Abstract
We report that breast cancer cells that infiltrate the lungs support their own metastasis-initiating ability by expressing tenascin C (TNC). We find that the expression of TNC, an extracellular matrix protein of stem cell niches, is associated with the aggressiveness of pulmonary metastasis. Cancer cell–derived TNC promotes the survival and outgrowth of pulmonary micrometastases. TNC enhances the expression of stem cell signaling components, musashi homolog 1 (MSI1) and leucine-rich repeat–containing G protein– coupled receptor 5 (LGR5). MSI1 is a positive regulator of NOTCH signaling, whereas LGR5 is a target gene of the WNT pathway. TNC modulation of stem cell signaling occurs without affecting the expression of transcriptional enforcers of the stem cell phenotype and pluripotency, namely nanog homeobox (NANOG), POU class 5 homeobox 1 (POU5F1), also known as OCT4, and SRY-box 2 (SOX2). TNC protects MSI1-dependent NOTCH signaling from inhibition by signal transducer and activator of transcription 5 (STAT5), and selectively enhances the expression of LGR5 as a WNT target gene. Cancer cell– derived TNC remains essential for metastasis outgrowth until the tumor stroma takes over as a source of TNC. These findings link TNC to pathways that support the fitness of metastasis-initiating breast cancer cells and highlight the relevance of TNC as an extracellular matrix component of the metastatic niche.
Many malignant tumors start releasing cancer cells into the circulation long before diagnosis1. To create a disseminated population that may eventually progress to overt metastasis, circulating cancer cells must first traverse endothelial capillary walls and then cope with the newly invaded parenchyma. At the distant site, cancer cells have to remain viable as metastasis-initiating entities to eventually grow out as overt metastatic lesions2. Specialized microenvironments called metastatic niches are thought to nurse the outgrowth of metastatic nodules from metastasis-initiating cells3. The niche components that support these functions remain mostly unknown, but the extracellular matrix may be relevant in this context. The extracellular matrix has a crucial role in developmental patterning, tissue organization and stem cell niches, and its composition is characteristically altered in tumors4, 5.
With a median survival of <2 years after diagnosis, lung metastasis from breast cancer remains a major clinical challenge6. We have identified a set of genes whose expression in breast tumors is associated with lung relapse7, 8. Several of these genes encode cytokines and other secreted products that enhance the transendothelial migration of breast cancer cells and their passage from circulation into the lung parenchyma9, 10. Another member of this lung metastasis gene set is TNC7. We find the possible involvement of TNC in metastasis intriguing, given its properties as an extracellular matrix protein. TNC is a hexameric glycoprotein that binds to fibronectin, periostin11, integrin cell adhesion receptors and syndecan membrane proteoglycans12. TNC is abundantly expressed in developing bone and cartilage, and in neural crest cells. In adults, TNC is present in stem cell niche regions and at sites of epithelial-mesenchymal interaction. TNC expression is elevated by mechanical stress and inflammation during wound healing and in tumor-associated connective tissue12, 13. On the basis of these clues, we wondered whether cancer cells expressing TNC have a selective advantage in the lung microenvironment. Here we provide evidence for such a role, and uncover new links of TNC to the critical stem and progenitor cell pathways NOTCH and WNT14, 15, and to the viability of metastatic cancer cells in the lungs.
RESULTS
TNC expression associates with aggressive lung metastasis
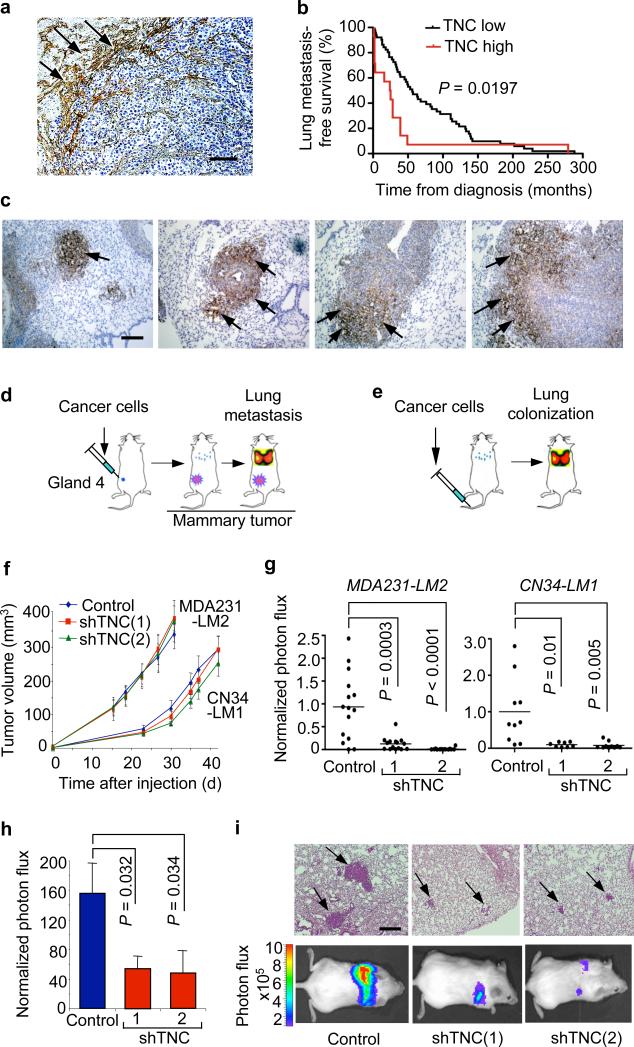
TNC is among a set of genes whose mRNA level in breast tumors is associated with relapse in the lungs7. To investigate this association, we analyzed TNC by immunostaining in human breast cancer tissue samples. In lung nodules, TNC staining was particularly evident at the invasive front (Fig. 1a). In lung metastases and in primary tumor samples, a high level of TNC staining was associated with a shorter progression to lung relapse (Fig. 1b and Supplementary Fig. 1). The median time from primary tumor diagnosis to lung metastasis was 24 months in cases with high expression of TNC in metastatic nodules versus 56 months in cases with low TNC (Fig. 1b). Moreover, high TNC expression in lung metastatic nodules predicted poor overall survival, as the median time from metastasis diagnosis to death was 7 months in cases with high TNC expression and 34 months for cases with low TNC.
Figure 1. TNC expression in lung metastatic foci and association with lung relapse.
(a) Heterogeneous TNC expression in human lung metastasis. TNC immunostaining (arrows) on lung metastasis section from an individual with breast cancer. Scale bar, 50 μm. (b) Kaplan-Meier analysis of lung metastasis–free survival in subjects with breast cancer, on the basis of TNC expression in dissected lung metastases. TNC immunostaining of tissue sections was used to classify metastases into two groups, TNC low (n = 51) or TNC high (n = 15). P value was determined by log-rank test. (c) Immunohistochemical analysis of TNC expression in lung metastatic foci of various sizes formed by MDA231-LM2 cells in mice. TNC accumulation at the invasive front in larger metastatic foci. Arrows, TNC expression. Scale bar, 50 μm. (d) Schematic of orthotopic metastasis assay in mice. MDA231-LM2 or CN34-LM1 cells were injected bilaterally into the fourth mammary fat pads leading to spontaneous lung metastasis. (e) Schematic of lung colonization assay by intravenous injection of cancer cells. (f) Growth curves of mammary tumors arising from MDA231-LM2 or CN34-LM1 cells and their TNC knockdown counterparts. Mammary tumor growth was measured with a digital caliper. Values are means ± s.e.m. (g) Lung metastasis in mammary tumor–bearing mice determined at week 6 by ex vivo luciferase luminescence. MDA231-LM2 control, n = 15; shTNC(1), n = 15; shTNC(2), n = 14. CN34-LM1 control, n = 10; shTNC(1), n = 8; shTNC(2), n = 10. P values were obtained by two-tailed Student's t test. (h) Lung colonization, as determined by bioluminescence, in mice injected intravenously with control and shTNC-transduced CN34LM1 cells . Values are mean ± s.e.m. P values were determined by Student's t test. (i) Representative luminescence images and H&E-stained lung sections from each group. Arrows, metastatic lesions. Scale bar, 100 μm.
Prompted by these results, we investigated the functional role of TNC in experimental models of breast cancer metastasis to lung. We used two breast cancer cell lines, MDA231-LM2 and CN34-LM1, which have high capacity to colonize the lungs in mice7, 8. The corresponding parental cell lines, MDA-MB-231 and CN34, were derived from malignant pleural fluids of patients with advanced metastatic disease. MDA231-LM2 and CN34-LM1 had greater TNC expression than their parental counterparts (Supplementary Fig. 2) and readily colonized the lungs when inoculated into the tail vein of immunodeficient mice (Fig. 1c). The resulting colonies showed a marked pattern of cancer cell–derived (human) TNC deposition, as determined by immunostaining with antibodies to human TNC (Fig. 1c and Supplementary Fig. 3). TNC was uniformly distributed throughout the micrometastases in early stages of lung seeding (Fig. 1c). As these colonies grew, cancer cell–derived TNC became progressively segregated to the invasive front (Fig. 1c), similar to its distribution in human metastasis nodules (Fig. 1a). Another lung metastasis gene product, fascin-1 (refs. 7,10), remained expressed throughout the metastatic nodules (Supplementary Fig. 4a), indicating that the absence of TNC expression outside the invasive front was not due to a general extinction of metastatic gene expression. The invasive front hosts presumptive cancer stem cells and dynamic interactions with mesenchymal, vascular and inflammatory components of the tumor stroma16, 17. Indeed, the TNC-positive areas in macrometastases were infiltrated with myofibroblasts (α-smooth muscle actin–staining cells; Supplementary Fig. 4b) and macrophages (CD68+ cells; Supplementary Fig. 4c).
Autocrine TNC initiates micrometastasis outgrowth
When we used shRNAs against TNC, the expression of TNC was reduced by about 90% in MDA231-LM2 and CN34-LM1 cells (Supplementary Fig. 5a). Control and TNC knockdown cells were injected orthotopically into the mammary fat pad (Fig. 1d) or directly to the lungs through the lateral tail vein (Fig. 1e). Reduced TNC expression did not diminish the ability of the cancer cells to grow as tumors when 5 × 105 cells were implanted in the mammary fat pad of female mice (Fig. 1f). However, TNC deficiency resulted in 90% inhibition of mammary tumors to metastasize to the lungs (Fig. 1g). TNC knockdown similarly caused a statistically significant decrease in lung colonization by these cells inoculated via the tail vein, as determined by bioluminescence imaging of a marker luciferase gene, and by H&E staining (Fig. 1h,i and Supplementary Fig. 5b,c).
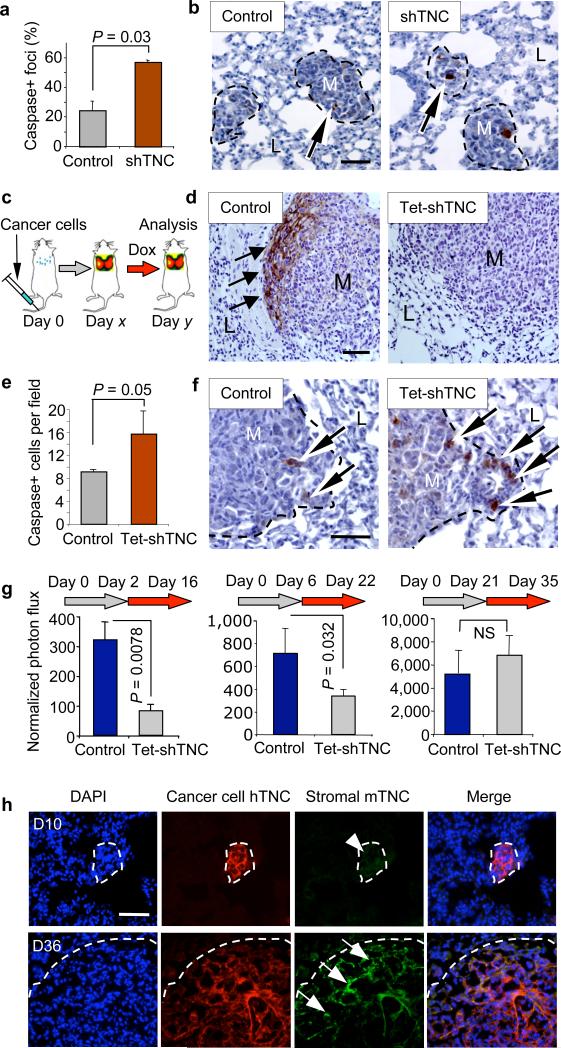
TNC influences cell migration in vitro18, and its knockdown in the breast cancer cells moderately decreased the cells' ability to migrate through membranes and matrigel (Supplementary Fig. 6 and ref. 8). This decrease was, however, much smaller than the 90% decrease in metastasis observed in vivo. Therefore, we investigated the effect of TNC knockdown on metastatic cell proliferation and survival. At 10 d after cancer cell inoculation into mice, the lung micrometastases showed no statistically significant effect of TNC knockdown on the proliferation marker phosphohistone H3 (Supplementary Fig. 7a,b). However, the TNC-depleted colonies showed a marked increase in the apoptosis marker, cleaved caspase 3 (Fig. 2a,b).
Figure 2. Cancer cell–derived TNC mediates resistance to apoptosis in micrometastasis.
(a,b) Apoptosis in early metastatic foci analyzed by immunohistochemistry. Average number (a) and representative images (b) of MDA231-LM2 apoptotic foci containing cleaved caspase 3+ cells at day 10 of lung colonization. n = 3 mice per condition; values are mean ± s.e.m. Arrows, apoptotic cells. M, metastatic foci. L, lung parenchyma. Scale bar, 20 μm. (c) Schematic of conditional TNC knockdown experiment. Lung colonization of MDA231-LM2 cells containing doxycycline (Dox)-inducible TNC shRNA (Tet-shTNC) was allowed for different lengths of time (day x) before TNC knockdown was induced by administration of doxycycline to the mice. Lung colonies were then analyzed (day y). (d) TNC staining in lung metastatic foci in control or Tet-shTNC MDA231-LM2. Cells were injected intravenously and allowed to colonize the lungs for 25 d. Thereafter, TNC knockdown was induced with doxycycline for 10 d, and lung sections were analyzed by TNC immunohistochemistry (arrows). Scale bar, 20 μm. (e) Analysis of apoptosis in established lung foci. Cleaved caspase 3 was quantified in control and Tet-shTNC (day 25 of lung colonization) mice after doxycycline administration for an additional 10 d. n = 3 mice per condition; values are mean ± s.e.m. (f) Representative examples of apoptotic cells at the invasive edge of established metastatic foci. Arrows, cleaved caspase 3+ cells. Scale bar, 20 μm. (g) Bioluminescence analysis of MDA231-LM2–mediated lung colonization in which TNC was conditionally knocked down at different time points. Left, TNC knocked down by 2-week-long doxycycline treatment starting 2 d after tail vein injection. Middle, doxycycline treatment at day 6. Right, doxycycline induction at day 21, maintained for 2 weeks. Two-tailed Student's t test was used to determine all P values. (h) Immunofluorescence analysis of TNC expression in MDA231-LM2 metastatic foci at different time points during lung colonization. Time points analyzed were days 10 and 36 (D10 and D36, respectively) after intravenous inoculation. Antibodies to stromal derived mouse TNC (mTNC) and tumor cell–derived human TNC (hTNC) were used. DAPI was used for nuclear staining. Scale bar, 20 μm.
To temporally map the effect of cancer cell–derived TNC during the course of lung colonization, we generated an MDA231-LM2 line that expresses TNC shRNA under the conditional control of doxycycline (Supplementary Fig. 8). We intravenously inoculated these cells and allowed them to reside in the lungs for different lengths of time before administering doxycycline to suppress TNC expression (Fig. 2c). A 10 d treatment with doxycycline caused an almost complete clearing of human TNC from established lung nodules (Fig. 2d). This was accompanied by an increase in apoptosis marker, cleaved caspase 3, at the invasive front (Fig. 2e,f). The administration of doxycycline starting 2 d or 6 d after cancer cell inoculation inhibited the outgrowth of the incipient micrometastases (Fig. 2g). When treatment was started later, doxycycline was less effective at curbing metastasis (Supplementary Fig. 9a). Administration of doxycycline at day 21, when the metastatic nodules had already become large, had no effect on further increase of the metastatic load (Fig. 2g). However, stromal myofibroblasts at the invasive front of large nodules (Supplementary Fig. 4b) produce TNC19. Indeed, large metastatic nodules stained positive for both cancer cell–derived (human) and stromal (mouse) TNC, whereas the micrometastases expressed only cancer cell–derived TNC (Fig. 2h). In agreement with this, α-smooth muscle actin, a marker of activated fibroblasts (myofibroblasts) was expressed in late metastatic foci but not in micrometastases (Supplementary Fig. 9b). Therefore, cancer cells in micrometastases act as their own main source of TNC until the tumor stroma takes over as a source of TNC.
TNC expression and metastasis-initiating characteristics
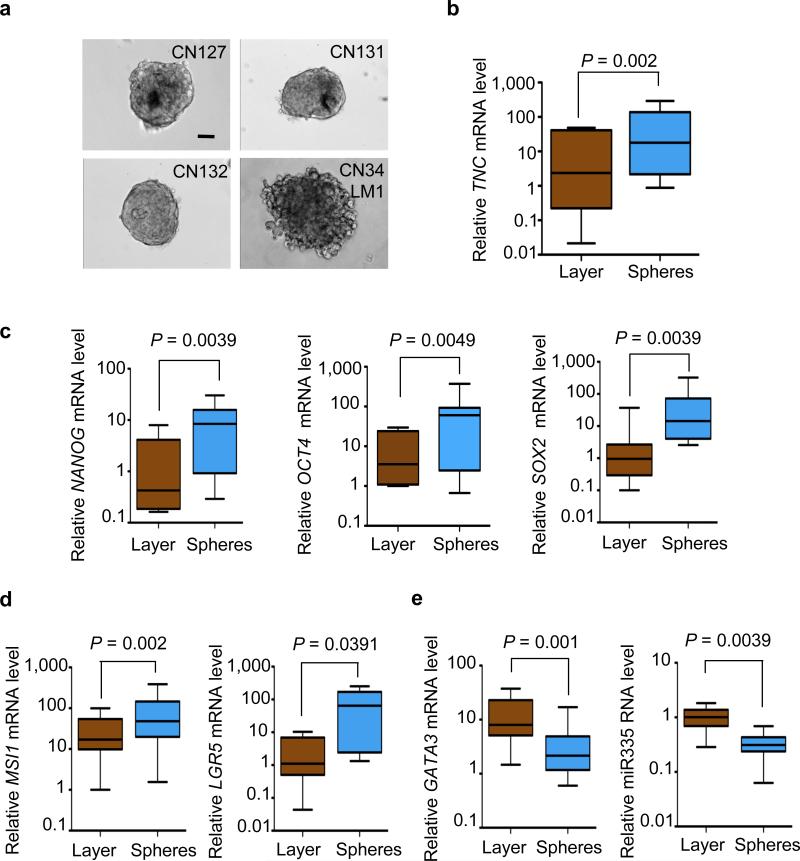
TNC expression has been detected in certain stem cell niches20, 21 and is abundantly expressed by mammary stem and progenitor cell populations22. Pluripotent mammary epithelial cells and metastasis-initiating breast cancer cell populations can grow as floating aggregates, called mammospheres and oncospheres, respectively, when placed on nonadhesive plates in serum-free conditions22, 23. We investigated the link between TNC and oncosphere formation in breast cancer cells from malignant pleural effusions obtained from human subjects. Both the recently isolated breast cancer cells and the CN34-LM1 cell line readily formed oncospheres under these culture conditions (Fig. 3a). Notably, TNC was upregulated in the oncospheres compared to monolayer cultures of the same samples (Fig. 3b), and was further upregulated after a cycle of oncosphere dissociation and regrowth (Supplementary Fig. 10a). The pluripotency markers NANOG24, OCT4 (refs. 25,26) and SOX2 (ref. 27) were upregulated in the oncospheres, and so were the adult stem cell markers MSI1 (also known as Musashi1)28 and LGR5 (ref. 29; Fig. 3c,d). Moreover, the mammary differentiation marker GATA3 (ref. 30) and the TNC-suppressor and metastasis-suppressor microRNA (miRNA) miR-335 (ref. 8) were expressed at low levels in oncospheres compared to monolayers (Fig. 3e). Thus, TNC expression was associated with progenitor markers in metastatic breast cancer cell populations.
Figure 3. Expression of TNC and stem cell markers in oncospheres.
(a) Examples of tumor spheres from various pleural effusion samples from subjects with breast cancer, grown in serum-free medium on nonadhesive plates (scale bar, 20 μm). (b–e) Gene expression analysis of oncospheres derived from pleural effusion samples compared to monolayer cultures of the same samples. Gene expression was analyzed by quantitative RT-PCR (qRT-PCR). TNC expression (b) was upregulated in oncospheres together with the expression of embryonic stem cell markers NANOG, OCT4 and SOX2 (c) and adult stem cell markers MSI1 and LGR5 (d). In contrast, the mammary differentiation marker GATA3 and the TNC- and metastasis-suppressor miRNA miR-335 were downregulated in oncospheres (e). n = 9–10 pleural fluid–derived cell samples. All P values were calculated by paired Wilcoxon signed rank test (two tailed). Whiskers represent minimum and maximum values.
Regulation of oncosphere growth MSI1 and LGR5 by TNC
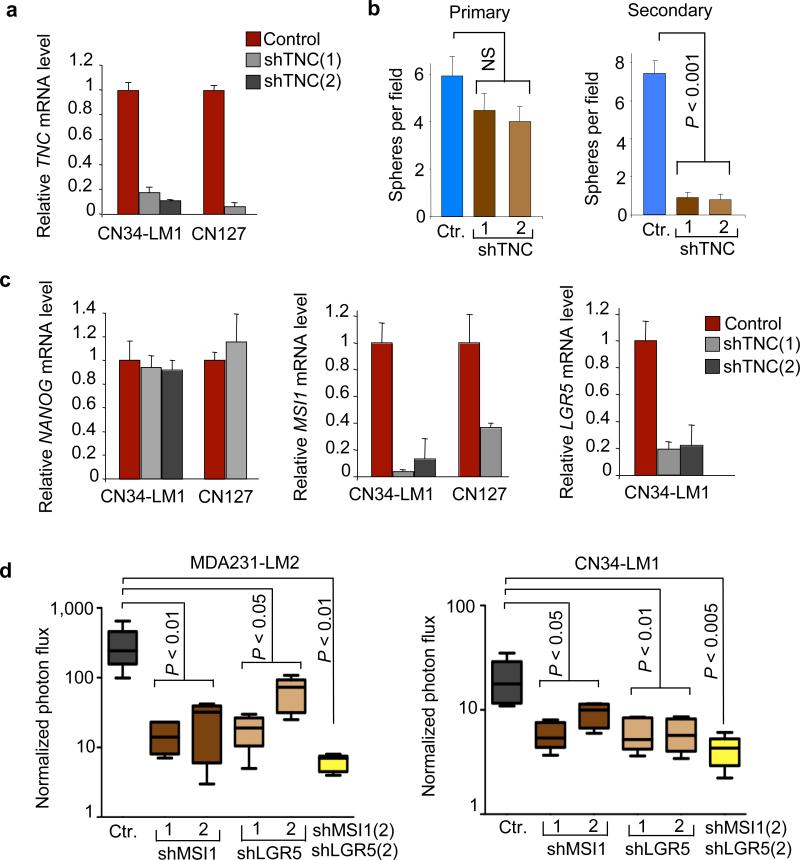
To test the functional implications of this link, we knocked down TNC in CN34-LM1 and in a primary culture of metastatic breast cancer cells from another individual with breast cancer, CN127, and generated oncospheres. TNC depletion (Fig. 4a) did not inhibit the formation of primary oncospheres but severely inhibited the formation of secondary oncospheres under standard culture conditions, and of primary oncospheres under growth factor–deprived conditions (Fig. 4b and Supplementary Fig. 10b). This suggests that TNC is required for the fitness of oncosphere initiating cells, particularly under stress.
Figure 4. Regulation of oncosphere growth MSI1 and LGR5 by TNC.
(a) TNC expression in oncospheres derived from CN34-LM1 cells and CN127 breast cancer cells expressing two independent TNC shRNAs. (b) Quantitative analysis of oncosphere formation in TNC-deficient CN34-LM1 cells. (c) Gene expression analysis in TNC-deficient oncospheres. Expression was analyzed by qRT-PCR; values are mean ± s.d. of triplicate experiments. For all genes, the expression in knockdown lines was normalized to control. (d) Lung colonization of MDA231-LM2 and CN34-LM1 cells transduced with short hairpins against MSI1 and LGR5 individually and in combination. Ctr, control. Student's t test was used to determine P values. n = 5 mice per condition. Whiskers represent minimum and maximum values.
Breast cancer cell populations with a CD44+CD24− surface marker phenotype are enriched in tumor-initiating cells31. In the parental CN34 and MDA231 cell lines and in their lung metastatic derivatives, ~90% of the population is CD44+CD24− (Supplementary Fig. 11 and ref. 32). TNC depletion did not change the CD44+CD24− population ratio in these cells (Supplementary Fig. 11). Moreover, TNC depletion did not affect the expression of NANOG, OCT4 and SOX2 (Fig. 4c and Supplementary Fig. 12). These genes encode transcription factors that are essential for the maintenance of pluripotency in stem cells24, 27. However, TNC depletion strongly suppressed the expression of LGR5 and MSI1 (Fig. 4c). LGR5 (ref. 29) and MSI1 (ref. 28) encode signal transduction components of the WNT and NOTCH pathways, respectively, which provide essential signals to stem and progenitor cells.
Knocking down MSI1 or LGR5 (Supplementary Fig. 13a,b) significantly reduced the lung metastatic efficiency of MDA231-LM2 and CN34-LM1 cells (Fig. 4d and Supplementary Fig. 13c). Moreover, the combined knockdown of both genes led to even further reduction in lung colonization. However, depletion of LGR5 and MSI1 did not alter the CD44+CD24− profile of the cells (Supplementary Fig. 14). Together, these results suggest that TNC is dispensable for the expression of pluripotency transcriptional enforcers in breast cancer cells but is required for expression of stem or progenitor cell signal transducers that support metastasis initiation by these cells.
TNC enhances WNT and NOTCH signaling in breast cancer cells
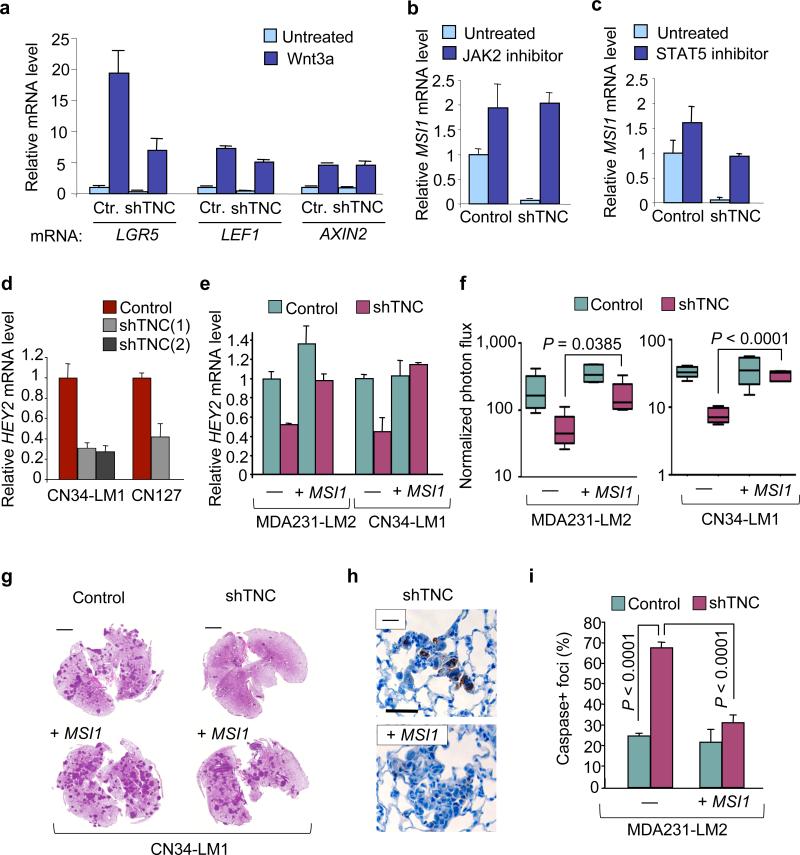
TNC has a complex molecular architecture consisting of a radial hexamer in which each monomer comprises an N-terminal TNC assembly domain, followed by 14 epidermal growth factor (EGF)-like repeats, up to 17 alternatively spliced fibronectin-like repeats and a C-terminal fibrinogen-like domain. The TNC hexabrachion binds to extracellular matrix fibronectin and periostin11, and to integrins, EGF receptors, syndecan-4 and other membrane proteins12, many of which are expressed in the CN34 and MDA231 breast cancer cells (Supplementary Table 1). These interactions have the potential to affect key pathways in the cell. We analyzed LGR5 and MSI1 expression in response to WNT3A in oncospheres. LGR5 is a key target of the WNT–T cell factor (TCF) pathway in adult stem cells29. Whereas addition of WNT3A did not alter the expression of MSI1 (data not shown), it strongly increased the expression of LGR5 and two other typical WNT target genes, those encoding lymphoid enhancer–binding factor 1 (LEF1) and axis inhibition protein 2 (AXIN2), which encode feedback regulators of WNT signaling (Fig. 5a). This effect was blocked by the WNT inhibitor dickkopf homolog 1 (DKK1) recombinant protein (Supplementary Fig. 15). Notably, the knockdown of TNC markedly reduced the response of LGR5 to WNT3A but had little or no effect on the response of LEF1 and AXIN2 (Fig. 5a). These results suggest that TNC specifically modulates a component of the LGR5 response to WNT.
Figure 5. TNC enhances WNT and NOTCH signaling in breast cancer cells.
(a) WNT3A induced LGR5, LEF1 and AXIN2 expression in CN34-LM1 oncospheres. Ctr, control. (b,c) MSI1 expression induced in TNC knockdown CN34-LM1 oncospheres by treatment with JAK2 inhibitor AG490 (b) or STAT5 inhibitor nicotinoyl hydrazone (c). (d) Expression of NOTCH target gene HEY2 in TNC knockdown oncospheres. (e) HEY2 expression in control or TNC knockdown CN34-LM1 and MDA231-LM2 cells transduced with MSI1 expression vector. (f) Lung luminescence in mice intravenously injected with CN34-LM1 or MDA231-LM2 cells. Control and shTNC knockdown cells were transduced with vector expressing MSI1 or control vector. Normalized photon flux was measured 1 week after tail vein injection. n = 5 mice per condition. P values were obtained using a two-tailed Student's t test. Whiskers represent minimum and maximum values. (g) Representative images of H&E-stained lung sections from experiment in f at 6 weeks. (h) Immunohistochemical analysis of cleaved caspase 3 in metastatic foci 10 d after tail vein injection. Representative images from MDA231-LM2 shTNC cells with or without ectopic MSI1 expression. Scale bar, 20 μm. (i) Number of cleaved caspase 3+ foci formed by MDA231-LM2 cells at day 10 of lung colonization. Control and shTNC cells were analyzed with or without overexpression of MSI1. n = 4 mice per condition. P values were obtained by two-tailed Student's t test. Error bars represent standard deviation.
MSI1 is an RNA-binding protein that inhibits translation of the NOTCH pathway inhibitor NUMB33. NOTCH signaling is implicated in the expansion of the luminal progenitor compartment of the mammary epithelium34, which is the presumptive origin of basal breast carcinomas35, 36. In contrast to the role of NOTCH, activation of Janus kinase 2 (JAK2) and STAT5 by prolactin leads to alveolar differentiation and β-casein production37. Notably, TNC inhibits β-casein production in mammary epithelial cells38. On the basis of these clues, we tested the hypothesis that TNC may counteract JAK2-STAT5–dependent inhibition of NOTCH signaling. Treatment with the JAK2 inhibitor AG490 (ref. 39) or the STAT5 inhibitor nicotinoyl hydrazone40 led to upregulation of MSI1 in the oncospheres (Fig. 5b,c). This effect was particularly prominent in TNC knockdown cells in which the proportional increase was significantly higher than in controls. The expression of the NOTCH target genes HEY2, encoding a basic-helix-loop-helix protein that functions as downstream effector of NOTCH signaling and DTX1, encoding an E3-ubiquitin ligase that regulates NOTCH activity (also known as deltex1) in oncospheres (Supplementary Fig. 16) was decreased by TNC depletion (Fig. 5d and Supplementary Fig. 17), and rescued by MSI1 overexpression in TNC-knockdown cells (Fig. 5e and Supplementary Fig. 18). Furthermore, MSI1 expression partially rescued the lung colonization capacity (Fig. 5f,g) and survival (Fig. 5h,i) of TNC-depleted breast cancer cells in the lungs. Thus, TNC suppressed JAK2-STAT5 signaling in breast cancer cells to enhance the expression of the NOTCH pathway component MSI1 and the prometastatic function of this pathway.
Association of TNC with MSI1 in clinical metastasis
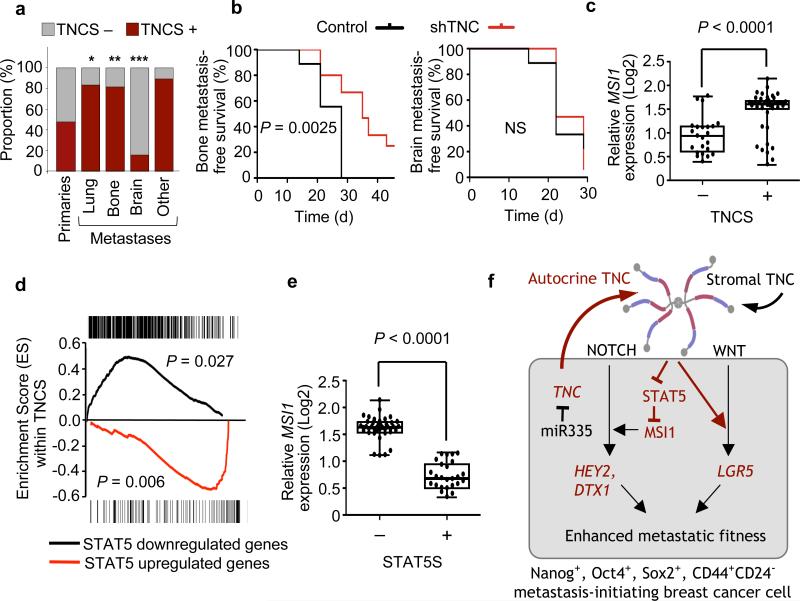
To seek clinical evidence for the links among TNC, STAT5 and MSI1 uncovered in our experimental system, we applied a bioinformatics classifier based on a set of genes whose expression denotes cell stimulation by TNC41. When applied to gene expression data from a large cohort of primary breast tumors, this TNC gene signature (TNCS) classifier separated breast tumors into two groups (Supplementary Fig. 19). When applied to metastatic tissue samples, a majority (>80%) of metastatic samples from lung, bone and other sites scored positive for TNCS, whereas 48% of the primary tumors and only 16% of the brain metastases did (Fig. 6a). This further strengthens the association of TNC with lung metastatic progression in patients, and suggested a role of TNC in metastasis to other sites. Intrigued by these results, we knocked down TNC in bone metastatic (MDA231-BoM1-1833, ref. 42) and brain metastatic (MDA231-BrM2, ref. 43) breast cancer cells (Supplementary Figs. 20a and 21a) and determined their metastatic efficiency. TNC depletion significantly inhibited bone metastasis but not brain metastasis (Fig. 6b and Supplementary Figs. 20b,c and 21b,c).
Figure 6. TNC association with STAT5 and Musashi in breast cancer metastasis.
(a) Proportion of TNCS+ cases in primary breast tumors and metastatic nodules from different organ sites. Primary breast tumors (n = 344), lung metastases (n = 18), bone metastasis (n = 16), brain metastasis (n = 19) and other metastasis (5 liver, 7 ovary, 1 chest wall and 1 duodenum). *P = 0.0033, **P = 0.01, ***P = 0.008, calculated by Fisher's exact tests. (b) Kaplan-Meier analysis of bone metastatic free survival (left) and brain metastatic survival (right) in a xenograft mouse model. MDA231-BoM1 and MDA231-BrM2 transduced with shTNC were injected intracardially and metastasis was determined by bioluminescence. Bone metastasis experiment: control, n = 9; shTNC, n = 15 (pooled samples from two TNC hairpins). Brain metastasis experiment: control, n = 9; shTNC, n = 17 (pooled samples from two TNC hairpins). P values were determined by log-rank test. (c) Relative levels of MSI1 in TNCS+ and TNCS− human metastasis samples. P value was calculated by two tailed Student's t test. (d) GSEA. Analysis of genes either up- or downregulated in the STAT5 signature (STAT5S) and their enrichment within TNCS+ metastases. P values were determined by a random permutation test45. (e) Relative expression of MSI1 within STAT5+ or STAT5− human breast cancer metastases. (f) Summary model of the role of TNC in the outgrowth of micrometastasis. Metastasis-initiating breast cancer cells with low expression of the metastasis suppressor miR-335 highly express TNC, supporting the survival and outgrowth of these cells in the pulmonary parenchyma. TNC interaction with cancer cells at the invasive front enhances signaling by the NOTCH and WNT pathways, promoting the viability of these cells and the reinitiation of metastatic outgrowth.
We applied the TNC signature and compared the level of MSI1 expression in TNCS− versus TNCS+ metastatic samples. MSI1 expression was significantly higher in TNCS+ metastatic samples than in TNCS− metastases (Fig. 6c). We also analyzed the correlation of the TNCS and a STAT5-dependent gene expression signature44 (Supplementary Tables 2–4). Unsupervised clustering of human metastasis samples showed that expression of this STAT5 signature was negatively correlated with expression of the TNCS (Supplementary Fig. 22). This negative correlation was even more apparent when gene set enrichment analysis (GSEA)45 was applied to determine the representation of STAT5 signature genes within the TNCS+ samples. Genes upregulated in the STAT5 signature were significantly (P = 0.006) underrepresented in the TNCS signature, and those downregulated in the signature significantly enriched (P = 0.027) (Fig. 6d). Moreover, MSI1 expression was significantly lower in STAT5 signature–positive human metastases (Fig. 6e). These results are in line with the functional evidence that TNC supports MSI1 expression in breast cancer cells by counteracting STAT5.
DISCUSSION
Our results place TNC on a metastasis-sustaining axis that engages the NOTCH and WNT pathways to support the fitness of initiating breast cancer cells during the establishment of lung metastases (Fig. 6f). Cancer cell–derived TNC provides a head start for the viability of breast cancer cells in this setting, and remains critical until TNC from infiltrating myofibroblasts or other stromal sources accumulates in the expanding nodules. Circulating cancer cells that extravasate into the lungs may thus be selected for expression of TNC as an advantage trait. Cancer cell–derived TNC was not essential for the bulk growth of cells implanted as mammary tumors in our model systems. However, the presence of TNC at the invasive front46 raises the possibility that TNC also supports breast cancer cells that locally migrate away from the bulk tumor mass as it supports cancer cells that reach the lung parenchyma or the bone marrow.
Our evidence suggests that TNC promotes the fitness of metastasis-initiating breast cancer cells by enhancing the expression of LGR5 and MSI1. Through these effects, TNC improves the performance of the NOTCH pathway and selectively enhances the LGR5 response to WNT in cancer cells. However, TNC seems dispensable for the expression of NANOG, OCT4 and SOX2, three transcription factors that are master enforcers of pluripotency and self-renewal in embryonic stem cells24, 25, 26, 27. TNC depletion also had no effect on the CD44+CD24− antigen profile, which is characteristic of breast cancer–initiating cells31. Thus, our evidence suggests that TNC specifically supports NOTCH and WNT signaling functions that are essential for the fitness of metastasis-initiating cells, but does not affect the expression of core transcriptional determinants of the stem cell phenotype. The differential effect of TNC on these two sets of molecular mediators not only sheds light on the contribution of an extracellular matrix component of the metastatic niche but also exemplifies how metastasis-initiating fitness and the metastasis-initiator phenotype are different, albeit interconnected, functions.
The complex molecular architecture of TNC and its ability to bind multiple types of cell adhesion receptors are thought to provide a coordinated set of environmental cues to cells in TNC-rich sites12. We show that the engagement of breast cancer cells by TNC protects NOTCH signaling from inhibition by JAK2-STAT5, and augments the ability of WNT to activate the expression of LGR5. The LGR5 effect may involve a TNC-dependent input on the unidentified factor that licenses stem cells to specifically activate this particular WNT target gene47. These links between TNC, NOTCH and WNT pathways are consistent with earlier reports observing a correlation of disease outcome with some of the components mechanistically brought together in our findings. Activity of the WNT pathway regulator β-catenin is associated with poor prognosis in breast cancer48. The accumulation of NOTCH intracellular domain predicts early recurrence in ductal carcinoma in situ49. High expression of MSI1, NOTCH1 and the NOTCH ligand Jagged1 predicts poor overall survival50, 51. The NOTCH inhibitor NUMB is frequently underexpressed in breast cancer52. Furthermore, loss of STAT5 activity has been linked to poorly differentiated breast tumors53, and poor breast cancer survival54. These observations reinforce the pathway delineated in this study, in which TNC promotes metastasis by inhibiting JAK2-STAT5 signaling in breast cancer cells, relieving MSI1 from repression and thus enhancing NOTCH signaling activity. A possible explanation for the negative selection of TNCS and the lack of effect of TNC depletion on brain metastasis is that extracellular matrix molecules other than TNC43 provide a metastasis niche in the unique microenvironment of the brain.
By interfering with TNC production in our preclinical models, we suppressed the ability of micrometastatic colonies to survive and expand. Notably, TNC-deficient mice are viable and do not show severe developmental defects55. As an extracellular matrix component, TNC may be required for the viability of stem or progenitor-like cells mainly under stress conditions56.
However, TNC may not be necessary for stem cell homeostasis otherwise. Considering that TNC is essential for the survival and outgrowth of disseminated cancer cells, but not essential in unstressed normal stem cell niches, therapeutically targeting TNC may provide a strategy for eliminating residual disease with minimal adverse effects and an approach for the combined inhibition of the NOTCH and WNT pathways in micrometastatic breast cancer.
ONLINE METHODS
Cell culture
We cultivated MDA231-LM2 (ref. 7), MDA231-BoM1 (ref. 42), MDA231-BrM2 (ref. 43) and 293T cells in DMEM supplemented with 10% vol/vol FBS, 2 mM L-Glutamine, 100 IU ml−1 penicillin, 100 μg ml−1 streptomycin and 1 μg ml−1 amphotericin B. We maintained CN34-LM1 cells8 and all primary pleural effusion samples in M199 medium containing 2.5% vol/vol FBS, 10 μg ml−1 insulin, 0.5 μg ml−1 hydrocortisone, 20 ng ml−1 EGF, 100 ng ml−1 cholera toxin, 0.5 μg ml−1 amphotericin B, 2 mM L-Glutamine, 100 IU ml−1 penicillin and 100 μg ml−1 streptomycin.
Pleural effusion samples
All procedures with human pleural effusion samples were approved by MSKCC Institutional Review Board. Procedures were done as described57. Briefly, we centrifuged the cells from pleural fluid at 300g for 10 min and treated them with ACK lysis buffer (LONZA) according to the manufacturer's instructions. We washed samples in PBS and plated cells.
Oncosphere formation
Oncospheres were grown by cultivating cancer cells derived from pleural effusions on ultralow-adhesive plates (Costar). We plated cells at 25,000 cells ml−1 in HuMEC medium (Invitrogen), supplemented with 5 μg ml−1 insulin, 20 ng ml−1 EGF, 10 ng ml−1 basic fibroblast growth factor and 2% vol/vol B27 (Invitrogen). Oncospheres were cultivated for 2 weeks.
Generation of various knockdown cells and MSI1 rescue cells
We knocked down TNC, MSI1 and LGR5 with Mission TRC lentiviral vectors expressing shRNA against these gene products (Sigma-Aldrich). To generate MSI1 rescue cells, we amplified MSI1 by PCR from a cDNA clone (clone ID 100014977, Open Biosystems) and subcloned it into pBabe-hygromycin retroviral vector via BamH1 and Sal1 restriction sites. Production of retroviral particles is described in Supplementary Methods.
mRNA and miRNA expression analysis
We isolated whole RNA from cancer cells using PrepEase RNA spin kit (USB). RNA (400 ng) was used to generate cDNA with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene expression was analyzed using Taqman gene expression assays (Applied Biosystems). For miRNA analysis, we isolated RNA enriched for small RNA molecules using the mirVana miRNA Isolation Kit (Ambion). We used Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems) to generate cDNA specific for miR-335 and RNU6B snRNA control from 10 ng of RNA. We carried out a quantitative PCR reaction on ABI 7900HT Fast Real-Time PCR System and used the software SDS2.2.2 (Applied Biosystems) for analysis. Statistical analysis was done using Graphpad Prism 5 software.
Immunostaining
We stained frozen, paraformaldehyde-fixed tissue sections with antibodies to human tenascin (clone BC-24, Sigma-Aldrich), mouse tenascin (clone MTn-12, Sigma-Aldrich), α-smooth muscle actin (clone 1A4, Dako) and CD68 (clone FA-11, AbD Serotec). Paraffin-embedded, paraformaldehyde-fixed tissue sections were stained with antibodies to human tenascin (clone BC-24, Sigma-Adrich) and fascin-1 (Chemicon). Cleaved caspase 3 and phosphohistone H3 stainings were carried out by the MSKCC Molecular Cytology Core Facility. We used antibodies to cleaved caspase 3 (Cell Signaling) and phosphohistone H3 (Upstate).
Mouse studies
All experiments using mice were done in accordance with a protocol approved by MSKCC Institutional Animal Care and Use Committee. We used nonobese diabetic–severe combined immunodeficiency female mice aged 6–8 weeks for all mouse experiments unless specified otherwise. The orthotopic metastasis assay has been previously described8. Briefly, we injected MDA231-LM2 or CN34-LM1 cells bilaterally into the fourth mammary fat pad of anesthetized mice (100 mg kg−1 ketamine and 10 mg kg−1 xylazine). Cells (500,000) were injected in 50-μl volume Matrigel (BD Biosciences) diluted with PBS (1:1). We monitored mammary tumor growth by regular measurements using a digital caliper. After 6 weeks, we killed mice and determined metastasis in lungs by ex vivo imaging. We carried out lung colonization assays by injecting 200,000 MDA231-LM2 or CN34-LM1 cells (suspended in 100 μl PBS) into the lateral tail vein. Lung colonization was studied and determined by in vivo bioluminescence imaging. Anesthetized mice were injected retro-orbitally with D-luciferin (150 mg kg−1) and imaged with IVIS Spectrum Xenogen machine (Caliper Life Sciences). Bone metastases and brain metastases were induced in athymic nude mice by intracardiac injections of 30,000 MDA231-BoM1 and 10,000 MDA231-BrM2 cells, respectively. Metastasis occurrence was determined by in vivo luminescence. For doxycyline-inducible knockdown experiments, we treated mice by adding doxycycline hyclate (Sigma-Aldrich) to the drinking water (2 mg ml−1). Bioluminescence analysis was carried out using Living Image software, version 2.50 (Caliper Life Sciences).
Human metastasis samples
We acquired paraffin-embedded tissue sections from lung metastases and tissue microarrays with multiple lung metastases or primary breast tumor cores from the MSKCC Department of Pathology in compliance with protocols approved by the MSKCC Institutional Review Board and after the subjects gave their informed consent. We stained sections for TNC expression using immunohistochemistry as described above. Total TNC immunoreactivity was evaluated and scored by a clinical pathologist (E.B.). We analyzed lung relapse–free survival as a function of TNC abundance in breast tumor samples (Supplementary Fig. 1c) or in metastasis samples (Fig. 1b).
Data set gene expression analysis
We obtained TNC signatures, comprised of 373 probe sets, as described41; gene expression of tumor cells was analyzed in response to TNC-coated surface. These TNC signature probes were used to cluster the microarray data of 344 primary breast tumors (GSE2603 and GSE5327 combined58) and 67 metastatic breast tumors from different distant sites (GSE14020; ref. 59). The heatmap.2 function in the R statistical package was used to obtain the clustering. TNC signature status among different tumor groups were then compared using Fisher's exact test. We derived a STAT5 gene expression classifier from a published gene expression profile44. This signature comprised 784 downregulated probe sets and 208 upregulated probe sets with fold change >10× and P < 0.05. STAT5 signature status (as a categorical variable) was determined using unsupervised clustering, the same approach as described above for TNCS. We calculated a Pearson correlation coefficient between STAT5 score and TNC score. P value was determined by a Student's t test.
Gene set enrichment analysis
We carried out GSEA as previously described45. Briefly, all genes were ranked on the basis of their correlation with TNCS status. Thereafter, genes from the STAT5 signature (either upregulated or downregulated) were aligned to the ranked list and the running sum was calculated, leading to an enrichment score. The P values were calculated on the basis of random permutations.
Statistical analysis
We determined differences in lung metastasis–free survival using log rank test. Comparison of means within groups in lung metastasis assays was analyzed using two-tailed unpaired Student's t test. We analyzed gene expression differences between primary breast cancer cells grown as a monolayer and those grown as oncospheres using paired Wilcoxon signed rank test (two tailed). P < 0.05 was considered significant.
Additional methods
Detailed methodology is described in Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Kim and E. Montalvo for technical support. This work was funded by grant CA94060 from the US National Institutes of Health (J.M.), the Hearst Foundation and the Alan and Sandra Gerry Metastasis Research Initiative. S.A. is supported by a Department of Defense Era of Hope postdoctoral fellowship. E.B is a recipient of the Exceptional Project Award from the Breast Cancer Alliance. J.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTIONS
T.O. and J.M. designed experiments, analyzed data and wrote the manuscript. J.M. supervised research. T.O. carried out experiments. S.A. carried out immunostaining and helped with pathway analyses. X.H.-F.Z. carried out bioinformatics analyses. S.V. carried out intracardiac injections and assisted with bone and brain metastasis assays. S.F.T. helped with miRNA analysis. P.G.M. and R.J.D. oversaw collection of clinical samples. K.M.-T. supervised histological staining and analysis. E.B. obtained and evaluated human breast cancer tissue sections. All authors discussed the results and commented on the manuscript.
Supplementary information is available on the Nature Medicine website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Hüsemann Y, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Cameron MD, et al. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res. 2000;60:2541–2546. [PubMed] [Google Scholar]
- 3.Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer. 2009;9:285–293. doi: 10.1038/nrc2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkan D, Green JE, Chambers AF. Extracellular matrix: a gatekeeper in the transition from dormancy to metastatic growth. Eur. J. Cancer. 2010;46:1181–1188. doi: 10.1016/j.ejca.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anan K, et al. Disparities in the survival improvement of recurrent breast cancer. Breast Cancer. 2010;17:48–55. doi: 10.1007/s12282-009-0103-2. [DOI] [PubMed] [Google Scholar]
- 7.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta GP, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- 10.Kim MY, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kii I, et al. Incorporation of tenascin-C into the extracellular matrix by periostin underlies an extracellular meshwork architecture. J. Biol. Chem. 2010;285:2028–2039. doi: 10.1074/jbc.M109.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orend G, Chiquet-Ehrismann R. Tenascin-C induced signaling in cancer. Cancer Lett. 2006;244:143–163. doi: 10.1016/j.canlet.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 13.von Holst A. Tenascin C in stem cell niches: redundant, permissive or instructive? Cells Tissues Organs. 2008;188:170–177. doi: 10.1159/000112848. [DOI] [PubMed] [Google Scholar]
- 14.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 15.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 16.Hermann PC, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deryugina EI, Bourdon MA. Tenascin mediates human glioma cell migration and modulates cell migration on fibronectin. J. Cell Sci. 1996;109:643–652. doi: 10.1242/jcs.109.3.643. [DOI] [PubMed] [Google Scholar]
- 19.Ishihara A, Yoshida T, Tamaki H, Sakakura T. Tenascin expression in cancer cells and stroma of human breast cancer and its prognostic significance. Clin. Cancer Res. 1995;1:1035–1041. [PubMed] [Google Scholar]
- 20.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcion E, Halilagic A, Faissner A, ffrench-Constant C. Generation of an environmental niche for neural stem cell development by the extracellular matrix molecule tenascin C. Development. 2004;131:3423–3432. doi: 10.1242/dev.01202. [DOI] [PubMed] [Google Scholar]
- 22.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponti D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 24.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 25.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 26.Adewumi O, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 28.Okano H, et al. Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 30.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imai T, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouras T, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 36.Molyneux G, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Wagner KU, Rui H. Jak2/Stat5 signaling in mammogenesis, breast cancer initiation and progression. J. Mammary Gland Biol. Neoplasia. 2008;13:93–103. doi: 10.1007/s10911-008-9062-z. [DOI] [PubMed] [Google Scholar]
- 38.Chammas R, Taverna D, Cella N, Santos C, Hynes NE. Laminin and tenascin assembly and expression regulate HC11 mouse mammary cell differentiation. J. Cell Sci. 1994;107:1031–1040. doi: 10.1242/jcs.107.4.1031. [DOI] [PubMed] [Google Scholar]
- 39.Sandberg EM, Sayeski PP. Jak2 tyrosine kinase mediates oxidative stress-induced apoptosis in vascular smooth muscle cells. J. Biol. Chem. 2004;279:34547–34552. doi: 10.1074/jbc.M405045200. [DOI] [PubMed] [Google Scholar]
- 40.Müller J, Sperl B, Reindl W, Kiessling A, Berg T. Discovery of chromone-based inhibitors of the transcription factor STAT5. ChemBioChem. 2008;9:723–727. doi: 10.1002/cbic.200700701. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz C, et al. Growth promoting signaling by tenascin-C. Cancer Res. 2004;64:7377–7385. doi: 10.1158/0008-5472.CAN-04-1234. [DOI] [PubMed] [Google Scholar]
- 42.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 43.Bos PD, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eilon T, Barash I. Distinct gene-expression profiles characterize mammary tumors developed in transgenic mice expressing constitutively active and C-terminally truncated variants of STAT5. BMC Genomics. 2009;10:231. doi: 10.1186/1471-2164-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ioachim E, et al. Immunohistochemical expression of extracellular matrix components tenascin, fibronectin, collagen type IV and laminin in breast cancer: their prognostic value and role in tumour invasion and progression. Eur. J. Cancer. 2002;38:2362–2370. doi: 10.1016/s0959-8049(02)00210-1. [DOI] [PubMed] [Google Scholar]
- 47.Casali A, Batlle E. Intestinal stem cells in mammals and Drosophila. Cell Stem Cell. 2009;4:124–127. doi: 10.1016/j.stem.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 48.Lin SY, et al. β-catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. USA. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farnie G, et al. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J. Natl. Cancer Inst. 2007;99:616–627. doi: 10.1093/jnci/djk133. [DOI] [PubMed] [Google Scholar]
- 50.Wang XY, et al. Musashi1 regulates breast tumor cell proliferation and is a prognostic indicator of poor survival. Mol. Cancer. 2010;9:221. doi: 10.1186/1476-4598-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reedijk M, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 52.Pece S, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cotarla I, et al. Stat5a is tyrosine phosphorylated and nuclear localized in a high proportion of human breast cancers. Int. J. Cancer. 2004;108:665–671. doi: 10.1002/ijc.11619. [DOI] [PubMed] [Google Scholar]
- 54.Nevalainen MT, et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J. Clin. Oncol. 2004;22:2053–2060. doi: 10.1200/JCO.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 55.Saga Y, Yagi T, Ikawa Y, Sakakura T, Aizawa S. Mice develop normally without tenascin. Genes Dev. 1992;6:1821–1831. doi: 10.1101/gad.6.10.1821. [DOI] [PubMed] [Google Scholar]
- 56.Mackie EJ, Tucker RP. The tenascin-C knockout revisited. J. Cell Sci. 1999;112:3847–3853. doi: 10.1242/jcs.112.22.3847. [DOI] [PubMed] [Google Scholar]
- 57.Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPβ at the core of the TGFβ cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 58.Smid M, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–3114. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 59.Zhang XH, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.