Abstract
The health of metazoan organisms requires an effective response to organellar and cellular damage – either by repair of such damage and/or by elimination of the damaged parts of the cells or the damaged cell in its entirety. Here we consider the progress that has been made in the last two decades in determining the fates of damaged organelles and damaged cells, through discrete, but genetically overlapping, pathways involving the selective autophagy and cell death machinery. We further discuss the ways in which the autophagy machinery may impact the clearance and consequences of dying cells for host physiology. Failure in the proper removal of damaged organelles and/or damaged cells by selective autophagy and cell death processes is likely to contribute to developmental abnormalities, cancer, aging, inflammation, and other diseases.
Introduction
As in all living things, each of our cells suffer the slings and arrows of outrageous fortune, facing damage from without and within. And, like the Prince of Denmark, each decides whether to be or not to be. To be, the cell must monitor and repair the damage. If not, it will “melt, thaw, and resolve itself into a dew,” dying and cleared from the body by other cells (with apologies to the bard for scrambling his immortal words).
Here, we consider how the molecular pathways of autophagy and cell death, and ultimately the clearance of dying cells, function in this crucial decision. While autophagy and cell death occur in response to a wide variety of metabolic and other cues, here our focus is restricted to those aspects of each that are directly concerned with the quality control of cells – the “garbage” (cellular or organellar) that must be managed for organismal function. And while there are many important functions of quality control mechanisms (e.g., DNA and membrane repair, cell growth and cell cycle control, unfolded protein and endoplasmic reticulum stress responses, innate and adaptive immunity, and tumor suppression), our discussion is limited to the selective disposal of damaged or otherwise unwanted organelles, and when necessary, damaged or excess cells, and how the autophagic and cell death mechanisms function in these processes. Overall, we focus on the overriding theme of waste management, but as we will see, many of the links between these elements remain largely unexplored. Further, while a great deal of what we know was delineated in yeast and invertebrate model systems, we largely restrict our consideration to what is known in mammals.
Engaging autophagy
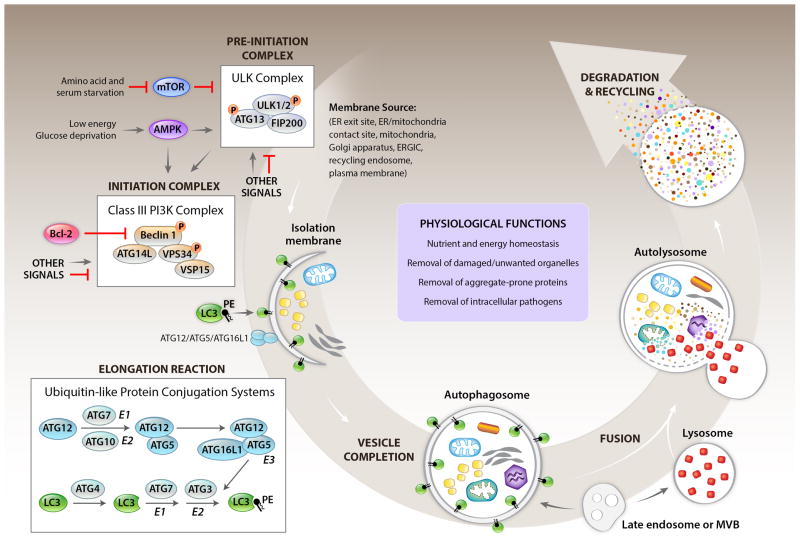
The process of macroautophagy (herein, autophagy) is best understood in the context of nutrient starvation (Kroemer et al., 2010; Mizushima and Komatsu, 2011). When energy in the form of ATP is limiting, AMP kinase (AMPK) becomes active, and this can drive autophagy. Similarly, deprivation from growth factors and/or amino acids leads to the inhibition of TORC1, which when active represses conventional autophagy. As a result of AMPK induction and/or TORC1 inhibition, autophagy is engaged, although other signals may bypass AMPK and TORC1 to engage autophagy (Figure 1).
Figure 1. Overview of the general autophagy pathway.
Shown are cellular events and selected aspects of the molecular regulation involved in the lysosomal degradation pathway of autophagy in mammalian cells. Several membrane sources may serve as the origin of the autophagosome and/or to contribute to its expansion. A “pre-initiation” complex (also called the ULK complex) is negatively and positively regulated by upstream kinases that sense cellular nutrient and energy status, resulting in inhibitory and stimulatory phosphorylations on ULK1/2 proteins. In addition to nutrient sensing kinases shown here, other signals involved in autophagy induction may also regulate the activity of the ULK complex. The pre-initiation complex activates the “initiation complex” (also called the Class III PI3K complex) through ULK-dependent phosphorylation of key components, and likely, other mechanisms. Activation of the Class III PI3K complex requires the disruption of binding of Bcl-2 anti-apoptotic proteins to Beclin 1, and is also regulated by AMPK, and a variety of other proteins not shown in figure. The Class III PI3K complex generates PI3P at the site of nucleation of the isolation membrane (also known as the phagophore) which leads to the binding of PI3P binding proteins (such as WIPI/II), and the subsequent recruitment of proteins involved in the “elongation reaction” (also called the ubiquitin-like protein conjugation systems) to the isolation membrane. These proteins contribute to membrane expansion, resulting in the formation of a closed double-membrane structure, the autophagosome, which surrounds cargo destined for degradation. The phosphatidylethanolamine-conjugated form of the LC3 (LC3-PE), generated by the ATG4-dependent proteolytic cleavage of LC3, and the action of the E1 ligase, ATG7, the E2 ligase, ATG3, and the E3 ligase complex, ATG12/ATG5/ATG16L, is the only autophagy protein that stably associates with the mature autophagosome. The autophagosome fuses with a lysosome to form an autolysosome; inside the autolysosome, the sequestered contents are degraded and released into the cytoplasm for recycling. Late endosomes or multivesicular bodies can also fuse with autophagosomes generating intermediate structures known as amphisomes, and they also contribute to the formation of mature lysosomes. Additional proteins (not depicted in diagram) function in the fusion of autophagosomes and lysosomes. The general autophagy pathway has numerous functions in cellular homeostasis (examples listed in box labeled “physiological functions”) which contribute to the role of autophagy in development and protection against different diseases.
The “goal” of the autophagy machinery is to deliver cytosolic materials to the interior of the lysosomes for degradation, thereby recovering sources of metabolic energy and requisite metabolites in times of starvation (general autophagy). Autophagy can similarly function to target damaged or otherwise unwanted organelles to lysosomes for removal (selective autophagy). While here we focus primarily on selective autophagy, it is useful to also consider general autophagy to highlight similarities and distinctions between the two processes.
In both cases a double-membrane structure, the autophagosome, fuses with lysosomes to deliver the contents for degradation, and this involves a proteolipid molecule, LC3-II, a component of the autophagosome comprised of a protein, LC3, and a lipid, phosphatidylethanolamine. LC3-II is generated by a process resembling ubiquitination, involving E1, E2, and E3 ligases (Figure 1). The parent molecule, LC3-I, is generated by the action of a protease, ATG4, which cleaves LC3 to produce LC3-I. This is bound by the E1, ATG7, and transferred to the E2, ATG3. The E3 ligase is a complex composed of ATG16L and ATG12-5; the latter is produced by another reaction in which ATG12 is bound by the E1, ATG7, transferred to a different E2, ATG10, and from there to ATG5. The process by which ATG12-5 is formed, and subsequently LC3-II (also known as LC3-PE) is generated, is referred to as the elongation reaction, and is required for the formation of the autophagosome.
While not entirely understood, the generation of the LC3-coupled autophagosome appears to originate through extension of intracellular membranes and several sources have been suggested including endoplasmic reticulum (ER), mitochondria, ER-mitochondrial contact sites, ER-Golgi intermediate compartment, the recycling endosome, and the plasma membrane (Hamasaki et al., 2013). The initiation process requires the action of the Class III PI3 kinase, VPS34, which converts phosphatidylinositol to phosphatylinositol 3-phosphate (PI3P); this is the only enzyme that performs this function in cells (Meijer and Klionsky, 2011). The activity of VPS34 requires VPS15 (which is myristoylated and binds to membranes), a requisite regulator, Beclin 1, and other proteins, including ATG14L, which bind to Beclin 1. The complex, termed the initiation complex, generates PI3P, which dictates the site at which the double membrane subsequently elongates by the E1-E2-E3 interaction described above (Figure 1).
During starvation-induced autophagy, the function of the Beclin 1-VPS34 initiation complex is controlled by a pre-initiation complex that includes a protein kinase, ULK1, and the function of this kinase activates the initiation complex to generate PI3P (Russell et al., 2013). This preinitiation complex is, in turn, activated by AMPK and inhibited by TORC1 (Wirth et al., 2013), and as we have discussed, starvation conditions result in AMPK activation and TORC1 inhibition. At least in the setting of glucose deprivation, AMPK can also act directly on the Beclin 1/VPS34 complex (Kim et al., 2013). For starvation-induced autophagy, it is also necessary to unleash negative regulators of the Beclin 1-VPS34 initiation complex, such as Bcl-2/Bcl-xL (Wirth et al., 2013).
Selective autophagic removal of organelles
As discussed in the Introduction, autophagy can function to remove damaged or otherwise unwanted organelles in a cell. By “unwanted” we mean organelles that are removed during differentiation (e.g., in maturing erythrocytes) or when environmental factors (e.g., hypoxia) disfavor some organelles in the cell. We refer to this process as selective autophagy. When considering selective autophagy, we are faced with two problems. First, how does the process “know” which structures or organelles to target for removal? And second, how does this occur even when the conventional autophagy machinery is suppressed (at least partially), such as in nutrient-rich conditions? With regard to the latter, the problem is confounded by the simple fact that lysosomal digestion of organelles will itself provide amino acids and other metabolites, presumably activating TORC1 and suppressing AMPK. As we have seen, such conditions inhibit the function of the pre-initiation complex. Nevertheless, animals lacking Ulk1 display a defect in at least one selective autophagic process, that of efficient removal of mitochondria during erythrocyte development (Kundu et al., 2008). Presumably, there are ways to bypass conventional inhibitory mechanisms to engage Ulk1 activity and promote selective autophagy in some settings. Alternatively, the pre-initiation complex may be bypassed in some situations. We will not fully resolve this paradox here, but perhaps provide clues as we consider the first problem, that of how specific cargoes are marked for clearance.
Before considering this issue, it may be useful to note that even in nutrient starved conditions, autophagy may be selective. Ribosomes represent a major portion of the biomass of many cells, and upon starvation, these are more rapidly removed than other structures in the cell (Cebollero et al., 2012). Similarly, there appears to be selective removal of peroxisomes during starvation (Hara-Kuge and Fujiki, 2008). The same may be the case for ER (reticulophagy), although it remains possible that in this case this is a consequence of developing the requisite autophagosomes for nutritional supplementation using the ER membrane (see above). Another possible selection during starvation is the preservation of functional mitochondria; since these are necessary for the catabolism of free fatty acids or amino acids and for the optimal generation of energy from glucose (all generated by lysosomal digestion), it simply does not make sense that inadvertent removal of mitochondria during starvation would be permitted. Such possible “anti-selection”, however, has not been fully documented nor adequately explored.
Targeting in selective organellar autophagy is perhaps best analyzed in the clearance of mitochondria (mitophagy) and peroxisomes (pexophagy). Tissues or cells lacking requisite components of the autophagy elongation machinery (e.g., ATG5, ATG7) often display greatly increased numbers of apparently damaged mitochondria (Mizushima and Levine, 2010) and peroxisomes (Till et al., 2012). That said, there is evidence that even in the absence of ATG5 and ATG7, some selective mitophagy continues via unknown mechanisms, perhaps via vesicular trafficking between mitochondria and lysosomes (Soubannier et al., 2012). Nevertheless, accumulated observations indicate that the autophagy elongation machinery and autophagosome formation is important for selective autophagy of damaged or otherwise unwanted organelles.
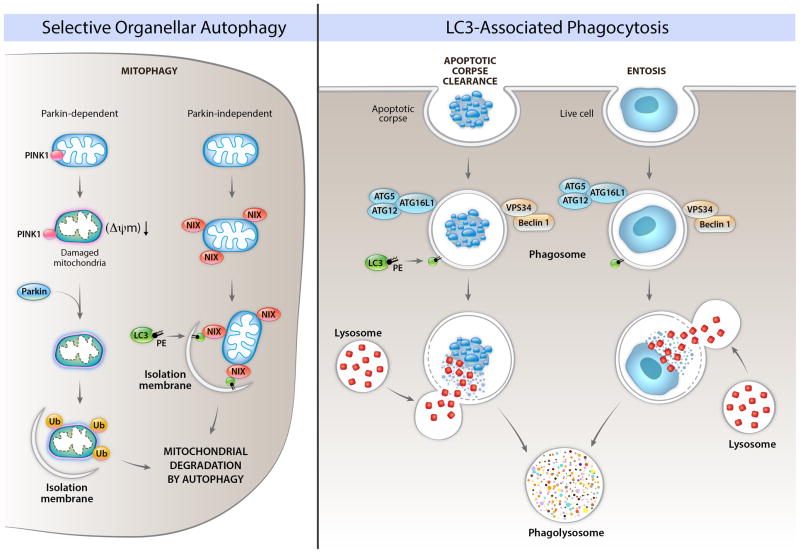
One way in which damaged mitochondria are removed by autophagy involves the action of two proteins, PINK1 and Parkin (Figure 2). PINK1 is a kinase that is constitutively imported into functional mitochondria and degraded by the rhomboid protease, PARL. As with most mitochondrial import, this requires the transmembrane potential of the inner mitochondrial membrane, ΔΨm. Loss of this potential, which can occur when the electron transport chain is damaged or if protons are allowed to pass freely across the inner membrane (i.e., due to the uncoupler protein, UCP, environmental protonophores, or in response to the mitochondrial permeability transition) causes active PINK1 to accumulate on the cytosolic face of the outer mitochondrial membrane. This then recruits and activates Parkin, which is a ubiquitin E3-ligase, which then ubiquitinates proteins on the mitochondria.
Figure 2. Roles of autophagy proteins in the removal of unwanted organelles and in the removal of cells.
The left panel shows Parkin-dependent and Parkin-independent mechanisms involved in the selective degradation of mitochondria by autophagy (mitophagy). In Parkin-dependent mitophagy, mitochondrial damage and loss of mitochondrial membrane potential (ΔΨm) leads to localization of the kinase, PINK1, on the cytoplasmic surface of the mitochondria, resulting in recruitment of the E3 ubiquitin ligase, Parkin, to the mitochondria, followed by the ubiquitination of mitochondrial proteins, and the formation of an isolation membrane that surrounds the damaged mitochondria. In Parkin-independent mitophagy, protein such as Nix (shown in figure), BNIP3, and FUNDC1 (not shown in figure) bind to LC3. Other autophagy proteins may be involved in Parkin-dependent and Parkin-independent mitophagy (discussed in text). The precise details of how an isolation membrane is formed around specific mitochondria earmarked for degradation are unclear. Other damaged/unwanted organelles such as ER, peroxisomes, and lipid droplets can also be degraded by selective autophagy; the molecular mechanisms of these forms of selective autophagy are not well understood in mammalian cells. The right panel depicts roles of LC3-associated phagocytosis (LAP) of apoptotic corpses and of live cells (entosis). In LAP, components of the autophagy initiation complex (Beclin 1, VPS34) are recruited to the phagosome, which leads to recruitment of LC3-PE, and facilitation of phagolyosomal fusion. This process requires other components of the elongation machinery, but – in contrast to general autophagy or selective autophagy – proceeds independently of the ULK pre-initiation complex.
While it is not clear how ubiquitination triggers mitophagy, several ubiquitin-binding proteins bear a motif that binds to LC3. These include p62/sequestosome1, NBR1, and optineurin, and the binding of p62 to LC3 has been implicated in mitophagy in some studies (Shaid et al., 2013). This leads to the notion that it is the binding of LC3 to ubiquitinated mitochondrial proteins that focuses the autophagy machinery on mitophagy (and perhaps other forms of selective autophagy). However, this idea has been challenged by studies showing that p62 is not required for Parkin-mediated mitophagy (Narendra et al., 2010). Moreover, AMBRA1 (a positive regulator of the Beclin 1/Class III PI3K initiation complex), binds to Parkin during mitophagy (Van Humbeeck et al., 2011), and several autophagy proteins, including ULK1, ATG14, ATG16L, and ATG9, are recruited to depolarized mitochondria in Parkin-expressing cells, independently of LC3 (Itakura et al., 2012). This suggests a model in which the damaged mitochondrion is not “recognized” by a preformed isolation membrane, but rather, recruits the machinery necessary for the de novo formation of an autophagosome.
Although it is understood that the activation of PINK1 and Parkin can trigger mitophagy, it is also likely that mitophagy proceeds via other mechanisms (Figure 2). One such mechanism involves either of two related proteins, BNIP3 and NIX (also known as BNIP3L). Animals lacking NIX fail to efficiently clear mitochondria in maturing erythrocytes, leading to anemia (Sandoval et al., 2008), an effect not observed in Parkin-deficient animals. Similarly, during hypoxia, BNIP3 expression is induced by HIF1, promoting mitophagy (Zhang et al., 2008). These proteins may act to directly recruit the autophagy machinery to the mitochondria (Zhang and Ney, 2009) and may also promote mitophagy through interaction with Bcl-2, an anti-apoptotic protein (see below) that resides on the mitochondrial outer membrane. Both BNIP3 and NIX promote the release of Beclin 1 from Bcl-2, and this may, in turn, promote mitophagy, although the details are unclear. Another mitochondrial outer membrane protein, FUNDC1, may also be required for mitophagy through interaction with LC3, in a process regulated by hypoxia and FUNDC1 dephosphorylation (Liu et al., 2012).
While pieces of the puzzle are beginning to emerge, a comprehensive understanding of how specific cargoes are targeted for autophagic degradation is still lacking. Clearly, the model of ubquitinated cargo binding to LC3-interacting proteins (through their ubiquitin-binding domains) which subsequently bind to LC3 (through their LC3-interaction region (LIR) motifs), and thereby, targeting the cargo for autophagy is – at least in many cases – an oversimplification. Even in this model, the relevant substrates for ubiquitination, the spectrum of ubiquitin ligases, the spectrum of ubiquitin-binding LC3-interacting autophagy “receptor” proteins, and the signals that trigger the initial ubiquitination of relevant substrates, are not completely known. In the future, we must look beyond established paradigms for additional signals and mechanisms of selective autophagic removal of damaged organelles.
One interesting alternative possibility is based on changes in lipid exposure, at least in the context of mitophagy. Damaged mitochondria expose cardiolipin, normally present predominantly on the inner mitochondrial membrane, on the outer membrane (Chu et al., 2013). Vertebrate LC3 orthologs bind to cardiolipin and this binding appears to be involved in the selective removal of the damaged mitochondria. If so, this may be specific to mitophagy in vertebrates, as the requisite docking site is not conserved in invertebrate or yeast orthologs of LC3.
While in this discussion we have focused on the selective removal of damaged organelles, we note that insights can be gleaned from examination of the process of xenophagy, the selective removal of infectious intracellular organisms by autophagic processes (Deretic et al., 2013), and this may inform this model further. For example, it has been shown that ATG16L is recruited to ubiquitin upstream of LC3 lipidation in xenophagic removal (Fujita et al., 2013), suggesting that ubiquitinated proteins on damaged organelles are potentially targeted by the ATG5-12-ATG16L E3 ligase to direct the process.
Of note, while lipid droplets are technically not organelles, their clearance is important for maintaining cellular health and their removal by selective autophagy (lipophagy) may bear some similarities to the selective removal of damaged organelles (Liu and Czaja, 2013). Lipophagy is a documented alternative form of lipid metabolism, and its failure in animals lacking efficient general or selective autophagy factors can lead to hepatic accumulation of intracellular lipid droplets (Singh et al., 2009; Orvedahl et al., 2011). However, the precise mechanisms by which lipid droplets are targeted for autophagic removal are underexplored.
Consequences of defective selective organellar autophagy
It is self-evident that the selective removal of damaged or excess organelles is a critical homeostatic process, but beyond this, our information on what happens when this goes wrong is somewhat limited. There is an accumulation of damaged organelles (including mitochondria, perixosomes, and ER) and organ degeneration in mice with tissue-specific knockout of core autophagy genes such as Atg5 and Atg7 in liver, neurons, heart, pancreatic acinar cells, muscle, podocytes, adipocytes, and hematopoietic stem cells (Mizushima and Levine, 2010). While it may not be possible to dissociate the effects of general autophagy from those of selective autophagy, it is reasonable to postulate that these phenotypes are partly related to defects in selective organellar autophagy – and at a minimum, such studies unequivocally establish a role for autophagy genes in the removal of damaged organelles in vivo.
The proper removal of excess or unwanted mitochondria is likely necessary for certain key aspects in development. As discussed above, the mitophagy factor, Nix, is required for mitochondrial clearance and erythroid maturation in vivo (Sandoval et al., 2008), and mouse erythrocytes lacking general autophagy factors such as Ulk1 and Atg7 fail to clear mitochondria and mature normally (Kundu et al., 2008; Mortensen et al., 2010). Reduction in mitochondrial number may also contribute to the role of core autophagy genes, such as Atg7, in white adipocyte differentiation (Zhang et al., 2009). An intriquing question is whether selective mitophagy – of paternal mitochondria – during embryonic development underlies mammalian maternal mitochondrial DNA (mtDNA) inheritance (Levine and Elazar, 2011). In C. elegans, several studies showed that paternal mitochondria and mtDNA are eliminated from the fertilized oocyte by autophagy (with surrounding membranous organelles but not the mitochondria themselves marked by ubiquitin) (Al Rawi et al., 2011; Sato and Sato, 2011; Zhou et al., 2011). In one of these studies (Al Rawi et al., 2011), p62 and LC3 were also found to colocalize with sperm mitochondria after fertilization in mice. However, a more recent study confirmed that sperm mitochondria colocalized with p62 and LC3 in mouse embryos, but concluded that this was not involved in their degradation (Luo et al., 2013). Thus, the question of whether selective mitophagy explains why our mitochondrial DNA comes mainly from our mothers remains to be resolved.
An emerging far-reaching biomedical paradigm is that defects in mitophagy – presumably through resulting abnormal mitochondrial function, abnormal mitochondrial biogenesis, and/or increased mitochondrial generation of reactive oxygen species (leading to genomic instability and enhanced pro-inflammatory signaling) – contributes to cancer, neurodegenerative diseases, myopathies, aging, and inflammatory disorders (reviewed in (Ding and Yin, 2012; Green et al., 2011; Lu et al., 2013; Narendra and Youle, 2011). This paradigm intuitively makes sense, and is consistent with a large body of literature in autophagy-deficient mice. Yet, it is difficult to establish a direct causal relationship between mitophagy defects and disease in mice lacking general autophagy factors. Presumably, phenotypes observed in mice lacking selective mitophagy factors may be more informative. For example, Parkin-deficient mice have cancer-prone phenotypes, including accelerated intestinal adenoma development (in the background of Apc mutation) (Poulogiannis et al., 2010) and the development of hepatocellular carcinoma (Fujiwara et al., 2008). However, these studies also do not provide direct evidence that Parkin-mediated mitophagy, rather than other potential effects of Parkin, contribute to its role in tumor suppression. Moreover, mice lacking Ulk1 (Kundu et al., 2008) or Nix (Sandoval et al., 2008) have progressive anemia with mature erythrocytes containing mitochondria, but no other obvious cancer-prone defects. In addition, Parkin null mice clear defective mitochondria normally in dopaminergic neurons in the substantia nigra (Sterky et al., 2011), even though PARKIN and PINK1 mutations in humans lead to overt degeneration of these neurons and Parkinson’s disease. It is not unlikely that there are several overlapping mechanisms for selective autophagy that compensate for such deficiencies. Another possible explanation for the lack of more striking phenotypes in mice lacking selective autophagy factors is that other processes help to mediate the damage that should accrue when damaged organelles are not effectively cleared from cells, including perhaps the removal of the cells themselves, considered next.
Removing excess or damaged cells
In multicellular organisms, the death of a cell is usually easily tolerated and indeed, part of normal development and homeostasis. Dying cells, regardless of the mode of cell death, are rapidly cleared from the body, either by shedding (e.g., skin, mucosa) or removal by phagocytosis (Figure 2). The latter in mammals is usually by “professional” phagocytes, such as macrophages, but can be mediated by other cells, such as epithelia.
For our discussion, it is useful to consider several distinctions between the many ways that cells can die (Green, 2011). First, cells can die actively or passively, that is, molecularly participating in their own demise, or not. Passive cell death occurs when a cell has accumulated so much acute damage that it cannot maintain its plasma membrane, and usually dies by a process that by morphology is classified as necrosis. Necrotic cells swell as water enters, expanding organelles and the nucleus (as we will see, however, morphological necrosis can also be active). Passive cell death can occur by environmental insult or by “murder,” such as via the action of complement, cytotoxic lymphocytes, or active engulfment of a living cell (e.g., when aged erythrocytes are cleared, or when epithelial cells lose contact with basal lamina). In contrast, active cell death involves intracellular processes that must be engaged for the cell to die. This is often referred to as “programmed cell death,” although the term was originally coined to indicate those cells that die at a defined (programmed) time in development. Here we do not distinguish between active and programmed, and use both to refer to those cell deaths that involve the participation of intracellular molecular machinery.
Active cell death can further be parsed into two general categories: cell suicide and cell sabotage (Green and Victor, 2012). Cell sabotage is akin to the conventional meaning of sabotage: just as a train must be moving quickly if it is to undergo destruction when a rail is dislodged, some disruptions of cellular functions may derail a cell to its destruction only if these functions are actively engaged. In cellular sabotage, the death is therefore molecularly active, but the process (presumably) was not selected during the evolution of multicellularity to transduce the death signal. Examples of cellular sabotage likely include the recently identified process of ferroptosis (Dixon et al., 2012), over-production of reactive oxygen species, and perhaps some forms of mitotic catastrophe.
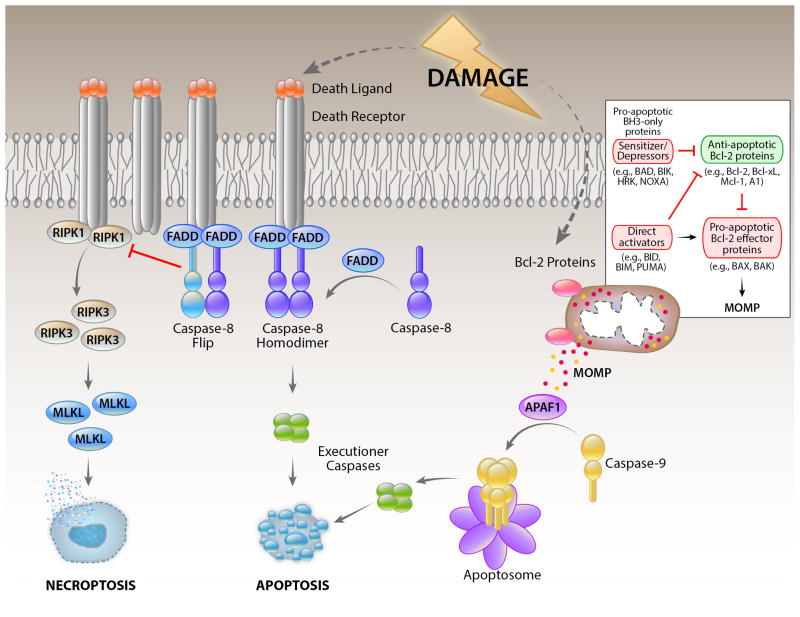
Cellular suicide, in contrast, involves the engagement of evolutionarily selected (again, presumably) molecular pathways that result in cell death (Figure 3). This is best exemplified by the process of apoptosis. There are multiple pathways of apoptosis, all of which involve the activation of caspase proteases which cleave many substrates within the cell, leading to its demise. Apoptosis is formally defined by morphology, in which the cell shrinks, the nucleus condenses, and the cell often fragments into smaller membrane-bound bodies, although more recent descriptions rely on the detection of caspase activation (Galluzzi et al., 2012). In most apoptotic pathways, caspase activation occurs by the formation of “caspase activation platforms” which bind and activate monomeric initiator caspases (e.g., caspase-8, caspase-9) which then cleave and thereby activate the executioner caspases (caspase-3, caspase-7) to orchestrate the death of the cell.
Figure 3. Cell death pathways engaged by cellular damage.
Cellular damage induces cell death by inducing expression and/or modification of pro-apoptotic BH3-only proteins of the Bcl-2 family (inset) which engage the mitochondrial pathway of apoptosis, in which MOMP releases proteins of the mitochondrial inter-membrane space. Among these is cytochrome c, which activates APAF1 to form a caspase-activation platform (the apoptosome) that binds and activates caspase-9. This then cleaves and thereby activates executioner caspases to promote apoptosis. Cellular damage can also induce the expression of death ligands of the TNF family, which bind their receptors to promote the activation of caspase-8 by FADD. The latter is antagonized by expression of c-FLIPL, and the caspase-8-FLIP heterodimer does not promote apoptosis, but instead blocks another cell death pathway engaged by death receptors, necroptosis. Necroptosis involves the activation of RIPK1 and RIPK3, resulting in phosphorylation and activation of the pseudokinase, MLKL, which promotes an active necrotic cell death.
While apoptosis occurs in a variety of settings and often via different pathways, our focus on the clearance of damaged and excess cells focuses much of our attention on only one such pathway which functions in this regard. In this, the mitochondrial (or intrinsic) pathway of apoptosis, the proteins of the Bcl-2 family control a process whereby the outer membranes of mitochondria become permeable (mitochondrial outer membrane permeabilization, MOMP), releasing the proteins of the intermembrane space to the cytosol. Cytochrome c, thus released, activates apoptosis activating factor-1 (APAF1) to form a caspase activation platform for the activation of the initiator caspase, caspase-9 (Figure 3).
Among the Bcl-2 proteins, the proapoptotic effector proteins, Bax and Bak, are responsible for the permeabilization of the outer membranes of mitochondria. These proteins, when activated, oligomerize and insert to cause permeabilization, probably causing the formation of lipidic pores that appear to be restricted to mitochondria. Active Bax and Bak are bound and inhibited by the anti-apoptotic Bcl-2 proteins (e.g., Bcl-2, Bcl-xL, Mcl-1), thereby blocking apoptosis. Another subfamily, the BH3-only proteins (so called because they carry only one of four Bcl-2 homology domains) function to antagonize the anti-apoptotic Bcl-2 proteins (the “sensitizer/de-repressors”) and/or to activate the pro-apoptotic effectors (the latter activity is a property of only a subset of these, the “direct activators,” notably Bid, Bim, and Puma) (Chipuk et al., 2010). Sequestration of the latter by anti-apoptotic Bcl-2 proteins is another way in which apoptosis can be blocked, rendering the cell “primed for death” if the sequestration is disrupted (Sarosiek et al., 2013). Cancers in which the cells are thereby primed are demonstrably more likely to respond to conventional therapies (Ni Chonghaile et al., 2011, Vo et al., 2012). This is the basis for the potential efficacy of BH3-mimetic drugs, such as Novatoclax and ABT-199.
Damage or other cues trigger the mitochondrial pathway of apoptosis via activation and/or expression of BH3-only proteins, which in turn de-repress the anti-apoptotic Bcl-2 proteins and activate the pro-apoptotic effectors. Structurally, the BH3 region of the BH3-only protein binds to a hydrophobic groove in the target Bcl-2 protein, blocking it in the case of anti-apoptotic proteins, or triggering conformational changes in the proapoptotic effector (Figure 3). The latter appears to occur by a “hit and run” mechanism, in which the BH3-only protein is subsequently displaced by the BH3 region of a neighboring, activated effector, to form a dimer. Higher order oligomers of the activated effector then form to perturb the membrane and effect MOMP.
Remarkably, the anti-apoptotic Bcl-2 proteins also bind to Beclin 1, via a bona fide BH3 domain in the latter, and this interaction inhibits the engagement of autophagy, discussed above. The roles of these proteins in controlling both mitochondrial apoptosis and autophagy represent a key node for cross-regulation, and may explain why cell death and autophagy often coincide as “autophagic cell death” (Gump and Thorburn, 2011). However, the latter is not well understood, and the precise interplay of the pathways remains unresolved. Further, the extent to which the Bcl-2 proteins regulate selective autophagy is unclear, and although the mitophagic proteins, Nix and BNIP3 are members of the Bcl-2 family and promote mitophagy, they do not do so via their BH3 domains, nor do they promote apoptosis. The lower affinity of anti-apoptotic proteins for the BH3 domain of Beclin 1 – versus those for the BH3 domains of pro-apoptotic proteins – may account for the early activation of autophagy in response to stress (which involves disruption of Bcl-2/Bcl-xL-Beclin 1 binding) which, if unsuccessful in keeping the cell alive, ultimately results in the cell’s transition to a pro-apoptotic state (which, as noted above, involves the release of pro-apoptotic molecules such as Bax and Bak) (Sinha and Levine, 2008). A crucial open question is how cells “know” whether autophagy will be successful in controlling cellular damage and therefore, they should make a decision in continuing “to be” or whether autophagy will be futile and they should decide “not to be” and to undergo apoptosis.
Other active cell deaths in response to damage
Programmed cell death does not necessarily proceed by apoptosis; there is extensive evidence for forms of programmed necrosis as well (Figure 3). These include excitotoxicity of neurons (Mehta et al., 2013), regulated necrosis by the mitochondrial permeability transition (MPT) (Rasola and Bernardi, 2011), and necroptosis, a form of programmed necrosis mediated by receptor interacting protein kinase 3 (RIPK3) and a pseudokinase, mixed lineage kinase like (MLKL) (Kaczmarek et al., 2013). While all have been implicated in pathological cell death under different conditions, and at least one (necroptosis) is potentially involved in responses to infection, there is currently no evidence that these are involved in physiological removal of damaged or unwanted cells. This view could change, however. Animals in which the intestinal epithelium lacks the pro-apoptotic effector proteins Bax and Bak, and therefore lack the mitochondrial pathway of apoptosis, nevertheless undergo death of intestinal cells in response to ionizing radiation (Kirsch et al., 2010). One or more of the programmed necrosis pathways may contribute to such cell death.
Further, systemic responses to environmental tissue damage can invoke a cytokine response, especially the production of tumor necrosis factor (TNF). TNF, as well as some other members of the TNF family (particularly CD95-ligand and TRAIL), the so-called “death ligands.” These are capable of engaging apoptosis via the death receptor (or extrinsic) pathway (Wilson et al., 2009). Ligation of the death receptors of the TNF-receptor (TNFR) family induces the formation of another caspase-activation platform, which in turn binds and activates the initiator caspase, caspase-8. Apoptosis promoted by caspase-8 is antagonized by a caspase-like molecule, cFLIPL (herein, FLIP) which can be induced by TNFR ligation and which forms a heterodimer with caspase-8, preventing apoptosis. In addition, the caspase-8-FLIP heterodimer functions as an active protease to block another pathway of cell death, that of necroptosis. Under conditions where TNFR1 is engaged but caspase-8-FLIP activity is blocked or disrupted, the RIPK3-MLKL interaction is promoted to cause necrotic cell death (Kaczmarek et al., 2013), through an effector mechanism that remains elusive, but is likely to involve the plasma membrane.
Clearance of dying cells
When dying cells are rapidly cleared from the body by phagocytosis, the process can be summarized as “find me, bind me, eat me, digest me”. As a result, regardless of how a cell has died, it is rapidly removed from the system.
If the plasma membrane of the dying cell is ruptured, signals are released that recruit and activate phagocytes such as macrophages. These recruitment signals, damage-associated molecular patterns (DAMPs), include ATP, UTP, and uric acid (Krysko et al., 2012). These and other DAMPs have additional consequences, considered below. However, cells that die by apoptosis usually do not release DAMPs prior to engulfment (although this has been contested (Kroemer et al., 2013), and other signals (“find me” signals) recruit macrophages. These include (depending on the dying cell) lysophosphatidylcholine, sphingosine-1-phosphate, fractalkine, and ATP (Ravichandran, 2011). In each case, the idea is that the release of find-me signals creates a local gradient that serves to bring nearby phagocytes to the dying cell for its clearance. Measurement and/or manipulation of such gradients in vivo for the elucidation of these find-me mechanisms are largely lacking, however.
An important consideration here is the negative regulation of the process of phagocytic clearance. While living cells can be engulfed by other cells (entosis, see below), this does not generally occur, leading to the concept of “don’t-eat-me” signals. One such signal appears to be CD47, widely expressed on living cells and lost prior to engulfment (Chao et al., 2011). CD47 ligates SIRP1α on phagocytic cells to generate the inhibitory signal (Willingham et al., 2012). Strikingly, neutralization of CD47 by antibodies or engineered SIRP1α promotes engulfment and destruction of living cancer cells in vivo, and represents a promising and novel avenue to cancer therapy.
Once recruited, phagocytes respond to the dying cell by binding and engulfing the cell. There are a large number of bind-me signals which serve to tether the phagocyte and enhance subsequent clearance, but these do not appear to functionally signal the phagocytic process. The eat-me signals function in this second event. The most prominent of the eat-me signals is the exposure of phosphatidylserine (PS) on the outer leaflet of the plasma membrane of the dying cell (Ravichandran, 2011).
PS in living cells is predominantly localized to the inner leaflet of the plasma membrane by the action of an ATP-dependent lipid translocase. If the plasma membrane is disrupted, however, lipid asymmetry is lost. Several proteins bind to PS, acting either as bridging molecules to phagocyte receptors, or as receptors on the phagocytes themselves (Ravichandran, 2011). Examples of the former are MFG-E8, a soluble molecule that binds PS on the dying cell and integrins on the phagocyte, and Gas6, which bridges PS and the MER kinase on the phagocyte. Phagocyte receptors that directly bind to PS include Tim-4, stabilin-2, and BAI-1. Each of these trigger pathways of actin reorganization and engulfment to bring the dying cell into the phagocyte. Which bridging molecules or receptors are most important appears to depend on the phagocyte that is engaged.
In apoptotic cells, PS is exposed on the intact plasma membrane to promote clearance of the dying cell prior to the release of DAMPs. Caspases, activated during apoptosis, promote this effect by cleaving the protein Xkr8 (Suzuki et al., 2013). In contrast, PS can also be exposed on living cells in response to calcium influx, via the action of another protein, TMEM16 (Suzuki et al., 2013). In the latter case, however, cells are not engulfed, and it is possible that don’t-eat-me signals are important in preventing these cells from being cleared.
Intriguingly, the autophagic process has been implicated in PS exposure on some, but perhaps not all cells (Qu et al., 2007; Mellén et al., 2008). In the absence of Beclin 1 or Atg5 (or with pharmacological inhibition of autophagy), the exposure of PS on apoptotic cells in differentiating mouse embryoid bodies or developing chick retina is impaired, as is their clearance, and this can be circumvented by increasing mitochondrial ATP production via addition of methylpyruvate. At present, a requirement for ATP in the function of Xkr8 is not established, and it is possible that another process functions (in an ATP-dependent manner) in these dying cells. One possibility is that fusion of PS-rich vesicles with the plasma membrane contributes to cell surface PS exposure in these cases (Lee et al., 2013), although a role for autophagy (and/or ATP) in this process has not been examined.
Once the cell is eaten, it must be efficiently digested, despite the simple fact that often an entire cell has been engulfed (effectively doubling the mass of the phagocyte). Upon uptake of the corpse into a phagosome, the phagosome then fuses with lysosomes (discussed in more detail below) and the cargo is digested. To handle the massive digestive activity, at least two changes to the phagocyte occur. The excess cholesterol is effluxed from the cell by the action of a transporter, the expression of which is induced by engagement of LXR signaling (Han and Ravichandran, 2011). In addition, the phagocyte expresses a protein, UCP2, which uncouples the electron transport chain of mitochondria from the proton gradient, causing the mitochondria to go into “over-drive,” consuming oxygen and free fatty acids (from the digested corpse) (Han and Ravichandran, 2011). Both are necessary for efficient clearance of the dying cell.
While the clearance of dying cells proceeds via the same mechanisms, regardless of how the cell died, the mode of cell death has different effects on the host. Dying cells release pro-inflammatory molecules, called damage-associated molecular patterns (DAMPs) when the plasma membrane is disrupted. These include uric acid, ATP/UTP, HMGB1, and other molecules. While necrotic cells release DAMPs, apoptotic cells, by virtue of their intact plasma membranes, generally do not, and therefore do not cause an inflammatory response. Apoptotic cells also often release lactoferin, which serves to inhibit neutrophil recruitment and activation (Bournazou et al., 2009) thereby contributing to the lack of inflammation often associated with apoptotic death.
In addition, molecules associated with dying cells can be targeted by the adaptive immune system, conditional on the mode of cell death, as the dead cells are taken up by dendritic cells to present the associated antigens to T lymphocytes (Green et al., 2009). Often, necrotic cells promote such adaptive immunity while apoptotic cells promote a state of active immune tolerance, blocking responses to the antigens. This is overly simplistic, however, and apoptotic cells can promote T cell immunity under some conditions (Kroemer et al., 2013). Such “immunogenic cell death” appears to depend on the autophagy pathway-dependent release of ATP/UTP and exposure of the endoplasmic reticulum protein, calreticulin, on the surface of the dying cell. In addition, the presentation of antigens from corpses engulfed by dendritic cells is dependent on autophagy in the dying cell (Perot et al., 2013). Thus autophagy plays roles in not only the processes of cell death, but also in its consequences. This may be particularly important in anti-cancer immunity.
Non-canonical autophagy pathway and clearance of dying cells
When dying cells are engulfed by a macrophage or other cell, the corpse-containing phagosome is rapidly decorated with the autophagic protein, LC3, which facilitates fusion with lysosomes and destruction of the cargo (Sanjuan et al., 2009). This LC3-associated phagocytosis (LAP) is dependent on the Beclin 1-VPS34 complex and the elongation machinery, but rather than generating a double-membrane autophagosome, LC3-II is generated on the single-membrane phagosome itself (Figure 2). In contrast to general or selective organellar autophagy, however, LAP appears to proceed independently of the ULK1 pre-initiation complex in mammalian cells (Henault et al., 2012). If LAP is defective (due, for example, to lack of the requisite autophagy machinery) the corpse is not digested, and macrophages produce high levels of pro-inflammatory cytokines (Martinez et al., 2011). This may have implications for disease. For example, systemic lupus erythematosis is often characterized by circulation of “LE cells” which have been identified as macrophages containing an undigested corpse. It is possible that pro-inflammatory signals emitted by such macrophages contribute to the disease.
LAP is also implicated in another process of “garbage disposal,” although in this case it involves clearance of a fragment rather than a dying cell. Every morning, the photoreceptor outer segments (POS) of the retina are shed and engulfed by the neighboring retinal pigment epithelia (RPE). LAP is engaged upon phagocytosis and facilitates degradation of the POS and recycling of the associated retinoic acid to the retina for new photoreceptor generation (Kim et al., 2013). A failure of LAP compromises this cycle and causes a progressive loss of vision.
The role of the autophagy pathway in LAP therefore presents a challenge to our interpretations of defective autophagy in disease processes. At least some diseases associated with age-related decline of expression of autophagy genes, or with polymorphisms in such genes (Choi et al., 2013) might arise as a consequence of defective LAP rather than conventional autophagy. The intimate relationships between the clearance of dying cells, LAP, and the inflammatory response support this idea.
The interface of autophagy and cell death in tissue homeostasis
LAP (see above) may also represent a link between autophagy components and a cell death process (and not only the clearance of dying cells), as engulfment of cells may restrict oncogenesis naturally. Immortalized mammary epithelial cells, upon loss of anchorage to basement membranes, engulf each other in a process called “entosis” (Florey et al., 2010). The engulfed cell dies by apoptosis due to nutrient deprivation. If the cell expresses an anti-apoptotic signal, such as Bcl-2, it is nevertheless killed as LAP in the engulfing cell promotes fusion with lysosomes (Figure 2). However, if cells resist apoptosis and fail to engage LAP (e.g. due to ablation of the autophagic machinery) such immortalized cells escape entosis and grow in an anchorage-independent manner. The implications of such an interplay between apoptosis and autophagy at the cellular level has obvious consequences for understanding oncogenesis.
In thinking about general autophagy (in response to metabolic stress), selective autophagy (in response to damaged organelles), and cell death (as a consequence of excessive damage), it is obvious that the pathways cross-talk at a superficial level. That is, a cell that is defective for autophagy will necessarily be more prone to die if faced with nutrient deprivation. Cells lacking selective autophagy will accumulate damaged organelles such as mitochondria, which can generate signals (e.g., ROS) that promote further damage and ultimately cell death. If cell death does not occur, the ensuing dysfunction may promote severe effects in the form of oncogenesis. And of course a cell that engages death pathways will circumvent any benefit that might arise from autophagy. In a recent study, the extent of autophagy of individual cells in a population inversely correlated with the likelihood that a cell would die in response to engagement of the death receptor pathway of apoptosis (Gump et al., 2013).
There are more fundamental molecular interactions between these pathways, but it is difficult to parse how specific interactions contribute to cross-regulation in the face of the over-arching effect of apoptotic defects on cellular health. For example, Beclin 1 is bound and inhibited by the anti-apoptotic Bcl-2 proteins (Pattingre et al., 2005) and pro-apoptotic BH3-only proteins appear to be capable of disrupting this interaction to promote autophagy (Maiuri et al., 2007). Other autophagy components also interact with apoptotic players, but again, it is unclear if these interactions, per se, influence cell fate. Examples include the binding and inhibition of Bax by UVRAG (Yin et al., 2011) and the binding and inhibition of Mcl-1 by ATG12 (Rubinstein et al., 2011), two interactions predicted to block or promote apoptosis, respectively.
Clearly, then, while it is highly likely that there are fundamental molecular interactions between the pathways of cell death and autophagy, it will be important to explore ways in which specific interactions can be disrupted without compromising the major pathways themselves, in order to separate specific and general effects. This is a challenge as we move forward in our understanding of these basic mechanisms of cellular quality control.
We began our discussion with what may be the most fundamental dichotomy in biology, to be or not to be. At the cellular level, the question might be less elegantly posed along these lines: should we (the cell) be functional or, if not, die? Or if we are dysfunctional and survive, do we risk compromising the life of the organism? Do we unite the fundamental pathways of garbage disposal, selective autophagy and active cell death, through complex (and largely unexplored) molecular interactions, or do we let the thresholds of damage dictate which pathway holds sway against the thousand natural shocks that flesh is heir to (again, with apologies to the Bard)? Those are the questions.
Acknowledgments
The authors thank Haley Harrington for assistance with manuscript preparation and Angela Diehl for expert scientific illustration. BL is supported by NIH R01 CA109618 and CPRIT grant RP120718-P1. DRG is supported by NIH R01 AI40646, AI47891, AI44828, CA169291, and GM 096208.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- Bournazou I, Pound JD, Duffin R, Bournazos S, Melville LA, Brown SB, Rossi AG, Gregory CD. Apoptotic human cells inhibit migration of granulocytes via release of lactoferrin. J Clin Invest. 2009;119:20–32. doi: 10.1172/JCI36226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebollero E, Reggiori F, Kraft C. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int J Cell Biol. 2012;2012:1–9. doi: 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, Majeti R, Weissman IL. Programmed cell removal: a new obstacle in the road to developing cancer. Nat Rev Cancer. 2011;12:58–67. doi: 10.1038/nrc3171. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The Bcl-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimiation signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15:1197–11205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Yin XM. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florey O, Krajcovic M, Sun Q, Overholtzer M. Entosis Curr Biol. 2010;20:R88–R89. doi: 10.1016/j.cub.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Fujita N, Morita E, Itoh T, Tanaka A, Nakaoka M, Osada Y, Umemoto T, Saitoh T, Nakatogawa H, Kobayashi S, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Marusawa H, Wang HQ, Iwai A, Ikeuchi K, Imai Y, Kataoka A, Nukina N, Takahashi R, Chiba T. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27:6002–6011. doi: 10.1038/onc.2008.199. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR. Means to an end. Apoptosis and other cell death mechanisms. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2011. [Google Scholar]
- Green DR, Ferguson T, Zitvogel L, Kroemer G. Immunogenic and tolerogenic cell death. Nat Rev Immunol. 2009;9:353–363. doi: 10.1038/nri2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR, Victor B. The pantheon of the fallen: why are there so many forms of cell death? Trends Cell Biol. 2012;22:555–556. doi: 10.1016/j.tcb.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gump JM, Staskiewicz L, Morgan MJ, Bamberg A, Riches DW, Thorburn A. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat Cell Biol. 2013;16:47–54. doi: 10.1038/ncb2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gump JM, Thorburn A. Autophagy and apoptosis: what is the connection? Trends Cell Biol. 2011;21:387–392. doi: 10.1016/j.tcb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol. 2013;25:455–460. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Han CZ, Ravichandran KS. Metabolic connections during apoptotic cell engulfment. Cell. 2011;147:1442–1445. doi: 10.1016/j.cell.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Kuge S, Fujiki Y. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp Cell Res. 2008;314:3531–3541. doi: 10.1016/j.yexcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012;37:986–997. doi: 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci. 2012;125:1488–1499. doi: 10.1242/jcs.094110. [DOI] [PubMed] [Google Scholar]
- Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity. 2013;38:209–223. doi: 10.1016/j.immuni.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Zhao H, Martinez J, Doggett TA, Kolesnikov AV, Tang PH, Ablonczy Z, Chan CC, Zhou Z, Green DR, et al. Noncanonical autophagy promotes the visual cycle. Cell. 2013;154:365–376. doi: 10.1016/j.cell.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch DG, Santiago PM, di Tomaso E, Sullivan JM, Hou WS, Dayton T, Jeffords LB, Sodha P, Mercer KL, Cohen R, et al. p53 controls radiation-induced gastrointestinal syndrome in mice independent of apoptosis. Science. 2010;327:593–596. doi: 10.1126/science.1166202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Immunol. 2013;31:51–72. doi: 10.1146/annurev-immunol-032712-100008. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer. 2012;12:860–875. doi: 10.1038/nrc3380. [DOI] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Meng XW, Flatten KS, Loegering DA, Kaufmann SH. Phosphatidylserine exposure during apoptosis reflects bidirectional trafficking between plasma membrane and cytoplasm. Cell Death Differ. 2013;20:64–76. doi: 10.1038/cdd.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Elazar Z. Development. Inheriting maternal mtDNA. Science. 2011;334:1069–1070. doi: 10.1126/science.1215480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20:3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- Lu H, Li G, Liu L, Feng L, Wang X, Jin H. Regulation and function of mitophagy in development and cancer. Autophagy. 2013;9:1720–1736. doi: 10.4161/auto.26550. [DOI] [PubMed] [Google Scholar]
- Luo SM, Ge ZJ, Wang ZW, Jiang ZZ, Wang ZB, Ouyang YC, Hou Y, Schatten H, Sun QY. Unique insights into maternal mitochondrial inheritance in mice. Proc Natl Acad Sci U S A. 2013;110:13038–13043. doi: 10.1073/pnas.1303231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, Geneste O, Kroemer G. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3:374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A. 2011;108:17396–17401. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mehta A, Prabhakar M, Kumar P, Deshmukh R, Sharma PL. Excitotoxicity: bridge to various triggers in neurodegenerative disorders. Eur J Pharmacol. 2013;698:6–18. doi: 10.1016/j.ejphar.2012.10.032. [DOI] [PubMed] [Google Scholar]
- Meijer AJ, Klionsky DJ. Vps34 is a phosphatidylinositol 3-kinase, not a phosphoinositide 3-kinase. Autophagy. 2011;7:563–564. doi: 10.4161/auto.7.6.14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén MA, de la Rosa EJ, Boya P. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ. 2008;15:1279–1290. doi: 10.1038/cdd.2008.40. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, Simon AK. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci U S A. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Youle RJ. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid Redox Signal. 2011;14:1929–1938. doi: 10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, del Moore VG, Deng J, Anderson KC, Richardson P, Tai YT, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. 2011;334:1129–1133. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Perot BP, Ingersoll MA, Albert ML. The impact of macroautophagy on CD8(+) T-cell-mediated antiviral immunity. Immunol Rev. 2013;255:40–56. doi: 10.1111/imr.12096. [DOI] [PubMed] [Google Scholar]
- Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, Luo F, Cantley LC, Wyllie AH, Adams DJ, et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci U S A. 2010;107:15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- Rasola A, Bernardi P. Mitochondrial permeability transition in Ca2+-dependent apoptosis and necrosis. Cell Calcium. 2011;50:222–233. doi: 10.1016/j.ceca.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Ravichandran KS. Beginnings of a good apoptotic meal: the find-me and eat-me signaling pathways. Immunity. 2011;35:445–455. doi: 10.1016/j.immuni.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan K-L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Milasta S, Green DR. Toll-like receptor signaling in the lysosomal pathways. Immunol Rev. 2009;227:203–220. doi: 10.1111/j.1600-065X.2008.00732.x. [DOI] [PubMed] [Google Scholar]
- Sarosiek KA, Ni Chonghaile T, Letai A. Mitochondria: gatekeepers of response to chemotherapy. Trends Cell Biol. 2013;23:612–619. doi: 10.1016/j.tcb.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–3. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH-3 only protein. Oncogene. 2008;1:S137–S148. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V, McLelland GL, Zunino R, Braschi E, Rippstein P, Fon EA, McBride HM. A vesicular transport pathway shuttles cargo from mitochondira to lysosomes. Curr Biol. 2012;22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- Sterky FH, Lee S, Wibom R, Olson L, Larsson NG. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc Natl Acad Sci U S A. 2011;108:12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science. 2013;341:403–406. doi: 10.1126/science.1236758. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Fujii T, Imao T, Ishihara K, Kuba H, Nagata S. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J Biol Chem. 2013;288:13305–13316. doi: 10.1074/jbc.M113.457937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till A, Lakhani R, Burnett SF, Subramani S. Pexophagy: the selective degradation of peroxisomes. Int J Cell Biol. 2012;2012:1–18. doi: 10.1155/2012/512721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B, Vandenberghe W. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci. 2011;31:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo TT, Ryan J, Carrasco R, Neuberg D, Rossi DJ, Stone RM, Deangelo DJ, Frattini MG, Letai A. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell. 2012;151:344–355. doi: 10.1016/j.cell.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, Wang J, Contreras-Trujillo H, Martin R, Cohen JD, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol. 2009;10:348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- Wirth M, Joachim J, Tooze SA. Autophagosome formation-The role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23:301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Yin X, Cao L, Kang R, Yang M, Wang Z, Peng Y, Tan Y, Liu L, Xie M, Zhao Y, et al. UV irradiation resistance-associated gene suppresses apoptosis by interfering with BAX activation. EMBO Rep. 2011;12:727–734. doi: 10.1038/embor.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li H, Xue D. Elimination of paternal mitochondria through the lysosomal degradation pathway in C. elegans. Cell Res. 2011;21:1662–1669. doi: 10.1038/cr.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]