Qu et al. provide new grist for the introns origin mill (1), adding one more chapter to what has become a very long narrative. Back in the day when spliceosomal introns in the protein-coding genes of eukaryotes were still very new to science (1977–1978), some of us argued that—in an evolutionary sense—they were actually very old (2, 3). According to our soon quite popular “introns-early” theory, pre-mRNA introns were actually the relics of ancient precellular gene assembly processes. We conjectured that these intervening sequences had been lost by “streamlining” in prokaryotes but retained in eukaryotes, where they might continue to play an evolutionary role in “exon shuffling” (4, 5). Despite the heroic efforts of Wally Gilbert to prove this notion right, it has largely fallen out of favor. Anticipated correlations between intron positions and protein module boundaries were never convincingly demonstrated, and the topology of the universal Tree of Life [especially now, with eukaryotes emerging from within archaea (6)] makes introns early unparsimonious.
What has taken its place is an “introns-late” view first articulated in 1987 by Tom Cavalier-Smith (7). According to this, spliceosomal introns are the descendants of group II introns introduced into eukaryotes via the genome of the α-proteobacterium that was to become the mitochondrion—a sort of cellular Trojan horse (Fig. 1). Phil Sharp’s aptly titled “Five easy pieces” (8) showed how one or more of the group II introns deposited in nuclear genomes might have over time degenerated into the five snRNAs that now, assisted by many proteins, are the agents of removal of what is left of all of the rest of the invaders. Everything that has happened in the quarter century since then serves to strengthen the belief that structural and functional similarities between group II introns and spliceosomal snRNAs are indeed true homologies. Almost certainly, the latter evolved from the former, by some sort of coming apart in the eukaryotic nuclear lineage.
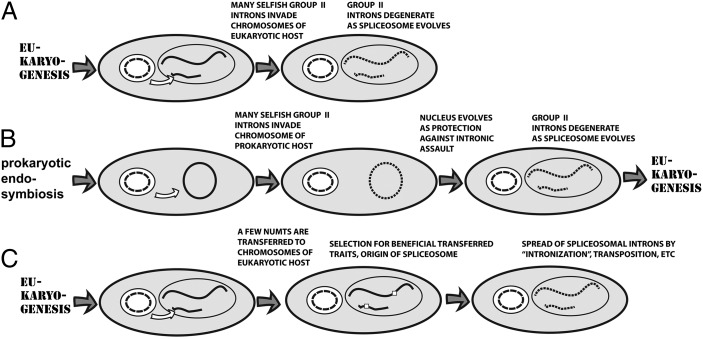
Fig. 1.
Three schemes for the evolution of spliceosomal introns. (A) In a relatively advanced eukaryotic cell, selfish group II introns from newly established mitochondria invade the nuclear genome and proliferate there, then degenerate into spliceosomal introns (7). (B) Selfish group II introns from an endosymbiotic bacterium invade the genome of a host archaean and harmfully proliferate there, forcing the evolutionary separation of nucleus and cytoplasm and the evolution of spliceosomes, and giving rise to eukaryotic cells (13). (C) In a relatively advanced eukaryotic cell, group II introns embedded in a relatively few numts that carry genes whose transfer to the nuclear genome confers selective advantage are established, degenerate into spliceosomal introns and spread through neutral processes.
The devil is in the details, of which early intron theorists—in their eagerness to explain why eukaryotic genes are now so beset with introns—were blissfully ignorant. If group II introns were fully self-splicing ribozymes (requiring no proteinaceous partners), then eukaryotic nuclear genomes would be an especially congenial place for them to proliferate wildly, as apparently they did, at the very beginning of eukaryote evolution (9). The spatial and temporal separation of eukaryotic transcription and translation would have obviated the awkward problem of ribosomes prematurely entering introns, before they are spliced. That “premature entry” is an issue for prokaryotes, in which transcription and translation are coupled, is indicated by the relative rarity of group II introns (compared with their presumed nuclear descendants) and their localization almost exclusively in mobile genetic elements or noncoding regions of bacterial genomes (10, 11).
However, unfortunately for advocates of this appealing scenario, group II introns are not fully self-splicing ribozymes. With a single, very recently discovered, apparent exception (12), all bacterial group II introns require the services of a protein [usually an intron-encoded protein (IEP)] to be spliced or to transpose. Eukaryote-type cytosolic ribosomes would not likely recognize the IEP coding sequence if it were presented to them, either in the unspliced pre-mRNA or excised intron, and the IEP produced would not likely find its way back to the nucleus. There would be no proliferation in the nucleus.
These barriers are not absolute, and evolution, ever the opportunist, might have worked around them. However, there is a radically different work-around, proposed in 2006 by Bill Martin and Eugene Koonin. They turned the Cavalier-Smith scenario on its head (13). Instead of group II introns invading a cell in which nucleus and cytoplasm were already separated, they ventured that the separation evolved later, as a way of accommodating the descendants of the invaders. In this schema, there was “a turbulent phase of genome evolution in the wake of mitochondrial origin, during which group II introns invaded the host’s chromosomes, spread as transposable elements to hundreds—perhaps thousands—of positions that have been conserved to the present, and fragmented into both mRNA introns and snRNA constituents of the spliceosome.” Subsequent fragmentation and increasing dependence on snRNAs and proteins acting in trans—for whatever reason they occurred—inevitably slowed splicing and exacerbated the premature ribosome entry problem. Elaboration of a nuclear envelope from the existing endomembrane system of the archaean-like cell that—according to Martin and Koonin—was the mitochondrion’s host was essential to save the day. Transcription (and splicing) became uncoupled from translation. Subsequent targeting of increasingly many proteinaceous spliceosome components to the nucleus, the elaboration of nonsense-mediated decay (NMD) and other surveillance systems (14, 15), and the development of alternative splicing and other intron-dependent regulatory devices ultimately made a good (or at least a necessary) thing out of a bad.
There are other possibilities of course. One, which has the advantage of not requiring a “turbulent phase” during which introns put survival of the nascent eukaryotic lineage at risk, would recognize the inevitability and potential value of numts (nuclear mitochondrial sequences) (16). These pieces of mitochondrial DNA, released by organellar lysis and deposited at random in nuclear chromosomes of many species, will sometimes carry group II introns. It will sometimes happen that having a nuclear copy of the gene in which such an intron is embedded confers some advantage. Because removal of the intron is necessary for expression, then both gene and intron will be spread (or maintained) within populations by positive (or purifying) selection—even as the intron’s splicing is taken over with increasing effectiveness by trans-acting snRNAs (derived from other numts) and proteins. There need have been very few such positively selected cases initially. Subsequent increases in intron number could have then been achieved early in eukaryote evolution by whatever mechanisms or processes currently create new introns (9). The existence of an effective system of splicing in trans makes at least one such process, “intronization” of parts of exons, selectively neutral and virtually inevitable (17). Also, there is good evidence for transposition in some systems. Although the preponderant mechanisms remain unclear, that intron gain occurs is an unavoidable inference of comparative genomics.
There may never be a principled way to verify any of the scenarios shown in Fig. 1. However, many observations are pertinent, and Qu et al. (1) go some way toward answering one of the questions that is relevant to any scheme. Namely, if the eukaryotic nucleus once housed group II introns (many, or even just a few essential ones), why are there none reported in any of the nearly 1,000 sequenced eukaryotic nuclear genomes? It is surely questionable whether group II introns (especially with nonfunctional or
Qu et al. provide new grist for the introns origin mill, adding one more chapter to what has become a very long narrative.
aberrant IEPs) would be easy to find in the noncoding “junk” DNA of many eukaryotes, where there would be much more room for them to locate and much less trouble for them to cause. However, the fact that none have been recorded seems significant.
In earlier work, this group (Marlene Belfort's) showed that when the Ll.LtrB group II intron of Lactococcus lactis is inserted into genes of Saccharomyces cerevisiae, it can (with the aid of its IEP encoded on another plasmid) be accurately excised, although mostly only after export to the cytoplasm (18). Before splicing, the pre-mRNA is subject to NMD, and after splicing, to some sort of translational repression. Therefore, they suggested that “a group II intron can splice from a nuclear transcript, but RNA instability and translational defects would have favored intron loss or evolution into protein-dependent spliceosomal introns, consistent with the bacterial group II intron ancestry hypothesis.”
In the current work, Qu et al. run the mechanism of repression to ground. It all turns out to be remarkably complex. First, because unspliced pre-mRNA, introns, and spliced mRNA are together at once in the cytoplasm, they can interact. Because of a complementarity characteristic of group II introns, base pairing occurs between exon-binding sequences in the intron and intron-binding sequences in the mRNA’s exons. Repression is due at least in part to this interaction, because in mutants with reduced complementarity, gene expression levels increase. Furthermore, immunoprecipitation showed that both spliced mRNA and unspliced pre-mRNA localize preferentially to cytoplasmic ribonucleoprotein particles: processing bodies and stress granules. If unspliced pre-mRNA is held up in the nucleus (by engineering an interaction between intron and small nucleolar RNAs), the gene silencing effect is somewhat ameliorated. They conclude that such results “provide a molecular basis for understanding why group II introns are absent from nuclear genomes, and they support the hypothesis that cytoplasm-nucleus partitioning contributed to the emergence of spliceosomal introns with the expulsion of group II introns from nuclear genomes,” consistent with Martin and Koonin’s conjecture (13).
Certainly, exclusion of group II introns from protein-coding genes is an important part of the story. However, reducing gene expression levels is not always selected against; indeed, it is sometimes selected for. More importantly, group II introns in noncoding regions would not have been subject to expulsion for the reasons invoked by Qu et al. (1). There are surely many more evolutionary forces at play in a story as complex as that of intron origins. The nucleus would still seem to be a great place for group II introns to spread and disport themselves, if their IEPs and spliced-out introns could relocalize there. So, maybe that’s the real problem. Maybe in some eukaryote yet to be sequenced, such an innovation has occurred. Stay tuned!
Footnotes
The author declares no conflict of interest.
See companion article on page 6612.
References
- 1.Qu G, et al. RNA–RNA interactions and pre-mRNA mislocalization as drivers of group II intron loss from nuclear genomes. Proc Natl Acad Sci USA. 2014;111:6612–6617. doi: 10.1073/pnas.1404276111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doolittle WF. Genes-in-pieces, were they ever together? Nature. 1978;272:581–582. [Google Scholar]
- 3.Darnell JE., Jr Implications of RNA-RNA splicing in evolution of eukaryotic cells. Science. 1978;202(4374):1257–1260. doi: 10.1126/science.364651. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert W. Why genes in pieces? Nature. 1978;271(5645):501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert W. The exon theory of genes. Cold Spring Harb Symp Quant Biol. 1987;52:901–905. doi: 10.1101/sqb.1987.052.01.098. [DOI] [PubMed] [Google Scholar]
- 6.Williams TA, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504(7479):231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 7.Cavalier-Smith T. The origin of eukaryotic and archaebacterial cells. Ann N Y Acad Sci. 1987;503:17–54. doi: 10.1111/j.1749-6632.1987.tb40596.x. [DOI] [PubMed] [Google Scholar]
- 8.Sharp PA. “Five easy pieces”. Science. 1991;254(5032):663. doi: 10.1126/science.1948046. [DOI] [PubMed] [Google Scholar]
- 9.Rogozin IB, Carmel L, Csuros M, Koonin EV. Origin and evolution of spliceosomal introns. Biol Direct. 2012;7:11. doi: 10.1186/1745-6150-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambowitz AM, Zimmerly S. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3(8):a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Candales MA, et al. Database for bacterial group II introns. Nucleic Acids Res. 2012;40(Database issue):D187–D190. doi: 10.1093/nar/gkr1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNeil BA, Simon DM, Zimmerly S. Alternative splicing of a group II intron in a surface layer protein gene in Clostridium tetani. Nucleic Acids Res. 2014;42(3):1959–1969. doi: 10.1093/nar/gkt1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440(7080):41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 14.Lynch M, Kewalramani A. Messenger RNA surveillance and the evolutionary proliferation of introns. Mol Biol Evol. 2003;20(4):563–571. doi: 10.1093/molbev/msg068. [DOI] [PubMed] [Google Scholar]
- 15.Koonin EV. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bensasson D, Zhang D, Hartl DL, Hewitt GM. Mitochondrial pseudogenes: Evolution’s misplaced witnesses. Trends Ecol Evol. 2001;16(6):314–321. doi: 10.1016/s0169-5347(01)02151-6. [DOI] [PubMed] [Google Scholar]
- 17.Gray MW, Lukes J, Archibald JM, Keeling PJ, Doolittle WF. Cell biology. Irremediable complexity? Science. 2010;330(6006):920–921. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
- 18.Chalamcharla VR, Curcio MJ, Belfort M. Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry. Genes Dev. 2010;24(8):827–836. doi: 10.1101/gad.1905010. [DOI] [PMC free article] [PubMed] [Google Scholar]