Abstract
Background
The Polo-like Kinase 1 (PLK1) protein regulates cell cycle progression and is overexpressed in many malignant tissues. Overexpression is associated with poor prognosis in several cancer entities, whereby expression of PLK1 shows high inter-individual variability. Although PLK1 is extensively studied, not much is known about the genetic variability of the PLK1 gene. The function of PLK1 and the expression of the corresponding gene could be influenced by genomic variations. Hence, we investigated the gene for functional polymorphisms. Such polymorphisms could be useful to investigate whether PLK1 alters the risk for and the course of cancer and they could have an impact on the response to PLK1 inhibitors.
Methods
The coding region, the 5′ and 3′UTRs and the regulatory regions of PLK1 were systematically sequenced. We determined the allele frequencies and genotype distributions of putatively functional SNPs in 120 Caucasians and analyzed the linkage and haplotype structure using Haploview. The functional analysis included electrophoretic mobility shift assay (EMSA) for detected variants of the silencer and promoter regions and reporter assays for a 3′UTR polymorphism.
Results
Four putatively functional polymorphisms were detected and further analyzed, one in the silencer region (rs57973275), one in the core promoter region (rs16972787), one in intron 3 (rs40076) and one polymorphism in the 3′untranslated region (3′UTR) of PLK1 (rs27770). Alleles of rs27770 display different secondary mRNA structures and showed a distinct allele-dependent difference in mRNA stability with a significantly higher reporter activity of the A allele (p < 0.01).
Conclusion
The present study provides evidence that at least one genomic variant of PLK1 has functional properties and influences expression of PLK1. This suggests polymorphisms of the PLK1 gene as an interesting target for further studies that might affect cancer risk, tumor progression as well as the response to PLK1 inhibitors.
Keywords: Polymorphism, PLK1, rs27770, 3′UTR
Background
Polo-like kinases (PLKs) belong to the family of serin/threonin kinases. They are involved in the regulation of cell division and centrosome cycle. Until now, four human PLKs have been identified. PLK1 is so far the best characterized polo-like kinase and a target for anticancer therapy [1,2].
PLK1 promotes proliferation by supporting mitotic entry and inhibits apoptosis by interaction with p53 [3-5]. PLK1 is up-regulated in many different tumour tissues like head and neck squamous cell carcinoma, oesophagus and stomach cancer, ovarian cancer, non-small cell lung cancer, liver cancer, cervical cancer and breast cancer [6-9]. Overexpression of PLK1 has been suggested as a biomarker for numerical chromosomal aberration [10,11]. Furthermore, overexpression is associated with poor prognosis in several cancer entities [9,12-15]. Consistent with these findings, different PLK1 inhibitors, i.e. small molecules as well as an siRNA-based formulation, are currently under preclinical and clinical evaluation as promising anticancer drugs [16-18].
The human PLK1 locus maps on chromosome 16p12.1, and the gene product comprises 10 exons and codes for a 67 kDa protein [19,20]. The 5′ region of PLK1 harbors three distinct regulatory regions. Next to the translation initiation site is the core promoter region of the gene. This region mediates up to 75% of the promoter activity and is followed by an extended promoter region with lower impact on activity. A silencer region which is able to suppress about 50% of the activity and an enhancer region are located distal to the promoter region (Figure 1A) [21]. Polymorphisms within the above mentioned gene regions can exert functional impact upon gene expression and protein function. In particular single nucleotide polymorphisms (SNPs) can be useful in association studies for studying complex genetic disorders by a candidate gene approach [22]. Functional polymorphisms are of interest in cancer research and treatment, because they could be used to analyze cancer risk and outcome as well as the response to therapeutic agents [23]. Until now, two reports indicate a possible impact of genetic variants on PLK1 function. In a genome wide bioinformatic approach a polymorphism of the PLK1 3′UTR (rs27770) was one of 117 variants that were predicted to be functional due to significant allele frequency deviations between HapMap (genomic level) and dbEST (mRNA level) data [24]. As part of a polymorphism panel, another polymorphism located within intron 3 of PLK1 (rs40076) has been suggested as an outcome predictor for Caucasian bladder cancer patients [25].
Figure 1.
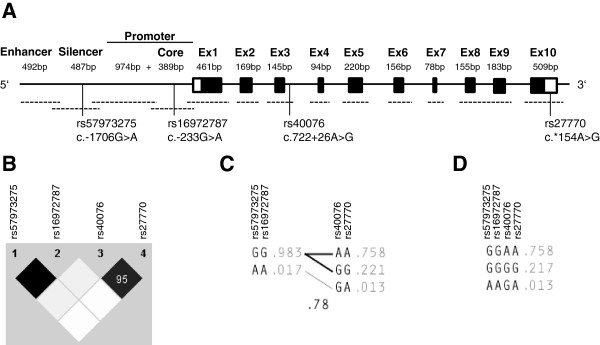
The PLK1 gene locus and genetic variants. Structure of the PLK1 gene (panel A). Black boxes represent exons, the size of exons and regulatory regions are given in base pairs (bp), they are not drawn to scale. The 5′ and 3′UTR are highlighted in white. Sequenced regions are depicted by dashed lines. Positions of PLK1 polymorphisms are given according to the nomenclature of the Human Genome Variation Society [26]. Haploview plot of the linkage disequilibrium analysis of PLK1 polymorphisms in 120 healthy Caucasians (panel B). Numbers in squares are D’ values in percent. If no number is given for a pairwise comparison D’ is 100 percent. Black color indicates an r2 of 1, shades of grey/black indicate an r2 between 1 and 0. White indicates an r2 of 0. Haplotypes of the two haplotype blocks (panel C) and across the PLK1 gene (panel D). The haplotype frequencies are shown to the right of each haplotype. Only haplotypes having a frequency > =1% are shown. Below the crossing area the multi-allelic D’, which indicates the level of recombination between the blocks, is shown. Connecting lines from one block to the other are shown for haplotypes with a frequency of at least 10% (thick lines) and with a frequency of at least 1% (thin lines), respectively.
PLK1 is an important oncogene and drug target in many cancer entities. Genetic variability of such proteins can have an impact upon the risk and the outcome of different cancer types as well as the response of an individual to drug treatments [27,28]. Until now, only very limited information about functionally relevant genetic variations of the PLK1 gene is available. The aim of this study was to systematically search for functional polymorphisms in the PLK1 gene, which could alter gene expression or protein function. We reviewed dbSNP and HapMap data and sequenced functionally relevant regions of PLK1. Retrieved polymorphisms were analyzed by in silico methods to predict functional polymorphisms. Four SNPs were selected for further evaluation and analyzed for linkage and haplotype structure. We identified rs27770 as a functional polymorphism that modulates the secondary structure and stability of PLK1 mRNA.
Results
Sequencing results of the PLK1 gene and linkage analysis of SNPs in regulatory regions
Data base analysis and PLK1 sequencing in healthy unrelated Caucasians revealed 49 SNPs with a minor allele frequency (MAF) of at least 1% but no other variations like insertions/deletions or repeats. The PLK1 coding region harbors no polymorphisms, whereas most of the SNPs where located in introns. Analysis of these intronic SNPs revealed that none of them is located within putatively functional regions (e.g. exon-intron boundaries or branch points), therefore, it is unlikely that they have functional properties, e.g. alteration of splicing. Three SNPs which occur in Caucasians were detected in regulatory regions and further investigated (Figure 1A). They are located in the silencer (rs57973275), in the promoter (rs16972787), and in the 3′UTR (rs27770). Additionally, we analyzed the genotype distribution and linkage of rs40076 in intron 3 based on the suggested association with survival of bladder cancer patients [25]. Table 1 shows the genotype distributions and allele frequencies of these polymorphisms. All distributions were compatible with Hardy-Weinberg equilibrium. The 5′-region SNPs rs57973275 and rs16972787 showed the same low MAF of 1.7%. They are in complete linkage to each other. Further linkage analysis using haploview (Figure 1B) revealed strong linkage disequilibrium between all four SNPs (D′ 0.95 to 1.0), whereas the correlation was high only for the pairs rs57973275/rs16972787 (r2 1.0) and rs40076/rs27770 (r2 0.89), but weak for other pairwise comparisons (r2 0.01 to 0.05). Analysis of the two haplotype blocks showed a D′ of 0.78 between blocks (Figure 1C). In line with these results, we identified 3 major haplotypes (GGAA, GGGG and AAGA) with a frequency above 1% (Figure 1D). These haplotypes represent 98.8% of all detected allele combinations of our Caucasian study population. Two rare haplotypes could be detected, one with a frequency of 0.8% (GGAG) and another with a frequency of 0.4% (AAGG).
Table 1.
Genotype distributions and allele frequencies in healthy Caucasians
| SNP | Genotype | n (%) | Allele frequency | HWE P | |
|---|---|---|---|---|---|
| rs57973275 |
GG |
116 (96.7) |
G |
0.98 |
|
| |
GA |
4 (3.3) |
A |
0.02 |
0.85 |
| |
AA |
0 |
|
|
|
| rs16972787 |
GG |
116 (96.7) |
G |
0.98 |
|
| |
GA |
4 (3.3) |
A |
0.02 |
0.85 |
| |
AA |
0 |
|
|
|
| rs40076 |
AA |
68 (56.7) |
A |
0.77 |
|
| |
AG |
48 (40.0) |
G |
0.23 |
0.20 |
| |
GG |
4 (3.3) |
|
|
|
| rs27770 |
AA |
71 (59.2) |
A |
0.77 |
|
| |
AG |
43 (35.8) |
G |
0.23 |
0.88 |
| GG | 6 (5.0) | ||||
rs57973275 (c.-1706 G > A) and rs16972787 (c.-233G > A) alleles generate different putative transcription factor binding sites
Functional polymorphisms in the silencer and promoter region may impair mRNA levels by changing transcription factor (TF) binding sites and concurrent modulation of the promoter activity. We predicted potential allele-dependent TF binding using MatInspector, Alibaba2 and Consite (Figure 2A) [29-31]. For rs57973275 a putative binding site of the TF Activator protein 1 (AP1) emerged for the G allele and C-Rel for the A allele. In the presence of the A allele, rs16972787 created binding sites for PAX-2 (Paired box gene 2), PAX-6 (Paired box gene 6) and Snail, as well as a MEF3 binding motif, whereas the same binding sites are not present for the G allele, which instead carries a putative binding site for KLF7 (Krueppel like transcription factor 7). We used these in silico results to design oligonucleotides which comprise the putative binding sites of the different TFs. To analyze differences between the alleles we performed electrophoretic mobility shift assays (EMSA) with these oligonucleotides. Nuclear extracts of three different cell lines (HEK293, HepG2 and HeLa) were used for both SNPs to cover the diversity of the predicted TFs. All three cell lines are known to express (HEK293) and overexpress (HepG2 and HeLa) PLK1 [32-34]. Figure 2B shows two representative results of these experiments. In contrast to the in silico results we were not able to detect any specific transcription factor binding to the alleles of both polymorphisms.
Figure 2.
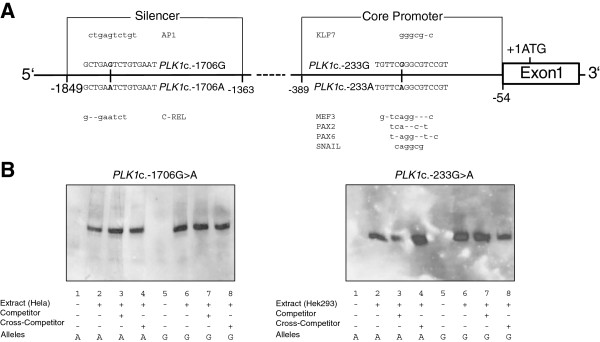
In silico transcription factor binding sites and electrophoretic mobility shift assay. Schematic representation of the PLK1 silencer and core promoter regions (panel A). Alleles and the surrounding sequence of the silencer SNP rs57973275 (c.-1706G > A) and the core promoter SNP rs16972787 (c.-233G > A) are shown. Transcription factors and their putative binding sites are shown above and below the corresponding PLK1 alleles, respectively. Representative electrophoretic mobility shift assays for both SNPs that failed to show genotype-dependent binding of nuclear extracts of different cell lines (panel B).
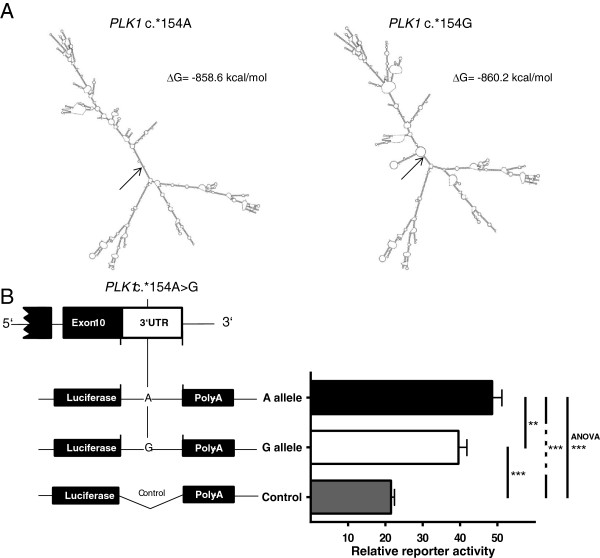
rs27770 (c.*154A > G) alleles generate different secondary mRNA structures and the A allele leads to increased expression in HEK293 reporter assays
To study whether microRNA binding, mRNA folding or regulatory RNA elements could be altered by rs27770, we performed bioinformatic analysis using RegRNA 2.0 and mfold web server [35,36]. According to the analysis of the PLK1 mRNA sequence [NCBI: NM005030] with and without the A to G substitution, the polymorphism is neither located within a microRNA binding site nor hampers or generates putative regulatory RNA elements like AU-rich motifs (data not shown). Mfold results strongly suggest that rs27770 in the 3′UTR has a marked effect on PLK1 mRNA structure (Figure 3A). The A and G alleles differ considerably in their predicted most favorable secondary mRNA structure. Under the assumption of a 5% suboptimality with regard to the minimum free energy, all additionally received secondary structures (A allele n = 30; G allele n = 39) were different between alleles. To study the functional impact of rs27770 alleles on PLK1 mRNA expression, 350 bp of the 3′UTR, containing either of the two alleles, were cloned into the pMIR-REPORT vector downstream of the Firefly luciferase coding region and co-transfected with the Renilla luciferase pGL4.74 reporter vector (Figure 3B). As expected, the 350 bp insert is functionally relevant and improves mRNA stability significantly in comparison with the empty vector. Furthermore, rs27770 alleles lead to significantly altered mRNA stability. In HEK293 cells, reporter activity of the A allele is about 25% higher compared with the G allele.
Figure 3.
Secondary PLK1 mRNA structure and reporter assays for allele-dependent mRNA stability. Mfold-predicted most favorable secondary PLK1 mRNA structure dependent on rs27770 (c.*154A > G) alleles (panel A). Prediction is based on PLK1 mRNA sequence according to NCBI accession number NM005030. Arrows indicate position of the polymorphism within the secondary structures. Allele-dependent activity of PLK1 3′UTR reporter constructs expressed in HEK293 cells (panel B). Structures of the constructs used for the reporter assay are depicted on the left side. Reporter activity was quantified by measuring Firefly luciferase activity normalized to Renilla luciferase activity. Data are mean ± SE of five independent experiments, ***P < 0.001, **P < 0.01, one-way ANOVA and Holm-Sidak’s post-hoc multiple comparisons test.
Discussion
According to our database review and sequencing results the coding region of PLK1 is conserved and polymorphisms are located in intronic and regulatory regions. This is in line with general findings with regard to the occurrence rate of genetic variations and especially of SNPs in different gene regions [22]. We considered database-derived polymorphisms with a MAF of at least 1% in Caucasians. Nevertheless, the analyzed databases comprise a relevant number of rare variants of the coding region with MAFs of less than 1% in Caucasian that could have a functional impact on PLK1. Some of these variants reach a MAF of more than 1% in other ethnicities (e.g. rs2230914). Due to the number of chromosomes investigated by sequencing, the probability to detect new, undescribed polymorphisms with a MAF of 1% was 33% only. An adequate probability of at least 90% for detecting new polymorphisms was only reached for polymorphisms with a MAF of more than 5%. Therefore, it is possible that the PLK1 gene still harbors undetected rare variants (most likely non-SNP variations). Furthermore, other databases that were not systematically analyzed for this study might contain additional variations with MAFs above 1% in Caucasians. For example, after completion of our experiments we became aware of a missense variant (rs45569335) with an overall MAF of 0.7%, but with a MAF of 1.2% within the Caucasian subset of the 1000 genome browser [37].
We selected four candidate SNPs for further investigation, which were either located within the regulatory regions of PLK1 (rs57973275, rs16972787 and rs27770) or showed an association with bladder cancer outcome in a previous study (rs40076). The results of the haplotype analysis implied that it is not necessary to genotype all 4 polymorphisms in future association studies [38] and that two tagging SNPs, either rs57973275 or rs16972787 in combination with rs27770 would be sufficient to represent the haplo- and diplotype structure of these PLK1 SNPs. Furthermore, the strong linkage disequilibrium suggests rs27770 as the underlying functional SNP in the detected association of the PLK1 intron 3 SNP rs40076 with bladder cancer outcome [39].
Since the two SNPs 5′ of the coding region are located within previously identified important regulatory regions of PLK1[21] they were considered eligible candidates for bioinformatic and experimental assessment [40], and because different software applications access different databases, we used three in silico tools to predict putative TF binding sites. Analysis revealed different allele-specific candidates for both SNPs. Computational approaches for identifying binding sites suffer from high error rates because binding motifs of TFs are typically short and degenerated [41], therefore, we performed EMSA experiments using three different cell lines to validate the in silico results. Unfortunately, the EMSA results clearly indicated no functional impact of these polymorphisms on TF binding, negating any further evaluation of the SNPs with regard to TF binding activity. Some authors reported that only 33% of all functional promoter variants were found in known consensus sequences or motifs [42], therefore, we cannot rule out an impact of the two SNPs on binding of other TFs which are not expressed by the selected cell lines. Another mechanism to regulate expression is methylation of CpG islands. Theoretically, the G allele of the promoter polymorphism rs16972787 could be a candidate for allele-specific methylation, however, changes of the methylation status in human malignant cells and tissues have been reported for PLK2 and PLK3 but not for PLK1[12,43,44]. A further study suggested that the PLK1 promoter is unmethylated in G0/G1 (PLK1 not expressed) as well as M phase (PLK1 expressed) and regulated during the cell cycle by transcription factors [45].
Alleles of the 3′UTR polymorphism rs27770 were analyzed with regard to different functional RNA motifs and microRNA binding sites. The analysis revealed neither motif nor target site differences. Until now at least six PLK1 mRNA-targeting microRNAs have been experimentally validated [46-51]. In line with our analysis, the predicted corresponding binding sites do not include the polymorphism. In consideration of the secondary mRNA structure, another key factor for microRNA target recognition is the accessibility of the binding site [52]. Different reports proposed altered microRNA binding because of allele-dependent changes of the secondary mRNA structure due to SNPs outside of the microRNA binding site [53,54]. Furthermore, alterations of the secondary structure itself can interfere with RNA-binding proteins, which can lead to altered mRNA stability [55]. We therefore investigated the secondary structure of the PLK1 mRNA dependence on rs27770 alleles. Although only one nucleotide was substituted, major changes of the secondary structure were predicted. Reporter assays of the 3′UTR of PLK1 consistently showed statistically significant allele-dependent differences in mRNA stability. In comparison to the G allele, the A allele showed 25% more reporter activity, which faithfully reflects mRNA levels [56]. Our results, as well as a previous bioinformatic comparison of HapMap and dbEST data, support a functional impact of rs27770. However, the results themselves are contradictory, because the previous report predicted an increased expression of the G allele [24]. This could have several reasons. First, an experimental validation of a subset of the predicted candidate SNPs confirmed only 36% of the results and rs27770 was not part of the validation subset. Most of the SNPs (59%) showed no differential allelic expression, but alleles corresponding to 5% of the SNPs were significantly associated with gene expression in the opposite direction. Second, according to the usual practice, we investigated mRNA stability of the 3′UTR by reporter assay, however the complete PLK1 mRNA contributes to the secondary structure. It is therefore possible that the hybrid mRNA of the Firefly luciferase coding region and the PLK1 3′UTR could lead to biased results because of other secondary structures. Third, both results could be genuine, if the effect of the SNP is context-dependent and tissue-related. This is a well-known phenomenon and occurs often in connection with regulatory SNPs, especially if the SNP effect depends on differentially expressed transcription factors and microRNAs respectively [57].
Finally, genetic variability of the PLK1 gene and rs27770 in particular are interesting candidates for additional studies. Because PLK1 plays an important role in the cell cycle and inhibits apoptosis, it should be investigated whether PLK1 polymorphisms have an impact on proliferation of malignant and non-malignant cells [3,5]. This would lead to altered expression profiles in cancer tissues and might partly explain the detected variability of PLK1 expression in different cancer entities [8]. In some malignancies like acute lymphoblastic leukemia (ALL) PLK1 expression is highly variable, but expression is not associated with any clinical or biological feature while ALL cell lines respond very well to PLK1 inhibitor treatment [58]. For these malignancies analysis of PLK1 polymorphisms would be an interesting approach to reanalyze genotype-dependent subsets with regards to expression patterns and clinical as well as biological features. PLK1 polymorphisms could be useful with regard to risk as well as outcome studies in Caucasian cancer patients but also in other ethnicities because, according to dbSNP data, rs27770 occurs in other ethnicities as well. Furthermore, functional SNPs in drug target genes may have an impact on targeted therapy. It would therefore be of interest to evaluate PLK1 inhibitor studies with regard to PLK1 polymorphisms, and, because of the predicted impact of the SNP on the secondary mRNA structure of PLK1 and the effects shown on mRNA stability, RNAi based PLK1 inhibitors would be of special interest in this case. It is also well-known that the target secondary structure has a major impact on siRNA and RNAi efficiency [59,60]. We have no evidence for an interaction of PLK1 polymorphisms with currently clinically evaluated RNAi-based PLK1 inhibitors [61], but it would be reasonable to analyze the respective binding sites with regard to allele-dependent target accessibility.
Conclusions
Altogether, our results contribute to reveal the functional impact of genetic variants on PLK1 function. Based on such results, we can speculate about a putative clinical impact: I. these variants may play a role in carcinogenesis and modulate the risk for cancer; II. variants may contribute to altered tumor growth which could lead to different disease courses; and III. PLK1 inhibitor response might be genotype dependent. Although our analyses were not exhaustive, data presented here strongly indicate that a relevant amount of the detectable inter-individual variability of the PLK1 expression with concomitant molecular changes is determined by genomic variants of PLK1.
Methods
Bioinformatic analyses
We have retrieved and analyzed data of genetic variations of the PLK1 gene region from NCBI dbSNP [62] and the HapMap database [63]. To further analyze HapMap data we used Haploview 4.2 [64] but none of the currently accessible HapMap versions contained data of all of the four polymorphisms of this study. Therefore, Haploview was used to analyze and visualize our own data only. Analysis of putative allele-dependent binding sites of transcription factors due to SNPs within regulatory regions of the PLK1 gene was performed with MatInspector [29], Consite [30] and Alibaba2.1 [31] using default settings. To study whether microRNA binding or regulatory RNA motifs could be altered by SNPs, we performed analysis of the 3′UTR using RegRNA 2.0 [36]. The mRNA sequences harboring the different rs27770 alleles were subjected to the web-tool mfold to predict secondary structures [35]. The full mRNA sequence of PLK1, [NCBI RefSeq NM005030], was used for analysis. Sequences were folded with mfold in a locally automated manner. The structures predicted to have the lowest energy were used to identify the folding state. For computing suboptimal foldings the percent suboptimality value that controls the free energy increment was set to 5%.
Sequencing of the PLK1 gene
DNA for sequencing (n = 20) and for genotyping (n = 100) were extracted from whole blood of healthy unrelated Caucasian blood donors, recruited at the local Department for Transfusion Medicine, University Hospital Essen, Germany, using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). This study was strictly performed according to the Declaration of Helsinki and was approved by the local ethics committee of the University Hospital Essen (073454). Informed consent was obtained from all study participants. Primers listed in Table 2 were used for PCR and subsequent sequencing. Primers for 5′UTR, coding regions and 3′UTR were designed to investigate the exons and the corresponding exon-intron boundaries. Relative positions of the enhancer, silencer and core promoter region of PLK1 have been adopted from Bräuninger et al. [21]. We used this information to design appropriate primers that generate overlapping PCR products (Figure 1A).
Table 2.
Primers for PCR and sequencing of PLK1
| Region | Sense (5′ – 3′) | Antisense (5′ – 3′) | Size (bp) |
|---|---|---|---|
| Enhancer |
GAGCAAGACTCCATCTCAACA |
AACCAGGTGTAAGCCTCCCA |
590 |
| Silencer |
CTTGTATACAACATTGCACATGG |
TCCTTCACCTGCCTTGCAGC |
603 |
| Promoter |
GCACTGCTCTGGGAGCTTGG |
TGATGCAACGAAGCTGTCTGG |
820 |
| Core Pro. |
TCTTCCAACCTTCCCTCCCTC |
GTCACTGCAGCACTCATGCTC |
690 |
| Exon1 |
GAGCGGTGCGGAGGCTCTGC |
CAGGGCTTTCCTCCTCTTGTGC |
1240 |
| Exon2 |
GCTGTGCTGGAGAAGGAATG |
ACAACCCACAAGTCAGTATCTTG |
455 |
| Exon3 |
CCTGGTTCTGGATGGTCAAA |
ATTGTCATATCTTTCCCTGTCAC |
409 |
| Exon4 |
CTGCTCAGTGGTCTTAGGGATT |
TATCCCACCTCTAAGGTAGCC |
359 |
| Exon5 |
AGTGGAGAACTTGGCATTG |
CTCTGTCCTTCAATCCGTG |
513 |
| Exon6 |
TCCCCAAAGCAGTGGTAGC |
TGTCTGCATAGGACCATTGGT |
385 |
| Exon7 |
CCCTGCTTTGCTCTTCTC |
GTTACAGACTCTGGCCTTTTTGAGC |
322 |
| Exon8 + 9 |
CTGGGCTCAAACAATCCTCCTCCCTCA |
GTGGGTTGAATGTGGAGTGAGCGGCT |
645 |
| Exon10 | TCTTCCCTCTACTCCCTAACA | GGGTTCTACAGCCTTGTCC | 473 |
Genotyping of PLK1 polymorphisms
The polymorphisms rs57973275, rs40076 and rs27770 were genotyped by restriction fragment length polymorphism analyses. For all polymerase chain reactions (PCR) the Taq DNA Polymerase Master Mix RED (Ampliqon, Herlev, Denmark) was used. The PCR for rs57973275 was performed with following primers: 5′-TCCCTGGACTTTGTCCATG-3′ and 5′-ACCACCTCCTAGTCTGATG-3′ resulting in a PCR product of 138 bp. Amplified fragments were digested with restriction enzyme DdeI (New England Biolabs, Beverly, MA, USA) by incubating for 4 hours at 37°C. DdeI specifically cuts PCR products that carry the G allele (98 + 40 bp). For rs40076 an 110 bp fragment was amplified from genomic DNA with the following primers: 5′-TGTGGTCCATTGGGTGTATC-3′ and 5′-AAGGTCCACAGAAAAGGTC-3′. The variant G allele generates a PsyI (Fisher Scientific, Schwerte, Germany) restriction site that leads to two bands (50 + 60 bp). Genotypes of rs27770 were determined using the primers 5′-CTCCCGCGGTGCCATGTCT-3′ and 5′-CCGAACATGTACAAAAATAACGTA-3′ and the restriction enzyme RsaI (New England Biolabs, Ipswich, MA, USA) which cuts the G allele (87 + 13 bp). For rs16972787 no appropriate allele-specific restriction enzyme was available and it was therefore genotyped by Pyrosequencing. PCR was performed using forward primer 5′-GGTCTCCGCATCCACGCCGG-3′ and biotinylated reverse primer 5′-TCCAAACC-CGCCCGCCGCGC-3′ resulting in a 150 bp fragment. The DNA amplification was carried out using Taq PCR Mastermix (Eppendorf, Hamburg, Germany). The biotinylated strand was captured on streptavidin coated beads, annealed with sequencing primer 5′-CCAGGCTATCCCACGTGTT-3′ and sequenced with a PyroMark Q96 MD (Qiagen, Hilden, Germany). Results were analyzed using the PSQ96 SNP software (Qiagen, Hilden, Germany). Adequate negative and positive controls were used for genotyping of all SNPs. Accuracy of genotyping was additionally validated by direct sequencing of 10% randomly selected samples and of the samples harboring rare haplotypes with a frequency under 1%. This revealed complete concordance with previous results.
Electrophoretic mobility shift assays (EMSA)
Nuclear extracts from HEK293, HeLa and HepG2 cells were prepared using the NuCLEAR™ extraction kit (Sigma, Deisenhofen, Germany) and stored at −80°C until use. EMSAs were done with the DIG Gel Shift kit (Roche Applied Science, Mannheim, Germany) using digoxigenin (DIG)-labeled double-stranded oligonucleotides. The double-stranded oligonucleotides were made of synthesized single-stranded oligonucleotides: rs57973275 G allele 5′-ACTACAGGCTGAGTCTGTGAATCTCC-3′ and 5′-GGAGATTCACAGACTCAGCCTGTAGT-3′, A allele 5′-ACTACAGGCTGAATCTGTGAATCTCC-3′ and 5′- GGAGATTCACAGATTCAGCCTGTAGT-3′; and for rs16972787 G allele 5′-CCACGTGTTCGGGCGTCCGTGTCAAT-3′ and 5′-ATTGACACGGACGCCCGAACACGTGG-3′, A allele 5′-CCACGTGTTCAGGCGTCCGTGTCAAT-3′ and 5′-ATTGACACGGACGCCTGAACACGTGG-3′. Single-stranded oligonucleotides (200 pmol) were mixed in TEN buffer (10 mM Tris, 1 mM EDTA, 0.1 M NaCl, pH 8.0), incubated at 95°C for 10 min and chilled on ice to let oligonucleotides anneal. Double-stranded oligonucleotides (3.85 pmol) were DIG labeled and equal labelling efficiency was verified by dot blot analysis. Probes were incubated with 10 μg nuclear extracts for 20 min at room temperature followed by non-denaturating 6% polyacrylamide gel electrophoresis with 0.5-fold TBE running buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.0). Controls contained labeled probe alone and competition experiments were performed with an additional 250-fold molar excess of unlabeled probe. EMSAs were performed in triplicate for every cell line and both polymorphisms. DNA-protein complexes were electroblotted to positively charged nylon membranes (Roche, Mannheim, Germany) and the band shifts were visualized according to the user’s manual for the DIG Gel Shift kit.
Transient transfection of HEK293 cells and luciferase reporter assay
The 3′UTR of PLK1 was PCR-amplified from genomic DNA and cloned in the pGEM-T Easy Vector (Promega, Madison, WI, USA). The amplified 3′UTR was restricted from the pGEM-T Vector and cloned downstream of the Firefly luciferase coding region into the pMIR-REPORT™ vector (Applied Biosystems, Foster City, CA). HEK293 cells were plated into 96-well plates at a density of 1,5 × 104 cells/well in 100 μl of DMEM medium with 10% FBS. After 24 h co-transfections were carried out in 50 μl of DMEM medium without serum using 150 ng of the respective pMIR reporter construct and 50 ng of Renilla luciferase control vector (pGL4.74, Promega, Madison, WI, USA) containing 0.5 μl of Lipofectamine 2000 (Invitrogen, Karlsruhe, Germany) per transfection according to the manufacturer’s instructions. After 6 h, the transfection mix was removed, and cells were incubated with new DMEM medium. 24 hours after treatment, cells were harvested and assayed for Firefly and Renilla luciferase activities using the Dual-Glo Luciferase Assay System (Promega, Madison, WI, USA) on a Lumat LB 9501 Luminometer (Berthold, Bad Wildbad, Germany). To correct for variable transfection efficiency Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Control for deviation from the Hardy–Weinberg equilibrium was conducted with a web-tool by Rodriguez et al. [65]. Linkage disequilibrium and haplotypes were assessed using Haploview [64]. One-way ANOVA was used to analyze the overall difference of reporter activities. To correct for multiple comparisons Holm-Sidak’s post-hoc multiple comparisons test was used for pairwise comparisons of reporter activities. All statistical analyses were performed using GraphPad Prism 6.0 (GraphPad Software, San Diego, CA, USA). Differences were regarded as significant at p < 0.05.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
All authors read and approved the final manuscript. KR, JW, WS and HSB contributed to conception and design of the study. NA, KR, JW, JH and HSB performed in silico analyses and functional assays. NA, WS and HSB contributed to data analysis and interpretation. NA, JH, WS and HSB drafted and revised the manuscript.
Contributor Information
Neval Akdeli, Email: neval.akdeli@gmx.de.
Kathrin Riemann, Email: kathrinriemann@gmx.de.
Jana Westphal, Email: janavogler@gmx.de.
Jochen Hess, Email: jochen.hess@uk-essen.de.
Winfried Siffert, Email: winfried.siffert@uk-essen.de.
Hagen S Bachmann, Email: hagen.bachmann@uk-essen.de.
References
- Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ji P, Liu J, Broaddus RR, Xue F, Zhang W. Centrosome-associated regulators of the G(2)/M checkpoint as targets for cancer therapy. Mol Cancer. 2009;8:8. doi: 10.1186/1476-4598-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Shao H, Huang Y, Yan F, Chu Y, Hou H, Zhu M, Fu C, Aikhionbare F, Fang G, Ding X, Yao X. PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J Biol Chem. 2011;286:3033–3046. doi: 10.1074/jbc.M110.165340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Kiyono T, Tomono Y, Kawajiri A, Urano T, Furukawa K, Nigg EA, Inagaki M. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat Cell Biol. 2006;8:180–187. doi: 10.1038/ncb1350. [DOI] [PubMed] [Google Scholar]
- Ando K, Ozaki T, Yamamoto H, Furuya K, Hosoda M, Hayashi S, Fukuzawa M, Nakagawara A. Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J Biol Chem. 2004;279:25549–25561. doi: 10.1074/jbc.M314182200. [DOI] [PubMed] [Google Scholar]
- Saxena M, Singh S, Negi MP, Srivastava AK, Trivedi R, Singh U, Pant MC, Bhatt ML. Expression profiling of G2/M phase regulatory proteins in normal, premalignant and malignant uterine cervix and their correlation with survival of patients. J Cancer Res Ther. 2010;6:167–171. doi: 10.4103/0973-1482.65242. [DOI] [PubMed] [Google Scholar]
- Winkles JA, Alberts GF. Differential regulation of polo-like kinase 1, 2, 3, and 4 gene expression in mammalian cells and tissues. Oncogene. 2005;24:260–266. doi: 10.1038/sj.onc.1208219. [DOI] [PubMed] [Google Scholar]
- Holtrich U, Wolf G, Brauninger A, Karn T, Bohme B, Rubsamen-Waigmann H, Strebhardt K. Induction and down-regulation of PLK, a human serine/threonine kinase expressed in proliferating cells and tumors. Proc Natl Acad Sci U S A. 1994;91:1736–1740. doi: 10.1073/pnas.91.5.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He ZL, Zheng H, Lin H, Miao XY, Zhong DW. Overexpression of polo-like kinase1 predicts a poor prognosis in hepatocellular carcinoma patients. World J Gastroenterol. 2009;15:4177–4182. doi: 10.3748/wjg.15.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieche I, Vacher S, Lallemand F, Tozlu-Kara S, Bennani H, Beuzelin M, Driouch K, Rouleau E, Lerebours F, Ripoche H, Cizeron-Clairac G, Spyratos F, Lidereau R. Expression analysis of mitotic spindle checkpoint genes in breast carcinoma: role of NDC80/HEC1 in early breast tumorigenicity, and a two-gene signature for aneuploidy. Mol Cancer. 2011;10:23. doi: 10.1186/1476-4598-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Matsuyama H, Kawauchi S, Matsumoto H, Nagao K, Ohmi C, Sakano S, Furuya T, Oga A, Naito K, Sasaki K. Overexpression of polo-like kinase 1 (PLK1) and chromosomal instability in bladder cancer. Oncology. 2006;70:231–237. doi: 10.1159/000094416. [DOI] [PubMed] [Google Scholar]
- Pellegrino R, Calvisi DF, Ladu S, Ehemann V, Staniscia T, Evert M, Dombrowski F, Schirmacher P, Longerich T. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology. 2010;51:857–868. doi: 10.1002/hep.23467. [DOI] [PubMed] [Google Scholar]
- Weichert W, Kristiansen G, Winzer KJ, Schmidt M, Gekeler V, Noske A, Muller BM, Niesporek S, Dietel M, Denkert C. Polo-like kinase isoforms in breast cancer: expression patterns and prognostic implications. Virchows Arch. 2005;446:442–450. doi: 10.1007/s00428-005-1212-8. [DOI] [PubMed] [Google Scholar]
- Kanaji S, Saito H, Tsujitani S, Matsumoto S, Tatebe S, Kondo A, Ozaki M, Ito H, Ikeguchi M. Expression of polo-like kinase 1 (PLK1) protein predicts the survival of patients with gastric carcinoma. Oncology. 2006;70:126–133. doi: 10.1159/000093003. [DOI] [PubMed] [Google Scholar]
- Wang R, Song Y, Xu X, Wu Q, Liu C. The expression of Nek7, FoxM1, and Plk1 in gallbladder cancer and their relationships to clinicopathologic features and survival. Clin Transl Oncol. 2013;15:626–632. doi: 10.1007/s12094-012-0978-9. [DOI] [PubMed] [Google Scholar]
- Garuti L, Roberti M, Bottegoni G. Polo-like kinases inhibitors. Curr Med Chem. 2012;19:3937–3948. doi: 10.2174/092986712802002455. [DOI] [PubMed] [Google Scholar]
- Korzeniewski N, Hohenfellner M, Duensing S. The centrosome as potential target for cancer therapy and prevention. Expert Opin Ther Targets. 2013;17:43–52. doi: 10.1517/14728222.2013.731396. [DOI] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay FJ, McEwen SJ, Bertoncello I, Wilks AF, Dunn AR. Identification and cloning of a protein kinase-encoding mouse gene, Plk, related to the polo gene of Drosophila. Proc Natl Acad Sci U S A. 1993;90:4882–4886. doi: 10.1073/pnas.90.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka R, Maloid S, Smith MR, O’Connell CD, Longo DL, Ferris DK. Cloning and characterization of human and murine homologues of the Drosophila polo serine-threonine kinase. Cell Growth Differ. 1994;5:249–257. [PubMed] [Google Scholar]
- Brauninger A, Strebhardt K, Rubsamen-Waigmann H. Identification and functional characterization of the human and murine polo-like kinase (Plk) promoter. Oncogene. 1995;11:1793–1800. [PubMed] [Google Scholar]
- Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3:391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. Silent polymorphisms speak: how they affect pharmacogenomics and the treatment of cancer. Cancer Res. 2007;67:9609–9612. doi: 10.1158/0008-5472.CAN-07-2377. [DOI] [PubMed] [Google Scholar]
- Ge B, Gurd S, Gaudin T, Dore C, Lepage P, Harmsen E, Hudson TJ, Pastinen T. Survey of allelic expression using EST mining. Genome Res. 2005;15:1584–1591. doi: 10.1101/gr.4023805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew AS, Gui J, Sanderson AC, Mason RA, Morlock EV, Schned AR, Kelsey KT, Marsit CJ, Moore JH, Karagas MR. Bladder cancer SNP panel predicts susceptibility and survival. Hum Genet. 2009;125:527–539. doi: 10.1007/s00439-009-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- Wheeler HE, Maitland ML, Dolan ME, Cox NJ, Ratain MJ. Cancer pharmacogenomics: strategies and challenges. Nat Rev Genet. 2013;14:23–34. doi: 10.1038/nrg3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliepaard M, Nofziger C, Papaluca M, Zineh I, Uyama Y, Prasad K, Grimstein C, Pacanowski M, Ehmann F, Dossena S, Paulmichl M. Pharmacogenetics in the evaluation of new drugs: a multiregional regulatory perspective. Nat Rev Drug Discov. 2013;12:103–115. doi: 10.1038/nrd3931. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 2002;2:S1–15. [PubMed] [Google Scholar]
- Lan R, Lin G, Yin F, Xu J, Zhang X, Wang J, Wang Y, Gong J, Ding YH, Yang Z, Lu F, Zhang H. Dissecting the phenotypes of Plk1 inhibition in cancer cells using novel kinase inhibitory chemical CBB2001. Lab Invest. 2012;92:1503–1514. doi: 10.1038/labinvest.2012.114. [DOI] [PubMed] [Google Scholar]
- Vitour D, Dabo S, Ahmadi PM, Vilasco M, Vidalain PO, Jacob Y, Mezel-Lemoine M, Paz S, Arguello M, Lin R, Yang J, Chen L, Zhao X, Wei Y. Polo-like kinase 1 (PLK1) regulates interferon (IFN) induction by MAVS. J Biol Chem. 2009;284:21797–21809. doi: 10.1074/jbc.M109.018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Jiang Q, Yang Y, Zhang S, Ma Y, Xie G, Chen X, Qian Z, Wen Y, Li J, Yang J, Chen L, Zhao X, Wei Y. Intravenous liposomal delivery of the short hairpin RNAs against Plk1 controls the growth of established human hepatocellular carcinoma. Cancer Biol Ther. 2011;11:401–409. doi: 10.4161/cbt.11.4.14178. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14(Suppl 2):S4. doi: 10.1186/1471-2105-14-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stram DO. Tag SNP selection for association studies. Genet Epidemiol. 2004;27:365–374. doi: 10.1002/gepi.20028. [DOI] [PubMed] [Google Scholar]
- Garner C, Slatkin M. On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci. Genet Epidemiol. 2003;24:57–67. doi: 10.1002/gepi.10217. [DOI] [PubMed] [Google Scholar]
- Chorley BN, Wang X, Campbell MR, Pittman GS, Noureddine MA, Bell DA. Discovery and verification of functional single nucleotide polymorphisms in regulatory genomic regions: current and developing technologies. Mutat Res. 2008;659:147–157. doi: 10.1016/j.mrrev.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S. Eukaryotic transcription factor binding sites–modeling and integrative search methods. Bioinformatics. 2008;24:1325–1331. doi: 10.1093/bioinformatics/btn198. [DOI] [PubMed] [Google Scholar]
- Buckland PR, Hoogendoorn B, Coleman SL, Guy CA, Smith SK, O’Donovan MC. Strong bias in the location of functional promoter polymorphisms. Hum Mutat. 2005;26:214–223. doi: 10.1002/humu.20207. [DOI] [PubMed] [Google Scholar]
- Benetatos L, Dasoula A, Hatzimichael E, Syed N, Voukelatou M, Dranitsaris G, Bourantas KL, Crook T. Polo-like kinase 2 (SNK/PLK2) is a novel epigenetically regulated gene in acute myeloid leukemia and myelodysplastic syndromes: genetic and epigenetic interactions. Ann Hematol. 2011;90:1037–1045. doi: 10.1007/s00277-011-1193-4. [DOI] [PubMed] [Google Scholar]
- Syed N, Coley HM, Sehouli J, Koensgen D, Mustea A, Szlosarek P, McNeish I, Blagden SP, Schmid P, Lovell DP, Hatzimichael E, Crook T. Polo-like kinase Plk2 is an epigenetic determinant of chemosensitivity and clinical outcomes in ovarian cancer. Cancer Res. 2011;71:3317–3327. doi: 10.1158/0008-5472.CAN-10-2048. [DOI] [PubMed] [Google Scholar]
- Kelly TK, Miranda TB, Liang G, Berman BP, Lin JC, Tanay A, Jones PA. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell. 2010;39:901–911. doi: 10.1016/j.molcel.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Song Y, Bi N, Shen J, Liu W, Fan J, Sun G, Tong T, He J, Shi Y, Zhang X, Lu N, He Y, Zhang H, Ma K, Luo X, Lv L, Deng H, Cheng J, Zhu J, Wang L, Zhan Q. DNA methylation-mediated repression of miR-886-3p predicts poor outcome of human small cell lung cancer. Cancer Res. 2013;73:3326–3335. doi: 10.1158/0008-5472.CAN-12-3055. [DOI] [PubMed] [Google Scholar]
- He J, Wu J, Xu N, Xie W, Li M, Li J, Jiang Y, Yang BB, Zhang Y. MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucleic Acids Res. 2013;41:498–508. doi: 10.1093/nar/gks995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lu KH, Liu ZL, Sun M, De W, Wang ZX. MicroRNA-100 is a potential molecular marker of non-small cell lung cancer and functions as a tumor suppressor by targeting polo-like kinase 1. BMC Cancer. 2012;12:519. doi: 10.1186/1471-2407-12-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagioni F, Bossel Ben-Moshe N, Fontemaggi G, Canu V, Mori F, Antoniani B, Di Benedetto A, Santoro R, Germoni S, De Angelis F, Cambria A, Avraham R, Grasso G, Strano S, Muti P, Mottolese M, Yarden Y, Domany E, Blandino G. miR-10b*, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol Med. 2012;4:1214–1229. doi: 10.1002/emmm.201201483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Sato F, Kan T, Cheng Y, David S, Agarwal R, Paun BC, Jin Z, Olaru AV, Hamilton JP, Selaru FM, Yang J, Matsumura N, Shimizu K, Abraham JM, Shimada Y, Mori Y, Meltzer SJ. Polo-like kinase 1 regulates cell proliferation and is targeted by miR-593* in esophageal cancer. Int J Cancer. 2011;129:2134–2146. doi: 10.1002/ijc.25874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Alajez NM, Bastianutto C, Hui AB, Mocanu JD, Ito E, Busson P, Lo KW, Ng R, Waldron J, O’Sullivan B, Liu FF. Significance of Plk1 regulation by miR-100 in human nasopharyngeal cancer. Int J Cancer. 2010;126:2036–2048. doi: 10.1002/ijc.24880. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Abrahams Y, Laguette MJ, Prince S, Collins M. Polymorphisms within the COL5A1 3′-UTR that alters mRNA structure and the MIR608 gene are associated with Achilles tendinopathy. Ann Hum Genet. 2013;77:204–214. doi: 10.1111/ahg.12013. [DOI] [PubMed] [Google Scholar]
- Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci U S A. 2007;104:13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Quon G, Lipshitz HD, Morris Q. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA. 2010;16:1096–1107. doi: 10.1261/rna.2017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheneval D, Kastelic T, Fuerst P, Parker CN. A review of methods to monitor the modulation of mRNA stability: a novel approach to drug discovery and therapeutic intervention. J Biomol Screen. 2010;15:609–622. doi: 10.1177/1087057110365897. [DOI] [PubMed] [Google Scholar]
- Fu J, Wolfs MG, Deelen P, Westra HJ, Fehrmann RS, Te Meerman GJ, Buurman WA, Rensen SS, Groen HJ, Weersma RK, van den Berg LH, Veldink J, Ophoff RA, Snieder H, van Heel D, Jansen RC, Hofker MH, Wijmenga C, Franke L. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira JC, Pezuk JA, Brassesco MS, Morales AG, Queiroz RG, Scrideli CA, Tone LG. PLK1 expression and BI 2536 effects in childhood acute lymphoblastic leukemia. Pediatr Blood Cancer. In press. [DOI] [PubMed]
- Chen J, Zhang W. Kinetic analysis of the effects of target structure on siRNA efficiency. J Chem Phys. 2012;137:225102. doi: 10.1063/1.4769821. [DOI] [PubMed] [Google Scholar]
- Shao Y, Chan CY, Maliyekkel A, Lawrence CE, Roninson IB, Ding Y. Effect of target secondary structure on RNAi efficiency. RNA. 2007;13:1631–1640. doi: 10.1261/rna.546207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan RK, Hamburg SI, Borad MJ, Seetharam M, Kundranda MN, Lee P, Fredlund P, Gilbert M, Mast C, Semple SC, Judge AD, Crowell B, Vocila L, MacLachlan I, Northfelt DW. A phase I dose escalation study of TKM-080301, aRNAi therapeutic directed against PLK1, in patients with advanced solid tumors. Proc 104th Annu Meet AACR. 2013;2013:LB-289. [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505–514. doi: 10.1093/aje/kwn359. [DOI] [PMC free article] [PubMed] [Google Scholar]