-phyto-072910-095242. [DOI] [PubMed] [Google Scholar]
- 3.Bender CL, Alarcón-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63(2):266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindeberg M, Cunnac S, Collmer A. Pseudomonas syringae type III effector repertoires: Last words in endless arguments. Trends Microbiol. 2012;20(4):199–208. doi: 10.1016/j.tim.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Lindgren PB, Peet RC, Panopoulos NJ. Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J Bacteriol. 1986;168(2):512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peñaloza-Vázquez A, Preston GM, Collmer A, Bender CL. Regulatory interactions between the Hrp type III protein secretion system and coronatine biosynthesis in Pseudomonas syringae pv. tomato DC3000. Microbiology. 2000;146(Pt 10):2447–2456. doi: 10.1099/00221287-146-10-2447. [DOI] [PubMed] [Google Scholar]
- 7.Oh HS, Park DH, Collmer A. Components of the Pseudomonas syringae type III secretion system can suppress and may elicit plant innate immunity. Mol Plant Microbe Interact. 2010;23(6):727–739. doi: 10.1094/MPMI-23-6-0727. [DOI] [PubMed] [Google Scholar]
- 8.Crabill E, Joe A, Block A, van Rooyen JM, Alfano JR. Plant immunity directly or indirectly restricts the injection of type III effectors by the Pseudomonas syringae type III secretion system. Plant Physiol. 2010;154(1):233–244. doi: 10.1104/pp.110.159723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salmeron JM, Staskawicz BJ. Molecular characterization and hrp dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato. Mol Gen Genet. 1993;239(1-2):6–16. doi: 10.1007/BF00281595. [DOI] [PubMed] [Google Scholar]
- 10.Rahme LG, Mindrinos MN, Panopoulos NJ. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992;174(11):3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huynh TV, Dahlbeck D, Staskawicz BJ. Bacterial blight of soybean: Regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245(4924):1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- 12.Tang X, Xiao Y, Zhou JM. Regulation of the type III secretion system in phytopathogenic bacteria. Mol Plant Microbe Interact. 2006;19(11):1159–1166. doi: 10.1094/MPMI-19-1159. [DOI] [PubMed] [Google Scholar]
- 13.Aldon D, Brito B, Boucher C, Genin S. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 2000;19(10):2304–2314. doi: 10.1093/emboj/19.10.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khokhani D, et al. Discovery of plant phenolic compounds that act as type III secretion system inhibitors or inducers of the fire blight pathogen, Erwinia amylovora. Appl Environ Microbiol. 2013;79(18):5424–5436. doi: 10.1128/AEM.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, et al. The plant phenolic compound p-coumaric acid represses gene expression in the Dickeya dadantii type III secretion system. Appl Environ Microbiol. 2009;75(5):1223–1228. doi: 10.1128/AEM.02015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schulte R, Bonas U. A Xanthomonas pathogenicity locus is induced by sucrose and sulfur-containing amino acids. Plant Cell. 1992;4(1):79–86. doi: 10.1105/tpc.4.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang S, et al. Type III secretion system genes of Dickeya dadantii 3937 are induced by plant phenolic acids. PLoS ONE. 2008;3(8):e2973. doi: 10.1371/journal.pone.0002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson JC, et al. Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J. 2011;67(2):258–268. doi: 10.1111/j.1365-313X.2011.04588.x. [DOI] [PubMed] [Google Scholar]
- 19.Sory MP, Cornelis GR. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14(3):583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 20.Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J Bacteriol. 2004;186(2):543–555. doi: 10.1128/JB.186.2.543-555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keshavarzi M, et al. Basal defenses induced in pepper by lipopolysaccharides are suppressed by Xanthomonas campestris pv. vesicatoria. Mol Plant Microbe Interact. 2004;17(7):805–815. doi: 10.1094/MPMI.2004.17.7.805. [DOI] [PubMed] [Google Scholar]
- 22.Durrett TP, Gassmann W, Rogers EE. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007;144(1):197–205. doi: 10.1104/pp.107.097162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White MC, Decker AM, Chaney RL. Metal complexation in xylem fluid I. Chemical composition of tomato and soybean stem exudate. Plant Physiol. 1981;67(2):292–300. doi: 10.1104/pp.67.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125(4):749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 25.Cai R, et al. The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog. 2011;7(8):e1002130. doi: 10.1371/journal.ppat.1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke CR, et al. Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol. 2013;200(3):847–860. doi: 10.1111/nph.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi K, et al. Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. J Bacteriol. 2003;185(22):6658–6665. doi: 10.1128/JB.185.22.6658-6665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brencic A, Winans SC. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol Mol Biol Rev. 2005;69(1):155–194. doi: 10.1128/MMBR.69.1.155-194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mo YY, Gross DC. Plant signal molecules activate the syrB gene, which is required for syringomycin production by Pseudomonas syringae pv. syringae. J Bacteriol. 1991;173(18):5784–5792. doi: 10.1128/jb.173.18.5784-5792.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuppels DA. Chemotaxis by Pseudomonas syringae pv. tomato. Appl Environ Microbiol. 1988;54(3):629–632. doi: 10.1128/aem.54.3.629-632.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XZ, Starratt AN, Cuppels DA. Identification of tomato leaf factors that activate toxin gene expression in Pseudomonas syringae pv. tomato DC3000. Phytopathology. 1998;88(10):1094–1100. doi: 10.1094/PHYTO.1998.88.10.1094. [DOI] [PubMed] [Google Scholar]
- 32.Sreedharan A, Penaloza-Vazquez A, Kunkel BN, Bender CL. CorR regulates multiple components of virulence in Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2006;19(7):768–779. doi: 10.1094/MPMI-19-0768. [DOI] [PubMed] [Google Scholar]
- 33.Bulgarelli D, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488(7409):91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 34.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rico A, Preston GM. Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol Plant Microbe Interact. 2008;21(2):269–282. doi: 10.1094/MPMI-21-2-0269. [DOI] [PubMed] [Google Scholar]
- 36.Kim YM, et al. Formation of dehydroalanine from mimosine and cysteine: Artifacts in gas chromatography/mass spectrometry based metabolomics. Rapid Commun Mass Spectrom. 2011;25(17):2561–2564. doi: 10.1002/rcm.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiller K, et al. MetaboliteDetector: Comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal Chem. 2009;81(9):3429–3439. doi: 10.1021/ac802689c. [DOI] [PubMed] [Google Scholar]
- 38.Kind T, et al. FiehnLib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81(24):10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sumner LW, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fiehn O, et al. Quality control for plant metabolomics: Reporting MSI-compliant studies. Plant J. 2008;53(4):691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 41.Shan L, He P, Zhou JM, Tang X. A cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Mol Plant Microbe Interact. 2000;13(6):592–598. doi: 10.1094/MPMI.2000.13.6.592. [DOI] [PubMed] [Google Scholar]
- 42.Kunze G, et al. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16(12):3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.