Significance
The mechanisms that enable Mycobacterium tuberculosis, the causative agent of tuberculosis, to resist drug treatment and survive the immune response are poorly understood. In this study we discovered that M. tuberculosis produces the protein channel protein with necrosis-inducing toxin (CpnT), which forms a channel in the outer membrane and releases a toxic domain into the extracellular milieu. This toxin has no similarity to known bacterial toxins and kills eukaryotic cells by necrosis, suggesting that it is required for escape of M. tuberculosis from macrophages and for dissemination. The channel domain of CpnT is used for uptake of nutrients across the outer membrane. Taken together, CpnT is a protein with functions in two fundamental processes in M. tuberculosis physiology: nutrient acquisition and control of host cell death.
Keywords: transport, pore, secretion
Abstract
The ability to control the timing and mode of host cell death plays a pivotal role in microbial infections. Many bacteria use toxins to kill host cells and evade immune responses. Such toxins are unknown in Mycobacterium tuberculosis. Virulent M. tuberculosis strains induce necrotic cell death in macrophages by an obscure molecular mechanism. Here we show that the M. tuberculosis protein Rv3903c (channel protein with necrosis-inducing toxin, CpnT) consists of an N-terminal channel domain that is used for uptake of nutrients across the outer membrane and a secreted toxic C-terminal domain. Infection experiments revealed that CpnT is required for survival and cytotoxicity of M. tuberculosis in macrophages. Furthermore, we demonstrate that the C-terminal domain of CpnT causes necrotic cell death in eukaryotic cells. Thus, CpnT has a dual function in uptake of nutrients and induction of host cell death by M. tuberculosis.
Toxins were recognized more than a century ago to play a major role in bacterial infectious diseases (1). Subsequently, hundreds of toxins from pathogenic bacteria have been characterized. Based on bioinformatic analysis of its genome, toxins appear to be absent from Mycobacterium tuberculosis (2–4), the causative agent of tuberculosis, a devastating disease with nine million new cases every year (5). Survival within host macrophages is a key trait enabling M. tuberculosis to persist in the human body (6), where it can reactivate after decades of quiescence (7). This so-called latent infection is poorly understood and is one of the reasons why tuberculosis remains a global public health problem. Alveolar macrophages engulf inhaled M. tuberculosis, contribute to killing of the bacteria, reduce inflammation of lung tissue, and limit uptake of M. tuberculosis by migratory dendritic cells to prevent bacterial dissemination (8). However, M. tuberculosis has evolved effective strategies to subvert this bactericidal response (6, 9). Death of M. tuberculosis-infected macrophages is caused by two processes: necrosis and apoptosis. Necrosis is characterized by metabolic collapse and loss of membrane integrity and is used by M. tuberculosis to exit destroyed cells, evade host defenses, and disseminate to other tissues and eventually to new hosts (10). By contrast, apoptosis of the infected macrophages helps the host to control the bacterial infection (11). Virulent M. tuberculosis strains induce a necrosis-like cell death and concomitantly suppress apoptosis of macrophages (12). Although M. tuberculosis is known to secrete virulence factors that interfere with phagosome maturation (9), it is unknown how M. tuberculosis kills macrophages.
Gram-negative bacterial pathogens use complex nanomachines such as type I–VI secretion systems to secrete effector proteins mediating host cell death and subverting immune responses (13). Similarly, proteins secreted by M. tuberculosis need to cross both an inner and an outer membrane (14), a barrier of notoriously low permeability in M. tuberculosis (15). However, the only known secretion systems capable of translocating proteins across both M. tuberculosis membranes are the type VII secretion systems encoded by esx operons. Although inner membrane proteins of ESX secretion systems have been characterized (16), channel proteins that are required for protein translocation across the outer membrane are currently unknown in M. tuberculosis. We hypothesized that deletion or inactivation of outer membrane channel proteins in M. tuberculosis may result in increased antibiotic resistance, as has been described for Gram-negative bacteria and Mycobacterium smegmatis (17, 18). Here we show that this approach identified a protein that enables uptake of small, hydrophilic molecules via its N-terminal pore domain and induces host cell necrosis by its secreted toxic C-terminal domain (CTD).
Results
CpnT Is Required for Efficient Growth on and Uptake of Glycerol by Mycobacterium bovis Bacillus Calmette–Guérin and M. tuberculosis.
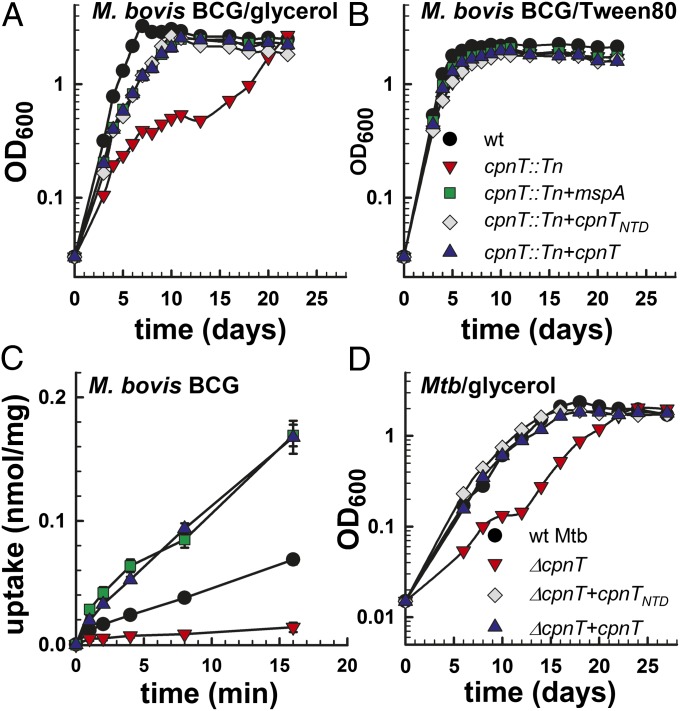
In a previous study, we identified an ampicillin-resistant mutant in a transposon library of M. bovis bacillus Calmette–Guérin with an insertion in the bcg3960c gene (19). The bcg3960c gene encodes a protein identical to Rv3903c (channel protein with necrosis-inducing toxin, CpnT) of M. tuberculosis (SI Appendix, Fig. S1A). Growth of the cpnT::Tn mutant in defined medium was impaired compared with wild-type (WT) M. bovis bacillus Calmette–Guérin when glycerol was the sole carbon source (Fig. 1A). No growth difference was observed when the detergent Tween-80 was the sole carbon source, indicating that the cpnT::Tn mutant did not have a general growth defect (Fig. 1B). Although glycerol rapidly accumulated in the WT strain, only minimal uptake of glycerol was detected in the cpnT::Tn mutant (Fig. 1C). These data show that CpnT is required for normal growth on, and uptake of, glycerol in M. bovis bacillus Calmette–Guérin. Deletion of cpnT (SI Appendix, Fig. S2) also reduced growth of M. tuberculosis on glycerol and glucose as sole carbon sources (Fig. 1D and SI Appendix, Fig. S3). The phenotype of the ΔcpnT M. tuberculosis mutant was less pronounced compared with the M. bovis bacillus Calmette–Guérin mutant, indicating that other pore proteins might be involved in uptake of small, hydrophilic nutrients in M. tuberculosis. Starvation-induced expression of silent porin genes (20) might explain the ability of the M. tuberculosis and M. bovis bacillus Calmette–Guérin cpnT mutants to eventually reach optical densities of the WT strains after an initial growth delay (Fig. 1 A and D). Importantly, both uptake and growth defects were rescued by expression of cpnT or of the porin gene mspA of M. smegmatis (Fig. 1 A and C and SI Appendix, Fig. S4), indicating that CpnT has the same outer membrane localization and a similar channel function as MspA (21, 22).
Fig. 1.
CpnT is required for efficient growth on and uptake of glycerol by M. bovis bacillus Calmette–Guérin (BCG) and M. tuberculosis. Growth of WT M. bovis BCG (black circle), cpnT::Tn (red inverted triangle), cpnT::Tn complemented with mspA (green square), cpnTNTD (gray diamond), or cpnT (blue triangle) in minimal Hartmans-de Bont (HdB) medium supplemented with 0.1% glycerol (A) and 1% Tween-80 (B). Experiments were carried out at least three times. Representative growth curves are shown. (C) [14C]Glycerol uptake by M. bovis BCG. The uptake rate is expressed as nanomole of glycerol per milligram of cells. Uptake experiments were done in triplicate and mean values are shown with SDs. The P value determined by Student t test was less than 0.05 for WT versus the cpnT::Tn mutant for all time points. (D) Growth of WT M. tuberculosis (black circle), ∆cpnT (red inverted triangle), and ∆cpnT complemented with cpnTNTD (black diamond) or cpnT (blue triangle) in 7H9 medium with 0.2% glycerol. Experiments were carried out three times. A representative growth curve is shown.
CpnT Is an Outer Membrane Protein of M. tuberculosis.
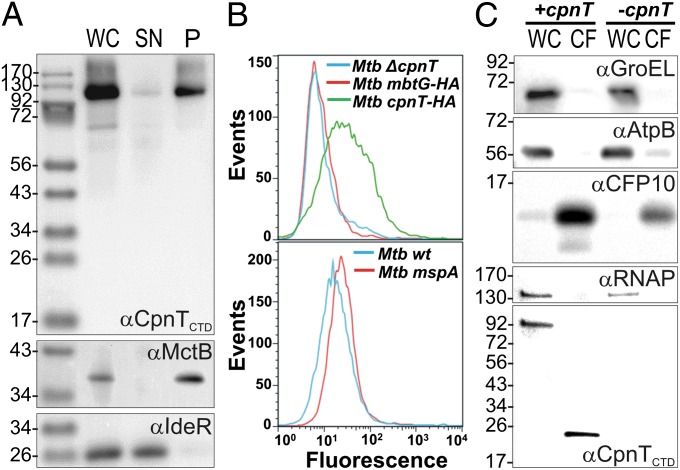
CpnT has no sequence similarity to any protein of known function and is unusually large for a porin (846 aa versus 184–505 aa of known porins). Subcellular fractionation of M. tuberculosis revealed that CpnT is membrane-associated (Fig. 2A). Furthermore, the N-terminal domain (NTD; aa 1–443) was sufficient for membrane localization of CpnT (SI Appendix, Fig. S5). To distinguish between inner and outer membrane proteins, we used flow cytometry analysis of M. tuberculosis. This approach relies on the detection of surface antigens by protein-specific antibodies (23). M. tuberculosis overexpressing cpnTHA with a C-terminal HA-tag displayed increased fluorescence compared with the strain lacking cpnT when stained with an anti-HA antibody (Fig. 2B). Importantly, surface staining of M. tuberculosis cells with the same antibody did not detect MbtGHA, a protein associated with the inner membrane (24), whereas the outer membrane porin MspA (22) was recognized using an MspA-specific antibody (Fig. 2B). These experiments indicate that the C terminus of CpnT is accessible to antibodies on the cell surface of M. tuberculosis. The membrane localization and surface accessibility of CpnT in combination with the observation that the outer membrane channel protein MspA complements the glycerol uptake defect of the cpnT mutant indicate that CpnT is localized in the outer membrane of M. bovis bacillus Calmette–Guérin and M. tuberculosis.
Fig. 2.
Subcellular localization of CpnT in M. tuberculosis. (A) Immunoblot analysis of whole-cell lysates (WC), water-soluble supernatant (SN), and membrane-associated pellet (P) protein fractions obtained by ultracentrifugation of WT M. tuberculosis mc26206. IdeR and MctB were used as controls for soluble and membrane-associated proteins, respectively. CpnT was detected using an antibody recognizing the CTD. (B) Surface accessibility of CpnT. Cells of M. tuberculosis ΔcpnT and of M. tuberculosis overexpressing cpnTHA and mbtGHA were incubated with an anti-HA antibody followed by detection with anti-rabbit AlexaFluor 488-labeled antibody (Upper). WT M. tuberculosis and M. tuberculosis expressing the porin gene mspA of M. smegmatis were incubated with a monoclonal anti-MspA antibody (P2) and an anti-mouse FITC-labeled antibody (Lower). The fluorescence of surface-stained M. tuberculosis cells was measured by flow cytometry and is displayed as histograms (MFI, mean fluorescence intensity). (C) Secretion of the CTD of CpnT. Immunoblot analysis of whole cell lysates (WC) and culture filtrates (CF) of the M. tuberculosis ΔcpnT mutant grown in vitro carrying integrative vectors with (+cpnT) or without (-cpnT) the cpnT gene. The cytoplasmic proteins GroEL and RNA polymerase (RNAP), the inner membrane protein AtpB, and the secreted protein CFP-10 served as markers for the subcellular fractions.
The NTD of CpnT Is an Outer Membrane Channel Protein of M. tuberculosis.
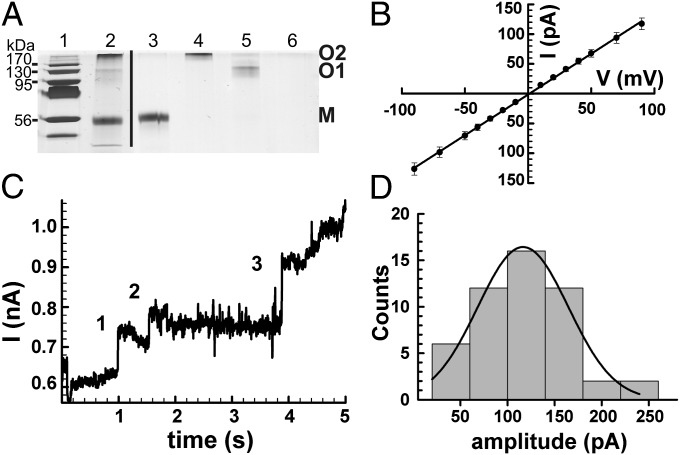
The NTD of CpnT (CpnTNTD) was sufficient to restore the ability of the cpnT mutants of M. tuberculosis and M. bovis bacillus Calmette–Guérin to take up (Fig. 1D and SI Appendix, Fig. S4B) and grow on glycerol (Fig. 1A and SI Appendix, Figs. S3 and S4), indicating that the pore activity of CpnT is mediated by this domain. Therefore, we sought to investigate the channel-forming properties of CpnTNTD in lipid bilayer experiments. The CpnTNTD–HA-His6 protein was purified from the M. smegmatis strain ML1910, which lacks all known endogenous porin genes and conditionally expresses cpnTNTD, to avoid contamination with M. smegmatis and M. tuberculosis pores. The porin-deletion mutant ML1910 is not viable without expression of cpnTNTD, indicating that it does not have outer membrane proteins with significant channel activity other than the Msp porins (25). The CpnTNTD protein was extracted from the M. smegmatis porin-deletion mutant with SDS and purified by Ni(II)-affinity followed by anion-exchange chromatography. The purified fraction contained a predominant 56 kDa band, the predicted size of the CpnTNTD monomer (Fig. 3A). Proteins with apparent molecular weights of 130 and 200 kDa also reacted with an HA-specific antibody (SI Appendix, Fig. S6), demonstrating that CpnTNTD exists in different oligomeric forms that are at least partially resistant to SDS. Proteoliposomes containing the purified protein sample with all three forms of CpnTNTD were added to solvent-free planar membranes and resulted in a step-wise current increase (SI Appendix, Fig. S7A) characteristic of water-filled membrane channels in a lipid bilayer setup (26), whereas the membrane current remained unchanged when empty liposomes were added. The average conductance of two individual CpnTNTD channels was 1.36 ± 0.01 nS as determined from the slopes of current–voltage (I/V) curves (Fig. 3B).
Fig. 3.
The oligomeric NTD of CpnT forms water-filled membrane channels. (A) To preserve disulfide bridges, proteins were mixed with nonreducing loading buffer and samples were loaded without boiling. Lane 1, molecular weight marker; lane 2, CpnTNTD after anion exchange chromatography; lane 3, gel-purified monomer of CpnTNTD (M); lane 4, gel-purified oligomer O2 of CpnTNTD; lane 5, gel-purified oligomer O1 of CpnTNTD (O1); lane 6, “protein-free” control gel. The gel was stained with silver. (B) Average I/V characteristics of two CpnTNTD channels. The I/V curves were recorded for two individual channels reconstituted into lipid bilayers in two separate experiments. The single channel conductance was calculated from the slope of the fitted line and was determined as 1.36 ± 0.01 nS. (C) Current trace of the O2 oligomer of CpnTNTD recorded at +100 mV and filtered by a Gaussian low-pass filter with a bandwidth of 100 Hz. The protein concentration was ∼0.3 µg/mL. The current trace represents a 5-s recording of a continuous multichannel experiment. Events 1, 2, and 3 had current amplitudes (conductances) of 95 pA (1 nS), 65 pA (0.7 nS), and 152 pA (1.5 nS), respectively. (D) Distribution of the current amplitudes of 50 channels of the CpnTNTD O2 oligomer recorded from 25 experiments at +100 mV. The dotted line represents a Gaussian fit for the data with a mean of 116 ± 48 pA. The average single-channel conductance was 1.2 ± 0.5 nS.
To examine whether the channel activity of CpnTNTD requires subunit association, we separated the monomer (∼56 kDa) and the two oligomeric forms O1 (∼130 kDa) and O2 (∼200 kDa) of purified CpnTNTD by electrophoresis and excised the corresponding protein bands from the gel (Fig. 3A and SI Appendix, Fig. S6B). Multichannel experiments with planar lipid membranes showed channel activity only with the sample containing the O2 oligomer (Fig. 3C), indicating that oligomer formation is required for channel activity of CpnTNTD. The average single-channel conductance of 50 channels of the CpnTNTD O2 oligomer was 1.2 ± 0.5 nS (Fig. 3D), which is consistent with the conductance of the unfractionated CpnTNTD sample but distinct from other mycobacterial pore proteins (27–30). The fast flickering events in the current traces of oligomeric CpnTNTD involve opening and closing of complete channels and of subconductance states (Fig. 3D) and might result from voltage gating as observed with other outer membrane pores (31). Neither the monomer nor the O1 oligomer had channel-forming activity when the same amount of protein was used in lipid bilayer experiments (SI Appendix, Fig. S7). The monomeric and oligomeric forms of CpnTNTD were in equilibrium with each other, as shown by the interconversion of the isolated forms after 1 wk of incubation at room temperature in the presence of the mild detergent n-octylpolyoxyethylene to promote refolding (SI Appendix, Fig. S6C). Taken together, the lipid bilayer experiments show that the NTD of CpnT is an integral membrane protein that forms water-filled channels in lipid membranes. The channel activity of CpnTNTD in vitro in combination with the observations that CpnTNTD and the outer membrane pore MspA complement the uptake defect of the M. bovis bacillus Calmette–Guérin cpnT mutant and the membrane association and the surface accessibility of CpnT in M. tuberculosis collectively demonstrate that CpnTNTD is a channel-forming protein in the outer membrane of M. tuberculosis.
The CTD of CpnT Is Toxic in Prokaryotic and Eukaryotic Cells.
Next, we examined possible functions of the C terminus of CpnT. Bioinformatic analysis suggested that the C terminus of CpnT (aa 720–846) belongs to the uncharacterized protein family DUF4237, comprising almost 200 bacterial and fungal proteins (SI Appendix, Fig. S8). Furthermore, homologs of CpnT in Mycobacterium marinum (MMAR_5464) and in other pathogenic mycobacteria have similar N but different C termini. Notably, the MMAR_5464 C terminus contains the ADP ribosyltranferase domain VIP2, often found in bacterial and viral toxins (SI Appendix, Fig. S1). However, the C terminus of CpnT has no sequence similarity to the VIP2 domain. These findings suggested a two-domain structure of CpnT with the N terminus common to mycobacteria and a species-specific C terminus of unknown function. To determine the domain borders of CpnT, we constructed several genes encoding various lengths of its C terminus. The longest fragment comprised aa 651–846 (defined as CpnTCTD) and corresponds to the size of the VIP2 domain of MMAR_5464. Attempts to express cpnTCTD failed in M. smegmatis, Escherichia coli, and Saccharomyces cerevisiae and yielded only minute amounts of protein in cell-free expression systems. By contrast, expression of DNA fragments encoding shorter CpnTCTD variants were not toxic in E. coli, suggesting that the fragment defined as CpnTCTD comprises an independent domain.
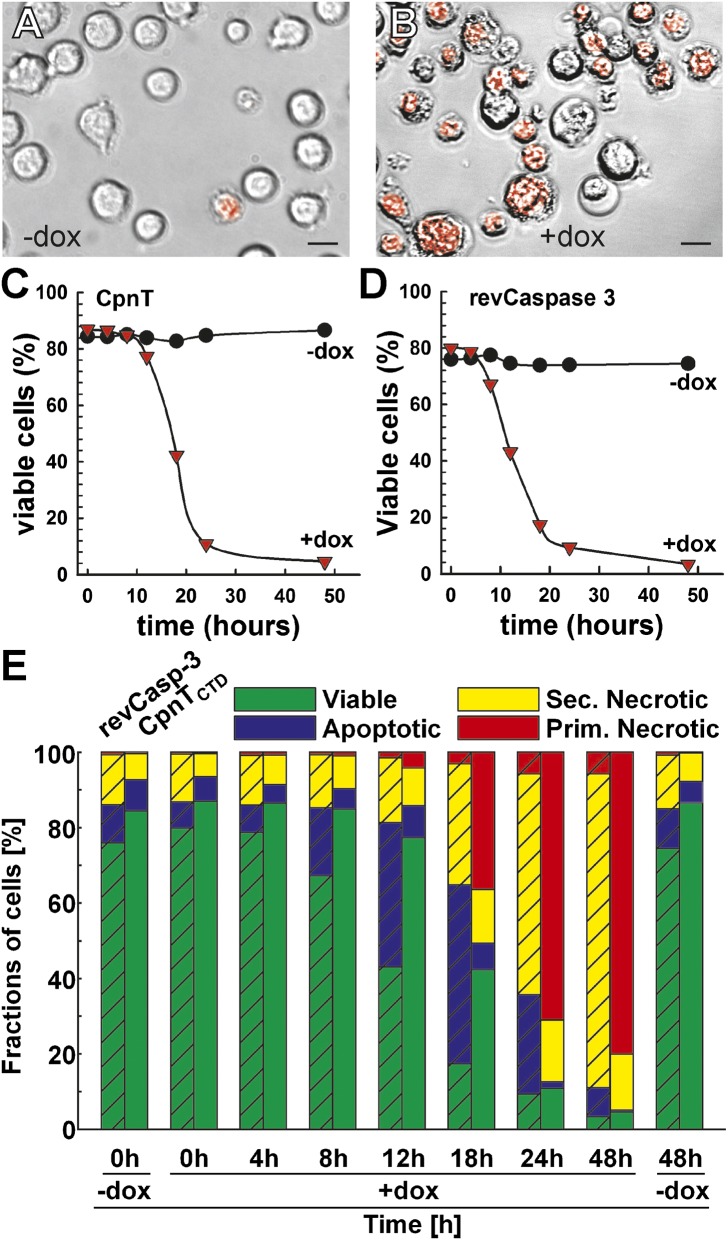
To test whether CpnTCTD is toxic in mammalian cells, we stably integrated the cpnTCTD gene under the control of a tetracycline-regulated promoter in the Jurkat T-cell line J644, which has been previously used for regulated expression of toxic genes (SI Appendix, Fig. S9A). To visualize CpnTCTD-induced morphological changes, Jurkat cells expressing cpnTCTD were examined by fluorescence microscopy after staining with 7-amino-actinomycin D, a marker of cells with damaged plasma membranes. Although only ∼5% of the cells were dead in the control culture (Fig. 4A), more than 80% of the cells had died 16 h after induction of cpnTCTD expression with doxycycline (Fig. 4B). To exclude the possibility that cell death induced by cpnTCTD was specific for the Jurkat cell line, we transiently transfected two other cell lines: the human embryonic kidney cell line 293T (HEK293T) and the C2C12 mouse myoblast cell line (SI Appendix, Fig. S10). In both cases, the majority of the cells were dead 24 h after transfection, as indicated by the disrupted monolayer. This suggested that cell death induced by CpnTCTD is not restricted to particular eukaryotic cell types. The absence of cell death after transfection with a cpnTCTD gene encoding the nontoxic CpnTCTD G818V mutant (SI Appendix, Fig. S10) further indicated that cell death was not an overexpression artifact. The G818V mutation was identified in a screen for nontoxic mutants in E. coli (SI Appendix) and modifies a glycine residue highly conserved in all DUF4237 family members (SI Appendix, Fig. S8). The elimination of CpnTCTD toxicity by a single point mutation argues against nucleic acids or unfolded protein as potential causes of toxicity in the cytosol of cells.
Fig. 4.
The CTD of CpnT induces necrotic cell death in eukaryotic cells. The Jurkat T-cell line J644 containing an integrated Tet-regulated cpnTCTD expression cassette was uninduced (A) or induced with 100 ng/mL doxycycline (B) for 16 h, and subsequently stained for cell viability with 7-amino-actinomycin D (7AAD). Bright field images were merged with the fluorescence image visualizing stained, nonviable cells in red. Magnification, 40×. (Scale bar, 20 µm.) (C and D) Death kinetics of J644-derived cells after induction of expression of cpnTCTD (C) or a gene encoding activated caspase-3 (revCasp-3) (D) with doxycycline (1 µg/mL). Viable and dead cells were distinguished by flow cytometry according to their position in forward scatter and log side scatter. (E) Characterization of the cell death induced by CpnTCTD and activated caspase-3. Cell death kinetics of the J644-derived cell lines either uninduced or induced with 1 µg/mL doxycycline. Cells were grouped into viable, apoptotic, secondary, and primary necrotic using a six-parameter classification scheme (cell size, cell granularity, membrane integrity, phosphatidyl serine exposure, mitochondrial potential, and nuclear degeneration) (32). All bars of one column sum up to 100% with the cells gated from the living and dead cells at each time point.
The CTD of CpnT Causes Necrotic Death in Jurkat T Cells.
To further examine cell death caused by CpnTCTD, we used a flow-cytometry–based six-parameter classification scheme (32) to compare Jurkat T-cell lines containing integrated plasmids stably expressing cpnTCTD (SI Appendix, Fig. S9) or a gene encoding an activated form of caspase-3 (revCasp-3), a key executioner caspase known to trigger apoptotic cell death (33) (SI Appendix, Figs. S11 and S12). Both cell lines displayed similar cell death kinetics in a doxycycline-dependent manner. Approximately 90% of the cells were dead 24 h after induction of their respective gene with doxycycline (Fig. 4 C and D). In the presence of doxycycline, cells expressing activated caspase-3 displayed the annexin-V-positive, propidium iodide-negative staining pattern typical for apoptotic cells (SI Appendix, Fig. S11). By contrast, cells expressing cpnTCTD were double-positive—a hallmark of necrotic cell death (Fig. 4E and SI Appendix, Fig. S12). The low mitochondrial potential and bright Hoechst 33342 staining of the cpnTCTD-expressing cells and the morphology of dead cells confirmed this classification (SI Appendix, Fig. S12). Consistent with these results was the finding that the pancaspase inhibitor z-d-CH2-DCB prevented only cell death induced by caspase-3 but not by CpnTCTD (SI Appendix, Table S4). Due to the fact that the pancaspase inhibitor also inactivates caspase-1 (34), combined with the lack of DNA fragmentation (SI Appendix, Fig. S13), another hallmark of inflammasome-mediated cell death (35), we can rule out pyroptosis as the mechanism of cell death. However, Necrostatin-1 did not inhibit death of cells expressing cpnTCTD (SI Appendix, Table S4), indicating a RipK1-independent (36), unknown type of cell death involving rupture of the plasma membrane without nuclear disintegration and chromatin cleavage. Based on these results, we conclude that the CpnTCTD of M. tuberculosis and likely the DUF4237 domains of other organisms (SI Appendix, Fig. S8) have necrosis-inducing activity in eukaryotic cells.
The CTD of CpnT Is Present in Culture Filtrates of M. tuberculosis.
Because many bacterial toxins are secreted, we examined the culture filtrate of M. tuberculosis using a purified CpnT antiserum produced against the C terminus of the protein. Although full-length CpnT was detected in whole cells of M. tuberculosis, a ∼24 kDa cleaved protein was present in the culture filtrate (Fig. 2C), suggesting that CpnT is translocated to the outer membrane as a full-length protein and that the CTD is released into the extracellular milieu. Further studies are needed to determine the mechanisms of translocation of CpnT to the outer membrane and the cleavage of the CTD.
CpnT Is Required for Survival, Replication, and Cytotoxicity of M. tuberculosis in Macrophages.
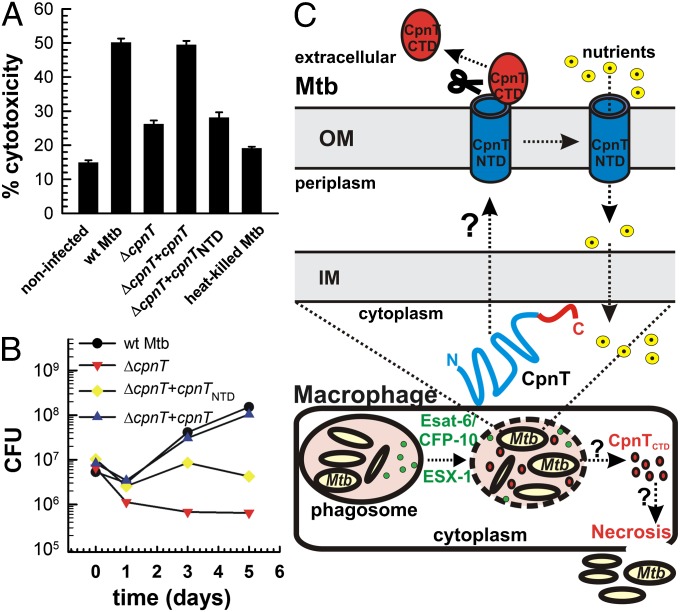
To assess the role of CpnT for virulence of M. tuberculosis, we examined the survival of macrophages after infection with WT M. tuberculosis and the ΔcpnT mutant. Loss of CpnT reduced the level of M. tuberculosis-induced cell death in differentiated THP-1 macrophages by 70%. Cytotoxicity was completely restored by expression of full-length cpnT, but was not complemented with a truncated gene encoding the N-terminal pore-forming domain (Fig. 5A). Furthermore, the ΔcpnT mutant did not replicate in differentiated THP-1 macrophages in contrast to WT M. tuberculosis (Fig. 5B). Complementation with the full-length cpnT gene restored the growth of the ΔcpnT mutant to the WT level, whereas the NTD of CpnT enhanced survival of the ΔcpnT mutant only until day 3 after infection (Fig. 5B). Thus, growth of M. tuberculosis in macrophages appears to benefit from the pore activity of CpnT during the early phase of infection, but the toxic C terminus is required for replication at a later phase. Interpretation of the loss of cytotoxicity of the M. tuberculosis ΔcpnT mutant is complicated by the fact that the ΔcpnT mutant does not replicate in macrophages in contrast to WT M. tuberculosis, leading to a reduced bacterial load. However, although the NTD of CpnT increased the bacterial load 12-fold by day 3 after infection (SI Appendix, Fig. S14), the cytotoxicity in macrophages remained unchanged compared with the ΔcpnT mutant (Fig. 5A). This indicated that the observed difference in cytotoxicity is likely not a consequence of fewer bacteria, but is dependent on the CpnTCTD activity. Taken together, these results show that CpnT is required for survival and replication of M. tuberculosis in macrophages. Transcriptional profiling showed that cpnT expression is induced under hypoxic conditions (37) and during growth in macrophages (38), but is repressed during nonreplicating persistence (39) and in the presence of reactive oxygen and nitrogen species (40), consistent with the dual role of CpnT in survival of M. tuberculosis in macrophages and in regulating the outer membrane permeability of M. tuberculosis.
Fig. 5.
CpnT is required for cytotoxicity and survival of M. tuberculosis in macrophages. (A) Differentiated THP-1 macrophages were infected with the indicated strains at an MOI of 20 for 2 d. Cytotoxicity was determined by flow cytometry after staining with the live/dead stain. (B) Differentiated THP-1 macrophages were infected with WT M. tuberculosis (black circle), ∆cpnT (red inverted triangle), and ∆cpnT complemented with cpnTNTD (yellow diamond) or cpnT (blue triangle) at an MOI of 10. Macrophages were lysed at the indicated time points, and the number of viable bacteria was counted as colony forming units (CFU) on agar plates. Bars represent mean ± SD. (C) Model of CpnT secretion and CpnT-induced cell death of macrophages. Secretion of CpnT by M. tuberculosis is mediated by unknown inner membrane (IM) and outer membrane (OM) components. We suggest that the C-terminal toxin is cleaved after integration of CpnT into the outer membrane. This leaves the N-terminal channel domain (CpnTNTD) in the outer membrane to enable uptake of nutrients. Esat-6 and CFP10 are secreted by ESX-1 and perforate the phagosomal membrane. Then, the secreted CTD of CpnT (CpnTCTD) probably gains access to the macrophage cytoplasm to induce necrosis by an unknown mechanism. These events enable M. tuberculosis to escape from the phagosome and eventually from the destroyed macrophage.
CpnT Is Not Required for Virulence of M. tuberculosis in Mice.
To further examine the role of CpnT in M. tuberculosis pathogenesis, we performed infection experiments in C57BL/6 mice. No significant difference was observed between WT and the cpnT mutant for up to 120 d after infection (SI Appendix, Fig. S15), indicating that cpnT is not required during acute and chronic infection in mice. Instead, CpnT might play a role in dissemination and reactivation from distant sites such as adipocytes (41), which cannot be easily assessed in an animal model. Furthermore, M. tuberculosis may have another protein with porin and/or toxin activity in addition to CpnT, masking the functional importance of these activities in vivo. This result is reminiscent of the toxin VacA, which is a key toxin of Helicobacter pylori with numerous in vivo effects (42). Nevertheless, the H. pylori vacA mutant is not attenuated in a mouse infection model (43).
Discussion
The outer membrane is of utmost importance for survival of M. tuberculosis under harsh conditions in vivo (44), yet the proteins that functionalize this membrane remain enigmatic. To date, only a handful of proteins have been suggested to be outer membrane proteins (15), but their functions have not been confirmed by phenotypes of the corresponding mutants of M. tuberculosis. This study, to our knowledge, represents the first example in which the phenotype of a gene deletion mutant—that is, an uptake defect for a small molecule—corresponds with the channel-forming activity of a purified M. tuberculosis protein.
The organization of CpnT is unusual for a channel-forming outer membrane protein. CpnT does not appear to have a classical Sec signal sequence, as do porins of Gram-negative bacteria (45) and MspA, the only known mycobacterial porin (46). In addition, CpnT is unusually large for a porin and seems to have at least two domains. Here, we show that the NTD of CpnT (aa 1–443) is sufficient for channel formation in the outer membrane of M. tuberculosis. Homologs of this domain are present in all mycobacteria with a sequenced genome, but they are connected to various CTDs of unknown functions (SI Appendix, Fig. S1). CpnT consists of an N-terminal outer membrane channel fused to a secreted CTD. This protein architecture resembles the intimin/invasin family of autotransporters from Gram-negative bacteria (47). Autotransporters facilitate translocation of a passenger domain via an integral outer membrane β-barrel domain (47–50) and have been identified in virtually all pathogenic Gram-negative bacteria. They frequently translocate toxic passenger domains (51). Furthermore, oligomer formation is required for channel activity of the NTDs of both CpnT and intimins (49). Thus, CpnT might represent the first autotransporter-like protein of M. tuberculosis. However, the mechanism of CpnT assembly and outer membrane integration is likely different from that of autotransporters of Gram-negative bacteria that are dependent on Omp85 family proteins such as the β-barrel assembly machinery (50) or the autotransporter translocation and assembly module (52).
The toxic CTD of CpnT challenges the paradigm that M. tuberculosis is one of the few bacterial pathogens that does not produce toxins (2). Based on our results, we propose the following model for the biological function of CpnT (Fig. 5C): After uptake of M. tuberculosis by macrophages, ESX-1–dependent permeabilization of the phagosome results in the mixing of phagosomal and cytosolic contents (53, 54). CpnT is integrated into the outer membrane and the toxic CTD is released from the cell surface of M. tuberculosis by proteolytic cleavage, whereas the NTD remains in the outer membrane and mediates uptake of small, hydrophilic molecules such as glycerol and ampicillin. Perforation of the phagosomal membrane by ESX-1 substrates (53, 54) might enable diffusion of CpnTCTD into the cytosol of macrophages to induce necrosis. Following necrotic disintegration of infected macrophages, M. tuberculosis escapes the phagosome and ultimately the macrophage. This model explains why ESX-1–dependent phagosomal escape of M. tuberculosis (55) is often observed in cells with a dispersed cytoplasm, destroyed plasma membranes, and no signs of apoptosis, which are, therefore, classified as necrotic (56). The proposed mechanism for escape of M. tuberculosis from the macrophage (Fig. 5C) is also consistent with the finding that ESX-1–mediated phagosomal rupture is followed by host cell necrosis within 48 h after infection (54). The ability of M. tuberculosis to enter and survive in macrophages and dendritic cells (6), and to escape from the phagosome, alleviates CpnT from the necessity of carrying an additional receptor-binding domain, as found in A–B-type toxins (57), to gain access to the cytosol of target cells.
Our study suggests that CpnT is the first example, to our knowledge, of an autotransporter-like protein in M. tuberculosis. CpnT is a multifunctional protein that increases nutrient uptake across the outer membrane by its channel-forming NTD. In addition, CpnT transports its toxic CTD to the cell surface of M. tuberculosis, where the toxin is cleaved to induce necrotic cell death of host cells.
Materials and Methods
Chemicals and reagents, bacterial strains, media and growth conditions, and detailed procedures are described in SI Appendix.
Glycerol Uptake Experiments.
Glycerol uptake experiments were carried out as described previously (58) with some modifications.
Channel Activity of CpnTNTD.
Lipid bilayer experiments were performed using a Port-a-Patch automated planar patch clamp system (Nanion Technologies GmbH) (26). Experiments were performed using 1 M KCl, 10 mM Hepes, pH 6.0 buffer. A diphytanoylphosphatidylcholine/cholesterol mixture was used to prepare giant unilamellar vesicles by electroformation using a Nanion Vesicle Prep Pro setup (Nanion Technologies). Two sets of experiments were performed to measure channel-forming properties of CpnTNTD. First, we used proteoliposomes containing purified CpnTNTD. Second, different oligomeric forms of CpnTNTD with electrophoretic mobilities of 56 kDa, 130 kDa, and 200 kDa were analyzed using planar lipid bilayers.
Cell Culture Experiments.
The Jurkat-derived cell line J644 stably expressing a doxycycline-controlled combined repressor/activator switch featuring the second-generation Tet-transregulators rtTA2S-M2 and tTSD-PP was used to create a stable cpnTCTD-expression cell line. Cell death was induced by expression of cpnTCTD and analyzed by flow cytometry.
Role of CpnT in Virulence of M. tuberculosis in Macrophages.
THP-1 macrophages differentiated with 12-phorbol, 13-myristate acetate were used for infection with M. tuberculosis strains at an MOI of 20 or 10 to assess cytotoxicity and intracellular survival, respectively.
Supplementary Material
Acknowledgments
We thank Chris Sassetti for the nitrile inducible expression vector and Nancy Mah and Miguel Andrade for help with the bioinformatic analysis. This work was supported by fellowships from the American Lung Association (to O.D.) and the Carmichael Fund of the University of Alabama at Birmingham (to M.P.) and by the National Institutes of Health Grants AI063446 (to S.E.) and AI63432, AI083632, and AI074805 (to M.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400136111/-/DCSupplemental.
References
- 1.Collier RJ. Understanding the mode of action of diphtheria toxin: A perspective on progress during the 20th century. Toxicon. 2001;39(11):1793–1803. doi: 10.1016/s0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- 2.Gordon SV, Bottai D, Simeone R, Stinear TP, Brosch R. Pathogenicity in the tubercle bacillus: Molecular and evolutionary determinants. Bioessays. 2009;31(4):378–388. doi: 10.1002/bies.200800191. [DOI] [PubMed] [Google Scholar]
- 3.Henkel JS, Baldwin MR, Barbieri JT. Toxins from bacteria. EXS. 2010;100:1–29. doi: 10.1007/978-3-7643-8338-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukhopadhyay S, Nair S, Ghosh S. Pathogenesis in tuberculosis: Transcriptomic approaches to unraveling virulence mechanisms and finding new drug targets. FEMS Microbiol Rev. 2012;36(2):463–485. doi: 10.1111/j.1574-6976.2011.00302.x. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization 2013. Global Tuberculosis Control, WHO report 2012 (World Health Organization)
- 6.Russell DG. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev. 2011;240(1):252–268. doi: 10.1111/j.1600-065X.2010.00984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn JL, Chan J. Tuberculosis: Latency and reactivation. Infect Immun. 2001;69(7):4195–4201. doi: 10.1128/IAI.69.7.4195-4201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilliams M, Lambrecht BN, Hammad H. Division of labor between lung dendritic cells and macrophages in the defense against pulmonary infections. Mucosal Immunol. 2013;6(3):464–473. doi: 10.1038/mi.2013.14. [DOI] [PubMed] [Google Scholar]
- 9.Poirier V, Av-Gay Y. Mycobacterium tuberculosis modulators of the macrophage’s cellular events. Microbes Infect. 2012;14(13):1211–1219. doi: 10.1016/j.micinf.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Divangahi M, Behar SM, Remold H. Dying to live: How the death modality of the infected macrophage affects immunity to tuberculosis. Adv Exp Med Biol. 2013;783:103–120. doi: 10.1007/978-1-4614-6111-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behar SM, et al. Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal Immunol. 2011;4(3):279–287. doi: 10.1038/mi.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Repasy T, Papavinasasundaram K, Sassetti C, Kornfeld H. Mycobacterium tuberculosis induces an atypical cell death mode to escape from infected macrophages. PLoS ONE. 2011;6(3):e18367. doi: 10.1371/journal.pone.0018367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlach RG, Hensel M. Protein secretion systems and adhesins: The molecular armory of Gram-negative pathogens. Int J Med Microbiol. 2007;297(6):401–415. doi: 10.1016/j.ijmm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci USA. 2008;105(10):3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H. Mycobacterial outer membranes: In search of proteins. Trends Microbiol. 2010;18(3):109–116. doi: 10.1016/j.tim.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houben EN, Korotkov KV, Bitter W. Take five—Type VII secretion systems of mycobacteria. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbamcr.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Pagès JM, James CE, Winterhalter M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. 2008;6(12):893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 18.Stephan J, Mailaender C, Etienne G, Daffé M, Niederweis M. Multidrug resistance of a porin deletion mutant of Mycobacterium smegmatis. Antimicrob Agents Chemother. 2004;48(11):4163–4170. doi: 10.1128/AAC.48.11.4163-4170.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danilchanka O, Mailaender C, Niederweis M. Identification of a novel multidrug efflux pump of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2008;52(7):2503–2511. doi: 10.1128/AAC.00298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blasband AJ, Schnaitman CA. Regulation in Escherichia coli of the porin protein gene encoded by lambdoid bacteriophages. J Bacteriol. 1987;169(5):2171–2176. doi: 10.1128/jb.169.5.2171-2176.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stahl C, et al. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol Microbiol. 2001;40(2):451–464. doi: 10.1046/j.1365-2958.2001.02394.x. and correction (2001) 457:1509. [DOI] [PubMed] [Google Scholar]
- 22.Faller M, Niederweis M, Schulz GE. The structure of a mycobacterial outer-membrane channel. Science. 2004;303(5661):1189–1192. doi: 10.1126/science.1094114. [DOI] [PubMed] [Google Scholar]
- 23.Song H, Sandie R, Wang Y, Andrade-Navarro MA, Niederweis M. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2008;88(6):526–544. doi: 10.1016/j.tube.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells RM, et al. Discovery of a siderophore export system essential for virulence of Mycobacterium tuberculosis. PLoS Pathog. 2013;9(1):e1003120. doi: 10.1371/journal.ppat.1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephan J, et al. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol Microbiol. 2005;58(3):714–730. doi: 10.1111/j.1365-2958.2005.04878.x. [DOI] [PubMed] [Google Scholar]
- 26.Kreir M, Farre C, Beckler M, George M, Fertig N. Rapid screening of membrane protein activity: Electrophysiological analysis of OmpF reconstituted in proteoliposomes. Lab Chip. 2008;8(4):587–595. doi: 10.1039/b713982a. [DOI] [PubMed] [Google Scholar]
- 27.Alahari A, et al. The N-terminal domain of OmpATb is required for membrane translocation and pore-forming activity in mycobacteria. J Bacteriol. 2007;189(17):6351–6358. doi: 10.1128/JB.00509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niederweis M. Mycobacterial porins—New channel proteins in unique outer membranes. Mol Microbiol. 2003;49(5):1167–1177. doi: 10.1046/j.1365-2958.2003.03662.x. [DOI] [PubMed] [Google Scholar]
- 29.Molle V, et al. pH-dependent pore-forming activity of OmpATb from Mycobacterium tuberculosis and characterization of the channel by peptidic dissection. Mol Microbiol. 2006;61(3):826–837. doi: 10.1111/j.1365-2958.2006.05277.x. [DOI] [PubMed] [Google Scholar]
- 30.Siroy A, et al. Rv1698 of Mycobacterium tuberculosis represents a new class of channel-forming outer membrane proteins. J Biol Chem. 2008;283(26):17827–17837. doi: 10.1074/jbc.M800866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baslé A, Iyer R, Delcour AH. Subconductance states in OmpF gating. Biochim Biophys Acta. 2004;1664(1):100–107. doi: 10.1016/j.bbamem.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Munoz LE, et al. Colourful death: Six-parameter classification of cell death by flow cytometry—Dead cells tell tales. Autoimmunity. 2013;46(5):336–341. doi: 10.3109/08916934.2012.755960. [DOI] [PubMed] [Google Scholar]
- 33.Danke C, et al. Adjusting transgene expression levels in lymphocytes with a set of inducible promoters. J Gene Med. 2010;12(6):501–515. doi: 10.1002/jgm.1461. [DOI] [PubMed] [Google Scholar]
- 34.Dostert C, et al. Malarial hemozoin is a Nalp3 inflammasome activating danger signal. PLoS ONE. 2009;4(8):e6510. doi: 10.1371/journal.pone.0006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergsbaken T, Cookson BT. Macrophage activation redirects yersinia-infected host cell death from apoptosis to caspase-1-dependent pyroptosis. PLoS Pathog. 2007;3(11):e161. doi: 10.1371/journal.ppat.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandenabeele P, Grootjans S, Callewaert N, Takahashi N. Necrostatin-1 blocks both RIPK1 and IDO: Consequences for the study of cell death in experimental disease models. Cell Death Differ. 2013;20(2):185–187. doi: 10.1038/cdd.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198(5):705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Homolka S, Niemann S, Russell DG, Rohde KH. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: Delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 2010;6(7):e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voskuil MI. Mycobacterium tuberculosis gene expression during environmental conditions associated with latency. Tuberculosis (Edinb) 2004;84(3-4):138–143. doi: 10.1016/j.tube.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Voskuil MI, Bartek IL, Visconti K, Schoolnik GK. The response of mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front Microbiol. 2011;2:105. doi: 10.3389/fmicb.2011.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neyrolles O, et al. Is adipose tissue a place for Mycobacterium tuberculosis persistence? PLoS ONE. 2006;1:e43. doi: 10.1371/journal.pone.0000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boquet P, Ricci V. Intoxication strategy of Helicobacter pylori VacA toxin. Trends Microbiol. 2012;20(4):165–174. doi: 10.1016/j.tim.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 43.Guo BP, Mekalanos JJ. Rapid genetic analysis of Helicobacter pylori gastric mucosal colonization in suckling mice. Proc Natl Acad Sci USA. 2002;99(12):8354–8359. doi: 10.1073/pnas.122244899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barry CE., 3rd Interpreting cell wall ‘virulence factors’ of Mycobacterium tuberculosis. Trends Microbiol. 2001;9(5):237–241. doi: 10.1016/s0966-842x(01)02018-2. [DOI] [PubMed] [Google Scholar]
- 45.de Keyzer J, van der Does C, Driessen AJ. The bacterial translocase: A dynamic protein channel complex. Cell Mol Life Sci. 2003;60(10):2034–2052. doi: 10.1007/s00018-003-3006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niederweis M, et al. Cloning of the mspA gene encoding a porin from Mycobacterium smegmatis. Mol Microbiol. 1999;33(5):933–945. doi: 10.1046/j.1365-2958.1999.01472.x. [DOI] [PubMed] [Google Scholar]
- 47.Oberhettinger P, et al. Intimin and invasin export their C-terminus to the bacterial cell surface using an inverse mechanism compared to classical autotransport. PLoS ONE. 2012;7(10):e47069. doi: 10.1371/journal.pone.0047069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- 49.Saurí A, et al. Autotransporter β-domains have a specific function in protein secretion beyond outer-membrane targeting. J Mol Biol. 2011;412(4):553–567. doi: 10.1016/j.jmb.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 50.Leyton DL, Rossiter AE, Henderson IR. From self sufficiency to dependence: Mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol. 2012;10(3):213–225. doi: 10.1038/nrmicro2733. [DOI] [PubMed] [Google Scholar]
- 51.Benz I, Schmidt MA. Structures and functions of autotransporter proteins in microbial pathogens. Int J Med Microbiol. 2011;301(6):461–468. doi: 10.1016/j.ijmm.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 52.Selkrig J, et al. Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol. 2012;19(5):506–510, S501. doi: 10.1038/nsmb.2261. [DOI] [PubMed] [Google Scholar]
- 53.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11(5):469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simeone R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8(2):e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Wel N, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129(7):1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 56.Welin A, Lerm M. Inside or outside the phagosome? The controversy of the intracellular localization of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2012;92(2):113–120. doi: 10.1016/j.tube.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 57.De Haan L, Hirst TR. Cholera toxin: A paradigm for multi-functional engagement of cellular mechanisms (review) Mol Membr Biol. 2004;21(2):77–92. doi: 10.1080/09687680410001663267. [DOI] [PubMed] [Google Scholar]
- 58.Danilchanka O, Pavlenok M, Niederweis M. Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob Agents Chemother. 2008;52(9):3127–3134. doi: 10.1128/AAC.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.