Abstract
Spatial distribution of components of nuclear metabolism provides a significant impact on regulation of the processes of gene expression. While distribution of the key nuclear antigens and their association with the defined nuclear domains were thoroughly traced in mammalian somatic cells, similar data for the preimplantation embryos are scanty and fragmental. However, the period of cleavage is characterized by the most drastic and dynamic nuclear reorganizations accompanying zygotic gene activation. In this minireview, we try to summarize the results of studies concerning distribution of major factors involved in RNA polymerase II-dependent transcription, pre-mRNA splicing mRNA export that have been carried out on early embryos of mammals.
1. Introduction
The main feature of the eukaryotic cell is the nucleus that holds the genetic information and realizes DNA replication and the processes of gene expression. Being the place where transcription and the related events occur, the nucleus demarcates nuclear processes from translation in cytoplasm, providing new additional levels of the regulation of gene expression that cannot be realized in prokaryotes.
Interchromatin space of the nucleus contains different nuclear domains. In mammalian somatic cells, molecular composition of major functional nuclear domains has been studied in detail [1]. Initially, the nuclear distribution of pre-mRNA splicing factors and the structures enriched with these factors have been explored with the use of antibodies against the Sm antigen (now known as symmetric dimethylarginine, sDMA) [2], 2,2,7-trimethylguanosine (TMG) cap of snRNAs, and some “signature” proteins of nuclear domains (coilin, SR-proteins) [3]. As a result, interchromatin granule clusters (IGCs), known as nuclear splicing speckles in terms of fluorescent microscopy, and also Cajal bodies have attracted a special attention as evolutionary conserved and “universal” nuclear domains [4].
IGCs/speckles are nuclear domains enriched in pre-mRNA splicing factors (snRNPs and SR-proteins), located in the interchromatin space of the nucleus. These domains are dynamic nuclear organelles, and their constituents can exchange continuously with the nucleoplasm and other nuclear locations, including active transcription sites [5]. IGCs not only serve as transient reservoirs for pre-mRNA splicing factors, but are also involved in many other functions (for reviews, see [5, 6]). The Cajal bodies have been implicated in RNA-related metabolic processes such as snRNP biogenesis, maturation and recycling, histone mRNA processing, and telomere maintenance (for reviews, see [7, 8]). Deciphering the fundamental rules that impact nuclear functions in the whole seems to be impossible without a large-scale comparative approach. As compared with traditionally used experimental objects including tissue-culture somatic cells, early mammalian embryos have been explored with a lesser extent.
The nuclei of early mammalian embryos are characterized not only by the unique functional status, but also by a peculiar nuclear ultrastructure [9]. Stepwise physiological reactivation of chromosomal transcription activity during early embryogenesis, known in literature as zygotic gene activation (ZGA), suggests that the embryos could be attractive experimental models in order to analyze nuclear distribution of gene expression factors that virtually may associate with the universal and/or specific nuclear domains. ZGA is one of the key points of the maternal-to-zygotic transition (MZT) that involves a set of structural, molecular, and biochemical rearrangements [10–14]. However, the majority of studies concerned first of all the changes of gene expression patterns [15–17] as well as the processes of chromatin remodeling during realization of ZGA events (for reviews, see [18, 19]). The relationships between these processes and the distribution of the essential components of nuclear metabolism were described with a lesser extent.
Here we review available literary data on nuclear distribution of some essential molecular components related to RNA polymerase II-dependent transcription and mRNA processing at different stages of ZGA in mammals. The dynamics of the nucleolar components in mammalian embryos was studied better [20–22] and remains beyond the scope of the present review.
2. RNA Polymerase II and Basal Transcription Factors
The association of RNA polymerase II (RNAP II) with many factors to form a polyfunctional holoenzyme is required for transcription [47, 48]. Besides core RNAP II enzyme, this complex includes basal transcription factors, mediator complexes, and transcription coactivators [49–51]. The large RNAP II subunit (RPB1) contains the carboxy-terminal domain (CTD) consisting of multiple heptapeptide repeats with the consensus sequence YSPTSPS. Dynamic phosphorylation of the CTD determines the progression of the RNAP II transcription cycle from initiation through elongation to termination [52–54].
Posttranslational modifications of RNAP II holoenzyme were studied in mouse and rabbit preimplantation embryos with the use of western blot analysis and immunofluorescent microscopy [23]. In this study, a set of the following monoclonal antibodies was applied: 8WG16 revealing the unphosphorylated CTD, CC-3 directed against a phosphorylated epitope of the CTD, and Pol3/3 against an internal epitope distinct from the CTD, thus recognizing two forms of RPB1 simultaneously. The authors have eventually suggested that phosphorylation of the CTD might control ZGA in the embryos of mammals with different ZGA chronologies. Several hours after fertilization, the CTD was found dephosphorylated. Dephosphorylation of the CTD occurs before the onset of a period characterized by a weak transcriptional activity (minor ZGA). Then, the CTD lacks immunological and drug-sensitivity characteristics related to its phosphorylation status, and RNAP II gradually translocates into the nuclei. At the major ZGA phase corresponding the 2-cell stage in mouse and the 8–16-cell stage in rabbit, phosphorylation pattern of the CTD was close to that observed in somatic cells. The authors also reported that actinomycin D does not prevent a new phosphorylation pattern of RNAP II at the onset of the major ZGA. As a result, a phosphorylated embryogenesis-specific RNAP II isoform was identified. This RNAP II isoform is insensitive to DRB, a CTD-kinase inhibitor, since it lacks a phosphoepitope generated by TFIIH-associated kinase phosphorylation. Finally, the authors have concluded that nuclear translocation of RNAP II and CTD phosphorylation might be major determinants of ZGA. The possible association of different RNAP II isoforms with intranuclear structures of mouse and rabbit preimplantation embryos was not explored in detail in this study.
In our immunocytochemical experiments, we used an affinity purified polyclonal serum to reveal the hyperphosphorylated form of RNAP II in mouse embryos. This form of RNAP II was detected before the major ZGA phase, namely, in early 2-cell embryos [24]. While ZGA proceeds, RNAP II was found in association with perichromatin fibrils (PFs) [25], which are referred to as the ultrastructural “in situ forms” of nascent pre-mRNA transcripts [55]. Besides, the hyperphosphorylated RNAP II was detected in nuclear speckles identified by the presence of the SR protein SC35. The intensity of anti-RNAP II immunostaining in speckles was being increased towards the end of ZGA [24] (Figure 1). If ZGA is delayed, for example, in embryos under the so-called “2-cell block in vitro” (for details, see [56, 57]), the hyperphosphorylated form of RNAP II begins to accumulate in enlarged nuclear speckles [26] (Figure 2).
Figure 1.
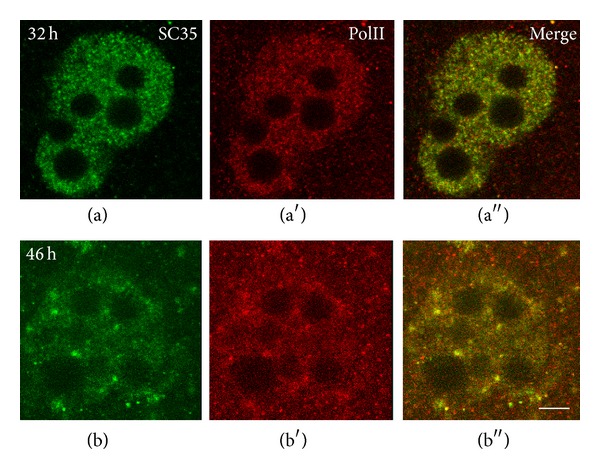
Double immunolocalization of SC35 (a), (b) and hyperphosphorylated RNA polymerase II (PolII) (a′), (b′) in mouse embryos. Fluorescence of nuclei begins to be detected only at the early 2-cell stage (line a). Association of RNAP II with SC35 domains (speckles) is observed already at this stage and is increased when ZGA finishes (line b). Bar is 10 μm, according to [24], open access.
Figure 2.
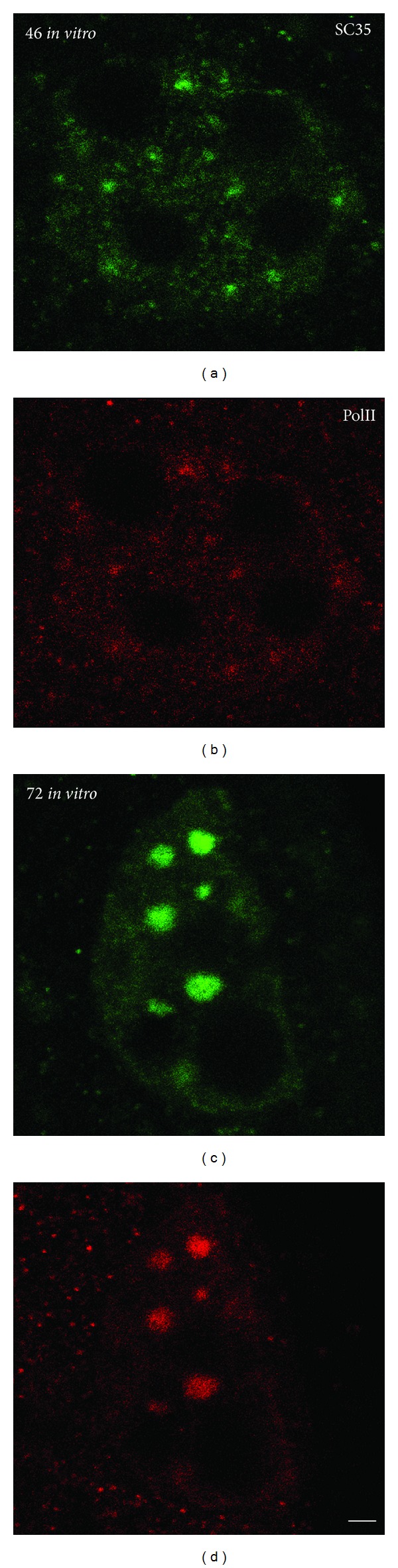
The distribution of SR protein SC35 (a), (c), hyperphosphorylated RNA polymerase II (PolII) (b), (d) in control (a), (b), and arrested in vitro 2-cell mouse embryos (c), (d). Note the large SC35 speckles enriched in hyperphosphorylated RNA polymerase II in the nucleus of blocked embryo. Scale bar is 5 μm, according to [26], reprinted from Tissue and Cell; Bogolyubova. Transcriptional activity of nuclei in 2-cell blocked mouse embryos 2011; 43: 262-265 [26], with permission from Elsevier.
Worrad et al. [29] have explored the concentration of the transcription factors Sp1 and TBP (TATA-binding subunit of TFIID) in 1-cell mouse embryos. The authors showed that concentration of both factors drastically increases in a time-dependent fashion after fertilization during the 1-cell stage. Six hours following the formation of pronuclei, this increase continued, and by the G2 phase the pronuclear concentration of Sp1 and TBP was very similar to that observed in 2-cell embryos. In addition, concentration of both transcription factors was greater in the male pronucleus and the difference was more pronounced for TBP.
Later, TBP and TBP-associated factor 1 (TAF1) were revealed in mouse zygotes [30]. TAF1 was not detected just after fertilization in transcriptionally silent cells, but it is expressed in pronuclei, reaching the maximum before the onset of ZGA. TAF1 and TBP shared similar dynamics of expression patterns. In 4 h after fertilization, anti-TAF1 immunofluorescent signal was not registered. The signal became obvious in a portion of embryos in 6 h after fertilization, in the majority of embryos in 9 h, and in all embryos in 11 h. TBP was revealed in some embryos for the first time in 4 h after fertilization and in almost all pronuclei 1 h later. In male pronuclei, transcriptional activity can be registered earlier. Respectively, both factors initially appear in male rather than in female pronuclei. The authors have supposed that the deficiency of transcription machinery might be a reason for the limitation of transcription at the beginning of embryogenesis.
In addition to TBP, Gazdag et al. [31] have revealed the related protein TBP2, also called TRF3 (TBP-related factor 3), in mouse 1-2-cell embryos. They also reported that TBP is expressed in the oocytes at the beginning of folliculogenesis, but cannot be detected during further stages of oocyte development and becomes abundant again only after fertilization. In contrast to TBP, TBP2 was almost undetectable after fertilization until the 2-cell stage.
In mouse embryo pronuclei, basal transcription factor TFIID has been revealed before the onset of ZGA in association with nuclear speckles already at the zygote stage [24]. During realization of ZGA, the presence of TFIID in the speckles became more evident (Figure 3). Localization data of RNAP II and TFIID in the IGCs of mouse embryos are in accordance with the results that came from somatic cell studies [58, 59] including IGC proteome analysis [60].
Figure 3.
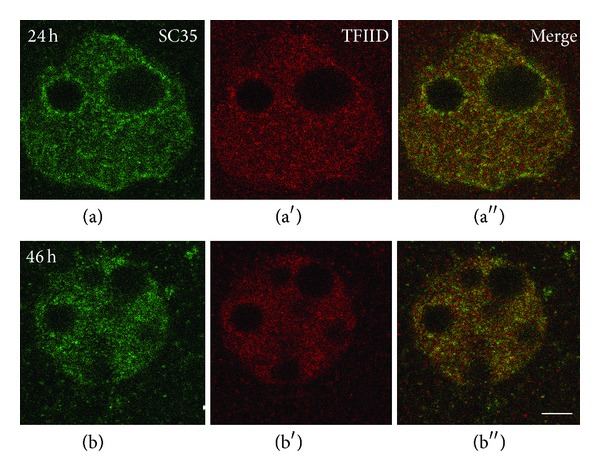
Double immunolocalization of SC35 (a), (b) and transcription factor TFIID (a′), (b′) in mouse embryos. TFIID is revealed in the nuclei at all studied stages. Colocalization of SC35 and TFIID is intensified during ZGA. Bar is 10 μm, according to [24], open access.
Interestingly, TFIID was also clearly detectable at the periphery of the nucleolar precursor bodies (NPBs) at the earliest stages of mouse cleavage [24]. The functional significance of this finding is ambiguous. Further studies are required to clarify the molecular composition and functions of NPBs, enigmatic structures of embryo nuclei.
Synoptic data on the revealing of RNAP II and transcription factors in mammalian embryos are presented in Table 1. At least a portion of RNAP II transcription machinery is detected in mammalian embryo nuclei already before ZGA. However, the final intranuclear patterns of RNAP II and transcription factors are being formed for the whole period of ZGA.
Table 1.
RNA polymerase II, mRNA processing factors, and mRNA export factors revealed in early mammalian embryos.
| Group of factors | Animal | Detected antigen | Method | Stage of embryogenesis | Reference |
|---|---|---|---|---|---|
| RNAP II | Mouse, rabbit | core of the RPB1 RNAPIIa, RNAPIIo |
IF*, WB | 1-cell–16–32 cell | [23] |
| Mouse | RNAPIIa, RNAPIIo | IF, IEM | 1-cell-2-cell | [24–27] | |
|
| |||||
| mRNA transcription factors | Mouse | TBP | IF | 1-cell | [28] |
| Sp1, TBP | IF Microinjection of Sp1-dependent luciferase reporter gene |
1-cell-2-cell | [29] | ||
| TBP, TAF1 | IF | 1-cell | [30] | ||
| TBP, TBP2 | IF, WB | 1-cell-2-cell | [31] | ||
| TFIID | IF | 1-cell-2-cell | [24] | ||
|
| |||||
| Splicing factors | Mouse | U1, U2, U4, U6 snRNAs snRNPs |
ISH IF |
1-cell-blastocyst | [32] |
| U1 snRNA U1 snRNP |
ISH IF |
1-cell-blastocyst | [33] | ||
| U1, U2 snRNAs snRNPs, SC35 |
ISH IF |
1-cell–8-cell | [34] | ||
| snRNPs, SC35 | IF, IEM | 1 cell-2-cell | [24, 35, 36] | ||
| snRNPs | IF | 1-cell-2-cell | [27] | ||
| snRNPs | IEM | 2-cell | [37] | ||
| hnRNPs, snRNPs | IEM | 1-cell–8-cell | [38] | ||
| Bovine | U2 snRNA snRNPs |
ISH, NB IF |
1-cell-blastocyst | [39] | |
| snRNPs, SC35 | IEM | 1-cell–16-cell | [40, 41] | ||
| Caprine | snRNPs, SC35 | IEM | 1-cell-2-cell | [41] | |
| Porcine | snRNPs | IF | 1-cell-blastocyst | [42] | |
| snRNPs, SC35 | IEM | 1-cell–4-cell | [43] | ||
| Hamster | U1, U2 snRNAs snRNPs SC35 |
ISH IF IF, IEM |
1-cell–8-cell | [44] | |
|
| |||||
| mRNA export-related factors | Mouse | hnRNPs | IF, WB | 1-cell–8-cell 1-cell |
[34] [28] |
| hnRNPs, Y14, Aly/REF, NXF1/TAP | IF, IEM | 1-cell-2-cell | [45, 46] | ||
| Hamster | hnRNPs | IF | 1-cell–8-cell | [44] | |
*Abbreviations: IF: immunofluorescence; IEM: immunoelectron microscopy; ISH: in situ hybridization; NB: northern blotting; RNAPIIa: hypophosphorylated form of RNA polymerase II; RNAPIIo: hyperphosphorylated form of RNA polymerase II; WB: western blotting.
3. Pre-mRNA Splicing Factors
Pre-mRNA splicing factors have been detected in the embryos of various mammalian species (Table 1). In the nuclei of early and late 2-cell mouse embryos, the distribution of snRNPs and non-snRNP protein SC35 was studied with the use of immunofluorescent and immunoelectron microscopy [35]. Following the activation of embryonic transcription in late 2-cell mouse embryos, splicing factors are revealed both in IGCs and in association with the PFs.
The Sm antigen of snRNPs and 2,2,7-trimethyl guanosine (TMG) cap characteristic for the mature snRNAs were localized in the nuclei of 1–4-cell porcine embryos at the ultrastructural level [43]. Surprisingly, unlike in bovine embryos (see below), immunoelectron microscopy also revealed the occurrence of snRNPs in the NPBs of porcine embryos. In bovine early preimplantation embryos, snRNPs were localized at different stages of ZGA [40]. Before the onset of transcription, up to the 4-cell stage, a diffuse labeling of the nucleoplasm was revealed. After the beginning of transcription, all 8-cell embryo nuclei were markedly stained, and mRNPs were shown to concentrate at the periphery of chromatin aggregates in association with the PFs. Interestingly, in vitro-produced 8-cell embryos showed a higher degree of chromatin condensation and a peripheral distribution of chromatin blocks as compared with the embryos produced in vivo. In 2- and 4-cell embryos, an intensive anti-snRNP labeling also characterized IGCs (nuclear speckles), which were often observed in the vicinity of the NPBs or even joined to them at the 4-cell stage. Using antibodies specific for the Sm epitope of snRNPs, the labeling was detected neither in the NPBs during embryonic nucleologenesis nor in the resulting nucleoli.
Analogous studies have been carried out later on bovine and caprine 2-cell embryos [41, 61]. The authors described several types of extrachromosomal nuclear bodies (NBs), 0.2–2.0 μm in diameter. The most striking feature of these NBs was the presence of proteins involved in pre-mRNA splicing. Some NBs having a rather dense finely fibrillar composition and, thus, named the dense bodies (DBs) were shown to contain the Sm-antigen. Moreover, more numerous NBs differed morphologically from the former by a much looser composition of fibrillogranular elements (called loose bodies, LBs). This type of the NBs, in addition to the Sm-antigen, contained the non-snRNP splicing factor SC35, a marker of IGCs/speckles. Both types of the NBs were distinguished clearly from the NPBs both morphologically and by the absence of NPB immunolabeling with antibodies against pre-mRNA splicing factors. Apart from the NBs described above, a high concentration of snRNPs was revealed in rather small, approximately 0.05 μm in diameter, morphologically and poorly defined domains named small snRNP-enriched areas (SSA). These domains housed a set of nuclear proteins typical for the CBs, including the CB signature protein coilin. However, it is still elusive whether the LBs correspond to the canonical IGCs of other cells. In the same way, it is unknown whether the DBs or SSA resemble the CBs in all respects.
With use of different approaches including light and electron immunocytochemistry and in situ hybridization, the dynamics of the CBs and/or IGCs has been studied in the early embryos of the mouse [24, 34, 62] and the golden hamster [44]. In the mouse, the CBs are already present in 1-cell embryos, before major transcription activation. On the contrary, in hamster embryos they appear during the 2-cell stage after the onset of transcription. On the other hand, hamster 1-cell embryos already display prominent IGCs, whereas typical IGCs appear only at the 2-cell stage in the mouse. Thus, the assembly of both CBs and IGCs seems to be independent of the onset of transcriptional activity in the early mammalian embryo [44]. Zatsepina et al. [62] have shown in early mouse embryos heterogeneity of CB-like (coilin-containing) nuclear bodies both in size and molecular composition. So, anti-Sm antibody was shown to stain small coilin-positive foci present in 1-cell and early 2-cell embryos and the periphery of significantly larger bodies present in middle and late 2-cell embryos.
Little is known about mammalian embryo nuclear structures that virtually could correspond to perichromatin granules (PGs). It is suggested that the structure of PGs arises due to the packaging of PFs, and the PGs participate in transient storage of mRNPs (in the form of heterogeneous nuclear (hn) RNPs) and/or also in mRNP nucleoplasmic transport [63]. Typical mammalian somatic PGs contain some splicing snRNAs [64] and snRNP proteins [65].
Numerous small PG-like granules, approximately 150 nm in diameter, resembling similar structures in the nucleus of antral oocytes [66] have been described in the nuclei of mouse 2-cell embryos [9]. Since the 4-cell stage, the number of the PGs decreases in the nuclei of mouse blastomeres and becomes comparable with their number in somatic cells. Taking into account the gradual increasing of embryonic RNA synthesis at this period, the authors believe that at least a part of the PGs in 2-cell mouse embryos contain maternal hnRNPs that will migrate into the cytoplasm or degrade. It cannot be excluded that some RNAs present in the PGs of mouse embryos may play a regulatory role.
The nature of larger RNP-containing granules, 30–50 nm in diameter, that have been described in the nuclei of 2–8-cell mouse embryos [9] remains unknown.
4. Some Proteins of the Exon-Exon Junction Complex
In our group, we also studied nuclear distribution of some representative proteins constituting the exon-exon junction complex (EJC) [45, 46, 67]. The EJC is a multiprotein complex that is loaded onto mRNA during splicing at a precise position upstream of exon-exon junctions [68]. Amongst other functions, the EJC enhances nuclear export of mRNA providing a platform to bind export factors [69]. Core EJC proteins including Y14 [70] provide stable association of the EJC with mRNA, whereas EJC shell proteins (e.g., Aly/REF) serve as adaptors providing a link between splicing and export [71].
In the nuclei of mouse embryos at the different stages of ZGA, the EJC core protein Y14, the shell protein Aly, and the essential mRNA export factor NXF1/TAP were detected in transcriptionally inert 1-cell embryos, but the intensity of fluorescence increased in late 2-cell embryos that are transcriptionally active [67]. Possible association of NXF1/TAP with nuclear speckles was also studied at the 2-cell stage [45]. In transcriptionally active embryos, NXF1/TAP was detected in the vicinity of the IGCs/speckles rather than inside these nuclear domains. Artificial inhibition of transcription by drugs resulted in significant accumulation of NXF1 in enlarged speckles.
We also studied possible spatial interactions between the EJC proteins and nuclear actin with the use of Förster resonance energy transfer (FRET) [46]. Two patterns of the FRET signal were detected in transcriptionally active nuclei of 2-cell embryos for the pairs: actin-Y14, actin-Aly/REF, and actin-NXF1/TAP. FRET areas were revealed both randomly distributed in the nucleoplasm and in association with NPBs. The means of FRET efficiency exceeded 25%–30% in separate areas. We supposed that FRET signals in the nucleoplasm correspond to transcriptionally active chromatin zones. The presence of Y14 and NXF1/TAP in the NPBs was confirmed at the ultrastructural level [46]; however, it is hard to explain detection of FRET in these nuclear structures of the embryos. The FRET pattern, typical for transcriptionally active embryos, was being retained after artificial suppression of transcription by drugs. In the specimens digested by RNAse, FRET efficiency was decreased significantly, showing the spatial interactions between EJC proteins and actin occur in an RNA-dependent manner.
5. Closing Remarks
The nucleus of early mammalian embryos is a highly dynamic system. The bulk of studies were devoted to the dynamics of intranuclear accumulation of the revealed antigens but not to their association with nuclear domains. Available data on the domain organization of mammalian embryo nuclei are not comprehensive, except for nucleolus; the genesis of the CBs and IGCs/speckles was described in the mouse and the golden hamster. As concerns the embryos of farm animals, real relationships between multifarious nuclear bodies known under various names and the canonical nuclear domains of somatic cell have to be ascertained. High rates of nuclear structure reorganization together with different chronologies that characterize early embryogenesis in various organisms make a comparative analysis of embryo nuclear domains difficult. Further studies in this field would be helpful to form an integral concept of the structural and functional compartmentalization of the nucleus as well as to extend our knowledge on the mechanisms of early embryogenesis in mammals.
Acknowledgment
This work was supported by the granting program “Molecular and Cell Biology” of Russian Academy of Sciences.
Conflict of Interests
The authors declare no conflict of interests in this study. No author of this paper has a direct financial relation with the commercial identities mentioned in the paper that might lead to a conflict of interests.
References
- 1.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends in Genetics. 2011;27(8):295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Lührmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. The Journal of Biological Chemistry. 2000;275(22):17122–17129. doi: 10.1074/jbc.M000300200. [DOI] [PubMed] [Google Scholar]
- 3.Spector DL, Fu X-D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. The EMBO Journal. 1991;10(11):3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogolyubov D, Stepanova I, Parfenov V. Universal nuclear domains of somatic and germ cells: some lessons from oocyte interchromatin granule cluster and Cajal body structure and molecular composition. BioEssays. 2009;31(4):400–409. doi: 10.1002/bies.200800100. [DOI] [PubMed] [Google Scholar]
- 5.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harbor Perspectives in Biology. 2011;3(2) doi: 10.1101/cshperspect.a000646.a000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anatomical Record A: Discoveries in Molecular, Cellular, and Evolutionary Biology. 2006;288(7):664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gall JG. Cajal bodies: the first 100 years. Annual Review of Cell and Developmental Biology. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- 8.Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harbor Perspectives in Biology. 2010;2(7) doi: 10.1101/cshperspect.a000653.a000653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fakan S, Odartchenko N. Ultrastructural organization of the cell nucleus in early mouse embryos. Biology of the Cell. 1980;37(3):211–218. [Google Scholar]
- 10.Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Experimental Cell Research. 1995;218(1):57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- 11.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Developmental Biology. 1997;181(2):296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 12.Minami N, Suzuki T, Tsukamoto S. Zygotic gene activation and maternal factors in mammals. Journal of Reproduction and Development. 2007;53(4):707–715. doi: 10.1262/jrd.19029. [DOI] [PubMed] [Google Scholar]
- 13.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136(18):3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Zheng P, Dean J. Maternal control of early mouse development. Development. 2010;137(6):859–870. doi: 10.1242/dev.039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamatani T, Carter MG, Sharov AA, Ko MSH. Dynamics of global gene expression changes during mouse preimplantation development. Developmental Cell. 2004;6(1):117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 16.Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Developmental Biology. 2005;283(1):40–57. doi: 10.1016/j.ydbio.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 17.Østrup O, Olbricht G, Østrup E, Hyttel P, Collas P, Cabot R. RNA profiles of porcine embryos during genome activation reveal complex metabolic switch sensitive to in vitro conditions. PLoS ONE. 2013;8(4) doi: 10.1371/journal.pone.0061547.e61547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tompson EM, Legouy E, Renard J-P. Mouse embryos do not wait for the MBT: chromatin and RNA Polymerase remodeling in genome activation at the onset of development. Developmental Genetics. 1998;22(1):31–42. doi: 10.1002/(SICI)1520-6408(1998)22:1<31::AID-DVG4>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 19.Østrup O, Andersen IS, Collas P. Chromatin-linked determinants of zygotic genome activation. Cellular and Molecular Life Sciences. 2013;70(8):1425–1437. doi: 10.1007/s00018-012-1143-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fléchon J-E, Kopečný V. The nature of the “nucleolus precursor body” in early preimplantation embryos: a review of fine-structure cytochemical, immunocytochemical and autoradiographic data related to nucleolar function. Zygote. 1998;6(2):183–191. doi: 10.1017/s0967199498000112. [DOI] [PubMed] [Google Scholar]
- 21.Hyttel P. Nucleolus formation in pre-implantation cattle and swine embryos. Italian Journal of Anatomy and Embryology. 2001;106(2) supplement 2:109–117. [PubMed] [Google Scholar]
- 22.Laurincik J, Maddox-Hyttel P. Nudeolar remodeling in nuclear transfer embryos. Advances in Experimental Medicine and Biology. 2007;591:84–92. doi: 10.1007/978-0-387-37754-4_6. [DOI] [PubMed] [Google Scholar]
- 23.Bellier S, Chastant S, Adenot P, Vincent M, Renard JP, Bensaude O. Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. The EMBO Journal. 1997;16(20):6250–6262. doi: 10.1093/emboj/16.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogolyubova I, Bogolyubov D. An immunocytochemical study of interchromatin granule clusters in early mouse embryos. BioMed Research International. 2013;2013:8 pages. doi: 10.1155/2013/931564.931564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogoliubova IO, Parfenov VN. Distribution of RNA polymerase II in nuclei of early murine embryos. Tsitologiia. 2002;44(2):175–180. [PubMed] [Google Scholar]
- 26.Bogolyubova IO. Transcriptional activity of nuclei in 2-cell blocked mouse embryos. Tissue and Cell. 2011;43(4):262–265. doi: 10.1016/j.tice.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 27.Vautier D, Besombes D, Chassoux D, Aubry F, Debey P. Redistribution of nuclear antigens linked to cell proliferation and RNA processing in mouse oocytes and early embryos. Molecular Reproduction and Development. 1994;38(2):119–130. doi: 10.1002/mrd.1080380202. [DOI] [PubMed] [Google Scholar]
- 28.Vautier D, Chesné P, Cunha C, Calado A, Renard J-P, Carmo-Fonseca M. Transcription-dependent nucleocytoplasmic distribution of hnRNP A1 protein in early mouse embryos. Journal of Cell Science. 2001;114(8):1521–1531. doi: 10.1242/jcs.114.8.1521. [DOI] [PubMed] [Google Scholar]
- 29.Worrad DM, Ram PT, Schultz RM. Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development. 1994;120(8):2347–2357. doi: 10.1242/dev.120.8.2347. [DOI] [PubMed] [Google Scholar]
- 30.Wang K, Sun F, Sheng HZ. Regulated expression of TAF1 in 1-cell mouse embryos. Zygote. 2006;14(3):209–215. doi: 10.1017/S0967199406003704. [DOI] [PubMed] [Google Scholar]
- 31.Gazdag E, Rajkovic A, Torres-Padilla ME, Tora L. Analysis of TATA-binding protein 2 (TBP2) and TBP expression suggests different roles for the two proteins in regulation of gene expression during oogenesis and early mouse development. Reproduction. 2007;134(1):51–62. doi: 10.1530/REP-06-0337. [DOI] [PubMed] [Google Scholar]
- 32.Dean WL, Seufert AC, Schultz GA, et al. The small nuclear RNAs for pre-mRNA splicing are coordinately regulated during oocyte maturation and early embryogenesis in the mouse. Development. 1989;106(2):325–334. doi: 10.1242/dev.106.2.325. [DOI] [PubMed] [Google Scholar]
- 33.Lobo SM, Marzluff WF, Seufert AC, et al. Localization and expression of U1 RNA in early mouse embryo development. Developmental Biology. 1988;127(2):349–361. doi: 10.1016/0012-1606(88)90321-1. [DOI] [PubMed] [Google Scholar]
- 34.Ferreira J, Carmo-Fonseca M. The biogenesis of the coiled body during early mouse development. Development. 1995;121(2):601–612. doi: 10.1242/dev.121.2.601. [DOI] [PubMed] [Google Scholar]
- 35.Bogoliubova IO, Parfenov VN. Pre-mRNA splicing factors in nuclei of two-cells murine embryos. Tsitologiia. 2000;42(9):884–890. (Rus). [PubMed] [Google Scholar]
- 36.Bogolyubova IO, Bogoliubova NA, Bogolyubov DS, Parfenov VN. Nuclear structure in early mouse embryos: a comparative ultrastructural and immunocytochemical study with special emphasis on the ‘2-cell block in vitro’. Tissue and Cell. 2006;38(6):389–398. doi: 10.1016/j.tice.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Baran V, Melcák I, Otcovský J, Landa V. Immunoelectron microscopic localization of small nuclear ribonucleoproteins and interchromatin granules in the 2-cell mouse embryo. Reproduction, Nutrition and Development. 1993;33(5):447–454. doi: 10.1051/rnd:19930505. [DOI] [PubMed] [Google Scholar]
- 38.Biggiogera M, Martin TE, Gordon J, Amalric F, Fakan S. Physiologically inactive nucleoli contain nucleoplasmic ribonucleoproteins: immunoelectron microscopy of mouse spermatids and early embryos. Experimental Cell Research. 1994;213(1):55–63. doi: 10.1006/excr.1994.1172. [DOI] [PubMed] [Google Scholar]
- 39.Watson AJ, Wiemer KE, Arcellana-Panlilio M, Schultz GA. U2 small nuclear RNA localization and expression during bovine preimplantation development. Molecular Reproduction and Development. 1992;31(4):231–240. doi: 10.1002/mrd.1080310402. [DOI] [PubMed] [Google Scholar]
- 40.Kopečný V, Fakan S, Pavlok A, et al. Immunoelectron microscopic localization of small nuclear ribonucleoproteins during bovine early embryogenesis. Molecular Reproduction and Development. 1991;29(3):209–219. doi: 10.1002/mrd.1080290302. [DOI] [PubMed] [Google Scholar]
- 41.Kopečný V, Biggiogera M, Pivko J, et al. The cell nucleus in early bovine and caprine preimplantation embryos: fine structural cytochemistry and immunoelectron microscopy. European Journal of Cell Biology. 1996;70(4):361–372. [PubMed] [Google Scholar]
- 42.Prather RS, Rickords LF. Developmental regulation of an snRNP core protein epitope during pig embryogenesis and after nuclear transfer for cloning. Molecular Reproduction and Development. 1992;33(2):119–123. doi: 10.1002/mrd.1080330202. [DOI] [PubMed] [Google Scholar]
- 43.Kopecny V, Biggiogera M, Laurincik J, et al. Fine structural cytochemical and immunocytochemical analysis of nucleic acids and ribonucleoprotein distribution in nuclei of pig oocytes and early preimplantation embryos. Chromosoma. 1996;104(8):561–574. doi: 10.1007/BF00352296. [DOI] [PubMed] [Google Scholar]
- 44.Ferreira J, Carmo-Fonseca M. Nuclear morphogenesis and the onset of transcriptional activity in early hamster embryos. Chromosoma. 1996;105(1):1–11. doi: 10.1007/BF02510033. [DOI] [PubMed] [Google Scholar]
- 45.Bogolyubova I, Bogolyubov D, Parfenov V. Localization of poly(A)+ RNA and mRNA export factors in interchromatin granule clusters of two-cell mouse embryos. Cell and Tissue Research. 2009;338(2):271–281. doi: 10.1007/s00441-009-0860-4. [DOI] [PubMed] [Google Scholar]
- 46.Bogolyubova I, Stein G, Bogolyubov D. FRET analysis of interactions between actin and exon-exon-junction complex proteins in early mouse embryos. Cell and Tissue Research. 2013;352(2):277–285. doi: 10.1007/s00441-012-1545-y. [DOI] [PubMed] [Google Scholar]
- 47.Greenblatt J. RNA polymerase II holoenzyme and transcriptional regulation. Current Opinion in Cell Biology. 1997;9(3):310–319. doi: 10.1016/s0955-0674(97)80002-6. [DOI] [PubMed] [Google Scholar]
- 48.Myer VE, Young RA. RNA polymerase II holoenzymes and subcomplexes. The Journal of Biological Chemistry. 1998;273(43):27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 49.Lee TIL, Young RA. Transcription of eukaryotic protein-coding genes. Annual Review of Genetics. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 50.Neish AS, Anderson SF, Schlegel BP, Wei W, Parvin JD. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Research. 1998;26(3):847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rachez C, Freedman LP. Mediator complexes and transcription. Current Opinion in Cell Biology. 2001;13(3):274–280. doi: 10.1016/s0955-0674(00)00209-x. [DOI] [PubMed] [Google Scholar]
- 52.Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. The Journal of Biological Chemistry. 1996;271(32):19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 53.Buratowski S. Progression through the RNA Polymerase II CTD Cycle. Molecular Cell. 2009;36(4):541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochimica et Biophysica Acta. 2013;1829(1):55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 55.Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends in Cell Biology. 1994;4(3):86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 56.Goddard MJ, Pratt HPM. Control of events during early cleavage of the mouse embryo: an analysis of the ‘2-cell block’. Journal of Embryology and Experimental Morphology. 1983;73:111–133. [PubMed] [Google Scholar]
- 57.Qiu JJ, Zhang WW, Wu ZL, Wang YH, Qian M, Li YP. Delay of ZGA initiation occurred in 2-cell blocked mouse embryos. Cell research. 2003;13(3):179–185. doi: 10.1038/sj.cr.7290162. [DOI] [PubMed] [Google Scholar]
- 58.Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. Journal of Cell Biology. 1995;129(2):287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mortillaro MJ, Blencowe BJ, Wei X, et al. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(16):8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic analysis of interchromatin granule clusters. Molecular Biology of the Cell. 2004;15(8):3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kopečný V, Biggiogera M, Pivko J, et al. Fine-structural cytochemical and immunocytochemical observations on nuclear bodies in the bovine 2-cell embryo. Zygote. 2000;8(4):315–328. doi: 10.1017/s0967199400001118. [DOI] [PubMed] [Google Scholar]
- 62.Zatsepina O, Baly C, Chebrout M, Debey P. The step-wise assembly of a functional nucleolus in preimplantation mouse embryos involves the cajal (coiled) body. Developmental Biology. 2003;253(1):66–83. doi: 10.1006/dbio.2002.0865. [DOI] [PubMed] [Google Scholar]
- 63.Daskal Y. The Cell Nucleus. Vol. 8. New York, NY, USA: Academic Press; 1981. Perichromatin granules; pp. 117–137. [Google Scholar]
- 64.Visa N, Puvion-Dutilleul F, Bachellerie J-P, Puvion E. Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. European Journal of Cell Biology. 1993;60(2):308–321. [PubMed] [Google Scholar]
- 65.Puvion E, Viron A, Assens C, Leduc EH, Jeanteur P. Immunocytochemical identification of nuclear structures containing snRNPs in isolated rat liver cells. Journal of Ultrastructure Research. 1984;87(2):180–189. doi: 10.1016/s0022-5320(84)80077-5. [DOI] [PubMed] [Google Scholar]
- 66.Palombi F, Viron A. Nuclear cytochemistry of mouse oogenesis. 1. Changes in extranucleolar ribonucleoprotein components through meiotic prophage. Journal of Ultrastructure Research. 1977;61(1):10–20. doi: 10.1016/s0022-5320(77)90003-x. [DOI] [PubMed] [Google Scholar]
- 67.Bogolyubova IO, Bogolyubov DS. Localization of mRNA export factors in early mouse embryos. HOAJ Biology. 2012;1(11) [Google Scholar]
- 68.le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. The EMBO Journal. 2000;19(24):6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. The EMBO Journal. 2001;20(17):4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tange TØ, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11(12):1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou Z, Luo M-J, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407(6802):401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]


