Significance
Bacteria use clustered regularly interspaced short palindromic repeats (CRISPRs) together with CRISPR-associated (Cas) proteins to defend themselves against viral infection. The CRISPR locus contains short segments acquired from viral genomes, and RNAs derived from these segments assemble with Cas proteins into programmable DNA-binding complexes that target DNA molecules complementary to the guide RNA for cleavage. In type I CRISPR-Cas systems, the CRISPR-associated complex for antiviral defense (Cascade) binds to target DNA sequences and then recruits the Cas3 enzyme to repeatedly cleave the bound DNA. In this study, we show how Cascade positions both the DNA and Cas3 to ensure DNA cleavage.
Abstract
In bacteria, the clustered regularly interspaced short palindromic repeats (CRISPR)–associated (Cas) DNA-targeting complex Cascade (CRISPR-associated complex for antiviral defense) uses CRISPR RNA (crRNA) guides to bind complementary DNA targets at sites adjacent to a trinucleotide signature sequence called the protospacer adjacent motif (PAM). The Cascade complex then recruits Cas3, a nuclease-helicase that catalyzes unwinding and cleavage of foreign double-stranded DNA (dsDNA) bearing a sequence matching that of the crRNA. Cascade comprises the CasA–E proteins and one crRNA, forming a structure that binds and unwinds dsDNA to form an R loop in which the target strand of the DNA base pairs with the 32-nt RNA guide sequence. Single-particle electron microscopy reconstructions of dsDNA-bound Cascade with and without Cas3 reveal that Cascade positions the PAM-proximal end of the DNA duplex at the CasA subunit and near the site of Cas3 association. The finding that the DNA target and Cas3 colocalize with CasA implicates this subunit in a key target-validation step during DNA interference. We show biochemically that base pairing of the PAM region is unnecessary for target binding but critical for Cas3-mediated degradation. In addition, the L1 loop of CasA, previously implicated in PAM recognition, is essential for Cas3 activation following target binding by Cascade. Together, these data show that the CasA subunit of Cascade functions as an essential partner of Cas3 by recognizing DNA target sites and positioning Cas3 adjacent to the PAM to ensure cleavage.
Bacteria and archaea target invasive DNA from viruses and plasmids using RNA-guided adaptive immune systems encoded by CRISPR (clustered regularly interspaced short palindromic repeat)-Cas (CRISPR-associated) loci (1). Short segments of foreign DNA are integrated into the host genome within the CRISPR locus, transcribed into long RNAs, and enzymatically processed into mature CRISPR RNAs (crRNAs). A defining feature of CRISPR-Cas systems is the use of Cas proteins in complex with crRNAs to identify and degrade target sequences (known as protospacers) that are complementary to the crRNA guide, preventing successful phage infection or plasmid transformation (2–4). Target recognition requires both crRNA–DNA base pairing and the presence of a conserved sequence element proximal to the target site called the protospacer adjacent motif (PAM), which enables discrimination between self and non-self DNA (5). In type I and III CRISPR systems, multisubunit Cas-targeting complexes assemble with crRNAs to recognize complementary DNA or RNA sequences (3, 6–9). In contrast, type II systems rely on a single crRNA-guided enzyme, Cas9, to both recognize and cleave DNA substrates (10, 11). Recently, Cas9 has been repurposed for genome-engineering applications in a variety of animals, plants, and other organisms (12).
In the extensively studied type I–E CRISPR-Cas system in Escherichia coli, five Cas proteins assemble along with the crRNA into a targeting complex known as Cascade (CRISPR-associated complex for antiviral defense) (3) (Fig. 1A). Recognition of double-stranded DNA (dsDNA) by Cascade leads to formation of an R loop as a result of base pairing between the crRNA and complementary DNA target strand and displacement of the non-target strand (6) (Fig. 1A). Following sequence recognition, Cascade recruits a transacting effector nuclease-helicase, Cas3, for target degradation (13–15). Cryoelectron microscopy (cryoEM) reconstructions of E. coli Cascade (16) revealed a helical backbone formed by six copies of the CasC subunit that cradles the crRNA and is capped on each end by unique interactions with the other Cas components. When bound to a complementary single-stranded RNA (ssRNA) oligonucleotide, Cascade undergoes a structural rearrangement as a result of base pairing along the length of the crRNA sequence (16). However, Cascade targets dsDNA rather than ssRNA in vivo (3), and must recruit the Cas3 nuclease for CRISPR-mediated interference. The positioning of dsDNA within the Cascade complex as well as the mechanism of subsequent Cas3 recruitment and activation are unknown.
Fig. 1.
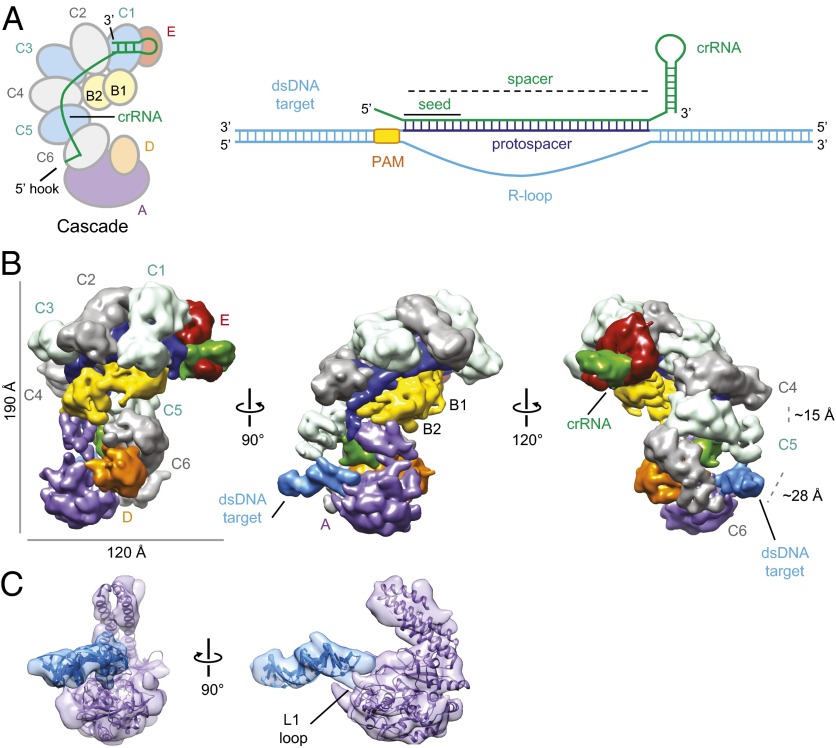
Cascade binding to dsDNA positions the PAM near CasA. (A) Subunit organization of Cascade and schematic of crRNA (green) and target DNA (sky blue). The PAM and protospacer of the target are depicted in orange and dark blue, respectively. The subunits of Cascade are colored as follows: purple, CasA (Cse1); yellow, CasB (Cse2); light blue and gray, CasC (Cas7); orange, CasD (Cas5e); and red, CasE (Cas6e). We use the Cas protein nomenclature from Brouns et al. (3), but the more recent protein names are included in parentheses (36). (B) CryoEM reconstruction of dsDNA target-bound Cascade at 9-Å resolution (0.5 FSC criterion) with subunits and target labeled and colored as in A. (Right) A larger distance between CasC5 (C5) and CasC6 (C6) relative to that between C4 and C5 allows the accommodation of the target DNA duplex. (C) Docking of the CasA crystal structure (24) (purple) and a modeled 17-bp B-form dsDNA (sky blue) into the EM density. The L1 loop of CasA is labeled.
We determined the cryoEM structure of Cascade bound to a 72-bp dsDNA target to visualize the process of natural target recognition. In addition, we investigated the mechanism of Cas3-mediated target degradation by both direct visualization of the Cas3 attachment site on Cascade and analysis of Cas3–Cascade‐mediated dsDNA cleavage using various DNA substrates and Cascade mutants. Along with recent structural studies of type II (17, 18) and III (7, 19, 20) CRISPR-Cas systems, these findings have important implications for our understanding of the emerging similarities and differences between CRISPR-associated surveillance complexes.
Results
The CasA Subunit of Cascade Directly Contacts the PAM Sequence in dsDNA.
We reconstituted a dsDNA-bound E. coli Cascade complex bearing the R44 crRNA (6, 21) with a 72-bp dsDNA target sequence containing the 32-bp R44 protospacer with 20-bp flanking sequences and an ATG PAM directly preceding the target sequence (Fig. 1A and Table S1). The binding reaction was followed by size-exclusion chromatography to remove unbound DNA (Fig. S1). Raw cryoEM micrographs of the dsDNA-bound complex embedded in ice showed monodisperse, easily identifiable particles (Fig. S2). Using the automated macromolecular microscopy system Leginon (22), we acquired ∼3,500 micrographs and automatically picked >280,000 target-bound Cascade particles using the Appion image-processing pipeline (23). After projection-matching refinement, we obtained a final target-bound Cascade structure at ∼9-Å resolution [using the 0.5 Fourier shell correlation (FSC) criterion], which shows clear structural features comparable to the previously determined Cascade structures (16) (Fig. 1B and Fig. S2).
Notably, an additional rod-shaped density not attributable to Cascade proteins projects out from one end of Cascade. A 17-bp B-form DNA duplex corresponding to the 17 bp preceding the PAM in the target dsDNA is readily accommodated within this additional density adjacent to the CasA subunit (Fig. 1C). This places the PAM directly adjacent to the L1 loop of CasA, which we previously linked to PAM recognition (24) (Fig. 1C). One helical end of the double-stranded target DNA extends into CasA and is stabilized by the distal domains of CasC5 (C5) and CasC6 (C6) (Fig. 1C). C5 and C6 have a different relative conformation (main body of C5 to C6 distance is ∼28 Å) compared with the other CasC subunits (C5 to C4 distance is ∼15 Å), as observed in the apo-Cascade structure (16). This arrangement places these subunits in the appropriate orientation and spacing to engage the target DNA duplex (Fig. 1B). We did not observe additional density for the other end of the target duplex, suggesting that this end may not be stably bound by Cascade.
Cascade maintains its overall superhelical morphology after dsDNA target binding and the density for the crRNA doubles in size relative to that of apo-Cascade, similar to what was observed in the previous ssRNA-bound Cascade structure. This crRNA–DNA hybrid density can be fit with discontinuous segments of 4–5 bp of double-stranded DNA (Fig. S3), as previously shown for the ssRNA-bound complex (16). Binding of the target dsDNA induces conformational changes in Cascade that are reminiscent of, but distinct from, those observed in the ssRNA-bound Cascade. The beak of the Cascade structure, consisting of CasE and the 3′ stem-loop of the crRNA, remains relatively unchanged between apo-Cascade and dsDNA-bound Cascade. The CasB subunits move ∼17 Å along the crRNA binding groove toward the tail (Fig. S3) and induce a ∼30° rotation of the four-helix bundle of CasA toward the tail, similar to that observed upon ssRNA binding (Fig. S3). However, in contrast to the ssRNA-bound structure, the position of the base of CasA in the DNA-bound complex remains similar to that occupied in apo-Cascade. Whereas ssRNA binding results in a concerted rigid-body rotation of the entire CasA subunit, dsDNA binding induces a ratcheting of the two major lobes of CasA relative to one another (Fig. S3). Thus, dsDNA binding triggers CasA to adopt a relative position within Cascade that is distinct from both apo-Cascade and ssRNA–Cascade. These differences are likely due to the extensive interactions between the target DNA and the base of CasA (Fig. 1B).
The Transacting Cas3 Nuclease Is Recruited Opposite the Target on CasA.
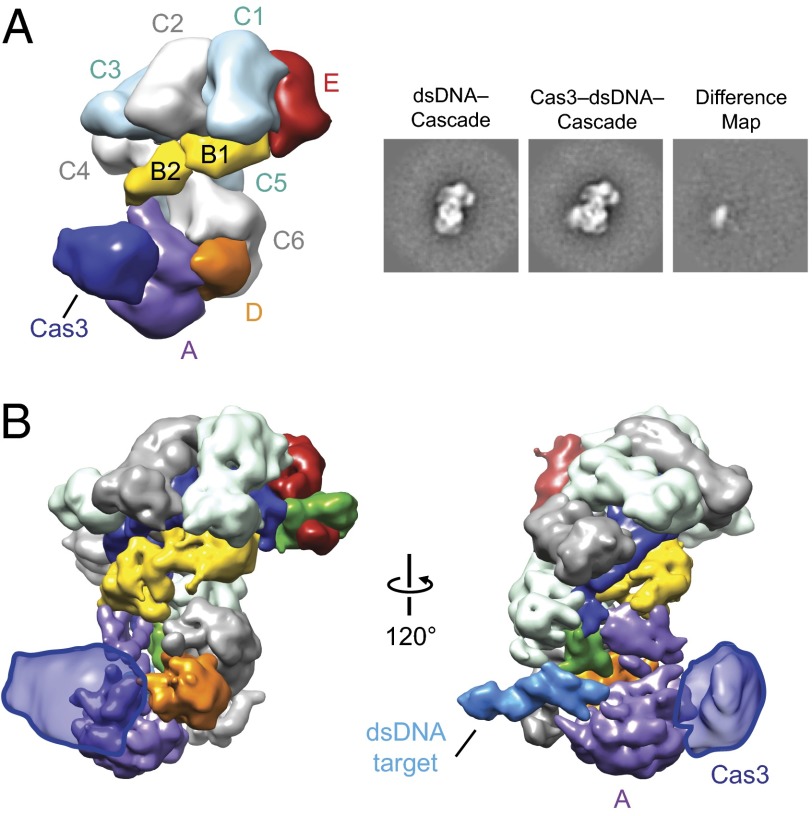
The manner in which Cascade recruits Cas3 has remained enigmatic in the CRISPR field. We formed Cas3–dsDNA–Cascade complexes and studied them by electron microscopy. Cas3–dsDNA–Cascade complexes dissociated under our initial conditions, with particles corresponding to Cascade and smaller particles with the predicted size of Cas3 easily recognizable in raw micrographs. Because of sample heterogeneity and the limiting amount of Cas3, we used negative-stain EM to provide the required signal to build a reliable reconstruction. To prevent target cleavage from occurring before observing the sample by EM, we kept all samples on ice to inhibit Cas3 activity (Fig. S4). To improve its stability, we examined the architecture of the complex after mild glutaraldehyde cross-linking following prebinding of the same 72-bp target and reconstitution with Cas3. This procedure resulted in the appearance of class averages with clear extra density that could be ascribed to Cas3 (Fig. 2A and Fig. S5). Importantly, we were able to localize Cas3 to the same region of Cascade in the absence of cross-linking agent by using a large (10×) molar excess of Cas3 over dsDNA-bound Cascade during the reconstitution (Fig. S5). To increase the proportion of stable complexes, we used the cross-linked sample for further structural studies. Using the 3D reconstruction of ssRNA–Cascade low-pass–filtered to 60 Å as a starting model for projection-matching refinement, we obtained a 3D reconstruction of Cas3–dsDNA–Cascade from ∼11,000 particles at a final resolution of ∼20 Å (Fig. 2B and Fig. S5). This structure features an extra globular density that can be assigned to Cas3 and is bound to CasA near the junction of its four-helix bundle and base (Fig. 2A). Interestingly, Cas3 interacts with CasA near the region of conformational changes induced by target DNA binding. Although nucleic acids cannot be directly visualized in EM structures of negatively stained complexes, we could easily map the Cas3 density onto our target-bound Cascade cryoEM structure (Fig. 2B). Based on the resulting model, Cas3 and the target duplex are engaged on opposite sides of CasA. The localization of Cas3 near CasA and CasD is consistent with naturally occurring fusions of Cas3 to both CasA (13) and CasD (UniProtKB accession no. D4CIN8). It is important to note that the density for Cas3 in our reconstruction is not large enough to account for the entire mass of the protein (Fig. S6). We hypothesize that this density likely corresponds to one domain of the multidomain Cas3.
Fig. 2.
Cas3 interacts with CasA. (A) (Left) Negative-stain reconstruction of TEV-cleaved Cas3-bound dsDNA–Cascade at 20-Å resolution (0.5 FSC criterion) with subunits and Cas3 labeled. (Right) Reference-free 2D class average of the dsDNA–Cascade complex, the corresponding reference-free 2D class average of Cas3–dsDNA–Cascade, and the difference map between these structures. The width of the boxes is ∼288 Å. (B) Cas3 density (outlined semitransparent blue surface) from the negative-stain reconstruction (A) mapped onto the cryoEM reconstruction of dsDNA-bound Cascade.
Cleavage of Cascade-Bound dsDNA by Cas3 Depends on Base-Pairing Potential of the PAM.
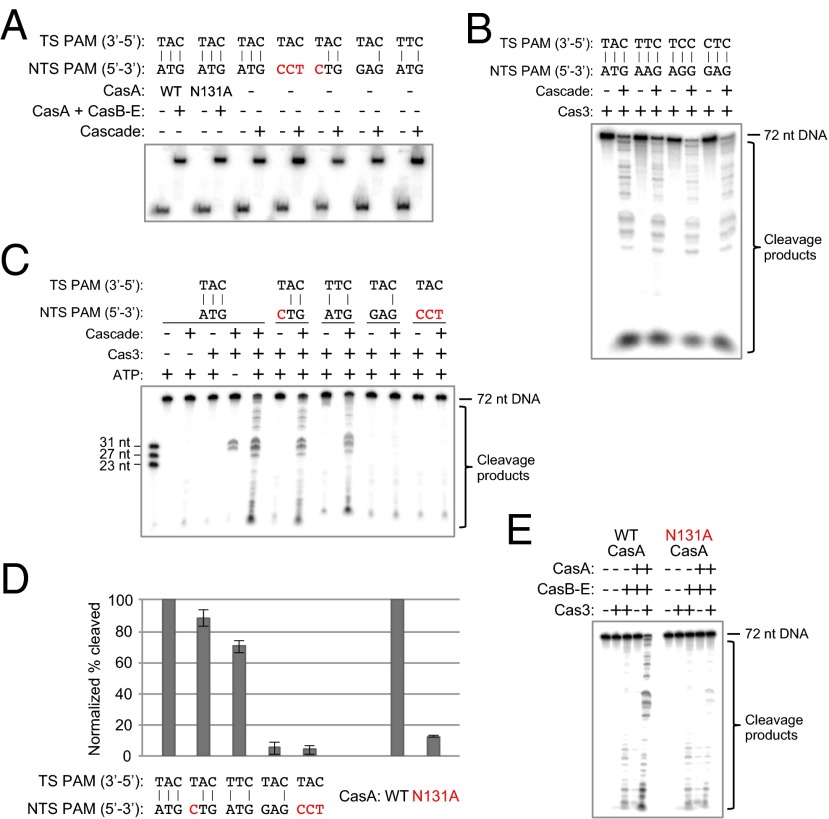
Because Cas3 localizes to CasA, the subunit responsible for PAM recognition during initial target binding, we wondered whether mutations to the PAM sequence might affect target cleavage. To test this, we performed in vitro Cas3 cleavage assays with Cascade and radiolabeled, double-stranded DNA substrates containing various mutations to the PAM region. Previous studies that tested cleavage of substrates with incorrect PAM sequences used low concentrations of Cascade and target DNAs that were bound either poorly or not at all under the test conditions (14, 15). Thus, any defects in cleavage were easily explained by the absence of Cascade targeting. We sought to investigate the effects of PAM mutations on cleavage in a manner independent of binding. To ensure that any observed cleavage defects were not simply a result of low Cascade affinity for a dsDNA target, we only made mutations to the non-target strand, which do not significantly affect binding affinity (13, 24) (Table S2). We used a saturating concentration of Cascade (1 μM) to ensure that all target DNA molecules were bound under cleavage conditions, as confirmed by native gel electrophoresis (Fig. 3A). We prebound the DNA targets with Cascade, added ATP and Cas3 to the indicated reactions, and allowed cleavage to proceed for 30 min before quenching and analysis. We found that alterations to the target strand PAM led to highly reduced binding by Cascade that cannot be saturated even at high concentrations, characteristic of the nonspecific binding behavior observed previously between Cascade and non-target DNA (24) (Fig. S7A), as well as negligible cleavage by Cas3 (Fig. S7B).
Fig. 3.
Target cleavage by Cas3 requires base pairing within the PAM sequence and integrity of the PAM-binding loop in CasA. (A) Native gel electrophoretic mobility-shift assay showing that all dsDNA substrates tested for Cas3-mediated degradation are fully bound by Cascade under the conditions used in the cleavage assays. Red text indicates a nucleotide mutation that changes the PAM to a sequence other than one of the four functional motifs. Cascade (1 µM) was incubated with 0.1–0.5 nM [32P]dsDNA at 37 °C for 30 min before gel-shift analysis by 10% native PAGE. NTS, non-target strand; TS, target strand. (B) All four PAM sequences that function in vivo also permit target cleavage in vitro. DNA cleavage reactions were performed using target dsDNAs with different PAM sequences at a final concentration of 1 nM in 1× reaction buffer. In the presence of 2 mM ATP, 1 μM targeting Cascade, and 500 nM MBP-tagged Cas3, all four targets are cleaved after incubation at 37 °C for 30 min. (C) PAM base-pair mismatches prevent efficient target cleavage by Cas3. Targeting Cascade was prebound to each dsDNA before addition of ATP and MBP-Cas3. (D) Quantified cleavage percentages for each condition in the presence of Cas3 and ATP are the average of three (E) or four (C) independent replicates; error bars represent ±1 SD. All values have been normalized to the WT cleavage efficiency. (E) Denaturing polyacrylamide gel depicting the cleavage defect caused by mutation of Asn131 in the L1 loop of CasA. Wild-type or N131A CasA (1 μM) was added back to 1 μM Strep-tag II–tagged CasB–E to reconstitute the Cascade complex and prebound to a 1 nM dsDNA target with a functional PAM before addition of ATP and MBP-Cas3.
Using 72-bp radiolabeled duplexes, we found that Cas3 cleaves targets with any of the four E. coli PAM sequences shown to be functional for in vivo interference: ATG, AAG, AGG, and GAG (13) (Fig. 3B). Consistent with previous work (14, 15), when a dsDNA target has a correct PAM and ATP is present, the action of Cas3 results in near-complete degradation, whereas in the absence of ATP, Cas3 nicks only the displaced, non-target strand between nucleotides 7 and 11 of the protospacer (Fig. 3C). We next mutated the sequence of the non-target strand PAM to prohibit Watson–Crick base pairing. Although single mismatches are somewhat tolerated, disrupting the base-pairing potential of two or all three PAM nucleotides prevents target cleavage by Cas3 (Fig. 3 C and D). Even when the non-target strand PAM consists of a sequence known to be sufficient for in vitro cleavage and in vivo interference (e.g., GAG, the fourth substrate tested in Fig. 3C), the target is not cleaved if it contains unpaired PAM nucleotides. Thus, the sequence on the non-target strand can be altered without greatly affecting binding affinity or cleavage efficiency, but base pairing in this region must be retained for the target to be degraded.
L1 Loop of CasA Is Required for Cas3-Mediated Cleavage of a Cascade Target.
The proximity of the PAM to CasA and Cas3 in the target-bound Cascade complex led us to hypothesize that the CasA subunit transmits information to Cas3 as a result of target recognition. To test this idea, we mutated the L1 loop of CasA and performed Cas3–Cascade cleavage assays as described above, using the same target DNA duplex used to generate our EM structures. We reconstituted the Cascade complex by incubating wild-type or N131A CasA with separately purified CasB–E, as described previously (24). Although the N131A mutation does not substantially impact Cascade’s affinity for a target, we prebound the reconstituted Cascade to the radiolabeled dsDNA target at a high concentration to ensure all targets were bound under cleavage conditions, which we confirmed by native gel-shift assays (Fig. 3A). We observed efficient DNA degradation with WT CasA, whereas the N131A mutation resulted in drastically diminished target cleavage efficiencies (Fig. 3 D and E). Thus, despite complete binding by Cascade, Cas3 is unable to cleave the DNA duplex when the CasA L1 loop is perturbed.
Discussion
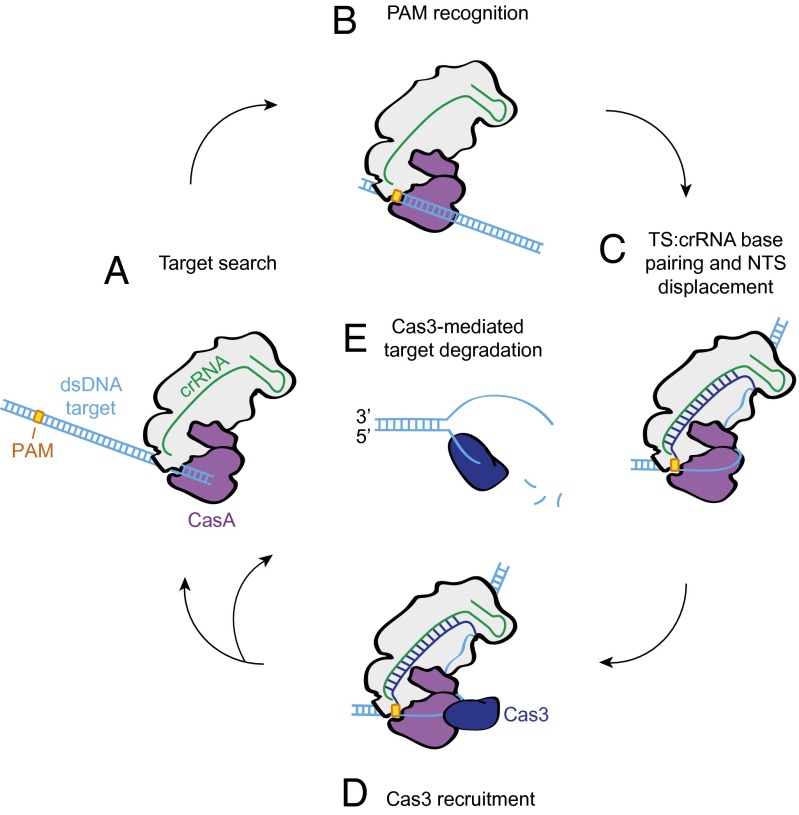
We propose a model for Cascade function in which binding to a dsDNA target with an intact PAM sequence drives critical rearrangements of the complex to enable productive encounters with Cas3 (Fig. 4). Our cryoEM reconstruction reveals that the target DNA duplex enters Cascade between CasC5 and C6 (Fig. 1B), in agreement with our current and previous observations (16) of a larger gap between these two proteins than between the other subunits of the helical backbone. Nonspecific interactions between CasA, these CasC subunits, and dsDNA may facilitate the initial target search before PAM recognition (Fig. 4A). The CasA L1 loop contacts the entering duplex at the approximate location of the PAM (Fig. 1C), where the target strand sequence is recognized (Fig. 4B). We hypothesize that in addition to initial target identification via PAM recognition, CasA is responsible for subsequent stabilization and arrangement of the displaced strand to ensure cleavage by Cas3 (Fig. 4C). The CasA L1 loop, as well as the base-pairing potential of the PAM nucleotides, is crucial for degradation to occur (Fig. 3).
Fig. 4.
Model for target recognition and Cas3 recruitment by Cascade. (A) Cascade searches dsDNA for PAM sites. (B) CasA recognizes the PAM sequence via the L1 loop. (C) Once CasA has located a bona fide target, a rearrangement of Cascade facilitates crRNA–protospacer DNA hybridization. (D) CasA positions the displaced, non-target strand for recruitment of the transacting Cas3 nuclease. (E) Following initial nicking of the displaced strand, Cas3 loads onto the newly formed ssDNA end and translocates along the substrate during processive degradation of the target dsDNA.
Moreover, our cryoEM structure of dsDNA-bound Cascade suggests the presence of single-stranded DNA that appears to be bound at the cleft between the four-helix bundle and the base domains of CasA immediately following the clearly visible double-stranded DNA (Fig. S6). Although the limited resolution of our present structure makes it hard to determine unambiguously whether this density is protein or DNA, the binding of ssDNA to this region of CasA would provide a structural link between the recognition of the PAM sequence and the site of Cas3 association. Higher-resolution studies will be required to confirm this idea.
Thus, we hypothesize that the non-target strand is looped around CasA such that binding of Cas3 at this site (Fig. 2) positions the nuclease domain to make its first nick ∼7–11 nt into the protospacer (Fig. 4D). Our laboratory has shown previously that the non-target strand is particularly susceptible to hydroxyl radical cleavage in this region (24). Intriguingly, this analysis revealed that the same CasA mutation that prevents target cleavage leads to a decrease in exposure of this particular site. Furthermore, it causes increased exposure of the nucleotides preceding the cut site, suggesting an overall weaker hold of the displaced strand. We predict that mutation of the L1 loop or disruption of PAM base pairing leads to improper binding of the non-target strand, inaccessibility of the Cas3 cleavage site, and failure of Cas3 to degrade the target DNA. When Cascade binds properly at target sites, Cas3 is recruited to the complex, nicks the displaced strand, and then processively unwinds and cleaves both strands to fully degrade the foreign DNA (Fig. 4E).
Together, these studies shed light on the mechanism by which Cascade binding to a dsDNA target leads to its degradation by the transacting nuclease-helicase Cas3. CasA links these two processes through physical interactions with DNA as well as with Cas3. We have shown that although target binding and degradation are linked, they have different requirements. Binding depends on sequence-specific recognition of the PAM on the target strand, whereas cleavage is influenced by the geometry of the PAM region, with a preference for a fully base-paired helix. Binding by Cascade alone does not necessarily lead to target degradation by Cas3, and previous observations indicate that reduced binding does not always lead to cleavage defects. Semenova et al. identified mutations to the protospacer sequence that decreased Cascade’s binding affinity substantially, but phage harboring these mutations were unable to escape interference in vivo (25). Our results complement this finding and underscore the complexity of target interference in the E. coli CRISPR system. The subtle interplay between DNA binding by Cascade and Cas3-mediated degradation awaits further elucidation and represents an exciting avenue of future CRISPR research.
Materials and Methods
Cloning and Protein Purification.
The Cas3 gene from E. coli K12 was amplified using PCR primers containing KasI and XhoI restriction sites (Table S1) and cloned into the expression vector pSV272, encoding an N-terminal His6-MBP (maltose-binding protein) tag followed by a tobacco etch virus (TEV) protease recognition site (Table S3).
Cas3 was overexpressed in E. coli BL21(DE3). Cells were grown at 20 °C to an OD600 of 0.3, at which point protein expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside, similar to the purification described previously (15). After overnight growth, the cells were harvested and lysed by sonication in 1× His (15.5 mM Na2HPO4, 4.5 mM NaH2PO4, 500 mM NaCl), 10 mM imidazole (pH 7.5), 5% (vol/vol) glycerol, 1 mM tris(2-carboxyethyl)phosphine (TCEP), Roche protease inhibitor tablets, 0.01% Triton X-100, and 0.5 mM PMSF. Lysate was clarified by centrifugation at 27,000 × g and purified using Ni-NTA affinity resin (QIAGEN). The resin was washed with buffer containing 1× His, 20 mM imidazole (pH 7.5), 5% (vol/vol) glycerol, and 1 mM TCEP, and protein was eluted with 1× His, 300 mM imidazole (pH 7.5), 5% (vol/vol) glycerol, and 1 mM TCEP. The protein was dialyzed for 3 h into buffer A [50 mM Hepes (pH 7.5), 50 mM KCl, 1 mM TCEP, 0.1 mM EDTA, and 5% (vol/vol) glycerol], loaded onto a 5-mL HiTrap Q HP anion-exchange column (GE Healthcare), and eluted using a linear gradient from buffer A to buffer B [50 mM Hepes (pH 7.5), 1 M KCl, 1 mM TCEP, 0.1 mM EDTA, and 5% (vol/vol) glycerol]. The protein was further purified on a HiLoad 16/60 Superdex 200 size-exclusion column (GE Healthcare) pre-equilibrated in gel-filtration buffer containing 50 mM Hepes (pH 7.5), 200 mM KCl, 1 mM TCEP, 0.1 mM EDTA, and 5% (vol/vol) glycerol. Most His6-MBP-Cas3 was flash-frozen and stored in gel-filtration buffer at −80 °C. The His6-MBP tag was removed from the remainder of the protein by incubation with TEV protease at 4 °C for 4 h. The cleaved sample was repurified by size-exclusion chromatography, concentrated, flash-frozen, and stored at −80 °C.
WT and N131A CasA proteins were purified as previously described (24). The CasB–E subcomplex and Cascade were initially purified using Strep-Tactin Superflow Plus resin (QIAGEN) using an N-terminal Strep-tag II on the CasB subunit, as previously described (16). The Strep-tag II peptide was removed from Cascade by cleavage with PreScission protease. The complexes were then purified by size-exclusion chromatography using a Superose 6 preparatory-grade column (GE Healthcare). CasB–E was stored in a buffer containing 20 mM Tris (pH 7.5), 100 mM NaCl, 5% (vol/vol) glycerol, and 1 mM TCEP, and Cascade was stored in 50 mM Hepes (pH 7.5), 100 mM KCl, 5% (vol/vol) glycerol, 1 mM TCEP, and 0.1 mM EDTA.
Proteins were analyzed for purity by SDS/PAGE (Fig. S8).
DNA–Cascade Complex Reconstitution for CryoEM.
DNA oligomers were ordered from Integrated DNA Technologies (Table S1). The dsDNA target used for DNA–Cascade complex reconstitution was formed by annealing the target strand (MLH-46; Table S1) with a 2× molar excess of non-target strand (MLH-47) in hybridization buffer containing 40 mM Tris (pH 8.0), 38 mM MgCl2, and 1 mM spermidine, heating at 95 °C for 2 min, and cooling at room temperature for 1 h. A 2× molar excess of the annealed dsDNA was incubated with Cascade at 37 °C for 30 min in a buffer containing 50 mM Hepes (pH 7.5), 100 mM KCl, 5% (vol/vol) glycerol, 0.1 mM EDTA, and 1 mM TCEP before running over a Superdex 200 10/300 size-exclusion column (GE Healthcare). Fractions from the first peak (DNA–Cascade) were pooled and concentrated for analysis by cryoEM.
CryoEM.
dsDNA-bound Cascade complexes were frozen in vitreous ice as described previously (16). Four-microliter droplets of sample (∼2.3 mg ml−1) were placed onto C-flat grids containing 2-μm holes with 2-μm spacing between holes (Protochips) immediately after the grids were glow-discharged for 60 s using an Edwards carbon evaporator. Grids were rapidly plunged into liquid ethane using an FEI Vitrobot maintained at 4 °C and 100% humidity after being blotted for 4–4.5 s using a blotting offset of −1 mm. Data were acquired using a Tecnai F20 Twin transmission electron microscope operating at 120 keV at a nominal magnification of 100,000× (1.15 Å at the specimen level) using low-dose exposures (∼20 e−⋅Å−2) with a random defocus ranging from −1.0 to −2.8 μm. A total of 3,580 images were automatically recorded on a Gatan 4k × 4k CCD camera using the MSI-Template application within Leginon (22).
Single-Particle Preprocessing.
All image processing and 2D classification were performed in Appion (23) as described previously (16). The contrast transfer function of each micrograph was estimated, and particles were selected during data collection using ACE2 (26) and a template-based particle picker (27) using apo E. coli Cascade reference-free 2D class averages as templates (16), respectively. Reference bias was assessed by an analysis of the similarities and differences between the reference-free 2D class average templates and the reference-free 2D class averages obtained in this study. Notably, many of the class averages from this study contain an additional density for the DNA duplex. Furthermore, because class averages were used as templates, the particle picking relied on low-resolution structural details. Phases of micrographs were corrected using ACE2 (26), and both the dsDNA-bound Cascade and negatively stained Cas3–dsDNA–Cascade particles were extracted using a 288 × 288-pixel box size. The particle stacks were binned by a factor of 2 for processing, and particles were normalized to remove pixels whose values were above or below 4.5-σ of the mean pixel value using XMIPP (28).
Three-Dimensional Reconstruction and Analysis.
Both 3D reconstructions were performed using iterative projection-matching refinement with libraries from the EMAN2 and SPARX software packages (30, 31). Refinement of the starting model [Cascade–ssRNA, Electron Microscopy Data Bank accession no. EMDB-5315 (16) low-pass–filtered to 60 Å] began using an angular increment of 25°, progressing down to 4° or 0.8° for the reconstruction of Cas3–dsDNA–Cascade or dsDNA–Cascade, respectively. The resolution was estimated by splitting the particle stack into two equally sized datasets and calculating the Fourier shell correlation between each of the back-projected volumes. The final reconstructions of dsDNA–Cascade and Cas3–dsDNA–Cascade showed structural features to ∼9- and ∼20-Å resolution, respectively, based on the 0.5 FSC criterion. Some α-helices can be seen as “sausages” in our ∼9-Å cryoEM map that agree with those in the crystal structure of CasA (Fig. 1C). This confirms that the resolution of this map is in the 9–10 Å range. Reprojections of the final 3D reconstruction showed excellent agreement with the reference-free class averages and displayed a large distribution of Euler angles, despite moderate preferential orientations of the particles on the carbon film (Fig. S5). The reconstruction of dsDNA–Cascade and Cas3–dsDNA–Cascade has an obvious additional density for the target DNA and Cas3, respectively. Additionally, the conformational changes we observe upon target binding for the dsDNA–Cascade structure are distinct from the previous reconstruction. These results suggest that there is not significant reference bias in our final models.
The final reconstruction was segmented using Segger (32) in Chimera (33). The crystal structure of Thermus thermophilus CasA [Protein Data Bank (PDB) ID code 4AN8] (24) is docked in both Fig. 1C and Fig. S6. The nuclease and helicase domains from a structural model of E. coli Cas3 generated by Phyre2 (34) based on PDB ID codes 4BUJ, 4BGD, 4F92, 2EYQ, 2VA8, 2XGJ, and 3SKD are also modeled separately in Fig. S6.
Cas3–Cascade Complex Reconstitution and Cross-Linking for Negative-Stain EM.
Cas3–dsDNA–Cascade complex reconstitution was performed in buffer containing 20 mM Hepes (pH 7.5), 150 mM KCl, 5% (vol/vol) glycerol, 1 mM NiCl2, and 1 mM MgCl2. A 1.5× molar excess of the gel-purified dsDNA target (MLH-46 + MLH-47) was preincubated with Cascade at 37 °C for 30 min before cooling on ice for 5 min. A 2× molar excess of TEV-cleaved Cas3 was added, and the reaction was kept on ice for 1 h. Glutaraldehyde was added to a final concentration of 0.15%, and the cross-linking reaction was incubated on ice for 30 min before dilution to a final protein concentration of 55–110 nM and application to grids for negative-stain EM analysis (Fig. 2).
Reactions were also performed in the absence of cross-linking agent. In this case, dsDNA–Cascade was prepared identically, but a 9.6× molar excess of Cas3 was incubated with Cascade on ice for 3 h in a buffer containing 20 mM Hepes (pH 7.5), 100 mM KCl, 5% (vol/vol) glycerol, 1 mM NiCl2, and 1 mM MgCl2. Samples were diluted to a final protein concentration of ∼85 nM (Fig. S5).
Negative-Stain Electron Microscopy.
Cas3–dsDNA–Cascade complexes in buffer containing 20 mM Hepes (pH 7.5), 150 mM KCl, 5% (vol/vol) glycerol, 1 mM NiCl2, and 1 mM MgCl2 were applied to glow-discharged 400-mesh continuous carbon grids. After adsorption for 1 min, we stained the samples consecutively with five droplets of 2% (wt/vol) uranyl acetate solution, blotted off the residual stain, and air-dried the sample in a fume hood. Data were acquired using a Tecnai F20 Twin transmission electron microscope operated at 120 keV at a nominal magnification of 80,000× (1.45 Å at the specimen level) using low-dose exposures (∼20 e−⋅Å−2) with a random defocus ranging from −0.5 to −1.3 μm. A total of 300–400 images of Cascade–dsDNA–Cas3 was automatically recorded on a Gatan 4k × 4k CCD camera using the MSI-Raster application within Leginon (22).
Localization of Cas3 by Difference Mapping.
Particles were subjected to five rounds of iterative MSA and MRA using the IMAGIC software package (29) to generate 2D class averages of each complex. The resulting set of class averages for each species was normalized using proc2d in EMAN (35). The EMAN classification program classesbymra was used to match the labeled class average to the best-matching unlabeled class average based on cross-correlation coefficients. The difference map was calculated by subtracting the class average of the dsDNA-bound Cascade from the class average of the dsDNA-bound Cascade containing Cas3, using proc2d in EMAN.
Oligoduplex Preparation.
DNA oligomers were ordered from Integrated DNA Technologies (Table S1). Single-stranded oligomers were purified on denaturing gels containing 10% (vol/vol) 29:1 polyacrylamide, 8 M urea, and 1× Tris/Borate/EDTA (TBE). DNA bands were visualized by UV light, excised, and eluted by soaking gel pieces in deionized H2O. Gel pieces were removed, and DNA was ethanol-precipitated and resuspended in deionized H2O. Duplexes were formed by mixing equimolar amounts of each strand in 20 mM Hepes (pH 7.5), 100 mM KCl, 5 mM MgCl2, and 5% (vol/vol) glycerol, heating at 95 °C for 2 min, and cooling at room temperature for 10 min. Duplexes were resolved on native gels containing 6% (vol/vol) 29:1 polyacrylamide and 1× TBE and purified as described above. DNA samples were 5′ labeled with [γ-32P]ATP using polynucleotide kinase (PNK; New England Biolabs) for 30 min at 37 °C. PNK was heat-denatured at 65 °C for 20 min, and excess ATP was removed using a G-25 spin column (GE Healthcare).
Electrophoretic Mobility-Shift Assays.
All binding assays were performed in 1× reaction buffer, which contained 20 mM Hepes (pH 7.5), 100 mM KCl, 5% (vol/vol) glycerol, 100 μM NiCl2, and 5 mM MgCl2. CasA and CasB–E were preincubated in 1× reaction buffer for 15 min at 37 °C to ensure complex formation before addition of DNA. After diluting all reaction components into 1× reaction buffer, DNA was added to a final concentration of 0.1–0.5 nM. Samples were incubated at 37 °C for 30 min and then resolved at 4 °C on a 6% 29:1 polyacrylamide gel containing 1× TBE. Gels were dried and DNA was visualized by phosphorimaging and quantified using ImageQuant software (GE Healthcare). The fraction of DNA bound (amount of bound DNA divided by the sum of free and bound DNA) was plotted versus the concentration of Cascade and fit to a binding isotherm using Kaleidagraph software (Synergy). Reported Kd values are the average of three independent replicates, and error bars represent the SD.
DNA Cleavage Assays.
All cleavage assays were performed in 1× reaction buffer. CasB–E and WT or N131A CasA were preincubated for 15 min at 37 °C to ensure complex formation before addition of DNA. Cascade components were prebound to DNA by incubating at 37 °C for 30 min. Samples were cooled on ice for 5 min before addition of ATP to a final concentration of 2 mM and His-MBP-Cas3 to 500 mM. The final concentration of DNA was 1 nM, Cascade was 1 μM, and CasA + CasB–E were 1 μM each. Reactions were incubated at 37 °C for 30 min and then resolved on denaturing gels containing 10% (vol/vol) 29:1 polyacrylamide, 8 M urea, and 1× TBE. Gels were dried and DNA was visualized by phosphorimaging and quantified using ImageQuant software (GE Healthcare). The percentage of DNA cleaved was determined by dividing the amount of cleaved DNA (the entirety of the lane below the 72-nt uncleaved band) divided by the sum of uncleaved and cleaved DNA. The background percentage cleaved in control lanes containing only Cascade components or only Cas3 (whichever value was higher) was subtracted to generate the corrected percent cleaved. Reported values are the average of at least three independent replicates, and error bars represent the SD. Values have been normalized to the WT percent cleaved (71 ± 3.0% for a substrate with WT PAM and 71 ± 1.1% when WT CasA is added back to CasB–E).
Supplementary Material
Acknowledgments
We thank G. Lander, P. Grob, Y. He, and T. Houweling for expert EM and image-processing assistance; K. Zhou and K. Condon for technical assistance; J. van der Oost, H.-W. Wang, B. Wiedenheft, and members of the E.N. and J.A.D. laboratories for helpful discussions. This project was funded by US National Science Foundation (NSF) Grant 1244557 (to J.A.D.). S.H.S. is an NSF and National Defense Science and Engineering Graduate Research Fellow. E.N. and J.A.D. are Howard Hughes Medical Institute Investigators.
Footnotes
The authors declare no conflict of interest.
Data deposition: The electron microscopy reconstructions of dsDNA–Cascade and Cas3–dsDNA–Cascade have been deposited in the Electron Microscopy Data Bank, www.emdatabank.org (EMDataBank accession nos. EMD-5929 and EMD-5930, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405079111/-/DCSupplemental.
References
- 1.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 5.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 6.Jore MM, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18(5):529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 7.Staals RH, et al. Structure and activity of the RNA-targeting type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 2013;52(1):135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139(5):945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hale CR, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45(3):292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terns RM, Terns MP. CRISPR-based technologies: Prokaryotic defense weapons repurposed. Trends Genet. 2014;30(3):111–118. doi: 10.1016/j.tig.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westra ER, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46(5):595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulepati S, Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem. 2013;288(31):22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinkunas T, et al. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 2013;32(3):385–394. doi: 10.1038/emboj.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477(7365):486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimasu H, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spilman M, et al. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell. 2013;52(1):146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouillon C, et al. Structure of the CRISPR interference complex CSM reveals key similarities with Cascade. Mol Cell. 2013;52(1):124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 22.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166(1):95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46(5):606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semenova E, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108(25):10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: Automated CTF estimation. Ultramicroscopy. 2005;104(1):8–29. doi: 10.1016/j.ultramic.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Roseman AM. FindEM—A fast, efficient program for automatic selection of particles from electron micrographs. J Struct Biol. 2004;145(1-2):91–99. doi: 10.1016/j.jsb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Sorzano CO, et al. XMIPP: A new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148(2):194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 29.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116(1):17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 30.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Hohn M, et al. SPARX, a new environment for cryo-EM image processing. J Struct Biol. 2007;157(1):47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170(3):427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 35.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 36.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.