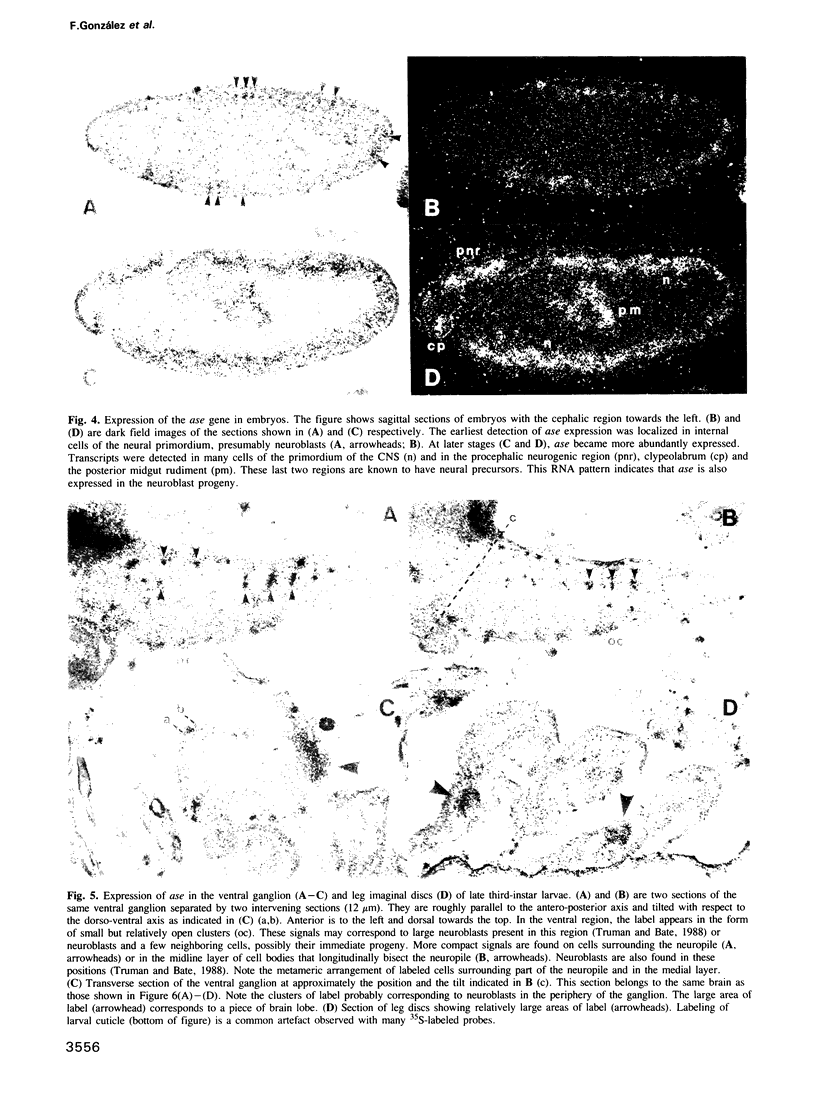
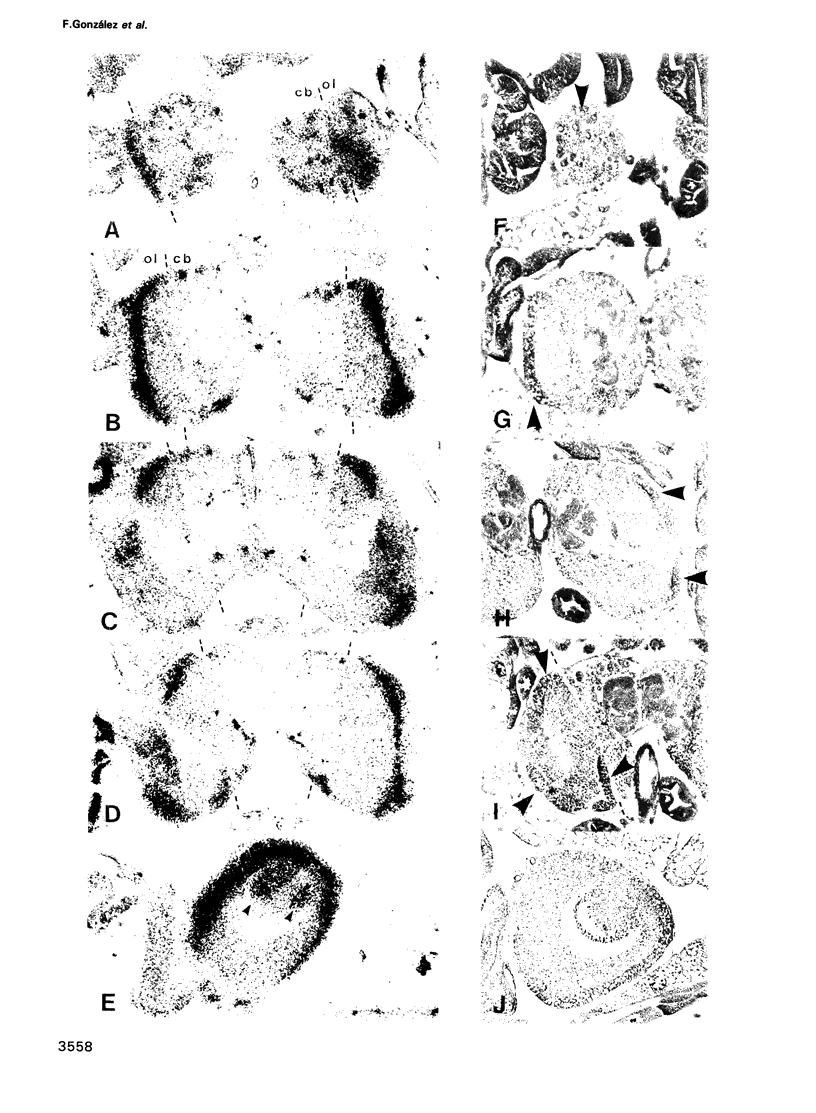
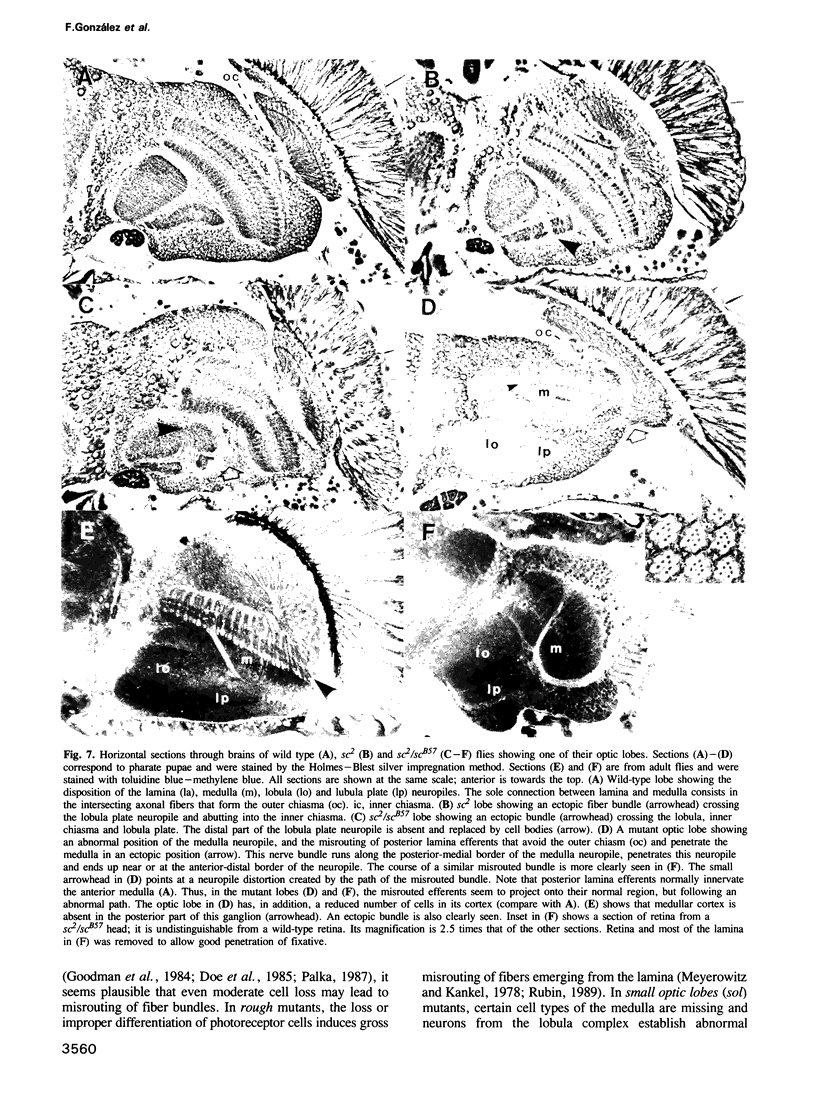
Abstract
The achaete-scute complex (AS-C) comprises five genetic regions: achaete, scute (sc) alpha, lethal of sc, sc beta and sc gamma. Each region promotes the determination and positional specification of different, but partially overlapping, subsets of neural elements of Drosophila. In this work, we report a molecular characterization of the sc gamma region. It comprises 22 kb of DNA and contains two transcription units, only one of which, named asense (ase), seems involved in neurogenesis. ase encodes a protein that shares with other three AS-C proteins a domain containing a helix--loop--helix motif characteristic of a group of DNA-binding proteins. In the embryo, ase is expressed in neural precursor cells, a pattern consistent with the known requirement of sc gamma for the development of the larval nervous system. In late third-instar larvae, the gene is expressed in developing structures of the central nervous system (CNS), namely the anlagen of the optic lobes and in many cells, including neuroblasts, of the central brain and ventral ganglia. Its removal leads to anatomical defects in the adult optic lobes. This is the first demonstration of a role for the AS-C in the development of the adult CNS.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alonso M. C., Cabrera C. V. The achaete-scute gene complex of Drosophila melanogaster comprises four homologous genes. EMBO J. 1988 Aug;7(8):2585–2591. doi: 10.1002/j.1460-2075.1988.tb03108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcells L., Modolell J., Ruiz-Gómez M. A unitary basis for different Hairy-wing mutations of Drosophila melanogaster. EMBO J. 1988 Dec 1;7(12):3899–3906. doi: 10.1002/j.1460-2075.1988.tb03276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamonte D., Modolell J. Search for Drosophila genes encoding a conserved domain present in the achaete-scute complex and myc proteins. Mol Gen Genet. 1989 Jan;215(2):281–285. doi: 10.1007/BF00339729. [DOI] [PubMed] [Google Scholar]
- Bensi G., Raugei G., Klefenz H., Cortese R. Structure and expression of the human haptoglobin locus. EMBO J. 1985 Jan;4(1):119–126. doi: 10.1002/j.1460-2075.1985.tb02325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Braun T., Buschhausen-Denker G., Bober E., Tannich E., Arnold H. H. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J. 1989 Mar;8(3):701–709. doi: 10.1002/j.1460-2075.1989.tb03429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera C. V., Martinez-Arias A., Bate M. The expression of three members of the achaete-scute gene complex correlates with neuroblast segregation in Drosophila. Cell. 1987 Jul 31;50(3):425–433. doi: 10.1016/0092-8674(87)90496-x. [DOI] [PubMed] [Google Scholar]
- Campuzano S., Balcells L., Villares R., Carramolino L., García-Alonso L., Modolell J. Excess function hairy-wing mutations caused by gypsy and copia insertions within structural genes of the achaete-scute locus of Drosophila. Cell. 1986 Jan 31;44(2):303–312. doi: 10.1016/0092-8674(86)90764-6. [DOI] [PubMed] [Google Scholar]
- Campuzano S., Carramolino L., Cabrera C. V., Ruíz-Gómez M., Villares R., Boronat A., Modolell J. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell. 1985 Feb;40(2):327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- Caudy M., Vässin H., Brand M., Tuma R., Jan L. Y., Jan Y. N. daughterless, a Drosophila gene essential for both neurogenesis and sex determination, has sequence similarities to myc and the achaete-scute complex. Cell. 1988 Dec 23;55(6):1061–1067. doi: 10.1016/0092-8674(88)90250-4. [DOI] [PubMed] [Google Scholar]
- Cavener D. R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987 Feb 25;15(4):1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia W., Howes G., Martin M., Meng Y. B., Moses K., Tsubota S. Molecular analysis of the yellow locus of Drosophila. EMBO J. 1986 Dec 20;5(13):3597–3605. doi: 10.1002/j.1460-2075.1986.tb04688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C. Q., Hiromi Y., Gehring W. J., Goodman C. S. Expression and function of the segmentation gene fushi tarazu during Drosophila neurogenesis. Science. 1988 Jan 8;239(4836):170–175. doi: 10.1126/science.2892267. [DOI] [PubMed] [Google Scholar]
- Doe C. Q., Kuwada J. Y., Goodman C. S. From epithelium to neuroblasts to neurons: the role of cell interactions and cell lineage during insect neurogenesis. Philos Trans R Soc Lond B Biol Sci. 1985 Dec 17;312(1153):67–81. doi: 10.1098/rstb.1985.0178. [DOI] [PubMed] [Google Scholar]
- Doe C. Q., Smouse D., Goodman C. S. Control of neuronal fate by the Drosophila segmentation gene even-skipped. Nature. 1988 May 26;333(6171):376–378. doi: 10.1038/333376a0. [DOI] [PubMed] [Google Scholar]
- Fischbach K. F., Heisenberg M. Structural brain mutant of Drosophila melanogaster with reduced cell number in the medulla cortex and with normal optomotor yaw response. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1105–1109. doi: 10.1073/pnas.78.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke W. W., Krien S., Brown R. M., Jr Simultaneous glutaraldehyde-osmium tetroxide fixation with postosmication. An improved fixation procedure for electron microscopy of plant and animal cells. Histochemie. 1969;19(2):162–164. doi: 10.1007/BF00281096. [DOI] [PubMed] [Google Scholar]
- García-Bellido A. Genetic Analysis of the Achaete-Scute System of DROSOPHILA MELANOGASTER. Genetics. 1979 Mar;91(3):491–520. doi: 10.1093/genetics/91.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghysen A., Dambly-Chaudière C. From DNA to form: the achaete-scute complex. Genes Dev. 1988 May;2(5):495–501. doi: 10.1101/gad.2.5.495. [DOI] [PubMed] [Google Scholar]
- Goodman C. S., Bastiani M. J., Doe C. Q., du Lac S., Helfand S. L., Kuwada J. Y., Thomas J. B. Cell recognition during neuronal development. Science. 1984 Sep 21;225(4668):1271–1279. doi: 10.1126/science.6474176. [DOI] [PubMed] [Google Scholar]
- Guarente L. UASs and enhancers: common mechanism of transcriptional activation in yeast and mammals. Cell. 1988 Feb 12;52(3):303–305. doi: 10.1016/s0092-8674(88)80020-5. [DOI] [PubMed] [Google Scholar]
- Ingham P. W. Genetic control of the spatial pattern of selector gene expression in Drosophila. Cold Spring Harb Symp Quant Biol. 1985;50:201–208. doi: 10.1101/sqb.1985.050.01.026. [DOI] [PubMed] [Google Scholar]
- Jiménez F., Campos-Ortega J. A. A region of the Drosophila genome necessary for CNS development. Nature. 1979 Nov 15;282(5736):310–312. doi: 10.1038/282310a0. [DOI] [PubMed] [Google Scholar]
- Jiménez F., Campos-Ortega J. A. Genes in subdivision 1B of the Drosophila melanogaster X-chromosome and their influence on neural development. J Neurogenet. 1987 Jun;4(4):179–200. [PubMed] [Google Scholar]
- Klämbt C., Knust E., Tietze K., Campos-Ortega J. A. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 1989 Jan;8(1):203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz E. M., Kankel D. R. A genetic analysis of visual system development in Drosophilia melanogaster. Dev Biol. 1978 Jan;62(1):112–142. doi: 10.1016/0012-1606(78)90096-9. [DOI] [PubMed] [Google Scholar]
- Modolell J., Bender W., Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Rogers S., Wells R., Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986 Oct 17;234(4774):364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Romani S., Campuzano S., Macagno E. R., Modolell J. Expression of achaete and scute genes in Drosophila imaginal discs and their function in sensory organ development. Genes Dev. 1989 Jul;3(7):997–1007. doi: 10.1101/gad.3.7.997. [DOI] [PubMed] [Google Scholar]
- Romani S., Campuzano S., Modolell J. The achaete-scute complex is expressed in neurogenic regions of Drosophila embryos. EMBO J. 1987 Jul;6(7):2085–2092. doi: 10.1002/j.1460-2075.1987.tb02474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M. Development of the Drosophila retina: inductive events studied at single cell resolution. Cell. 1989 May 19;57(4):519–520. doi: 10.1016/0092-8674(89)90120-7. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gómez M., Modolell J. Deletion analysis of the achaete-scute locus of Drosophila melanogaster. Genes Dev. 1987 Dec;1(10):1238–1246. doi: 10.1101/gad.1.10.1238. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., Stoetzel C., Gorostiza-Thisse C., Perrin-Schmitt F. Sequence of the twist gene and nuclear localization of its protein in endomesodermal cells of early Drosophila embryos. EMBO J. 1988 Jul;7(7):2175–2183. doi: 10.1002/j.1460-2075.1988.tb03056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman J. W., Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988 Jan;125(1):145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Villares R., Cabrera C. V. The achaete-scute gene complex of D. melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987 Jul 31;50(3):415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985 Jan;40(1):55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]
- White K., Kankel D. R. Patterns of cell division and cell movement in the formation of the imaginal nervous system in Drosophila melanogaster. Dev Biol. 1978 Aug;65(2):296–321. doi: 10.1016/0012-1606(78)90029-5. [DOI] [PubMed] [Google Scholar]
- Wright W. E., Sassoon D. A., Lin V. K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989 Feb 24;56(4):607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]