Abstract
Objective
In the past two decades, approximately 1,000 reports have been published regarding associations between genetic variants in candidate genes and risk of colorectal cancer (CRC). Study results are inconsistent. We aim to provide a synopsis of the current understanding of genetic factors for CRC risk through systematically evaluating results from previous studies.
Design
We searched PubMed and Google Scholar to identify papers that investigated associations between genetic variants and CRC risk and published through December 25, 2012. With data from 950 papers, we conducted 910 meta-analyses for 267 genetic variants in 150 candidate genes with at least three data sources. We used Venice criteria and false-positive report probability tests to grade levels of cumulative epidemiological evidence of significant associations with CRC risk.
Results
Sixty-two variants in 50 candidate genes showed a nominally significant association with CRC risk (p<0.05). Cumulative epidemiological evidence for a significant association with CRC risk was graded strong for eight variants in five genes (APC, CHEK2, DNMT3B MLH1, and MUTYH), moderate for two variants in two genes (GSTM1 and TERT), and weak for 52 variants in 45 genes. In addition, 40 variants in 33 genes showed convincing evidence of no association with CRC risk in meta-analyses including at least 5,000 cases and 5,000 controls.
Conclusion
Approximately 4% of genetic variants evaluated to date in candidate-gene association studies showed moderate to strong cumulative epidemiological evidence of an association with CRC risk. These genetic variants, if confirmed, may explain approximately 5% of familial CRC risk.
Keywords: colorectal cancer, meta-analysis, systematic review, genetic epidemiology, genome-wide association study, genetic variants, gene
Introduction
Colorectal cancer (CRC) is the third-most common cancer, and the second leading cause of cancer death worldwide (1). Genetic factors play an important role in CRC development (2-6). High-penetrance germline mutations in the APC, MUTYH, SMAD4, BMPR1A, STK11, and mismatch repair genes have been identified to account for about 6% of CRC cases (Table 1) (6-13). Since 2007, common genetic variants in approximately 21 loci have been identified through genome-wide association studies (GWAS) (Table 2) (14-24). GWAS-identified variants, however, are associated with weak to moderately elevated risk of CRC, and explain approximately 8% of the familial risk of CRC (20;21).
Table 1. Known high-penetrance mutations in genes contribute to familial colorectal cancer.
| Gene | Variants | Hereditary syndrome | Population frequency | References |
|---|---|---|---|---|
| APC | Nonsense or frameshift mutations | Familial adenomatous polyposis | 0.01-0.02% | 6, 7 |
| MLH1 | Truncating and missense mutations | Lynch syndrome | 0.10% | 6, 8 |
| MSH2 | Truncating and missense mutations | Lynch syndrome | <0.1% | 6, 8 |
| MSH6 | Truncating and missense mutations | Lynch syndrome | <0.05% | 6, 8 |
| PMS2 | Truncating and missense mutations | Lynch syndrome | <0.05% | 6, 8 |
| STK11 | Multiple mutations | Peutz-Jeghers syndrome | 0.0005-0.01% | 9, 10 |
| BMPR1A | Multiple mutations | Juvenile polyposis syndrome | <0.0005% | 10 |
| SMAD4 | Multiple mutations | Juvenile polyposis syndrome | <0.0005% | 10 |
| MUTYH | Nonsense and missense mutations | MUTYH-associated polyposis | <0.02% | 11, 12 |
Table 2. Low-penetrance loci associated with colorectal-cancer risk, identified by genome-wide association studies with p<5×10-8.
| Loci | Genes * | Variants | Alleles † | MAF (%) ‡ | OR (95% CI) | P value | Ethnicity | Reference |
|---|---|---|---|---|---|---|---|---|
| 1q41 | DUSP10 | rs6691170 | T/G | 36 | 1.06 (1.03-1.09) | 9.55 × 10-10 | European | 21 |
| 1q41 | DUSP10 | rs6687758 | G/A | 20 | 1.09 (1.06-1.12) | 2.27 × 10-9 | European | 21 |
| 3q26.2 | MYNN | rs10936599 | T/C | 25 | 0.93 (0.91-0.96) | 3.39 × 10-8 | European | 21 |
| 5q31.1 | PITX1 | rs647161 | A/C | 31 | 1.11 (1.08-1.15) | 1.22 × 10-10 | Asian | 24 |
| 6p21.31 | CDKN1A | rs1321311 | A/C | 23 | 1.10 (1.07-1.13) | 1.14 × 10-10 | European | 22 |
| 6q26-q27 | SLC22A3 | rs7758229 | T/G | 22 | 1.28 (1.18-1.39) | 7.92 × 10-9 | Asian | 23 |
| 8q23.3 | EIF3H | rs16892766 | C/A | 7 | 1.25 (1.19-1.32) | 3.30 × 10-18 | European | 19 |
| 8q24.21 | MYC | rs10505477 | A/G | 51 | 1.17 (1.12-1.23) | 3.16 × 10-11 | European | 14 |
| 8q24.21 | MYC | rs6983267 | G/T | 52 | 1.21 (1.15-1.27) | 1.27 × 10-14 | European | 15 |
| 8q24.21 | MYC | rs7014346 | A/G | 37 | 1.19 (1.14-1.24) | 8.60 × 10-26 | European | 18 |
| 10p14 | FLJ3802842 | rs10795668 | A/G | 33 | 0.89 (0.86-0.91) | 2.50 × 10-13 | European | 19 |
| 11q13.4 | POLD3 | rs3824999 | A/C | 50 | 0.93 (0.91-0.95) | 3.65 × 10-10 | European | 22 |
| 11q23 | Unknown | rs3802842 | C/A | 29 | 1.11 (1.08-1.15) | 5.82 × 10-10 | European | 18 |
| 12p13.32 | CCND2 | rs10774214 | T/C | 35 | 1.09 (1.06-1.13) | 3.06 × 10-8 | Asian | 24 |
| 12q13.13 | LARP4, DIP2 | rs7136702 | T/C | 35 | 1.06 (1.04-1.08) | 4.02 × 10-8 | European | 21 |
| 12q13.3 | DIP2B, ATF1 | rs11169552 | T/C | 28 | 0.92 (0.90-0.95) | 1.89 × 10-10 | European | 21 |
| 14q22.2 | BMP4 | rs4444235 | C/T | 46 | 1.11 (1.08-1.15) | 8.10 × 10-10 | European | 20 |
| 15q13.3 | GREM1 | rs4779584 | T/C | 18 | 1.26 (1.19-1.34) | 4.44 × 10-14 | European | 17 |
| 16q22.1 | CDH1 | rs9929218 | A/G | 29 | 0.91 (0.89-0.94) | 1.20 × 10-8 | European | 20 |
| 18q21.1 | SMAD7 | rs4939827 | C/T | 48 | 0.85 (0.81-0.89) | 1.00 × 10-12 | European | 16 |
| 19q13.1 | RHPN2 | rs10411210 | T/C | 10 | 0.87 (0.83-0.91) | 4.60 × 10-9 | European | 20 |
| 20p12.3 | HAO1 | rs2423279 | C/T | 30 | 1.10 (1.06-1.14) | 6.64 × 10-9 | Asian | 24 |
| 20p12.3 | BMP2 | rs961253 | A/C | 36 | 1.12 (1.08-1.16) | 2.00 × 10-10 | European | 20 |
| 20q13.33 | LAMA5 | rs4925386 | T/C | 32 | 0.93 (0.91-0.95) | 1.89 × 10-10 | European | 21 |
| Xp22.2 | SHROOM2 | rs5934683 | T/C | 33 | 1.07 (1.04-1.10) | 7.30 × 10-10 | European | 22 |
Candidate gene in the locus.
Minor (bold)/major alleles (per initial studies).
MAF=minor allele frequency in controls.
In addition to GWAS, approximately 1,000 papers have been published over the past 25 years investigating genetic variants in candidate genes in relation to CRC risk. Because of the limitation of SNP arrays used in GWAS, many genetic variants evaluated in candidate gene association studies have not been adequately investigated in GWAS. Results from previous candidate gene studies have been inconsistent and are difficult to interpret. Most findings from candidate gene association studies cannot be replicated. Furthermore, sample size from most previous candidate gene association studies was small, so these studies often do not have adequate power to detect a true association. Meta-analysis is a useful tool to systematically evaluate available results published to date to assess evidence for a true association. By pooling data from multiple studies, meta-analysis can increase statistical power and evaluate consistency of association, a major criterion for determining causality. Recently, an interim guideline, named Venice criteria, has been used to systematically grade the cumulative evidence of genetic associations (25;26). Systematic field synopses and meta-analyses have been utilized to evaluate the association of genetic variations in candidate genes with several diseases, including Alzheimer's disease (27), schizophrenia (28), breast cancer (29), cutaneous melanoma (30), and Parkinson's disease (31). Herein, we sought to systematically collect and comprehensively evaluate all candidate-gene association studies of CRC risk, perform meta-analyses for variants with at least three independent datasets, and provide a systematic synopsis of our current understanding of the genetic basis of CRC risk.
Methods
Search strategy and selection criteria
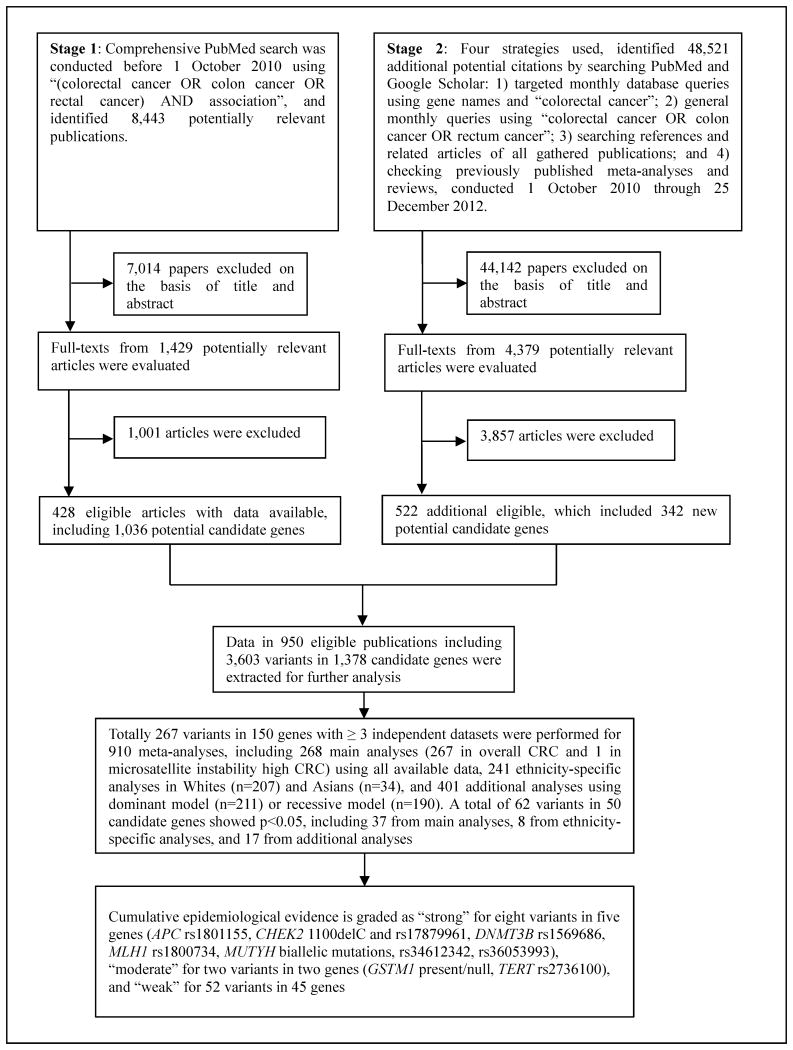
Literature searches were conducted through a two-stage strategy (Figure 1). In Stage 1, we searched the PubMed database using key terms “(colorectal cancer OR colon cancer OR rectal cancer) AND association” before October 1, 2010. This search yielded 8,443 potentially relevant articles which were screened for eligibility by title, abstract, or full text, as necessary – 428 reports, which included 1,036 potential candidate genes, then met eligibility criteria. In Stage 2, conducted October 1, 2010 through December 25, 2012, we used four supplementary approaches to query PubMed and Google Scholar: 1) monthly database queries for “colorectal cancer” and the 1,036 gene names identified in Stage 1 such as “MTHFR”; 2) monthly queries using “colorectal cancer OR colon cancer OR rectum cancer”; 3) searching references and related articles of all gathered papers; and 4) checking previously published meta-analyses and reviews. These four searches identified 48,521 additional reports, of which 522 met our inclusion criteria, adding genetic variants in 342 additional candidate genes. In Stages 1 and 2 combined, we screened a total of 56,964 articles, identifying 945 which reported 3,603 variants in 1,378 independent candidate genes which met our criteria for further analysis.
Figure 1. Profiles of literature search, meta-analysis and evaluation of cumulative evidence.
Studies were eligible for inclusion in this meta-analysis if they met the following criteria: 1) data were published in a peer-reviewed journal in English; 2) the study used a case-control, cohort, or a cross-sectional design in human beings; 3) the study provided sufficient information for the genotypic or allelic distribution of individual variants for both CRC cases and controls, and 4) CRC cases were diagnosed by pathological and/or histological examination. We did not include in the meta-analyses the following two groups of variants: 1) high-penetrance germline mutations in known CRC susceptibility genes, and 2) risk variants identified and confirmed in recent GWAS (Table 2). When multiple publications reported on the same or overlapping data, we used the most informative or most recent publication. Only data from original published papers were included in the present analysis. All variants, regardless of their minor allele frequency (MAF), were considered for meta-analyses when genotype counts or allelic counts were provided in the original studies.
Data extraction and management
All data were extracted by two authors (XM and BZ), and disagreement was resolved by discussion. We recorded first author, year of publication, study name, geographic location of study, ethnicity, PubMed identification number, study design, sample size, mean ages of cases and controls, sample source, genes, variants, major and minor alleles, genotype counts or allelic counts for cases and controls, and Hardy-Weinberg equilibrium (HWE) in controls. Ethnicity was classified as African descendants, Asian (East Asian descent), White (European descent), or Other (including mixed), based on ethnicity of at least 80% of the study population (32). If ethnicity was not reported, we considered ethnicity of the source population where the study was conducted (32). Finally, if a report included several sources or study populations, data were extracted separately.
Statistical analysis and evaluation of cumulative evidence
Statistical analyses were performed by STATA, version 11.0. All tests were two-sided, and p<0.05 was considered statistically significant unless otherwise stated.
Summary odds ratios (ORs) with 95% confidence intervals (CIs) for alleles and genotypes, were used to assess strength of associations between genetic variants and CRC risk by the random-effects method (33). Genotype counts or allelic counts for cases and controls from each original study were used to estimate summary ORs. We did not use adjusted ORs to estimate summary ORs since inconsistent covariates were used for adjustment in original studies included in this meta-analysis. In the primary analyses, we evaluated common variants (MAF≥0.05) using additive model and rare variants (MAF<0.05) using dominant model. For some common variants, a few original studies did not provide sufficient data for analyses with additive model, and thus dominant/recessive model was applied in the primary analyses. For some specific variants, we used the conventional comparisons in original studies, like GSTM1 ‘Present/Null’, NAT2 phenotype (predicted by genetic variants) and MUTYH rs36053993 in the primary analyses. We also conducted subgroup analyses by ethnicities. Dominant and recessive models were also used to assess associations between genetic variants and CRC risk, if available. Meta-analyses were performed only for variants with at least three independent datasets. Because major and minor alleles can be reversed in populations of different ethnicities, averaged MAFs across studies might be greater than 50%. When this occurred, the minor allele among White populations was used as the minor allele in all analyses. For genetic variants other than SNPs, the less prevalent variant or trait was evaluated for associated effects unless otherwise stated. HWE among control groups in each study was assessed by Fisher's exact test to compare observed and expected genotype frequencies (34). We conducted power analysis to evaluate the statistical power of meta-analyses in detecting an association (i.e., OR=1.15) with certain allele frequency (i.e., MAF=0.10) under the additive genetic model, assuming an alpha of 0.05 (35). We calculated the proportion of the familial risk of CRC based on the formula provided by Houlston et al (20).
To determine heterogeneity, we performed Cochran's Q test (36) and calculated the I2 statistic to quantify the proportion of total variation due to heterogeneity (37). Heterogeneity was considered significant if p<0.10. Generally, I2 values <25% correspond to no or little heterogeneity, values 25% – 50% correspond to moderate heterogeneity, and values >50% correspond to strong heterogeneity between studies. Potential small-study bias was assessed with a modified Egger test by Harbord et al. (38). We also evaluated if there was any excess in studies with positive findings than expected using the method described by Ioannidis and Trikalinos (39). To evaluate small-study bias and excessive significant findings, we used p<0.10 as the significant level, as recommended (38;39). For variants showing statistically significant association with CRC risk, sensitivity analyses were performed to determine if the association would be lost when the first published or first positive report was excluded, or when all studies deviated from HWE in controls were excluded.
For statistically significant associations identified by meta-analyses, Venice criteria were applied to assess cumulative evidence (Webappendix notes for Venice criteria). Venice criteria details are published elsewhere (25). For amount of evidence, we did not apply this criterion for rare variants with frequency<1% since an A grade is virtually unobtainable (29). For protection from bias, we also considered GWAS results for all common SNPs (MAF≥5%). If a common variant that can be adequately tagged by GWAS chips was not identified by GWAS, that variant would be downgraded for its evidence of association with CRC risk. Cumulative epidemiological evidence of significant associations in meta-analyses were considered strong if all three grades were A, moderate if all three grades were A or B, and weak if any grade was C. We also performed false-positive report probability (FPRP) analysis to determine if a significant association can be excluded as a false-positive finding. We used the approach developed by Wacholder et al (40) to calculate FPRP for the 62 significant associations. We used prior probability of 0.05 to estimate FPRP value for each of the 62 associations based on p-value and OR obtained from meta-analysis. FPRP<0.05, 0.2≤ FPRP≤0.05, and FPRP>0.2 were considered strong, moderate, and weak evidence of true association, respectively. We upgraded cumulative evidence from moderate to strong, and from weak to moderate, if evidence of true association based on the FPRP analysis was strong. We downgraded cumulative evidence from strong to moderate, and from moderate to weak if evidence of true association was weak. For the 25 significant associations derived from subgroup analysis of different ethnicities or under dominant or recessive model, we also assessed significance based on Bonferroni corrected p-value (5.49×10-5=0.05/910). Regardless of Venice criteria and FPRP grades, we assigned weak evidence of association credibility if p-value > 5.49×10-5.
Results
A total of 945 articles reporting 3,603 variants in 1,378 independent genes were eligible for our analysis (Figure 1). Most of these reports (n=884, 93.5%) were published since 2000. We conducted 910 meta-analyses for 267 variants (241 common and 26 rare) in 150 genes that had at least three data sources (Figure 1). For the 267 main meta-analyses with the use of all available data, mean sample size was 9,633 (range: 519-76,991) from a mean of seven (range: 3-68) independent studies (Webappendix Table 1).
Among the main meta-analyses, 37 (13.9%) variants within 28 genes showed nominally significant association (p<0.05) for CRC risk (Table 3; Webappendix Table 2: references used; Webappendix Table 3). The 37 variants are not in linkage disequilibrium (r2 < 0.1). Mean pooled sample size in the 37 meta-analyses that showed significant association was 15,912 (range: 1,730-51,971), drawn from an average of 11 independent studies (range: 3-56). Approximately 10-fold elevated risk of CRC risk showed association with MUTYH biallelic mutations. Strong associations with CRC (ORs 2.0-10.0) were detected for four rare variants (MLH1 rs121912963, OR=2.74; MLH1 rs63750447, OR=2.14; MUTYH rs34612342, OR=3.32; MUTYH rs36053993, OR=6.49). Moderate associations with CRC (ORs 1.5-2.0 or 0.50-0.67) were found for three rare variants (APC rs1801155, OR=1.96; CHEK2 rs17879961, OR=1.56; CHEK2 1100delC, OR=1.88) and two common variants (DNMT3B rs1569686, OR=0.57; MLH1 rs1800734, OR=1.51). Associations with CRC risk, ORs 0.67-1.50, were observed for the remaining 27 variants, of which most are common. Four of the 37 positive variants (MLH1 rs1800734; MUTYH biallelic mutations; CHEK2 rs17879961; DNMT3B rs1569686) showed highly significant association with CRC risk at p<5×10-7; 13 showed association with CRC risk at p<0.01, and the remaining 20 had p<0.05 (Table 3).
Table 3. Genetic variants nominally significantly associated with colorectal-cancer risk in meta-analyses of all available data.
| Number evaluated | Colorectal-cancer risk meta-analysis | Venice criteria garde‡ |
False-positive report probability§ |
Cumulative evidence of association |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||||
| Genes | Variants | Alleles * | Chromosome | Frequency (%)† | Ethinicity | Studies | Cases | Controls | Genetic models | OR (95% CI) | P value | I2 (%) | Pheterogeneity | |||
| APC | rs1801155 | A/T | 5 | 6.80 | Jewish | 3 | 804 | 6,188 | Dominant | 1.96 (1.37-2.79) | 1.99×10-4 | 0 | 0.84 | BAA | 0.007 | Strong |
| CHEK2 | 1100delC | 1100delC/– | 22 | 0.71 | White | 7 | 3,874 | 11,630 | Dominant | 1.88 (1.29-2.73) | 0.001 | 0 | 0.50 | ×AA | 0.036 | Strong |
| CHEK2 | rs17879961 | C/T | 22 | 3.91 | White | 6 | 6,042 | 17,051 | Dominant | 1.56 (1.32-1.84) | 1.22×10-7 | 0 | 0.76 | BAA | <0.001 | Strong |
| CYP1A1 | rs1048943 | G/A | 15 | 10.33 | All | 16 | 6,704 | 8,009 | Additive | 1.24 (1.05-1.47) | 0.014 | 74 | 0.00 | ACC | 0.338 | Weak |
| CYP2E1 | 96-bp insertion | 96-bp ins/– | 10 | 16.98 | All | 4 | 1,412 | 1,781 | Additive | 1.24 (1.03-1.49) | 0.023 | 35 | 0.20 | ABA | 0.462 | Weak |
| DNMT3B | rs1569686 | G/T | 20 | 16.99 | All | 4 | 1,054 | 1,224 | Additive | 0.57 (0.47-0.68) | 1.86×10-9 | 0 | 0.99 | BAA | <0.001 | Strong |
| GH1 | rs2665802 | A/T | 17 | 45.39 | All | 7 | 3,275 | 3,848 | Additive | 0.89 (0.80-0.99) | 0.025 | 49 | 0.07 | ABC | 0.508 | Weak |
| GSTM1 | Present/Null | NA | 1 | 50.64 | All | 56 | 20,552 | 31,419 | null vs present | 1.10 (1.04-1.17) | 0.001 | 48 | 0.00 | ABC | 0.046 | Moderate |
| GSTT1 | Present/Null | NA | 22 | 29.53 | All | 43 | 15,144 | 23,847 | null vs present | 1.15 (1.05-1.27) | 0.004 | 68 | 0.00 | ACC | 0.144 | Weak |
| IGFBP3 | rs2854746 | G/C | 7 | 46.08 | All | 5 | 4,282 | 7,365 | Additive | 1.07 (1.01-1.14) | 0.016 | 0 | 0.07 | AAC | 0.447 | Weak |
| MLH1 | rs121912963 | C/G | 3 | 0.43 | All | 3 | 1,412 | 1,508 | Additive | 2.74 (1.31-5.75) | 0.008 | 15 | 0.31 | ×AC | 0.231 | Weak |
| MLH1 | rs1800734 | A/G | 3 | 21.11 | White | 5 | 801 | 10,890 | Additive | 1.51 (1.34-1.69) | 6.74×10-12 | 46 | 0.11 | AAA | <0.001 | Strong |
| MLH1 | rs63750447 | A/T | 3 | 1.96 | Asian | 3 | 937 | 919 | Additive | 2.14 (1.12-4.12) | 0.022 | 41 | 0.18 | BBC | 0.458 | Weak |
| MMP1 | rs1799750 | 2G/1G | 11 | 39.63 | All | 8 | 1,477 | 1,751 | Additive | 0.76 (0.64-0.92) | 0.004 | 61 | 0.01 | ACC | 0.138 | Weak |
| MSH3 | rs184967 | A/G | 5 | 15.25 | White | 3 | 5,085 | 7,136 | Additive | 1.11 (1.03-1.20) | 0.005 | 0 | 0.38 | AAC | 0.182 | Weak |
| MSH3 | rs26279 | G/A | 5 | 28.13 | White | 4 | 5,691 | 7,665 | Additive | 1.1 (1.03-1.17) | 0.006 | 17 | 0.31 | AAC | 0.157 | Weak |
| MTHFD1 | rs1950902 | A/G | 14 | 18.78 | White | 3 | 3,822 | 5,452 | Additive | 0.90 (0.84-0.98) | 0.010 | 0 | 0.79 | AAC | 0.275 | Weak |
| MUTYH | Monoallelic mutation | NA | 1 | 1.69 | White | 17 | 25,981 | 18,811 | Carriers vs wild homozygotes | 1.17 (1.01-1.34) | 0.036 | 0 | 0.84 | BAC | 0.546 | Weak |
| MUTYH | Biallelic mutation | NA | 1 | 0.01 | White | 17 | 25,981 | 18,811 | Carriers vs wild homozygotes | 10.19 (5.00-22.04) | 5.30×10-10 | 0 | 0.88 | ×AA | <0.001 | Strong |
| MUTYH | rs34612342 | G/A | 1 | 0.01 | White | 17 | 27,041 | 19,641 | GG vs AA | 3.32 (1.13-9.81) | 0.030 | 0 | 1.00 | ×AA | 0.533 | Strong |
| MUTYH | rs36053993 | A/G | 1 | 0.00 | White | 17 | 26,957 | 19,870 | AA vs GG | 6.49 (2.57-1.35) | 7.49×10-5 | 0 | 0.85 | ×AA | 0.003 | Strong |
| NAT2 | Fast/slow | NA | 8 | 47.39 | All | 35 | 11,684 | 15,348 | Slow vs fast | 0.94 (0.89-0.99) | 0.023 | 1 | 0.45 | AAC | 0.47 | Weak |
| NOD2 | rs2066844 | T/C | 16 | 6.15 | White | 9 | 3,297 | 3,088 | Dominant | 1.35 (1.02-1.78) | 0.038 | 34 | 0.14 | BBC | 0.581 | Weak |
| NOD2 | rs2066847 | C/– | 16 | 6.21 | White | 11 | 4,337 | 5,395 | Dominant | 1.30 (1.02-1.65) | 0.032 | 33 | 0.13 | BBC | 0.546 | Weak |
| PTGS1 | rs5788 | A/C | 9 | 13.35 | White | 4 | 3,989 | 6,659 | Additive | 1.13 (1.04-1.22) | 0.004 | 0 | 0.64 | AAC | 0.113 | Weak |
| PTGS2 | rs689466 | G/A | 1 | 30.34 | All | 9 | 4,076 | 7,610 | Additive | 0.88 (0.80-0.98) | 0.018 | 56 | 0.02 | ACC | 0.405 | Weak |
| SCD | rs7849 | G/A | 10 | 18.49 | All | 3 | 2,011 | 2,580 | Additive | 0.85 (0.73-0.98) | 0.025 | 29 | 0.25 | ABC | 0.488 | Weak |
| TERT | rs2736100 | T/G | 5 | 49.34 | White | 8 | 16,176 | 18,135 | Additive | 1.07 (1.04-1.1) | 2.92×10-5 | 0 | 0.53 | AAC | 0.001 | Moderate |
| TGFB1 | rs1800469 | T/C | 19 | 38.74 | All | 10 | 4,405 | 5,383 | Additive | 0.88 (0.79-0.97) | 0.013 | 55 | 0.02 | ACC | 0.33 | Weak |
| TNF | rs1800629 | A/G | 6 | 13.78 | All | 11 | 2,296 | 2,283 | Additive | 1.28 (1-1.62) | 0.046 | 71 | 0.00 | ACC | 0.625 | Weak |
| TP73 | G4C14/A4T14 | NA | 1 | 24.02 | All | 4 | 858 | 1,168 | Additive | 1.20 (1.04-1.40) | 0.015 | 6 | 0.36 | AAC | 0.363 | Weak |
| UBD | rs2076485 | C/T | 6 | 26.07 | White | 3 | 4,281 | 6,157 | Additive | 1.07 (1.01-1.14) | 0.034 | 0 | 0.77 | AAC | 0.563 | Weak |
| VDR | rs11568820 | A/G | 12 | 36.61 | White | 4 | 3,228 | 3,455 | Dominant | 1.15 (1.04-1.27) | 0.005 | 0 | 0.93 | AAB | 0.165 | Weak |
| VDR | rs1544410 | A/G | 12 | 38.96 | All | 17 | 11,687 | 12,301 | Additive | 0.85 (0.72-0.99) | 0.040 | 93 | 93.40 | ACC | 0.87 | Weak |
| VEGF | rs3025039 | T/C | 6 | 19.43 | All | 6 | 1,925 | 1,884 | Additive | 1.19 (1.04-1.37) | 0.014 | 29 | 0.22 | ABC | 0.347 | Weak |
| XPA | rs1800975 | A/G | 9 | 36.46 | White | 3 | 593 | 1,137 | Additive | 0.82 (0.70-0.96) | 0.016 | 4 | 0.36 | AAC | 0.379 | Weak |
| XPC | rs2228001 | C/A | 3 | 37.38 | All | 9 | 2,978 | 5,204 | Additive | 1.08 (1.01-1.16) | 0.021 | 0 | 0.83 | AAC | 0.486 | Weak |
Minor alleles/major alleles (Per Caucasian); majors alleles were treated as reference alleles in the analyses;
Allelic ORs were estimated under the additive model. For dominant or recessive models, ORs were estimated for subjects who carry one or two minor alleles or subjects homozygous for the minor alleles,respectively.
Frequency of minor allele or effect genotype(s) in controls in primary meta-analysis
Venice criteria grades are for amount of evidence, replication of the association, and protection from bias.
False-positive report probability (FPRP) was determined based on OR and P value of each variant from meta-analysis and a prior probability of 0.05.
Cumulative epidemiological evidence as graded by combination of results from Venice criteria and FPRP for association with colorectal-cancer risk.
Of the 267 meta-analyses of all available data, 120 (44.9%) had little or no heterogeneity, 43 (16.1%) had moderate heterogeneity, and 104 (39.0%) had strong heterogeneity. The proportion of studies with strong heterogeneity was significantly lower for the 37 positive variants (Table 3) than the remaining 230 variants (19% vs 42%, Fisher's exact p < 0·01). Small-study bias was detected for 36 variants (13.5%), of which seven were positive variants. Of the 267 variants, 38 (14.2%) showed evidence of excess studies with significant findings including four positive variants. When considering all studies included in 267 meta-analyses as a whole, the number of studies with significant findings was also greater than that expected (666 vs 301, p < 0.0001).
In sensitivity analyses, nine SNPs (rs7849, rs1800469, rs3025039, rs1048943, rs689466, rs1544410, rs2854746, rs1800629, G4C14/A4T14) became non-significant after exclusion of HWE-violating studies, and 13 variants (rs2854746, rs121912963, rs63750447, rs26279, rs1950902, MUTYH monoallelic mutation, NAT2 Fast/slow, rs2066844, rs2066847, rs1800629, G4C14/A4T14, rs2076485, rs1544410) became non-significant after exclusion of the first positive or first published report.
We next calculated FPRP value at the prior probability, 0.05, to evaluate the probability of true association with CRC risk for the 37 positive variants from the main analyses. Associations with CRC risk had a FPRP value <0.05 for nine variants in seven genes (APC rs1801155, CHEK2 1100delC and rs17879961, DNMT3B rs1569686, GSTM1 deletion, MLH1 rs1800734, MUTYH biallelic mutations, rs36053993, TERT rs2736100), FPRP 0.05-0.2 for 6 variants in 5 genes (GSTT1 deletion, MMP1 rs1799750, MSH3 rs184967 and rs26279, PTGS1 rs5788, VDR rs11568820), and FPRP > 0.2 for the remaining 22 variants (Table 3).
Epidemiological credibility of significant associations was graded for the 37 positive variants identified through the main analyses (Table 3 and Webappendix Table 3). We first applied Venice criteria. Grades of A were given to 25, 22, and 9 meta-analyses for amount of evidence, replication of association, and protection from bias, respectively. Grades of B were given to 7, 8, and 1 meta-analyses for amount of evidence, replication of association, and protection from bias, respectively. Grades of C were given to 0, 7, and 27 meta-analyses for these three criteria, respectively. Next, strong, moderate, and weak for evidence of true association with CRC risk were assigned to 9, 6, and 22 variants, respectively, based on FPRP. For MUTYH rs34612342, we disregarded FPRP value (FPRP=0.533) when evaluating cumulative evidence because this mutation is pathogenic and has strong evidence to increase the risk of developing multiple adenomatous polyps and colorectal cancer (41). Altogether, eight variants in five genes (APC rs1801155, CHEK2 1100delC and rs17879961, DNMT3B rs1569686, MLH1 rs1800734, MUTYH biallelic mutations, rs34612342, rs36053993), were graded strong for evidence of association with CRC risk using combined Venice criteria and FPRP results. Two variants (GSTM1 Present/Null, TERT rs2736100) scored moderate for evidence of association with CRC risk. The remaining 27 variants scored C in one or more Venice criteria or were downgraded due to high FPRP. These variants were graded weak for cumulative evidence of association with CRC risk, based on combined Venice criteria and FRPR results.
Next, we performed stratified meta-analyses by ethnicity for 207 variants among Whites and 34 variants among Asians (Webappendix Table 5) and identified eight additional variants from eight genes to be nominally associated with CRC risk (p<0.05, Table 4 and Webappendix Table 3). Six of them (rs16260, rs28362491, rs1800566, rs1052133, rs1801394, rs7903146) were associated with CRC risk only in Whites; the other two (rs20417, rs1042522) were associated with CRC risk only in Asians. We also performed meta-analyses using dominant and recessive models to evaluate associations of genetic variants with CRC risk, identifying 17 additional variants across 17 genes showing significant association, although none were statistically significant in additive model (Table 5, and Webappendix Table 4). Similar to the 37 positive variants identified in the main analyses, we applied Venice criteria and FRRP to evaluate these 25 variants. We also considered Bonferroni corrected p-value. All were graded weak for cumulative evidence of association with CRC risk.
Table 4. Genetic variants nominally significantly associated with colorectal-cancer risk identified from additional analyses by ethnic group in additive model.
| Number evaluated | Colorectal-cancer risk meta-analysis | Venice criteria garde‡ |
False-positive report probability§ |
Cumulative evidence of association |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||||
| Genes | Variants | Alleles * | Chromosome | MAF (%)† | Ethinicity | Studies | Cases | Controls | OR (95% CI) | P value | I2 (%) | Pheterogeneity | |||
| CDH1 | rs16260 | A/C | 16 | 28.02 | White | 6 | 6,761 | 6,646 | 0.93 (0.87-1.00) | 0.048 | 23 | 0.26 | AAC | 0.642 | Weak |
| MTRR | rs1801394 | A/G | 5 | 44.65 | White | 10 | 6,430 | 9,746 | 0.98 (0.93-1.02) | 0.030 | 4 | 0.41 | AAC | 0.535 | Weak |
| NQO1 | rs1800566 | T/C | 16 | 17.88 | White | 8 | 6,293 | 6,566 | 1.09 (1.03-1.16) | 0.006 | 0 | 0.50 | AAC | 0.183 | Weak |
| OGG1 | rs1052133 | G/C | 3 | 21.59 | White | 14 | 5,908 | 7,355 | 1.15 (1.01-1.32) | 0.033 | 74 | 0.00 | ACC | 0.558 | Weak |
| PTGS2 | rs20417 | C/G | 1 | 2.76 | Asian | 4 | 1,285 | 3,040 | 1.44 (1.06-1.95) | 0.019 | 28 | 0.25 | BBC | 0.420 | Weak |
| NFKB1 | rs28362491 | –/ATTG | 4 | 41.05 | White | 6 | 1,199 | 3,134 | 1.29 (1.11-1.50) | 0.001 | 55 | 0.05 | ACB | 0.036 | Weak |
| TCF7L2 | rs7903146 | T/C | 10 | 29.08 | White | 3 | 1,960 | 14,290 | 1.12 (1.02-1.22) | 0.015 | 0 | 0.47 | AAC | 0.335 | Weak |
| TP53 | rs1042522 | C/G | 17 | 37.15 | Asian | 8 | 3,993 | 4,943 | 1.14 (1.02-1.27) | 0.021 | 60 | 0.02 | ACC | 0.430 | Weak |
Minor alleles/major alleles (Per Caucasian); majors alleles were treated as reference alleles in the analyses
Allelic ORs were estimated under the additive model. For dominant or recessive models, ORs were estimated for subjects who carry one or two minor alleles or subjects homozygous for the minor alleles,respectively.
MAF=minor allele frequency in controls.
Venice criteria grades are for amount of evidence, replication of the association, and protection from bias.
False-positive report probability (FPRP) was determined based on OR and P value of each variant from meta-analysis and a prior probability of 0.05.
Cumulative epidemiological evidence as graded by combination of results from Venice criteria and FPRP for association with colorectal-cancer risk.
Table 5. Additional genetic variants nominally significantly associated with colorectal-cancer risk in meta-analyses using dominant or recessive models.
| Number evaluated | Colorectal-cancer risk meta-analysis | Venice criteria grade‡ |
False-positive report probability§ |
Cumulative evidence of association |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||||
| Genes | Variants | Alleles * | Chromosome | MAF (%)† | Ethnicity | Studies | Cases | Controls | Genetic models | OR (95% CI) | P value | I2 (%) | Pheterogeneity | |||
| SELS | rs34713741 | T/C | 15 | 33.22 | All | 3 | 1,442 | 2,071 | Dominant | 1.21 (1.05-1.39) | 0.008 | 0 | 0.40 | AAC | 0.235 | Weak |
| SERPINE1/PAI-1 | rs1799889 | 5G/4G | 7 | 44.94 | White | 4 | 2,241 | 4,534 | Dominant | 0.87 (0.78-0.97) | 0.014 | 0 | 0.56 | AAC | 0.337 | Weak |
| EPHX1 | rs2234922 | G/A | 1 | 19.38 | All | 13 | 5,329 | 6,700 | Dominant | 0.91 (0.85-0.99) | 0.020 | 0 | 0.50 | AAC | 0.46 | Weak |
| ERCC5/XPG | rs17655 | C/G | 13 | 24.73 | All | 9 | 6,322 | 7,537 | Dominant | 1.13 (1.01-1.25) | 0.027 | 38 | 0.12 | ABC | 0.48 | Weak |
| RAD18 | rs373572 | C/T | 3 | 29.22 | All | 3 | 3,174 | 3,397 | Dominant | 1.18 (1.01-1.37) | 0.033 | 27 | 0.25 | ABC | 0.55 | Weak |
| CCND1 | rs9344 | A/G | 11 | 48.15 | All | 22 | 6,316 | 8,272 | Dominant | 1.13 (1.01-1.26) | 0.035 | 43 | 0.00 | ABC | 0.569 | Weak |
| IGF1 | rs35767 | T/C | 12 | 24.75 | All | 3 | 2,717 | 4,880 | Recessive | 0.75 (0.62-0.91) | 0.003 | 0 | 0.57 | BAC | 0.11 | Weak |
| MGMT | rs12917 | T/C | 10 | 12.99 | All | 7 | 4,127 | 7,284 | Recessive | 1.54 (1.14-2.08) | 0.005 | 0 | 0.47 | BAA | 0.158 | Weak |
| CRP | rs1800947 | C/G | 1 | 5.70 | All | 4 | 2,916 | 3,544 | Recessive | 3.84 (1.38-10.74) | 0.010 | 0 | 0.47 | CAC | 0.277 | Weak |
| HPGD | rs2612656 | G/A | 4 | 22.75 | White | 3 | 2,979 | 5,575 | Recessive | 1.31 (1.05-1.64) | 0.016 | 21 | 0.28 | BAC | 0.380 | Weak |
| FRZB | rs7775 | G/C | 2 | 8.77 | White | 3 | 1,256 | 3,000 | Recessive | 3.20 (1.17-8.73) | 0.023 | 64 | 0.06 | CCC | 0.468 | Weak |
| TGFBR1 | rs334354 | A/G | 9 | 26.71 | All | 4 | 1,226 | 2,776 | Recessive | 1.38 (1.04-1.84) | 0.029 | 8 | 0.35 | BAC | 0.516 | Weak |
| TGFB1 | rs4803455 | A/C | 19 | 47.48 | All | 3 | 2,786 | 3,516 | Recessive | 1.14 (1.01-1.28) | 0.030 | 0 | 0.37 | AAC | 0.536 | Weak |
| LIPC | rs6083 | A/G | 15 | 36.52 | All | 3 | 4,702 | 4,914 | Recessive | 0.85 (0.74-0.99) | 0.032 | 25 | 0.27 | AAA | 0.56 | Weak |
| MTHFR | rs1801133 | T/C | 1 | 33.50 | All | 68 | 32,608 | 44,383 | Recessive | 0.92 (0.85-1) | 0.036 | 52 | 0.00 | ACC | 0.61 | Weak |
| CYP2C9 | rs1799853 | T/C | 10 | 13.31 | White | 6 | 4,915 | 5,237 | Recessive | 1.36 (1.02-1.83) | 0.038 | 0 | 0.76 | BAA | 0.60 | Weak |
| MTRR | rs10380 | T/C | 5 | 9.31 | White | 4 | 3,869 | 5,141 | Recessive | 1.61 (1.02-2.52) | 0.039 | 6 | 0.36 | BAA | 0.597 | Weak |
Minor alleles/Major alleles (Per Caucasian); majors alleles were treated as reference alleles in the analyses
Allelic ORs were estimated under the additive model. For dominant or recessive models, ORs were estimated for subjects who carry one or two minor alleles or subjects homozygous for the minor alleles, respectively.
MAF=minor allele frequency in controls.
OR=odds ratio; CI=confidence interval.
Venice criteria grades are for amount of evidence, replication of the association, and protection from bias.
False-positive report probability (FPRP) was determined based on OR and P value of each variant from meta-analysis and a prior probability of 0.05.
Cumulative epidemiological evidence as graded by combination of results from Venice criteria and FPRP for association with colorectal-cancer risk.
The vast majority of meta-analyses performed in this project (205 variants in 130 genes) did not yield any evidence of significant association. These meta-analyses included a mean of six studies (range 3-34) and 7,916 participants (range 519-36,982). Table 6 shows results for 40 variants from 33 genes that showed no evidence of association with CRC risk in meta-analyses with a minimum of 5,000 cases and 5,000 controls.
Table 6. Genetic variants showing no relation to colorectal-cancer risk in meta-analyses with at least 5,000 cases and 5,000 controls in additive model.
| Number assessed | Colorectal cancer risk | Heterogeneity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| Genes | Variants | Comparisons* | Frequency (%)† | Ethnicity | Studies | Cases | Controls | OR (95% CI) | P value | I2 (%) | Pheterogeneity |
| ABCB1 | rs1202168 | T vs C | 41.25 | White | 4 | 6,318 | 5,805 | 1.05 (0.97-1.14) | 0.191 | 55 | 0.08 |
| ABCB1 | rs9282564 | G vs A | 8.89 | White | 4 | 5,792 | 5,234 | 1.21 (0.78-1.88) | 0.400 | 96 | 0.00 |
| ABCB1/MDR1 | rs1045642 | C vs T | 47.52 | All | 13 | 6,312 | 7,128 | 0.98 (0.89-1.07) | 0.611 | 58 | 0.00 |
| APC | rs459552 | A vs T | 22.27 | All | 8 | 6,654 | 7,117 | 0.96 (0.91-1.02) | 0.205 | 0 | 0.66 |
| CASP8 | rs3834129 | 6 bp ins vs del | 41.14 | All | 10 | 6,922 | 10,750 | 1.05 (0.92-1.20) | 0.441 | 82 | 0.00 |
| CASR | rs1042636 | G vs A | 8.52 | White | 4 | 6,298 | 7,839 | 1.00 (0.92-1.09) | 0.936 | 0 | 0.66 |
| CDH1 | rs16260 | A vs C | 28.09 | All | 9 | 7,220 | 7,045 | 0.94 (0.88-1.01) | 0.116 | 21 | 0.26 |
| COMT | rs4680 | A vs G | 48.89 | White | 5 | 5,074 | 5,239 | 1.05 (0.94-1.16) | 0.390 | 56 | 0.06 |
| CYP1A1 | rs4646903 | C vs T | 14.42 | All | 15 | 7,258 | 8,154 | 1.05 (0.92-1.19) | 0.500 | 65 | 0.00 |
| CYP1A2 | rs762551 | C vs A | 30.45 | All | 11 | 7,667 | 8,242 | 1.02 (0.94-1.10) | 0.664 | 55 | 0.01 |
| CYP1B1 | rs1056836 | G vs C | 43.27 | White | 9 | 8,709 | 9,097 | 1.02 (0.97-1.06) | 0.488 | 0 | 0.44 |
| CYP1B1 | rs1800440 | G vs A | 18.36 | White | 6 | 6,679 | 6,923 | 0.97 (0.88-1.07) | 0.580 | 53 | 0.06 |
| CYP2C9 | rs1057910 | G vs A | 7.16 | All | 8 | 8,538 | 9,182 | 1.00 (0.86-1.16) | 0.994 | 62 | 0.01 |
| EPHX1 | rs1051740 | C vs T | 30.95 | All | 18 | 10,478 | 12,372 | 1.02 (0.97-1.08) | 0.447 | 36 | 0.06 |
| ERCC2/XPD | rs13181 | C vs A | 30.25 | All | 17 | 6,039 | 8,749 | 0.99 (0.92-1.05) | 0.649 | 24 | 0.17 |
| ERCC2/XPD | rs1799793 | A vs G | 29.71 | All | 7 | 5,470 | 7,135 | 1.01 (0.96-1.07) | 0.674 | 0 | 0.68 |
| GSTP1 | rs1138272 | T vs C | 9.84 | All | 10 | 7,160 | 7,789 | 0.92 (0.80-1.06) | 0.234 | 56 | 0.02 |
| GSTP1 | rs1695 | G vs A | 27.54 | All | 33 | 9,986 | 15,562 | 0.98 (0.93-1.03) | 0.487 | 25 | 0.10 |
| IGF1 | (CA)n | non R19 vs R19 | 36.71 | All | 8 | 5,493 | 6,827 | 0.99 (0.89-1.09) | 0.769 | 70 | 0.00 |
| IGFBP3 | rs2854744 | C vs A | 46.30 | All | 9 | 6,872 | 10,606 | 1.03 (0.98-1.07) | 0.284 | 0 | 0.98 |
| IL6 | rs1800795 | C vs G | 38.55 | All | 14 | 6,952 | 8,657 | 1.01 (0.93-1.11) | 0.749 | 65 | 0.00 |
| IRS1 | rs1801278 | A vs G | 6.92 | White | 7 | 7,048 | 7,533 | 1.08 (0.96-1.22) | 0.219 | 39 | 0.13 |
| MLH1 | rs1799977 | G vs A | 29.56 | All | 10 | 6,384 | 8,972 | 1.01 (0.92-1.11) | 0.904 | 58 | 0.01 |
| MTHFD1 | rs2236225 | A vs G | 45.19 | White | 6 | 6,535 | 9,347 | 0.98 (0.90-1.07) | 0.603 | 67 | 0.01 |
| MTHFR | rs1801131 | C vs A | 30.14 | All | 34 | 14,965 | 22,017 | 0.99 (0.94-1.03) | 0.514 | 32 | 0.04 |
| MTR | rs1805087 | G vs A | 19.42 | All | 19 | 12,945 | 17,655 | 0.99 (0.94-1.05) | 0.717 | 33 | 0.09 |
| MTRR | rs1801394 | A vs G | 48.33 | All | 16 | 7,674 | 11,593 | 0.96 (0.91-1.01) | 0.110 | 37 | 0.12 |
| MUTYH | rs3219484 | A vs G | 8.20 | White | 3 | 5,391 | 5,222 | 0.95 (0.68-1.34) | 0.787 | 91 | 0.00 |
| MUTYH | rs3219489 | C vs G | 28.44 | All | 4 | 5,082 | 5,280 | 1.09 (0.92-1.28) | 0.317 | 81 | 0.00 |
| NAT1 | Phenotype | Fast vs slow | 44.37 | All | 15 | 7,336 | 9,825 | 1.03 (0.92-1.16) | 0.596 | 61 | 0.00 |
| NQO1 | rs1800566 | T vs C | 22.90 | All | 12 | 7,209 | 8,783 | 1.07 (0.99-1.15) | 0.090 | 32 | 0.13 |
| OGG1 | rs1052133 | G vs C | 25.79 | All | 18 | 6,654 | 8,599 | 1.10 (0.99-1.23) | 0.085 | 71 | 0.00 |
| PPARG | rs1801282 | G vs C | 9.10 | All | 18 | 13,758 | 20,300 | 0.97 (0.91-1.03) | 0.339 | 18 | 0.24 |
| PPARG | rs3856806 | T vs C | 11.39 | All | 10 | 6,189 | 8,707 | 1.03 (0.96-1.11) | 0.412 | 2 | 0.42 |
| PTGS2/COX2 | rs5275 | C vs T | 33.17 | All | 10 | 6,059 | 8,084 | 1.01 (0.97-1.07) | 0.579 | 0 | 0.98 |
| TGFBR1 | rs11466445 | 9 bp del vs ins | 9.19 | All | 10 | 6,338 | 6,689 | 1.04 (0.96-1.13) | 0.379 | 1 | 0.43 |
| TP53 | rs1042522 | C vs G | 31.28 | All | 31 | 10,515 | 12,909 | 1.00 (0.92-1.10) | 0.922 | 72 | 0.00 |
| VDR | rs2228570 | A vs G | 37.32 | All | 20 | 13,631 | 15,155 | 1.00 (0.94-1.06) | 0.959 | 53 | 0.00 |
| VDR | rs7975232 | A vs C | 43.08 | All | 9 | 5,421 | 5,377 | 1.08 (0.98-1.19) | 0.105 | 58 | 0.02 |
| XRCC1 | rs25487 | G vs A | 31.87 | All | 25 | 9,541 | 14,448 | 1.04 (0.97-1.11) | 0.281 | 49 | 0.00 |
OR=odds ratio; CI=confidence interval.
Genetic comparison used in meta-analysis: Minor allele vs Major allele.
Frequency of minor allele or effect genotype(s) in controls in primary meta-analysis
Discussion
To our knowledge, this study is the largest and most comprehensive assessment of the literature regarding candidate-gene association studies for CRC risk conducted to date. We systematically evaluated data for 3,603 variants in 1,378 independent candidate genes from 950 reports published in the past two decades. Several meta-analyses have been conducted to evaluate candidate-gene association studies of CRC risk for single gene or several genes. These early analyses, however, were limited to 52 variants in 34 genes (Webappendix Table 6). Recently, Theodoratou et al (42) evaluated genetic variants for CRC risk using data from 635 publications and conducted meta-analyses for 92 polymorphisms in 64 genes, including 18 variants identified from GWAS studies. We did not include GWAS-identified risk variants in this study since they have been robustly replicated and should be considered to have strong evidence of association. Our study not only provides an update of the variants meta-analyzed previously using data from more studies and a bigger sample size, but also assessed more than 193 variants that have not been assessed in any previous meta-analyses, including the meta-analysis conducted by Theodoratou, et al (42). Of the 267 variants in 150 genes summarized by our 910 meta-analyses, 62 variants in 50 genes showed nominally significant association with CRC risk. Using Venice criteria plus FPRP results, we graded eight variants strong for cumulative epidemiological evidence of association with CRC risk (APC rs1801155, CHEK2 1100delC and rs17879961, DNMT3B rs1569686, MLH1 rs1800734, MUTYH biallelic mutations, rs34612342, rs36053993), two variants moderate for cumulative evidence of association with CRC risk (GSTM1 Present/Null, TERT rs2736100), and the remaining 52 variants weak. Of the eight strong variants, MUTYH rs36053993 was also rated as having ‘strong’ evidence for association in Theodoratou's study (42). For 40 variants in 33 genes, we showed no evidence of association with CRC risk in meta-analyses with large sample sizes (10,000 individuals minimum). Our study provides a comprehensive research synopsis of candidate-gene association studies of CRC risk. Results from this study will be helpful for future studies to evaluate genetic risk factors for CRC.
The adenomatous polyposis coli (APC) gene, a tumor suppressor gene at chromosome 5q21, encodes a large multidomain protein including 2,843 amino acids that play a central role in the Wnt singling pathway (43). Germline pathogenic mutations in the APC gene result in autosomal dominant inherited familial adenomatous polyposis (FAP) in which more than 100 adenomatous polyps can develop (3;6). Our meta-analysis provides strong evidence of association for CRC risk with a heterozygous variant at codon 1,307 in exon 15 of the gene (rs1801155), with a 1.96-fold increased risk of CRC in Jews (including Ashkenazi and Israeli Jews). This variant is present in 7% of Ashkenazi Jews, while population frequency is very low in Europeans and Asians (based on HapMap data).
The CHEK2 gene maps to chromosome 22q12.1 and encodes a protein kinase that is activated in response to DNA damage and is involved in cell cycle arrest (44). Our meta-analysis revealed strong evidence of association with CRC risk for a truncating mutation at codon 381 in exon 10 (1100delC) and a missense polymorphism in exon 3 (rs17879961, Ile157Thr). The 1100delC mutation leads to kinase-deficient molecules due to protein truncation (45), while Ile157Thr results in a CHEK2 protein with deficient binding and phosphorylation of downstream substrates (46). Interestingly, in a previous meta-analysis, we found strong cumulative evidence of association for these two variants with breast-cancer risk (29), indicating the CHEK2 gene may play a role in both CRC and breast cancer.
Our meta-analyses revealed strong evidence for an association of CRC risk with three rare variants in the MUTYH gene based on data from 17 population-based studies excluding cases with MUTYH-associated polyposis (MAP). Biallelic mutations in the MUTYH gene mainly constitute either homozygotes (two same) or compound heterozygotes (two different) of Gly382Asp and Tyr165Cys. Gly382Asp and Tyr165Cys are located in exon 7 and exon 13 of the MUTYH gene, respectively, and have been predicted to be deleterious by SIFT (47) and confirmed to be pathogenic (41). However, the monoallelic mutation, including a heterozygous genotype of 12 mutations in the MUTYH gene showed only weak evidence for association with CRC risk in our study. Two common variants (MLH1 rs1800734, DNMT3B rs1569686) showed strong cumulative evidence of association with CRC risk. MLH1, which maps to chromosome 3p22.2, is a human homolog of the E. coli DNA mismatch repair gene mutL and is a locus frequently mutated in hereditary nonpolyposis colon cancer (HNPCC) (48). Approximately 85% of genetically defined HNPCC patients have germline mutations in the MLH1 gene (49). Interestingly, meta-analysis of five studies, comprised of 801 microsatellite instability high (MSI-H) cases and 10,890 controls, identified a highly significant association of rs1800734 (-93G>A) with MSI-H CRC (p=1.67×10-12). This promoter SNP showed a much stronger association with MSI-H CRC (OR=1.51) than overall CRC cases (OR=1.05, p=0.013) based on meta-analysis of six studies: 17,174 cases, 13,166 controls. The DNMT3B gene plays an important role in the generation of aberrant methylation in carcinogenesis (50). Although this gene was not identified as a susceptibility locus for CRC by GWAS, we still rated the SNP (rs1569686) in this gene as having strong evidence for association given the highly consistent results across studies included in our meta-analysis.
Two common variants (GSTM1 null, TERT rs2736100) scored moderate for cumulative evidence of association with CRC risk, and both of them were upgraded from ‘weak’ for having a low false-positive report probability (<0.05). Additional investigations of these variants are needed, particularly since sample sizes of studies for both variants are relatively small. Cumulative epidemiological evidence of association with CRC was weak for the remaining 52 variants, many of which are common and were identified through ethnicity-specific meta-analyses or meta-analyses using dominant or recessive models. Well-designed studies with large samples are warranted to clarify association with CRC for these variants.
Our meta-analysis provides no evidence for association with CRC risk for 205 of the 267 variants evaluated in our study, supporting the notion that the vast majority of genetic variants evaluated in candidate gene association studies may not be truly related to CRC risk. Methodological limitations in previous candidate gene studies, such as small sample size, may explain some of the null associations. However, of the 205 non-significant variants, 40 variants in 33 genes showed no association with CRC risk in meta-analyses including a minimum of 5,000 cases, 5,000 controls, which provides approximately 85% power to detect an OR of 1.15 under the additive model for a variant with MAF 0.10, Type 1 error 0.05. Thus, future epidemiological studies with a similar sample size are unlikely to be helpful in assessing effects of these variants.
There are several limitations of this study. First, although we have systematically searched the literature to identify eligible studies using two stages, it is possible that some studies might have been missed. PubMed was the main database we used for our literature search. To expand our search, we also queried Google Scholar which links multiple databases. Compared with previous meta-analyses which also used multiple databases (Webappendix Table 7), we yielded more studies with a bigger combined sample size for most variants included in our evaluation. Second, we did not assess gene-gene or gene-environment interactions. Additional studies specifically designed to identify these interactions are needed. Third, heterogeneity across studies, including differences in study populations, study designs and genotyping platforms, may have contributed to some of the null associations in this study. More than one-third of the meta-analyses had high heterogeneity, especially for variants with non-significant association. We attempted to address study heterogeneity through stratification analyses by ethnicity. Other sources of heterogeneity also exist and are difficult to address in this meta-analysis because of limited available data. Finally, Venice criteria use p-value<0.05 as significance level to determine association. However, we found most associations with a p-value 0.005-0.05 to have weak evidence for association with CRC in this study. Thus, a more stringent threshold of p-value would be helpful to evaluate evidence for a true-positive association. In addition, Venice criteria offer the advantage of evaluating multiple sources of potential bias, some of which, such as genotyping error, phenotype misclassification, and population stratification, are difficult to assess in meta-analyses.
In our meta-analyses, we identified ten genetic variants showing strong or moderate epidemiological evidence of associations with CRC risk. If all these 10 variants are confirmed to be associated with CRC risk, they could explain approximately 5% of familial CRC risk in European populations. Nevertheless, genetic risk factors identified to date account for less than 30% familial risk of CRC. Some of the missing heritability could be due to methylation markers, copy number variations, structural variants, and rare variants, for which conventional candidate gene association studies and GWAS are inadequate to investigate. Gene-gene and gene-environment interactions may also play a significant role in the etiology of CRC. Additional research, including those with a large sample size, use of higher density SNP arrays and next-generation sequencing technologies, imputation using data from the 1000 Genomes Project and better defined CRC subtypes, are needed to clarify the missing heritability of CRC. Our study, the largest field synopsis conducted to date for CRC candidate gene association studies, not only summarizes the current literature regarding genetic epidemiology of CRC, but also provides comprehensive data and helpful clues for designing future studies to further investigate genetic risk factors for CRC.
Supplementary Material
Significance of this study.
What is already known about this subject?
Colorectal cancer (CRC) is one of most commonly diagnosed cancers in the world.
Approximately 35% of CRC risk could be attributable to inheritable factors.
Many studies have been conducted to evaluate associations between genetic variants in candidate genes and risk of CRC over the past two decades – with inconsistent results.
What are the new findings?
This study is the largest, most comprehensive assessment of the literature to date regarding genetic association studies in CRC risk.
Of the 267 variants evaluated, 62 variants in 50 candidate genes showed a statistically significant association with CRC risk.
Eight variants in five genes showed strong cumulative evidence of association with CRC risk, and two variants in two genes showed moderate evidence.
This study provides clues for designing future studies to further investigate genetic risk factors for CRC.
How might it impact on clinical practice in the foreseeable future?
Genetic risk variants may be used to identify high-risk individuals for CRC screening and prevention.
Acknowledgments
We thank the authors of many original studies for clarification of data and providing additional information, and Mary Jo Daly for her help with manuscript preparation. This research is supported in part by NIH grant R37 CA070867 and Ingram Professorship funds.
The corresponding author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or nonexclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Gut editions and any other BMJPGL products to exploit all subsidiary rights, as set out in our licence.
Abbreviations
- CRC
colorectal cancer
- GWAS
genome-wide association studies
- MAF
minor allele frequency
- HWE
Hardy-Weinberg equilibrium
- ORs
odds ratios
- CIs
confidence intervals
- FPRP
false-positive report probability
- APC
adenomatous polyposis coli
- FAP
familial adenomatous polyposis
- MAP
MUTYH-associated polyposis
- HNPCC
hereditary nonpolyposis colon cancer
- MSI-H
microsatellite instability high
Footnotes
Conflicts of interest: We declare no conflict of interest.
Contributors: X Ma and B Zhang conducted literature searches, data extraction, quality assessment and analyses. X Ma and B Zhang drafted the manuscript with substantial contributions from W Zheng. W Zheng reviewed results and provided guidelines for presentation and interpretation.
Reference List
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 3.Jass JR. Familial colorectal cancer: pathology and molecular characteristics. Lancet Oncol. 2000;1:220–6. doi: 10.1016/s1470-2045(00)00152-2. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham D, Atkin W, Lenz HJ, et al. Colorectal cancer. Lancet. 2010;375(9719):1030–47. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 5.Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet. 2009;10(6):353–8. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 6.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4(10):769–80. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 7.Fearnhead NS, Britton MP, Bodmer WF. The ABC of APC. Hum Mol Genet. 2001;10(7):721–33. doi: 10.1093/hmg/10.7.721. [DOI] [PubMed] [Google Scholar]
- 8.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87(2):159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 9.van Lier MG, Wagner A, Mathus-Vliegen EM, et al. High cancer risk in Peutz-Jeghers syndrome: a systematic review and surveillance recommendations. Am J Gastroenterol. 2010;105(6):1258–64. doi: 10.1038/ajg.2009.725. [DOI] [PubMed] [Google Scholar]
- 10.Nagy R, Sweet K, Eng C. Highly penetrant hereditary cancer syndromes. Oncogene. 2004;23(38):6445–70. doi: 10.1038/sj.onc.1207714. [DOI] [PubMed] [Google Scholar]
- 11.Jasperson KW, Tuohy TM, Neklason DW, et al. Hereditary and familial colon cancer. Gastroenterology. 2010;138(6):2044–58. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubbe SJ, Di Bernardo MC, Chandler IP, et al. Clinical implications of the colorectal cancer risk associated with MUTYH mutation. J Clin Oncol. 2009;27(24):3975–80. doi: 10.1200/JCO.2008.21.6853. [DOI] [PubMed] [Google Scholar]
- 13.Aaltonen L, Johns L, Jarvinen H, et al. Explaining the familial colorectal cancer risk associated with mismatch repair (MMR)-deficient and MMR-stable tumors. Clin Cancer Res. 2007;13(1):356–61. doi: 10.1158/1078-0432.CCR-06-1256. [DOI] [PubMed] [Google Scholar]
- 14.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39(8):989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 15.Tomlinson I, Webb E, Carvajal-Carmona L, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39(8):984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 16.Broderick P, Carvajal-Carmona L, Pittman AM, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39(11):1315–7. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 17.Jaeger E, Webb E, Howarth K, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40(1):26–8. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 18.Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40(5):631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40(5):623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 20.Houlston RS, Webb E, Broderick P, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40(12):1426–35. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houlston RS, Cheadle J, Dobbins SE, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42(11):973–7. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunlop MG, Dobbins SE, Farrington SM, et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet. 2012;44(7):770–6. doi: 10.1038/ng.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui R, Okada Y, Jang SG, et al. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut. 2011;60(6):799–805. doi: 10.1136/gut.2010.215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia WH, Zhang B, Matsuo K, et al. Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat Genet. 2013;45(2):191–6. doi: 10.1038/ng.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37(1):120–32. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 26.Khoury MJ, Bertram L, Boffetta P, et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. Am J Epidemiol. 2009;170(3):269–79. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertram L, McQueen MB, Mullin K, et al. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 28.Allen NC, Bagade S, McQueen MB, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40(7):827–34. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B, Beeghly-Fadiel A, Long J, et al. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 2011;12(5):477–88. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatzinasiou F, Lill CM, Kypreou K, et al. Comprehensive field synopsis and systematic meta-analyses of genetic association studies in cutaneous melanoma. J Natl Cancer Inst. 2011;103(16):1227–35. doi: 10.1093/jnci/djr219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lill CM, Roehr JT, McQueen MB, et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: The PDGene database. PLoS Genet. 2012;8(3):e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ioannidis JP, Ntzani EE, Trikalinos TA. 'Racial' differences in genetic effects for complex diseases. Nat Genet. 2004;36(12):1312–8. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 33.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 34.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76(5):887–93. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skol AD, Scott LJ, Abecasis GR, et al. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38(2):209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 36.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127(9):820–6. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 37.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 38.Harbord RM, Egger M, Sterne JA. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med. 2006;25(20):3443–57. doi: 10.1002/sim.2380. [DOI] [PubMed] [Google Scholar]
- 39.Ioannidis JP, Trikalinos TA. An exploratory test for an excess of significant findings. Clin Trials. 2007;4(3):245–53. doi: 10.1177/1740774507079441. [DOI] [PubMed] [Google Scholar]
- 40.Wacholder S, Chanock S, Garcia-Closas M, et al. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Tassan N, Chmiel NH, Maynard J, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet. 2002;30(2):227–32. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 42.Theodoratou E, Montazeri Z, Hawken S, et al. Systematic meta-analyses and field synopsis of genetic association studies in colorectal cancer. J Natl Cancer Inst. 2012;104(19):1433–57. doi: 10.1093/jnci/djs369. [DOI] [PubMed] [Google Scholar]
- 43.Fodde R, Smits R, Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer. 2001;1(1):55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 44.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282(5395):1893–7. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 45.Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31(1):55–9. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 46.Kilpivaara O, Vahteristo P, Falck J, et al. CHEK2 variant I157T may be associated with increased breast cancer risk. Int J Cancer. 2004;111(4):543–7. doi: 10.1002/ijc.20299. [DOI] [PubMed] [Google Scholar]
- 47.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bronner CE, Baker SM, Morrison PT, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368(6468):258–61. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 49.Goecke T, Schulmann K, Engel C, et al. Genotype-phenotype comparison of German MLH1 and MSH2 mutation carriers clinically affected with Lynch syndrome: a report by the German HNPCC Consortium. J Clin Oncol. 2006;24(26):4285–92. doi: 10.1200/JCO.2005.03.7333. [DOI] [PubMed] [Google Scholar]
- 50.Robertson KD, Keyomarsi K, Gonzales FA, et al. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28(10):2108–13. doi: 10.1093/nar/28.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.