Abstract
Botulinum neurotoxins (BoNTs) are produced as progenitor toxin complexes (PTCs) by Clostridium botulinum. The PTCs are composed of BoNT and non-toxic neurotoxin-associated proteins (NAPs), which serve to protect and deliver BoNT through the gastrointestinal tract in food borne botulism. HA33 is a key NAP component that specifically recognizes host carbohydrates and helps enrich PTC on the intestinal lumen preceding its transport across the epithelial barriers. Here, we report the crystal structure of HA33 of type B PTC (HA33/B) in complex with lactose at 1.46 Å resolution. The structural comparisons among HA33 of serotypes A–D reveal two different HA33–glycan interaction modes. The glycan-binding pockets on HA33/A and B are more suitable to recognize galactose-containing glycans in comparison to the equivalent sites on HA33/C and D. On the contrary, HA33/C and D could potentially recognize Neu5Ac as an independent receptor, whereas HA33/A and B do not. These findings indicate that the different oral toxicity and host susceptibility observed among different BoNT serotypes could be partly determined by the serotype-specific interaction between HA33 and host carbohydrate receptors. Furthermore, we have identified a key structural water molecule that mediates the HA33/B–lactose interactions. It provides the structural basis for development of new receptor-mimicking compounds, which have enhanced binding affinity with HA33 through their water-displacing moiety.
Keywords: Botulinum neurotoxins, Progenitor toxin complex, Hemagglutinin, Carbohydrate receptor, Host–pathogen interaction
1. Introduction
Botulinum neurotoxins (BoNTs) are highly potent bacterial toxins, which are classified as the Tier 1 select agents by the Centers for Disease Control and Prevention (CDC). Eight serotypes of BoNTs have been identified that are designated as BoNT/A–H [1,2]. BoNTs attack motoneurons at neuromuscular junctions (NMJs) and cleave the soluble N-ethylmaleimide sensitive factor attachment protein receptors (SNAREs) complex, which subsequently inhibits the release of acetylcholine neurotransmitter and paralyzes the affected muscles [3–5]. This is the common mechanism underlying the life-threatening disease botulism in human and other animals.
It is well known that the acidic environment, the abundant digestive proteases, and the tightly sealed intestinal epithelial cell layers in the gastrointestinal (GI) tracts act as major barriers against effective oral delivery of therapeutic proteins or peptides. Therefore, it is remarkable that BoNT, a ~150 kDa protein, is able to survive the harsh environment of the GI tract and gain access to the general circulation in food borne botulism. It is believed that BoNTs manage to do so in the form of progenitor toxin complex (PTC), although the underlying molecular mechanism is still largely unknown [6]. This notion is strongly supported by the fact that the oral toxicity of PTCs of different BoNT serotypes is ~360–16,000-fold more potent than that of the free BoNT [7–10]. The PTC is a large multi-protein complex, which is composed of a BoNT molecule and several auxiliary proteins termed neurotoxin-associated proteins (NAPs). BoNT/A–D and G are produced in bacteria together with four NAPs, which include non-toxic nonhemagglutinin (NTNHA) protein and three hemagglutinins (HAs: HA33, HA17 and HA70, also known as HA1, HA2 and HA3, respectively) [6]. In contrast, BoNT/E and F do not have the ha genes. Instead, they contain genes of orfX1/X2/X3, whose expression and function are still unknown [1,11].
The structure of the PTC of serotype A consists of two relatively independent modules. NTNHA assembles with BoNT/A to form a ~290 kDa minimally functional PTC (M-PTC) that protects BoNT/A against the low pH condition and proteases present in the GI tract [12,13]. The HA proteins form a ~470 kDa 12-subunit complex (termed the HA complex) with HA70, HA17, and HA33 in a 3:3:6 stoichiometry [14,15]. The M-PTC and the HA complex together form the large PTC (L-PTC) that displays the most potent oral toxicity. The HA complex exhibits multiple carbohydrate-binding sites located on HA70 and HA33, which could help the L-PTC bind to glycans on the intestinal epithelial cell surface. In this regard, the HA complex may facilitate PTC absorption and enhance its oral toxicity [7–9,16], even though BoNT alone may be able to penetrate the GI barrier by transcytosis [17–19].
The HA33–glycan interaction is particularly important for the function of the HA complex, because each HA complex contains six molecules of HA33 to allow multivalent binding. Disrupting glycan binding to HA33 abolished the transport of the HA complex across epithelial cell layers [14]. Furthermore, the earlier structural studies on HA33/A, C and D have revealed serotype-specific HA33– glycan interactions [14,20,21], and raised the possibility that the HA33–glycan binding specificity may partly contribute to the host tropism of various BoNT serotypes. For example, BoNT/A and B are the major cause of human botulism, while BoNT/C and D predominantly cause botulism in cattle, poultry, and wild birds. Here we report a 1.46 Å resolution crystal structure of HA33/B in complex with a receptor-mimicking lactose. Together with the crystal structure of the apo form of HA33/B at 3.5 Å resolution reported recently [15], our structure completes the gallery of HA33 of serotypes A–D and provides a unique opportunity to investigate the serotype-specific HA33-mediated pathogen–host interactions.
2. Materials and methods
2.1. Construct design and cloning
The gene encoding full length HA33/B (residues E2–P294; Genbank ID YP_001693310.1) from Clostridium botulinum Okra producing BoNT/B1 was cloned into expression vector pQE30. This construct carries five point mutations (T6V, I288K, T290N, M291I, and S292R) as cloning artifacts. The gene encoding full length HA33/C (residues S2-I286; Genbank ID YP_398514.1) from C. botulinum phage Stockholm producing BoNT/C1 was cloned into expression vector pQE30. To facilitate protein purification, a 6 ×His tag was introduced to the N-terminus of HA33/B and C followed by a thrombin cleavage site and a Strep tag was added to the C-terminus of HA33/B and C.
2.2. Protein expression and purification
HA33/B and C were expressed in the Escherichia coli strain BL21-RIL (DE3) (Novagen). Protein expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) when OD600 of the bacterial culture had reached 0.8, and continued at 18 °C overnight. HA33/B and C were purified by Ni-NTA (nitrilotriacetic acid, Qiagen) affinity column in a buffer containing 50 mM Tris (pH 8.0) and 400 mM NaCl, and subsequently eluted in the same buffer containing 300 mM imidazole. The eluted fractions were dialyzed at 4 °C overnight against a buffer composed of 20 mM Tris (pH 8.0) and 50 mM NaCl. After His-tag was removed by thrombin protease, HA33/B and C were further purified by MonoQ ion-exchange chromatography in the buffer of 20 mM Tris (pH 8.0) and eluted with a NaCl gradient, followed by Superdex 200 size-exclusion chromatography in 20 mM Tris (pH 8.0) and 50 mM NaCl. The protein was concentrated to ~6 mg/ml using Amicon Ultra centrifugal filter (Millipore) for crystallization.
2.3. Isothermal titration calorimetry (ITC)
The calorimetry titration experiments were performed on an ITC200 calorimeter (GE Life Sciences). HA33/B or HA33/C (200 μM) was used as the titrand in the cell, while lactose was used at 40 mM and 100 mM for HA33/B and HA33/C, respectively, as the titrant in the syringe. The titrations were performed at pH 8 (20 mM Tris, pH 8.0, 100 mM NaCl) or pH 5 (20 mM Sodium citrate, pH 5.0, 100 mM NaCl). The data were analyzed using the Origin software package.
2.4. Crystallization, data collection, and structure determination
Initial crystallization screens were performed using a phoenix crystallization robot (Art Robbins Instruments) and commercial high-throughput screening kits. Manual optimization was performed at 18 °C using the hanging-drop vapor diffusion method with a 1:1 (v/v) ratio of protein and reservoir. The best crystals of the HA33/B–lactose complex were obtained using 0.1 M Hepes (pH 7.0), 5% MPD, 5% (w/v) PEG [poly(ethylene glycol)] 6k, and 20 mM lactose.
The crystals were cryo-protected in the original mother liquor supplemented with 20% (v/v) glycerol and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source (APS). The data were processed with HKL2000 [22]. Data collection statistics are summarized in Table 1. The structure of the HA33/B–lactose complex was determined by molecular replacement software Phaser [23] using HA33/A (PDB: 4LO2) [14] as the search model. The model building and refinements were performed in COOT [24] and PHENIX [25]. Lactose was modeled into the corresponding structure during the refinement based on the F0–Fc electron density maps. The refinement progress was monitored with the free R value using a 5% randomly selected test set [26]. MolProbity web server was used for structure validation [27]. Structural refinement statistics are listed in Table 1. The coordinate and diffraction data have been deposited in the Protein Data Bank under accession code 4OUJ. The conformational change of HA33 was measured by DynDom [28]. All structure figures were prepared with PyMol. (http://www.pymol.org).
Table 1.
Data collection and refinement statistics.
| HA33/B–lactose | |
|---|---|
| Data collection | |
| Space group | P1 21 1 |
| Cell dimensions | |
| a, b, c (Å) | 83.8, 36.9, 114.6 |
| α, β, γ (°) | 90, 102.1, 90 |
| Resolution range (Å) | 37–1.46 (1.54–1.46)a |
| Measured reflections | 374,338 |
| Unique reflections | 118,971 |
| Completeness (%) | 99.0 (97.1)a |
| R merge | 0.04 (0.413)a |
| I/σ(I) | 15.9 (2.3)a |
| Structure refinement | |
| Resolution (Å) | 19.80–1.46 |
| No. reflections | 118,909 |
| Rwork/Rfree (%) | 16.3/18.1 |
| Restraints (RMS observed) | |
| Bond length (Å) | 0.009 |
| Bond angle (°) | 1.209 |
| Average isotropic B–value (Å2) | 20.8 |
| Number of atoms | |
| Protein | 9001 |
| Ligand | 46 |
| Water | 635 |
Values for the highest resolution shell.
3. Results and discussion
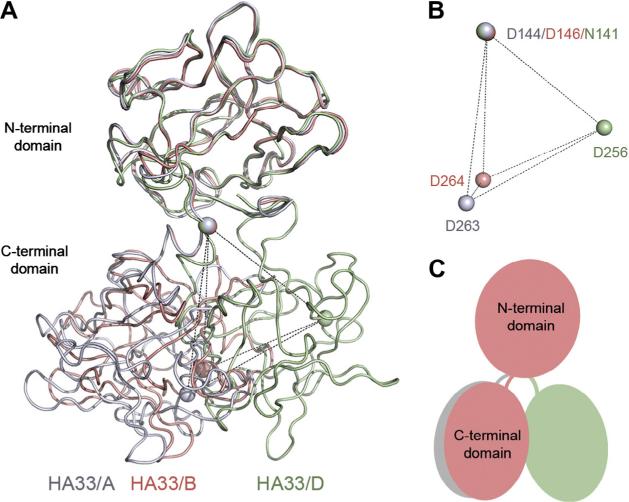
The complex of HA33/B and lactose was obtained by co-crystallization of HA33/B in the presence of 20 mM lactose, and the structure was determined by molecular replacement using the structure of HA33/A as the model [14] before the structure of apo HA33/B is available. HA33/B is composed of two β-trefoil domains linked by a short α-helix (residues Tyr147–Phe152) (Fig. 1A). The overall structure of each HA33 domain is highly conserved among HA33/ A–D. The root-mean-square deviation (RMSD) is ~0.53–1.08 Å and ~1.14–1.22 Å for the N- and C-terminal domains, respectively, in pair-wise structure superimposition among the four HA33 sero-types. However, we observed a different rotation via the α-helix linker between the N- and C-terminal b-trefoil domains of HA33/ B in comparison to other HA33 serotypes (Fig. 1). Using the structure of HA33/A as a reference, the two domains of HA33/B show an inter-domain twist of ~20°. In contrast, HA33/D shows an inter-domain twist of ~65° when compared with HA33/A, whereas HA33/C and D are highly similar [14]. The two independently solved structures of HA33/B, in complex with lactose or in the apo form [15], are almost identical, even though they are from crystals adopting completely different packing lattices. Therefore, the structure of HA33/B observed here is likely a faithful mimic of its physiological conformation. Taken together, we have observed two major conformations of HA33, which are represented by HA33/A–B and HA33/C–D, respectively. The potential functional role of the serotype-specific HA33 conformation awaits clarification in future studies.
Fig. 1.
HA33 of different serotypes adopt two major conformations. (A) The structures of HA33/B (this study, salmon), HA33/A (PDB: 4LO2, gray) [14], and HA33/D (PDB: 2E4M, green) [38] are superimposed based on Ca atoms in their N-terminal domains. (B) The equivalent residue D144HA33/A, D146HA33/B, and N141HA33/D, which is located in the linker connecting the two HA33 domains, is used as the origin for analysis. A key Gal-binding residue D263HA33/A, D264HA33/B, and D256HA33/D were used to quantify the inter-domain rotation of HA33. The corresponding Ca atoms are shown as spheres. (C) A cartoon showing the different inter-domain rotation of HA33/A (gray), HA33/B (salmon), and HA33/D (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
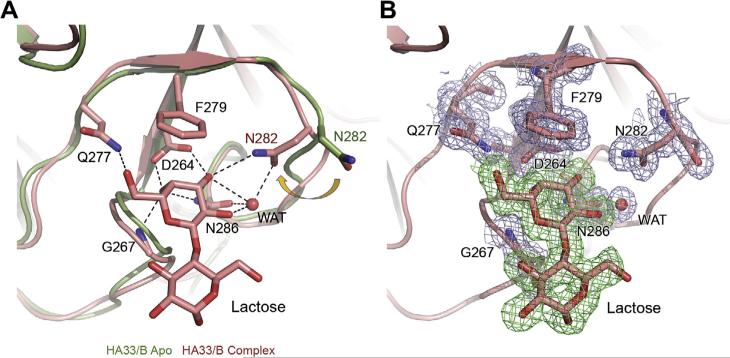
We next analyzed the HA33/B–lactose interactions. There are two commonly used strategies to pursue a protein–ligand complex, crystal soaking and co-crystallization. Crystal soaking starts with pre-formed protein crystals in the absence of ligand. Ligand is then added during the soaking procedure, which diffuses into the crystal and binds to the protein. In contrast, the ligand always binds to the protein during co-crystallization, thus the protein–ligand interaction is less likely to be biased by crystal packing. Here, we obtained the HA33/B–lactose complex by co-crystallization, complementing the previous structures of glycan-bound HA33/A or C obtained by crystal soaking [14,20,21]. We found that one lactose molecule binds at the glycan-binding site on HA33/B, and the binding mode is almost identical to that observed in the HA33/A–lactose complex [14]. The interactions are mediated by the galactose (Gal) moiety through seven pairs of hydrogen bonds as well as a crucial stacking interaction between Phe279 and the hexose ring of Gal (Fig. 2A). In contrast, the glucose (Glc) moiety is pointing away from HA33/B and not directly involved in protein interaction. This finding is consistent with the result of a recent glycan array study, which suggests that HA33/A does not have strict preference for saccharides other than the terminal Gal [29].
Fig. 2.
HA33/B specifically binds lactose. (A) The structures of the apo HA33/B (PDB: 3WIN, green) [15] and the HA33/B–lactose complex (this study, salmon) were superimposed based on Cα atoms in the C-terminal domain. The lactose-binding residues in the HA33/B–lactose complex are shown as sticks. The apo conformation of residue N282 is also shown for comparison. A structural water molecule in the binding pocket is shown as sphere. Hydrogen bonds are shown as dashed lines. (B) A sigma-A weighted 2F0–Fc electron density map (contoured at 1.0 σ) was shown for lactose-binding residues of HA33/B (light blue) and lactose (green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
It is worth noting that our high-resolution structure allows identification of a well-defined water molecule in the glycan-binding pocket, which is absent in the structure of apo HA33/B (Fig. 2). This water molecule contributes four pairs of hydrogen bonds, bridging O2 and O3 atoms of Gal and residues Asn282 and Asn286 of HA33/B. No such water molecule was observed in the HA33/A–lactose complex [14], probably because the water-binding residue Asn282 of HA33/B is replaced by residue His281 of HA33/A (Fig. 3). One strategy in small molecule lead optimization is to structurally modify lead compounds that display ordered water molecules in a protein-binding site to gain improved binding affinity [30,31]. Therefore, new carbohydrate-mimicking compounds with higher binding affinity to HA33/B could be designed by incorporating the equivalent of this key structural water molecule into the new compounds.
Fig. 3.
Structure-based sequence alignment of HA33 serotype A–D. The structure of HA33/B in the current work, HA33/A (PDB: 4LO2) [14], HA33/C (PDB: 3AJ5) [21], and HA33/D (PDB: 2E4M) [38] are used for analysis using PROMALS3D [39] and ESPript [40]. Identical residues are indicated with white letters on a red background, similarly conserved residues are in red letters, varied residues are in black letters, and dots represent gaps. The secondary structure of HA33/B studied in this work is shown on the top, and the lactose-binding residues are indicated by red ovals. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We next compared the structures of HA33/B in the apo and the complex forms and found that the lactose-binding site is largely pre-formed except for residue Asn282. Residue Asn282 adopts a large side-chain reorientation upon lactose binding and forms a hydrogen bond with O3 atom of Gal (Fig. 2A and B). Interestingly, Asn282 is the only Gal-interacting residue that is different between HA33/A and B, while HA33/A has a histidine residue (His281) at the equivalent position (Fig. 3). The pKa (Ka is the acid dissociation constant) of His281 of HA33/A is ~6.22 calculated based on its crystal structure [32]. Protonation of histidine has been reported to be involved in low pH-sensing in many proteins, such as KcsA potassium channel, pH-gated urea channel, sensor histidine kinase of Salmonella enterica, and viral membrane fusion proteins [33–36]. Therefore, we performed isothermal titration calorimetry (ITC) experiments at pH 8.0 and 5.0 to further examine the pH effect on HA33–lactose interactions (Fig. 4). HA33/B binds lactose with dissociation constant (Kd) of ~1.4 mM and ~1.6 mM at pH 8.0 and 5.0, respectively. In contrast, HA33/A binds to lactose with a ~ 3.7-fold lower affinity at pH 5.0 (~3.5 mM) than that at pH8.0 (~0.95 mM) [14]. Therefore, protonation of His281 of HA33/A in the acidic GI tract could weaken its interaction with the carbohydrate receptors, which may lead to decreased efficiency of intestinal binding and absorption of the L-PTC. Such difference between HA33/A and B could partly contribute to the observation that L-PTC/B is ~90-fold more lethal than L-PTC/A in a mouse oral intoxication model [37], although these two PTCs have similar overall architectures [14,15].
Fig. 4.
Thermodynamic studies of HA33–lactose interactions by isothermal titration calorimetry. The representative titration curves are shown for (A) HA33/B at pH 8.0, (B) HA33/B at pH 5.0, (C) HA33/A at pH 5.0, and (D) HA33/C at pH 8.0. The thermodynamic values reported are the average of three independent experiments (means ± SD).
HA33/C and D show very different glycan-binding features in comparison to HA33/A and B. Although the Gal-binding pockets are similarly configured among the four different HA33 serotypes, HA33/C binds lactose with ~7-fold lower affinity than that of HA33/B (Fig. 4). Structural analyses reveal two possible reasons for this difference. First, residue Phe278HA33/A/Phe279HA33/B, which provides an important packing interaction with the hexose ring of Gal, is replaced by residue Asp271HA33/C/Asp271HA33/D. Second, Gly267HA33/B and Gly266HA33/A, which is located very close to the Glc moiety of lactose, is replaced by Asn259HA33/C and Asn259-HA33/D (Fig. 3). As a result, the side chain of Asn259 of HA33/C and D has to take a large reorientation to avoid clashing with Glc. Besides Gal binding, HA33/C is reported to bind Neu5Ac in an adjacent pocket [20]. HA33/A and B do not bind Neu5Ac because they do not have the essential Neu5Ac-binding residues corresponding to Trp176 and Arg183 of HA33/C.
In summary, our results are a step towards further understanding of the host susceptibility of various BoNT serotypes, and have implications for the development of receptor-mimicking compounds, which could potentially inhibit BoNT oral intoxication upon pre-treatment.
Acknowledgments
This work was partly supported by National Institute of Allergy and Infectious Diseases (NIAID) Grant R01AI091823 to Rongsheng Jin and by the Swiss Federal Office for Civil Protection (BABS #353003325) to Andreas Rummel. This work is based upon research conducted at the Advanced Photon Source (APS) on the Northeastern Collaborative Access Team beamlines, which are supported by a grant from the National Institute of General Medical Sciences, United States (P41 GM103403). Use of the APS, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Abbreviations
- BoNT
botulinum neurotoxin
- PTC
progenitor toxin complex
- NAP
neurotoxin-associated protein
- NTNHA
non-toxic non-hemagglutinin
- HA
hemagglutinin
- NMJ
neuromuscular junction
References
- 1.Hill KK, Smith TJ. Genetic diversity within Clostridium botulinum serotypes, botulinum neurotoxin gene clusters and toxin subtypes. Curr. Top. Microbiol. Immunol. 2013;364:1–20. doi: 10.1007/978-3-642-33570-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Dover N, Barash JR, Hill KK, Xie G, Arnon SS. Molecular characterization of a novel botulinum neurotoxin type H gene. J. Infect. Dis. 2014;209:192–202. doi: 10.1093/infdis/jit450. [DOI] [PubMed] [Google Scholar]
- 3.Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta BR, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 4.Blasi J, Chapman ER, Link E, Binz T, Yamasaki S, De Camilli P, Sudhof TC, Niemann H, Jahn R. Botulinum neurotoxin A selectively cleaves the synaptic protein SNAP-25. Nature. 1993;365:160–163. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- 5.Schiavo G, Matteoli M, Montecucco C. Neurotoxins affecting neuroexocytosis. Physiol. Rev. 2000;80:717–766. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- 6.Collins MD, East AK. Phylogeny and taxonomy of the food-borne pathogen Clostridium botulinum and its neurotoxins. J. Appl. Microbiol. 1998;84:5–17. doi: 10.1046/j.1365-2672.1997.00313.x. [DOI] [PubMed] [Google Scholar]
- 7.Ohishi I, Sugii S, Sakaguchi G. Oral toxicities of Clostridium botulinum toxins in response to molecular size. Infect. Immun. 1977;16:107–109. doi: 10.1128/iai.16.1.107-109.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng LW, Onisko B, Johnson EA, Reader JR, Griffey SM, Larson AE, Tepp WH, Stanker LH, Brandon DL, Carter JM. Effects of purification on the bioavailability of botulinum neurotoxin type A. Toxicology. 2008;249:123–129. doi: 10.1016/j.tox.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 9.Ohishi I. Oral toxicities of Clostridium botulinum type A and B toxins from different strains. Infect. Immun. 1984;43:487–490. doi: 10.1128/iai.43.2.487-490.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakaguchi G. Clostridium botulinum toxins. Pharmacol. Ther. 1982;19:165–194. doi: 10.1016/0163-7258(82)90061-4. [DOI] [PubMed] [Google Scholar]
- 11.Lin G, Tepp WH, Pier CL, Jacobson MJ, Johnson EA. Expression of the Clostridium botulinum A2 neurotoxin gene cluster proteins and characterization of the A2 complex. Appl. Environ. Microbiol. 2010;76:40–47. doi: 10.1128/AEM.01882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gu S, Jin R. Assembly and function of the botulinum neurotoxin progenitor complex. Curr. Top. Microbiol. Immunol. 2013;364:21–44. doi: 10.1007/978-3-642-33570-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu S, Rumpel S, Zhou J, Strotmeier J, Bigalke H, Perry K, Shoemaker CB, Rummel A, Jin R. Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science. 2012;335:977–981. doi: 10.1126/science.1214270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K, Gu S, Jin L, Le TT, Cheng LW, Strotmeier J, Kruel AM, Yao G, Perry K, Rummel A, Jin R. Structure of a bimodular botulinum neurotoxin complex provides insights into its oral toxicity. PLoS Pathog. 2013;9:e1003690. doi: 10.1371/journal.ppat.1003690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amatsu S, Sugawara Y, Matsumura T, Kitadokoro K, Fujinaga Y. Crystal structure of Clostridium botulinum whole hemagglutinin reveals a huge triskelion-shaped molecular complex. J. Biol. Chem. 2013;288:35617–35625. doi: 10.1074/jbc.M113.521179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue K, Fujinaga Y, Watanabe T, Ohyama T, Takeshi K, Moriishi K, Nakajima H, Inoue K, Oguma K. Molecular composition of Clostridium botulinum type A progenitor toxins. Infect. Immun. 1996;64:1589–1594. doi: 10.1128/iai.64.5.1589-1594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couesnon A, Pereira Y, Popoff MR. Receptor-mediated transcytosis of botulinum neurotoxin A through intestinal cell monolayers. Cell. Microbiol. 2008;10:375–387. doi: 10.1111/j.1462-5822.2007.01051.x. [DOI] [PubMed] [Google Scholar]
- 18.Couesnon A, Shimizu T, Popoff MR. Differential entry of botulinum neurotoxin A into neuronal and intestinal cells. Cell. Microbiol. 2009;11:289–308. doi: 10.1111/j.1462-5822.2008.01253.x. [DOI] [PubMed] [Google Scholar]
- 19.Couesnon A, Molgo J, Connan C, Popoff MR. Preferential entry of botulinum neurotoxin A Hc domain through intestinal crypt cells and targeting to cholinergic neurons of the mouse intestine. PLoS Pathog. 2012;8:e1002583. doi: 10.1371/journal.ppat.1002583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura T, Tonozuka T, Ide A, Yuzawa T, Oguma K, Nishikawa A. Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin. J. Mol. Biol. 2008;376:854–867. doi: 10.1016/j.jmb.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura T, Tonozuka T, Ito S, Takeda Y, Sato R, Matsuo I, Ito Y, Oguma K, Nishikawa A. Molecular diversity of the two sugar-binding sites of the beta-trefoil lectin HA33/C (HA1) from Clostridium botulinum type C neurotoxin. Arch. Biochem. Biophys. 2011;512:69–77. doi: 10.1016/j.abb.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 27.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. cta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- 29.Yao G, Lee K, Gu S, Lam KH, Jin R. Botulinum neurotoxin A complex recognizes host carbohydrates through its hemagglutinin component. Toxins (Basel) 2014;6:624–635. doi: 10.3390/toxins6020624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel J, Tirado-Rives J, Jorgensen WL. Energetics of displacing water molecules from protein binding sites: consequences for ligand optimization. J. Am. Chem. Soc. 2009;131:15403–15411. doi: 10.1021/ja906058w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ladbury JE. Just add water! The effect of water on the specificity of protein–ligand binding sites and its potential application to drug design. Chem. Biol. 1996;3:973–980. doi: 10.1016/s1074-5521(96)90164-7. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 33.Thompson AN, Posson DJ, Parsa PV, Nimigean CM. Molecular mechanism of pH sensing in KcsA potassium channels. Proc. Natl. Acad. Sci. USA. 2008;105:6900–6905. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weeks DL, Sachs G. Sites of pH regulation of the urea channel of Helicobacter pylori. Mol. Microbiol. 2001;40:1249–1259. doi: 10.1046/j.1365-2958.2001.02466.x. [DOI] [PubMed] [Google Scholar]
- 35.Perez JC, Groisman EA. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol. Microbiol. 2007;63:283–293. doi: 10.1111/j.1365-2958.2006.05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller DS, Kampmann T, Yennamalli R, Young PR, Kobe B, Mark AE. Histidine protonation and the activation of viral fusion proteins. Biochem. Soc. Trans. 2008;36:43–45. doi: 10.1042/BST0360043. [DOI] [PubMed] [Google Scholar]
- 37.Cheng LW, Henderson TD., 2nd Comparison of oral toxicological properties of botulinum neurotoxin serotypes A and B. Toxicon. 2011;58:62–67. doi: 10.1016/j.toxicon.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasegawa K, Watanabe T, Suzuki T, Yamano A, Oikawa T, Sato Y, Kouguchi H, Yoneyama T, Niwa K, Ikeda T, Ohyama T. A novel subunit structure of Clostridium botulinum serotype D toxin complex with three extended arms. J. Biol. Chem. 2007;282:24777–24783. doi: 10.1074/jbc.M703446200. [DOI] [PubMed] [Google Scholar]
- 39.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]