Abstract
Carvacrol, thymol, and eugenol are naturally occurring phenolic compounds known to possess antimicrobial activity against a range of bacteria, as well as antioxidant activity. Biodegradable poly(anhydride-esters) composed of an ethylenediaminetetraacetic acid (EDTA) backbone and antimicrobial pendant groups (i.e., carvacrol, thymol, or eugenol) were synthesized via solution polymerization. The resulting polymers were characterized to confirm their chemical composition and understand their thermal properties and molecular weight. In vitro release studies demonstrated that polymer hydrolytic degradation was complete after 16 days, resulting in the release of free antimicrobials and EDTA. Antioxidant and antibacterial assays determined that polymer release media exhibited bioactivity similar to that of free compound, demonstrating that polymer incorporation and subsequent release had no effect on activity. These polymers completely degrade into components that are biologically relevant and have the capability to promote preservation of consumer products in the food and personal care industries via antimicrobial and antioxidant pathways.
Introduction
Prevention of microbial contamination in the personal care and food industries is of paramount importance for consumer protection. However, most commonly used synthetic preservatives, such as parabens, suffer from problems such as sensitization and toxicity.1 Another commonly used antimicrobial, triclosan, is thought to breed bacterial resistance. Triclosan can potentially disrupt the endocrine system and its level in the blood correlating to a consumer’s use has been discovered.2 Moreover, parabens, triclosan, and other preservatives have environmental bioaccumulation issues, with concentrations of these chemicals detected in aquatic life,3 leading to a call for safer alternatives. Alternatively, many naturally occurring phenolic compounds possess known activity against a range of Gram-positive and Gram-negative bacteria. Carvacrol, thymol, and eugenol are three such phenolic compounds that are major constituents of oregano, thyme, and clove essential oils, respectively.4,5 In addition to their antibacterial activity,6 these compounds are potent antioxidants;7 thus, they can potentially be used to prevent the bacterial spoilage8,9 and oxidation10,11 of perishable food items (e.g., meat, dairy, fruit) as well as personal care products.12
Currently, antimicrobials are physically mixed into formulations to prevent bacterial growth. The sustained release of these bioactive compounds can naturally preserve consumer products thereby increasing shelf life. To extend bioactive release, researchers have physically incorporated the aforementioned phenols into polymer matrices. Chitosan nanoparticles loaded with eugenol or carvacrol were successfully formulated but suffered from low antimicrobial content (ca. 3% w/w)13 or a lack of release studies.14 Likewise, carvarcol-containing polyethylene-co-vinylacetate films15 and thymol-containing polycaprolactone films16 suffered from low loading at 7 and 10 wt %, respectively. Eugenol has been physically incorporated into methacrylate polymers; however, release studies were conducted under nonphysiological conditions (i.e., ethanol).17 Additionally, poly(lactic-co-glycolic acid) (PLGA) films containing carvacrol within the polymer matrix exhibited antibiofilm and antibacterial activities,18 although the release is not quantified. Eugenol and carvacrol-loaded PLGA nanoparticles displaying antibacterial activity have been formulated by multiple groups but the nanoparticles exhibited a burst release; nearly 50% antimicrobial released in the first 8 h for eugenol19 and 60% released in 3 h for carvacrol.20
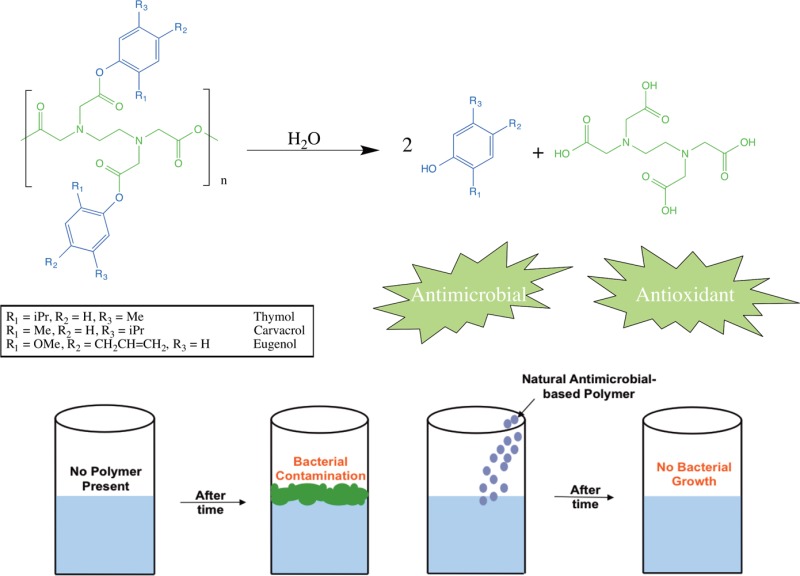
Chemical incorporation of bioactives into a biodegradable polymer backbone has multiple advantages (e.g., higher drug loading, sustained release, tunable release rates and geometry depending on desired application) over the aforementioned physical incorporation methods.21,22 Chemical incorporation of eugenol into a polymer backbone has also been achieved, along with antibacterial activity, but the polymethacrylate was not degradable, limiting its use in other applications.23 In this work, we present the synthesis of biodegradable poly(anhydride-esters) (PAEs) containing one of the three phenolic compounds (carvacrol, thymol, or eugenol) chemically linked and subsequently polymerized through EDTA, a widely used chelator known to prevent oxidation caused by metals,24 to achieve high drug loading (>50%). Previously, our group has synthesized polymers chemically, incorporating various bifunctional25,26 and monofunctional27,28 bioactives; however, in this research the polymers hydrolyze into the phenols and EDTA, both of which are known preservatives and safe to use in consumer products.29,30 Furthermore, all ultimate degradation products are found on the FDA’s generally regarded as safe (GRAS) list. Chemical structures, thermal properties, and molecular weights of polymer and precursors were determined and in vitro release studies confirmed that the polymer degraded into the free antimicrobials and EDTA. The release media antioxidant efficacy and antibacterial activity were determined using a free radical quenching assay and a disk diffusion assay with a Gram-positive bacterium (Staphylococcus aureus) and a Gram-negative bacterium (Escherichia coli), respectively, and compared to activities of the free phenolic compounds.
Experimental Section
Materials
Concentrated hydrochloric acid (HCl), 1 N HCl, 1 N sodium hydroxide (NaOH), poly(vinylidine fluoride), and poly(tetrafluoroethylene) syringe filters, and Wheaton glass scintillation vials were purchased from Fisher Scientific (Fair Lawn, NJ). All other reagents, solvents, and fine chemicals were obtained from Aldrich (Milwaukee, WI) and used as received.
Structural and Chemical Composition Characterization
1H NMR spectra of all products were recorded on a Varian 400 MHz spectrophotometer. Samples (5–10 mg) were dissolved in DMSO-d6, which was also used as the internal reference. Fourier-transform infrared (FT-IR) absorbance spectra were measured on a Thermo Nicolet/Avatar 360 FT-IR spectrometer. Samples (1 wt %) were ground with KBr and pressed into a disc. Each spectrum was an average of 32 scans. Elemental analyses were performed by QTI (Whitehouse, NJ).
Molecular Weight
Polymer precursors were analyzed by mass spectrometry (MS) to determine molecular weights. A Finnigan LCQ-DUO running Xcalibur software and an adjustable atmospheric pressure ionization electrospray source (API-ESI Ion Source) was used. Samples were dissolved in methanol (10 μg/mL) and injected with a glass syringe. During the experiment, the pressure was 0.8 × 10–5 Torr and the API temperature was 150 °C. Polymer weight-averaged molecular weights (Mw) and polydispersity indices (PDI) were determined by gel permeation chromatography (GPC) on a PerkinElmer liquid chromatography system consisting of a Series 200 refractive index detector, a Series 200 LC pump, and an ISS 200 autosampler. Automation of the samples and collection and processing of the data was done using a Dell OptiPlex GX110 computer running PerkinElmer TurboChrom 4 software using a PerkinElmer Nelson 900 Series Interface and 600 Series Link. Polymer samples were prepared for autoinjection by dissolving polymer in DCM (10 mg/mL) and filtering through 0.45 μm poly(tetrafluoroethylene) (PTFE) syringe filters. Samples were resolved on a Jordi divinylbenzene mixed-bed GPC column (7.8 × 300 mm, Alltech Associates, Deerfield, IL) at 25 °C, with DCM as eluent at a flow rate of 0.5 mL/min. Molecular weights were calibrated relative to polystyrene standards (Polymer Source Inc., Dorval, Canada).
Thermal Properties
Differential scanning calorimetry (DSC) was performed using a Thermal Analysis (TA) DSC Q200 to evaluate thermal transitions (i.e., melting points of polymer precursors and glass transition temperatures of polymers). TA Universal Analysis 2000 software was used for automation and data collection on an IBM ThinkCentre computer. Samples (5–10 mg) were heated under dry nitrogen gas from −10 to 200 °C at a heating rate of 10 °C/min and cooled to −10 °C at a rate of 10 °C/min with a two-cycle minimum. Melting temperatures were calculated at the peak of melting.
Thermogravimetric analyses (TGA) were performed on a PerkinElmer Pyris 1 system with TAC 7/DX instrument controller. PerkinElmer Pyris software running on a Dell Optiplex GX110 computer was used for automation and data collection and processing. Samples (5–10 mg) were heated under dry nitrogen gas from 25 to 400 °C at a heating rate of 10 °C/min. Decomposition temperatures were measured at the onset of thermal decomposition.
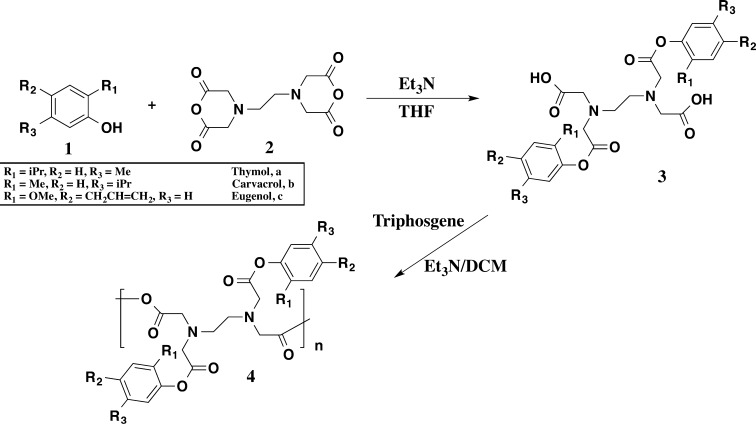
Polymer Precursor (3) Synthesis
Antimicrobial-containing diacids were synthesized using a modified previously developed procedure.27 Diacids (3) were prepared by reaction of ethylenediaminetetraacetic acid (EDTA) dianhydride (2) with the appropriate antimicrobial (1) in the presence of a base (triethylamine) (Scheme 1). The full characterization of the thymol-based system is presented as an example. The data for carvacrol- and eugenol-based systems can be found in the Supporting Information. Thymol (1a; 18 mmol) was dissolved in anhydrous THF (75 mL) and anhydrous triethylamine (Et3N, 64 mmol). EDTA dianhydride (5; 9 mmol) was added to the reaction mixture while stirring at room temperature to yield a suspension. The reaction stirred at room temperature under nitrogen overnight. Then, the reaction mixture was poured over DI water (∼500 mL) and acidified to pH 2 using concentrated HCl. The solid diacid (3a) that formed was isolated via vacuum filtration, washed with water (3 × 200 mL), and dried overnight under vacuum at room temperature.
Scheme 1. Synthesis of Antimicrobial-Containing PAEs (4) and Precursors (3).
Thymol-Based Diacid (3a)
Yield: 76% (off-white powder). 1H NMR (DMSO-d6, 400 MHz): δ 7.21 (d, 2H, Ar–H); 7.02 (d, 2H, Ar–H); 6.87 (s, 2H, Ar–H); 4.15 (s, 4H, CH2); 3.77 (s, 4H, CH2); 3.13 (s, 4H, CH2); 2.92 (m, 2H, CH); 2.23 (s, 6H, CH3); 1.08 (d, 12H, CH3). IR (KBr, cm–1): 3400 (OH, COOH), 1760 (C=O, ester), 1712 (C=O, COOH). MS m/z = 557.2 [M + 1], (C30H40N2O8)n (556.7) Calcd: C, 64.14; H, 7.4; N, 4.70. Found: C, 64.73; H, 7.24; N, 5.03. Tm: 113 °C.
Polyanhydride-ester (4) Synthesis
Diacid (3; 5.4 mmol) was dissolved in 20% (w/v) anhydrous DCM, and triethylamine (24 mmol) was added. Then, triphosgene (6 mmol) dissolved in anhydrous DCM (15 mL) was added dropwise at 0 °C to the stirring reaction mixture over 1 h using a syringe pump to yield a suspension. After stirring for 2 h at 0 °C under nitrogen, the reaction mixture was poured over diethyl ether (∼400 mL). The solid polymer (4) that formed was isolated by vacuum filtration, washed with acidic water (100 mL) and dried overnight under vacuum at room temperature.
Thymol-Based Polymer (4a)
Yield: 39% (brown powder). 1H NMR (DMSO-d6, 400 MHz): δ 7.22 (d, 2H, Ar–H); 7.03 (d, 2H, Ar–H); 6.82 (s, 2H, Ar–H); 4.41 (s, 4H, CH2); 3.68 (s, 4H, CH2); 2.92 (m, 5H, CH2, CH); 2.24 (s, 6H, CH3); 1.06 (d, 12H, CH3). IR (KBr, cm–1): 1815 and 1745 (C=O, anhydride), 1730 (C=O, ester). Mw = 23200 Da; PDI 1.3. Tg = 77 °C. Td = 223 °C.
In Vitro Bioactive Release
First, the release of diacid (3) from polymer (4) was evaluated to determine the amount of time required to hydrolyze anhydride bonds through in vitro degradation in phosphate buffered saline (PBS). Polymers were ground into powder using mortar and pestle to obtain particles of ∼300–500 μm, as determined by standard testing sieves (Aldrich, Milwaukee, WI). Powdered polymer samples (15 mg) were incubated in 10 mL of PBS (pH 7.4) in 20 mL Wheaton glass scintillation vials (Fisher, Fair Lawn, NJ) using a controlled environment incubator-shaker (New Brunswick Scientific Co., Edison, NJ) at 60 rpm at 37 °C. At predetermined time intervals, the media was replaced with fresh PBS and the spent media was analyzed by ultraviolet–visible (UV–vis) spectrophotometry (PerkinElmer Lambda XLS spectrophotometer, Waltham, MA) at λ = 270 nm. Additionally, the bioactive (1) release from diacid (3) was elucidated under the same conditions listed above, using powdered diacid samples (15 mg) incubated in 10 mL of PBS (pH 7.4) in 15 mL centrifuge tubes (BD Falcon, Franklin Lakes, NJ). After centrifugation at 6000 rpm for 4 min, the degradation media (5 mL) was collected at predetermined time points and fresh PBS (5 mL) replaced that which was removed. High performance liquid chromatography (HPLC) quantified the amount of free phenol in spent media released by comparison to calibration curves of standard solutions. Media was analyzed via HPLC using an XTerra RP18 3.5 μm 4.6 × 150 mm column (Waters, Milford, MA) on a Waters 2695 Separations Module equipped with a Waters 2487 Dual Absorbance Detector. All samples were filtered using 0.22 μm poly(vinylidine fluoride) syringe filters and subsequently injected (20 μL) using an autosampler. The mobile phase composed of methanol (55%) and 50 mM KH2PO4 with 1% formic acid in DI water at pH 2.5 (45%) run at 0.8 mL/min flow rate at ambient temperature. Absorbance was monitored at λ = 270. The degradation experiments were performed in triplicate.
Antioxidant Activity via Radical Scavenging
To determine the degradation media antioxidant activity, a 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging assay was used.31 This was performed by adding a sample (0.1 mL) to a 24 μg/mL DPPH solution in methanol (3.9 mL). Hour-24 polymer degradation media samples (0.1 mL) were incubated with the DPPH solution (3.9 mL) at room temperature with gentle shaking. After 1 h, solutions were analyzed by UV/vis spectrophotometry at λ = 517 nm. For comparison, a solution of freshly prepared bioactive (1) at the same concentration of the 24-h degradation media (as determined at HPLC) was prepared and analyzed via the same method. DPPH % radical reduction was calculated by [(Abst0 – Abst)/Abst0] × 100, where Abst0 is the initial absorbance, and Abst is the absorbance after 1 h. All radical quenching assays were performed in triplicate. Student’s t tests were used to determine the significant difference of the antioxidant activity between degradation media and free antimicrobial (significantly different if p < 0.05)
Antibacterial Testing
Carvacrol, thymol, and eugenol are well-known antimicrobial agents, exhibiting activity against a range of bacteria;6 thus, the activity of degradation media against Gram-positive and Gram-negative bacteria was tested using the disc diffusion method.32 First, polymer was completely hydrolyzed with 1 N NaOH, then acidified to pH 2 using concentrated HCl, after which bioactives were extracted with ethyl acetate. The organic layer was dried over MgSO4 and concentrated in vacuo. Upon solubilization in 1:1 PBS/DMSO, the concentration of all bioactives was 10 mg/mL. The three free bioactives were tested against equal concentrations of bioactives extracted from the degradation media (10 mg/mL) and tested as follows: Muller-Hinton agar (Becton Dickinson, Sparks, MD) was poured into sterile Petri dishes (Fisher, Fair Lawn, NJ) to an even thickness of 4 mm. Bacteria inocula (S. aureus or E. coli) were suspended in nutrient broth (EMD Chemicals, Gibbstown, NJ) and turbidity to give a bacterial count of approximately 108 colony forming units per mL. The agar plate was inoculated with bacteria broth culture using a sterile cotton swab (Fisher, Fair Lawn, NJ). Sterile paper discs (6 mm diameter, Becton Dickinson, Franklin Lakes, NJ) were impregnated with 25 μL of test solutions (one for each free phenol, one for each phenol degradation media, one for EDTA, and one for PBS/DMSO). Discs were then placed onto an agar plate and gently pressed down. Plates were incubated at 37 °C for 24 h, after which zones of inhibition were measured with a ruler and rounded to the nearest millimeter.
Results and Discussion
Synthesis and Characterization
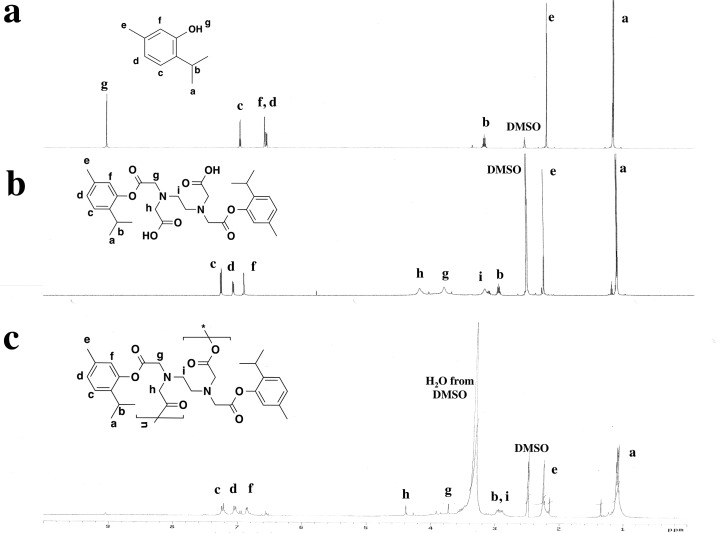
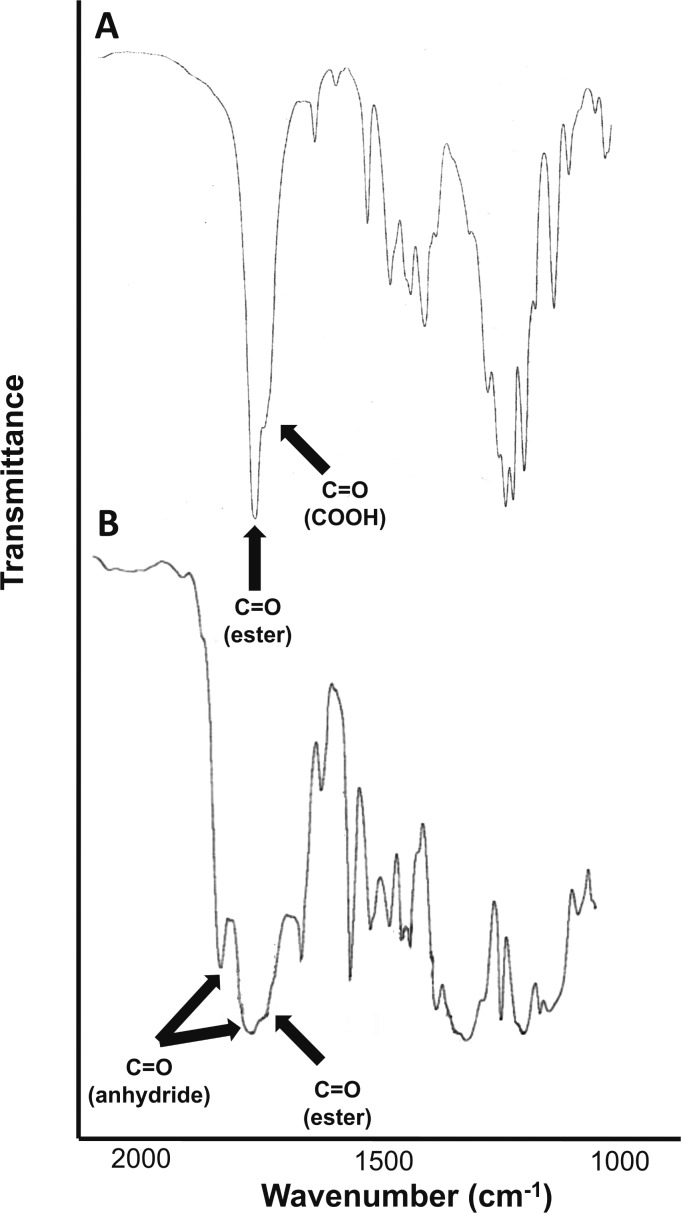
The phenolic antimicrobials (1) are reacted with EDTA dianhydride (2) in the presence of triethylamine to yield diacid (3) via a ring-opening transesterification (Scheme 1). The diacids, 3, were successfully prepared in high yields (76–85%) with only minor purification necessary. Diacid structures were confirmed by 1H NMR and FT-IR spectra, while DSC, MS, and elemental analysis were used for melting point, molecular weight, and chemical composition determination, respectively. In the 1H NMR spectrum of 3a, which is provided as an example (Figure 1), the disappearance of the phenol signal of 1a at 9.02 ppm and the appearance of EDTA linker peaks at 4.15, 3,77, and 3.13 ppm demonstrate successful ring-opening esterification to generate 3a. The diacids 3 were polymerized via solution polymerization techniques,33,34 using triphosgene as the coupling reagent in the presence of triethylamine at 0 °C. Solution polymerization was chosen instead of melt-condensation to prevent potential ring closure and regeneration of the EDTA dianhydride, 5.34 Polymers were characterized by 1H NMR to confirm structure. Additionally, FT-IR (Figure 2) confirmed the synthesis of a poly(anhydride-ester) through the disappearance of the carboxylic acid stretch at 1712 cm–1 in diacid 4 and the appearance of the C=O anhydride stretches at 1815 and 1745 cm–1 in polymer 5.
Figure 1.
1H NMR spectra of thymol (a), corresponding diacid (b), and polymer (c).
Figure 2.
FTIR spectra of thymol-containing diacid 3a (A, top) and polymer 4a (B, bottom).
The polymers (4) displayed moderate molecular weights typical for solution polymerization techniques, ranging from 11000 to 23000 Da.33,34 PDI values were narrow (1.3–1.5) following isolation from the reaction mixture, indicating high homogeneity. The polyanhydride with the bulkiest antimicrobial, eugenol (4c), was the most difficult to polymerize and displayed the lowest molecular weight. All polymers (4) are amorphous with no indication of melting temperatures (up to 200 °C), exhibiting only glass transition temperatures in the range of 65 to 86 °C. With thermal decomposition occurring at temperatures >220 °C, it is anticipated that processing via melt-extrusion will be a viable method for film production and processing at elevated temperatures.
In Vitro Bioactive Release
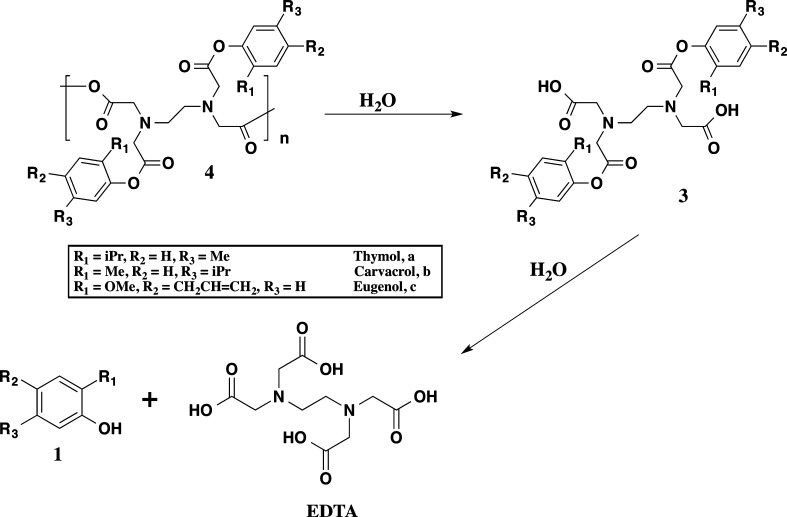
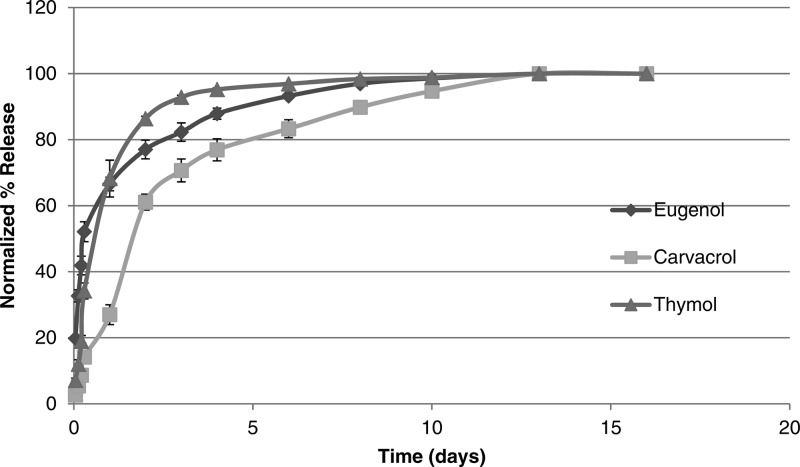
Bioactive release was carried out on powdered samples in PBS at physiological conditions (37 °C, pH 7.4). The degradation rate of polymer into bioactive via anhydride and ester bond hydrolysis is an important factor in obtaining controlled antimicrobial release. Based upon previous knowledge of PAEs, the anhydride bonds are expected to hydrolyze first, followed by the ester bonds (Scheme 2).35 To obtain polymer degradation lag time (i.e., the time during which little degradation takes place) under physiological conditions, degradation of polymer into diacid was first determined by UV–vis spectrophotometry. All polymers (4) exhibited a brief lag time (<6 h) before any degradation was detected, as polyanhydrides commonly exhibit a degradation lag time due to their surface eroding properties.35,36 To study complete degradation, bioactive release was monitored under identical conditions with HPLC. The studies were concluded upon complete degradation into respective bioactives and EDTA. Small amounts of diacid were also observed in degradation media, with the amounts of thymol increasing over time (e.g., thymol retention time of 14.45 min, thymol diacid retention time of 4.54 min), thus, proving that the anhydride bonds hydrolyze first, followed by ester bonds. All chemically incorporated bioactive was completely released after 16 days. At the end of the study, a mass balance was performed (>97% mass accounted for) and the release data was normalized (Figure 3)
Scheme 2. Proposed Hydrolytic Degradation Scheme of Antimicrobial-Containing PAEs (4) into Diacid (3) and Free Antimicrobial (1) and EDTA.
Figure 3.
Normalized release of antimicrobials (1) from polymers (4) is a result of in vitro hydrolytic degradation.
Antioxidant Activity
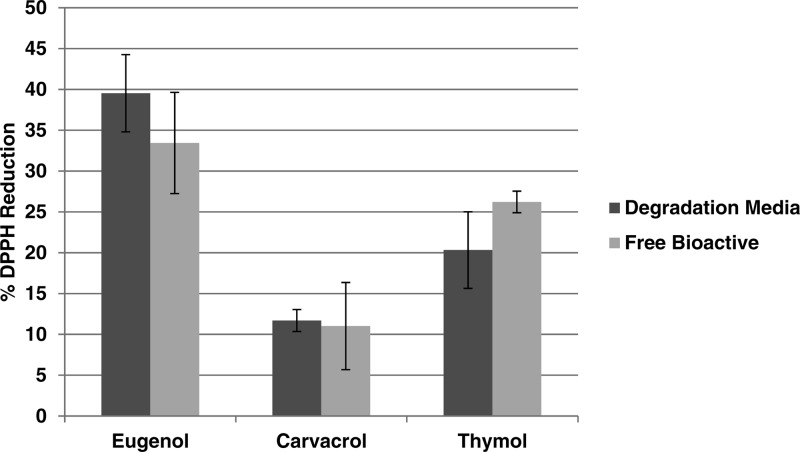
To ensure bioactive released from polymers exhibited the same efficacy as free bioactive, a DPPH radical quenching assay was used.31 DPPH is often used to assess antioxidant activity by determining the change in absorbance at 517 nm via UV–vis spectrophotometry; the solution color turns from deep purple to light yellow upon free radical quenching, thus reducing absorbance at 517 nm. Given that the assay is dependent on antioxidant concentration,37 degradation media at 24 h were analyzed (Figure 4) and compared to freshly prepared solutions of free bioactives of the same concentrations (52 μg/mL for carvacrol, 74 μg/mL for eugenol, and 100 μg/mL for thymol). Student’s t tests were performed to ascertain significant differences (p < 0.05) between the released and freshly prepared samples. The observed activities displayed no significant differences between released samples and free bioactive for all three phenols; thus the intermediate degradation products (i.e., diacid) and EDTA present in the degradation media had a negligible effect on free radical quenching ability. The DPPH quenching efficacy of the concentrations tested are consistent when compared to literature values of 500 μg/mL solutions of eugenol, carvacrol, and thymol (93, 79, and 82%, respectively).7
Figure 4.
DPPH reduction results comparing bioactive released from polymer at a 24 h time point to free bioactive.
Antibacterial Testing
To ensure that polymer degradation products diffusing from the discs would cause clear zones of growth inhibition, the polymer was completely hydrolyzed and a specific bioactive concentration was prepared from extracted products and compared to that of equivalent concentrations of free bioactives; the concentration for all bioactives was kept at 10 mg/mL (greater than the minimum inhibitory concentrations (MICs) to ensure a clear zone is observed) in 1:1 PBS/DMSO. S. aureus, a Gram-positive bacterium, and E. coli, a Gram-negative bacterium, were both evaluated, since both strains are commonly encountered and often responsible for contamination of products leading to spoilage.38 As shown in Figure 5, the free phenols diffused from the discs and prevented bacterial growth on the agar. Both the free bioactives (e−g) and the extracted bioactives (b–d) exhibited nearly the same zones of inhibition for both strains (Table 1). Neither EDTA (h) nor PBS/DMSO (a) controls exhibited any activity against either strain. Carvacrol and thymol displayed greater activity compared to eugenol, which is expected owing to eugenol’s higher MICs against both strains.6 Overall, this assay shows that the methods used to process the bioactives used did not alter their antibacterial activity upon release from polymer.
Figure 5.
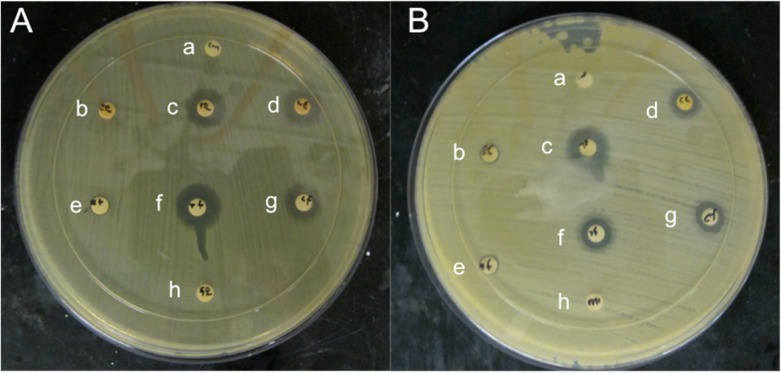
Disk diffusion assay results for E. coli (A) and S. aureus (B) showing zones of growth inhibition for 1:1 PBS/DMSO (a), extracted eugenol (b), extracted thymol (c), extracted carvacrol (d), free eugenol (e), free thymol (f), free carvacrol (g), and EDTA (h).
Table 1. Zones of Growth Inhibition for Extracted and Free Phenols.
| E. coli | S. aureus | |
|---|---|---|
| compound | zone of inhibition (mm) | zone of inhibition (mm) |
| PBS/DMSO | ||
| extracted eugenol | 4 | 4 |
| free eugenol | 4 | 4 |
| extracted thymol | 8 | 7 |
| free thymol | 8 | 6 |
| extracted carvacrol | 7 | 6 |
| free carvacrol | 7 | 6 |
| EDTA |
Conclusion
PAEs containing naturally occurring antimicrobials found in plant extracts were synthesized using solution polymerization methods, resulting in products with high drug loading (>50%). Polymers (4) hydrolytically degraded after 16 days, releasing free phenolic antimicrobials and EDTA in a controlled manner. Final polymer degradation products, phenolic antimicrobials and EDTA, are both commonly used as preservatives and are found on the FDA GRAS list. These polymers are unique in that the polymer completely breaks down into useful products: EDTA and an antimicrobial. A sustained release of compounds with antimicrobial, antioxidant, and chelation abilities can be beneficial for consumer product protection; the spoilage of products caused by bacterial contamination and oxidation can be prevented by polymer degradation products. Furthermore, bioactives released from polymer display similar antioxidant and antibacterial activities. PAE properties allow for future formulations, such as films and microspheres, in addition to tunable release rates and thermal properties by changing the dianhydride used.
Acknowledgments
The authors thank the National Institutes of Health and Center for Advanced Food Technology (Rutgers University) for financial support, Michelle Moy (Chemistry, Rutgers University) for assistance with the experiments, and Dr. Susan Skelly (Microbiology, Rutgers University) for assistance with the antimicrobial studies.
Supporting Information Available
Characterization data for carvacrol- and eugenol-based polymers and precursors are included. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Soni M. G.; Carabin I. G.; Burdock G. A. Food Chem. Toxicol. 2005, 43, 985–1015. [DOI] [PubMed] [Google Scholar]
- Dann A. B.; Hontela A. J. Appl. Toxicol. 2011, 31, 285–311. [DOI] [PubMed] [Google Scholar]
- Brausch J. M.; Rand G. M. Chemosphere 2011, 82, 1518–1532. [DOI] [PubMed] [Google Scholar]
- Chaieb K.; Hajlaoui H.; Zmantar T.; Ben K.-N. A.; Rouabhia M.; Mahdouani K.; Bakhrouf A. Phytother. Res. 2007, 21, 501–506. [DOI] [PubMed] [Google Scholar]
- Lee S.-J.; Umano K.; Shibamoto T.; Lee K.-G. Food Chem. 2004, 91, 131–137. [Google Scholar]
- Burt S. Int. J. Food Microbiol. 2004, 94, 223–253. [DOI] [PubMed] [Google Scholar]
- Lin C.-W.; Yu C.-W.; Wu S.-C.; Yih K.-H. Yaowu Shipin Fenxi 2009, 17, 386–395. [Google Scholar]
- Cleveland J.; Montville T. J.; Nes I. F.; Chikindas M. L. Int. J. Food Microbiol. 2001, 71, 1–20. [DOI] [PubMed] [Google Scholar]
- Holley R. A.; Patel D. Food Microbiol. 2005, 22, 273–292. [Google Scholar]
- Baron C. P.; Andersen H. J. J. Agric. Food Chem. 2002, 50, 3887–3897. [DOI] [PubMed] [Google Scholar]
- Soliva-Fortuny R. C.; Biosca-Biosca M.; Grigelmo-Miguel N.; Martin-Belloso O. J. Sci. Food Agric. 2002, 82, 1490–1496. [Google Scholar]
- Varvaresou A.; Papageorgiou S.; Tsirivas E.; Protopapa E.; Kintziou H.; Kefala V.; Demetzos C. Int. J. Cosmet. Sci. 2009, 31, 163–175. [DOI] [PubMed] [Google Scholar]
- Woranuch S.; Yoksan R. Carbohydr. Polym. 2013, 96, 586–592. [DOI] [PubMed] [Google Scholar]
- Chen F.; Shi Z.; Neoh K. G.; Kang E. T. Biotechnol. Bioeng. 2009, 104, 30–39. [DOI] [PubMed] [Google Scholar]
- Nostro A.; Scaffaro R.; D’Arrigo M.; Botta L.; Filocamo A.; Marino A.; Bisignano G. Appl. Microbiol. Biotechnol. 2012, 96, 1029–1038. [DOI] [PubMed] [Google Scholar]
- Sanchez-Garcia M. D.; Ocio M. J.; Gimenez E.; Lagaron J. M. J. Plast. Film Sheeting 2008, 24, 239–251. [Google Scholar]
- Peppas N. A.; am E. D. J. J. Appl. Polym. Sci. 1997, 66, 509–513. [Google Scholar]
- Zodrow K. R.; Schiffman J. D.; Elimelech M. Langmuir 2012, 28, 13993–13999. [DOI] [PubMed] [Google Scholar]
- Gomes C.; Moreira R. G.; Castell-Perez E. J. Food Sci. 2011, 76, N16–N24. [DOI] [PubMed] [Google Scholar]
- Iannitelli A.; Grande R.; Di S. A.; Di G. M.; Sozio P.; Bessa L. J.; Laserra S.; Paolini C.; Protasi F.; Cellini L. Int. J. Mol. Sci. 2011, 12, 5039–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrich K. E.; Cannizzaro S. M.; Langer R. S.; Shakesheff K. M. Chem. Rev. (Washington, D. C.) 1999, 99, 3181–3198. [DOI] [PubMed] [Google Scholar]
- Prudencio A.; Schmeltzer R. C.; Uhrich K. E. Macromolecules 2005, 38, 6895–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo L.; Vazquez B.; Parra J.; Lopez B. A.; Deb S.; San R. J. Biomacromolecules 2006, 7, 2751–2761. [DOI] [PubMed] [Google Scholar]
- Cannarsi M.; Baiano A.; Sinigaglia M.; Ferrara L.; Baculo R.; Del N. M. A. Int. J. Food Sci. Technol. 2008, 43, 573–578. [Google Scholar]
- Erdmann L.; Uhrich K. E. Biomaterials 2000, 21, 1941–1946. [DOI] [PubMed] [Google Scholar]
- Rosario-Meléndez R.; Harris C. L.; Delgado-Rivera R.; Yu L.; Uhrich K. E. J. Controlled Release 2012, 162, 538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudencio A.; Carbone A. L.; Griffin J.; Uhrich K. E. Macromol. Rapid Commun. 2009, 30, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Rosario-Melendez R.; Yu W.; Uhrich K. E. Biomacromolecules 2013, 14, 3542–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brul S.; Coote P. Int. J. Food Microbiol. 1999, 50, 1–17. [DOI] [PubMed] [Google Scholar]
- Whittaker P.; Vanderveen J. E.; Dinovi M. J.; Kuznesof P. M.; Dunkel V. C. Regul. Toxicol. Pharmacol. 1993, 18, 419–427. [DOI] [PubMed] [Google Scholar]
- Scherer R.; Godoy H. T. Food Chem. 2008, 112, 654–658. [Google Scholar]
- Manual of Clinical Microbiology, 7th ed.; Murray P. R., Baron E. J., Pfaller M. A., Tenover F. C., Yolken R. H.. ASM Press: Washington, DC, 1999. [Google Scholar]
- Domb A. J.; Ron E.; Langer R. Macromolecules 1988, 21, 1925–1929. [Google Scholar]
- Schmeltzer R. C.; Johnson M.; Griffin J.; Uhrich K. J. Biomater. Sci., Polym. Ed. 2008, 19, 1295–1306. [DOI] [PubMed] [Google Scholar]
- Whitaker-Brothers K.; Uhrich K. J. Biomed. Mater. Res., Part A 2006, 76A, 470–479. [DOI] [PubMed] [Google Scholar]
- Gopferich A.; Tessmar J. Adv. Drug Delivery Rev. 2002, 54, 911–931. [DOI] [PubMed] [Google Scholar]
- Brand-Williams W.; Cuvelier M. E.; Berset C. Food Sci. Technol. (London) 1995, 28, 25–30. [Google Scholar]
- Cosentino S.; Tuberoso C. I. G.; Pisano B.; Satta M.; Mascia V.; Arzedi E.; Palmas F. Lett. Appl. Microbiol. 1999, 29, 130–135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.