Abstract
The focus here is on incorporating electronic polarization into classical molecular mechanical force fields used for macromolecular simulations. First, we briefly examine currently used molecular mechanical force fields and the current status of intermolecular forces as viewed by quantum mechanical approaches. Next, we demonstrate how some components of quantum mechanical energy are effectively incorporated into classical molecular mechanical force fields. Finally, we assess the modeling methods of one such energy component—polarization energy—and present an overview of polarizable force fields and their current applications. Incorporating polarization effects into current force fields paves the way to developing potentially more accurate, though more complex, parameterizations that can be used for more realistic molecular simulations.
1. Introduction
One grand challenge in computational chemistry, in the context of molecular modeling, is to develop accurate force fields. The notion of the force field comprises a mathematical formula and a set of parameters used to calculate the energy of molecular systems. Despite more than 40 years of efforts devoted to force field development, many problems still need to be addressed. Usually, force fields are specialized, that is they are not intended for all types of organic, inorganic and biologically relevant molecules. Instead, they are devoted either to proteins and nucleic acids or to other organic molecules such as sugars or lipids, though more general force fields have been proposed, such as the universal force field (UFF) (Rappe et al 1992) and the generalized AMBER force field (GAFF) (Wang et al 2004). The force fields usually suffer from not being able to reproduce properties of molecules under different temperature, pressure and environmental conditions, such as pH, ion concentration and type of solvent. All force fields can be improved either by reparameterizing the existing set of parameters or extending the energy formula by adding terms and then parameterizing the entire force field anew. Force field parameters are usually selected in such a way that, for as large a set of test molecules as possible, they reproduce experimental or quantum mechanically determined geometries, conformational energies, binding energies, vibrational modes and many other properties that characterize a gas phase or condensed state. Numerous attempts have been made to modify known force fields to more reliably model the way a molecule responds to changing conditions, such as a dielectric environment. Most popular modifications include incorporating more interacting points (e.g. lone pairs or other extra points), point multipoles and polarizabilities. A major focus in research currently underway is to reliably model molecular responses to changes in a dielectric environment by including the effects of electronic polarization.
Polarization refers to the redistribution of a molecule’s electron density due to an electric field exerted by other molecules (Rick and Stuart 2002). If more than two molecules are involved, polarization leads to nonadditivity, since any two molecules will interact differently when polarized by a third molecule than if the third molecule was not present. For over 30 years, many attempts have been made to include the effects of polarization in simulations of molecular systems. In 1976, Warshel and Levitt (1976) implemented one of the first methods that included polarization effects in molecular mechanics. Yet only recently has substantial effort been devoted to developing consistently parameterized, polarizable force fields suitable for protein and nucleic acid simulations, as well as for ligands and organic molecules. In addition, attempts have been made to improve force fields by including other than nonadditive polarization energy components in intermolecular interactions. However, this review’s central focus is the polarization energy component of force fields. Here we evaluate work mainly published after 2000.
2. Classical molecular mechanical force fields
Molecular mechanical force fields can be divided into two core groups. The first group, Class I or diagonal force fields, includes AMBER (Cornell et al 1995, Wang et al 2000, Duan et al 2003), CHARMM (MacKerell et al 1998a), OPLS (Jorgensen et al 1996), GROMOS (Schuler et al 2001), ECEPP (Zimmerman et al 1977) and several other less popular force fields. The analytical formula used to calculate energy is defined as the sum of bonded (e.g. short range) and nonbonded terms, which describes the interactions between atoms separated by more than two bonds. The same nonbonded terms are used to determine interactions between atoms belonging either to the same or different molecules. For example, in the AMBER force field (assisted model building with energy refinement) (Cornell et al 1995), energy is calculated as the sum of intramolecular bonded terms (bonds, angles, torsions), intra- and intermolecular nonbonded terms (e.g. van der Waals and electrostatic terms, as modeled by the Coulomb formula, which describes interactions between partial charges on atoms):
| (1) |
| (2) |
| (3) |
The symbols represented are as follows: req and θeq are equilibrium values for bond length and angles between atoms; n is the dihedral multiplicity; γ is the dihedral angle phase; and Kr, Kθ and Vn are force constants for bonds, angles and dihedral terms. The following mixing rules for A and B van der Waals coefficients are often used when well depth (εi) and atomic radii (Ri), which describe interactions for the same atom types, are known:
| (4) |
| (5) |
Usually, the so-called 1–4 van der Waals and electrostatic interactions (e.g. interactions between atoms separated by three covalent bonds) are treated separately and their magnitude is scaled down. Small variations in a number of energy terms and their functional forms are found in Class I force fields.
The second group, Class II force fields, comprises the CFF (Niketic and Rasmussen 1977), CVFF (Dauber-Osguthorpe et al 1988), MMFF (Halgren 1999), MM3 (Allinger et al 1989), MM4 (Allinger et al 2003) and UFF (Rappe et al 1992). Functional forms of these force fields are more complicated and contain higher-order terms for calculating the bond and valence angle terms as well as nondiagonal, mixing terms in the Ebonded part of the energy. Such terms increase force field accuracy when calculating relative energies and geometries, as well as performing vibrational analyses.
Class I force fields have been developed mainly to reproduce condensed state properties (e.g. molecules in a liquid or crystal environment), which can be simulated by Monte Carlo or Molecular Dynamics methods. These force fields are ‘effective’ potentials because intermolecular two-body interactions implicitly absorb many energy contributions in an effective way. On the other hand, Class II force fields have been parameterized, with the focus on more precisely reproducing molecular structures, conformational equilibria, accurate descriptions of molecular vibrations (IR spectra) and intermolecular interactions of dimers in a vacuum. Thus, Class II force fields typically fail when applied to condensed state simulations (Halgren and Damm 2001).
The Coulomb formula describing interactions between point charges is typically employed for most force fields to evaluate electrostatic interactions. Interactions between higher-order multipoles are generally not included. In most cases, point charges are distributed at the nuclei. However, electrostatic interactions in MM3 force fields are described in terms of point dipoles located at chemical bonds (Allinger et al 1989). The nonbonded van der Waals interactions are modeled as the sum of attractive and repulsive terms that are proportional to R−n, where R is the distance between two interacting points. Usually, interacting points coincide with atom position but additional points, located at lone electronic pairs, have also been employed (Dixon and Kollman 1997). Most force fields apply a standard Lennard-Jones 12-6 (R−12 - R−6) potential (Cornell et al 1995, Jorgensen et al 1996, MacKerell et al 1998a); however, other R function potentials are also used, such as 9–6 (Dauber-Osguthorpe et al 1988), buffered 14–7 (Halgren 1999) or Buckingham-type exp(−α R) − 6 (Stone 1996).
How well do the above potentials correctly match the description of intermolecular interactions? To answer this question, we consider the current status of intermolecular interaction theories as formulated by quantum mechanics.
3. Quantum mechanical theories for intermolecular interactions
In 1930, London (1937) laid the foundation for today’s theories of intermolecular interactions. He proposed that the interaction energy between molecules is the sum of four basic components: electrostatic, induction, dispersion and exchange (Margenau and Kestner 1971, Stone 1996). All are caused by electrostatic interactions between all particles in the molecules (e.g. electrons and nuclei). These components can be associated with molecular properties, such as permanent multipole moments and static and dynamic polarizabilities. Each energy component has a specific interpretation: electrostatic energy results from interactions between each molecule’s permanent electric multipole moments; induction energy is defined by one molecule’s permanent multipoles interacting with multipole moments induced in another molecule; dispersion energy originates from interactions between mutually polarized electronic charge distributions, which are often approximated by interactions between each molecule’s instantaneously induced multipole moments; and exchange energy, which arises from repulsion between overlapping electron densities at short distances.
Two general quantum mechanical approaches are used to calculate intermolecular interactions: the supermolecular theory and the perturbation theory. An exhaustive introduction to these modern theories is found in reviews published by Chalasinski and Szczesniak (1994) and Jeziorski et al (1994), as well as in books published by Stone (1996) and Margenau and Kestner (1971).
According to the supermolecular approach, the interaction energy is defined as the difference between the energy of the dimer (EAB) and the energies of the two monomers (EA and EB):
| (6) |
Intermolecular interaction between two molecules measures about several kcal mol−1 and is five to six orders of magnitude less than the quantum mechanical energies of the dimer AB; therefore, the quality of the basis set and level of applied theory affects the calculation results. Chalasinski and Szczesniak (1994) and others have proposed an approach to decompose supermolecular energies into well-defined contributions. One problem with the supermolecular approach is the presence of basis set superposition error (BSSE) (Boys and Bernardi 1970). It is caused by a nonphysical lowering of the monomer’s energy in a dimer’s calculations, since each monomer uses a partner’s basis set to lower its own energy.
For perturbation theory, the simplest approach is based on applying standard Rayleigh–Schrödinger perturbation theory (RSPT), which is also called the ‘polarization approximation’. Here, the intermolecular interaction potential operator V is treated as a perturbation operator to the unperturbed Hamiltonian H0 = HA + HB for the dimer AB. In the polarization approximation, the total dimer wavefunction is a product of monomer wavefunctions: ΦAB = ΦAΦB. The Schrödinger equation for a dimer is as follows:
| (7) |
The formal parameter ξ is introduced to define orders of the perturbation expansion. The total dimer’s energy and wavefunction depend implicitly on ξ. When the value of the formal parameter ξ is equal to 1, the complete intermolecular interactions in equation (7) are recovered. As a result of the perturbation expansion, with respect to the parameter ξ, the total intermolecular interaction energy is defined as an infinite series (in ξ) of energy corrections called polarization energies:
| (8) |
The first order of polarization energy is the electrostatic interaction energy. In deriving , multipole expansion of the V operator can be applied, which leads to energy components depending on the powers of intermolecular separation, R. can be interpreted as an interaction between permanent multipole moments of monomers A and B. At the short intermolecular distance, an additional term, , becomes important. This term originates from mutual penetration of monomer electron clouds. Thus, the first-order interaction energy is expressed as follows:
| (9) |
In practical applications, electrostatic energy is often calculated as the interaction between point multipoles distributed over monomers at a set of points, which usually coincides with atom positions. Two formulations have been proposed: Stone’s distributed point multipole analysis (DMA) (Stone 1981) and Sokalski’s cumulative atomic multipole moments (CAMM) (Sokalski and Poirier 1983).
Polarization energies of the second and third order are sums of induction and dispersion energies (see the first paragraph of this section for interpretation of these components):
| (10) |
| (11) |
Induction and dispersion energy components can be expanded in terms of the power of intermolecular distance R, which yield well-interpreted terms that describe interactions between permanent and induced multipole moments and between mutually induced multipole moments, respectively.
One problem with polarization perturbation theory is that total dimer wavefunction is not antisymmetric with respect to electron exchange between monomers. To alleviate the problem, Jeziorski et al (1994) developed the symmetry-adapted perturbation theory (SAPT), in which the antisymmetrized product of monomer wavefunctions AΦAB = AΦAΦB is used for the zeroth-order approximation to the dimer wavefunction, instead of applying ΦAB, which is used in the polarization approximation. The A represents the intermolecular antisymmetrization operator. By applying this type of wavefunction, additional energy terms, such as Eexch, can be defined at each order of energy correction. The Eexch components make the perturbation energy expansion a rapidly converged series with correct asymptotic behavior, as compared to the polarization approximation. The perturbation expansion is defined as follows:
| (12) |
| (13) |
The most important exchange term contribution belongs to because it accounts for about 90% of the total exchange energy (Jeziorski et al 1994).
To obtain correct interaction energies for many-electron systems, intra-monomer electron-correlation effects need be taken into account (Jeziorski et al 1994). This problem can be solved by applying double-perturbation theory to each term of the polarization energy expansion (Jeziorski et al 1994). With this approach, each component of the nth-order interaction energy is expanded into a series with respect to the ith and jth orders of the intra-monomer correlation operator of molecules A and B, respectively:
| (14) |
As a result of this expansion, we can derive additional energy corrections to each , such as Eelst-corr, Eind-corr and Eexch-corr. The symmetry-adapted perturbation theory program (SAPT) (Jeziorski et al 1994) can be used here to conduct numerical calculations for the most important components of the interaction energy. Calculations can be practically performed for molecular systems containing 20–30 atoms. This is sufficient for accurately determining intermolecular potentials for molecular systems containing representative chemical groups important in biomolecular modeling. As an example, Bukowski et al developed the ‘first-principles’ accurate potential for water (Bukowski et al 2007).
Besides the above classification, intermolecular interaction energy for many-body systems may be expressed as a series of two-, three-, … n-body terms, which depend on the presence of two molecules, three molecules or more:
| (15) |
Three- and higher-body terms contain nonadditive contributions to the interaction energy. In table 1, we summarize the properties of key contributions to the total interaction energy and the presence of nonadditivity effects.
Table 1.
Intermolecular distance dependence, sign of energy, additivity characteristics and per cent contribution at R corresponding to the van der Waals (vdW) minimum and for larger distances, for the most important components of the intermolecular interaction energy for polar systems (Jeziorski et al 1994, Chalasinski and Szczesniak 1994).
| R dependence | Sign | Additive? | % Contribution in vdW minimum (%) | ||
|---|---|---|---|---|---|
|
|
R−1, R−2, R−3, … | +/− | Yes | 50–70 | |
|
|
exp(−a R) | + | Yes | 1–5 | |
|
|
exp(−a R) | + | No | 5–10 | |
|
|
R−4, R−6, … | − | No (nonadditivity could be +/−) | 10–20 | |
|
|
R−6, R−8, R−10, … | − | Yes | 15–30 | |
|
|
R−9, … | +/− | No (nonadditivity: could be: +/−) | 1–2 | |
| Various Ecorrel contributions | <5 |
Though popular among chemists, the charge-transfer term (van der Vaart and Merz 1999) is not included in table 1. Commonly accepted in the field of experimental spectroscopy and for partitioning interaction energies in the supermolecular approach, it is not easily defined within the scope of perturbation theory. More rigorously, charge-transfer energy is considered to be a part of the Einduction energy component. It is usually heavily contaminated by a basis set superposition error but is, apparently, less dependent on a basis set (see Stone (1996), pp 102–104). When the charge-transfer term is extracted from Einduction, it turns out to be relatively small, often negligible, and exponentially dependent on molecular separation. Stone and Misquitta (2009) demonstrated that the charge-transfer term is small, if proper handling of exchange repulsion is taken into account. The charge-transfer term is also attractive and nonadditive, as is the rest of Einduction.
All empirical force fields employ simplified formulae for calculating intermolecular interactions that are intended to reproduce complicated energy hypersurfaces; in this sense, they are effective potentials with various approximation levels and error compensations. But the question is, what is missing, and what can be improved on in those molecular mechanical force fields, given the present knowledge of rigorous theories? Most force fields utilize intermolecular potentials consisting of electrostatic, repulsion and attraction terms. Electrostatic interaction is modeled using simple Coulomb terms involving interactions between distributed partial atomic charges on atoms or on other points (~R−1, where R is the distance between interacting centers). Less often, electrostatic energy is calculated as the interactions between point dipoles on chemical bonds (~R−3) (Allinger et al 1989, 2003) or between higher-order multipoles, usually coinciding with the positions of nuclei (Ren and Ponder 2003). Repulsion between atoms is modeled by a single term that is proportional to either the power of R(~R−9, ~R−12, ~ R−14) or less often, but more accurate, by the term that is proportional to ~ exp(α R). The latter expression is more accurate because the magnitude of quantum mechanical exchange-repulsion energy depends on the overlapping of molecular electronic densities and, so, exhibits an exponential dependence on R. In the majority of cases, the attractive component of intermolecular interaction is represented by a dispersion term that is proportional to ~R−6, which neglects the higher-order powers of R.
Other ways used to improve the accuracy of each energy component include higher-order permanent multipoles (terms ~R−n, n > 1), multipole polarizabilities, higher-order dispersion terms and nonadditive terms, as well as an increasing number of interacting points in molecules, but these methods are much less often used (Rick and Stuart 2002). The attractive terms, which are proportional to greater than the (−6) power of R, are also rarely used. Such terms originate from the multipole expansion of Eind and Edisp energy components (see table 1). Thus, while the basic, most popular terms to describe intermolecular interactions, such as R−1, R−6 and R−12 (equation (2)), have been adopted by those working in classical force fields, they actually absorb many of the energy components in an effective, yet not always correct, way.
The charge-transfer term is explicitly treated in the SIBFA (sum of interactions between fragments ab initio computed) polarizable molecular mechanics force field (Gresh et al 2007) and in the CTPOL force field (Sakharov and Lim 2009). Merz et al (Ababou et al 2007) have also demonstrated the charge-transfer term’s importance in quantum mechanical simulations using semiempirical approaches, though its contribution is small and depends on energy-decomposition schemes.
Current force fields rarely include other than polarization energy terms that explicitly describe nonadditive interactions, but such terms could be important in various applications. For example, it was shown that an explicit triple exponential term employed to describe the repulsion between two water molecules and an ion improved the agreement between calculated and experimental enthalpies of formation for the ion–(H2O)n cluster as a function of n (Clementi et al 1980, Cieplak et al 1987). This term describes three-body nonadditive repulsion, which could be associated with some components that comprise the term.
Several force fields have been developed which incorporate the effects of electronic polarization in intra- and intermolecular interaction models. This is a step in the right direction, because Einduction, which is partially modeled by a polarization term used in the classical force fields, is an important component, contributing 10%–20% of the total interaction energy. Using a polarization term to some extent introduces a nonadditive, n-body effect. It is suggested that, by including polarization energy and other nonadditive terms in classical molecular mechanics, only a single set of parameters may be required to correctly describe both gas and condensed-phase environments (Ponder and Case 2003). In a number of applications it has already been demonstrated that polarization energy improves the agreement of molecular mechanics with experimental or high-level ab initio calculations. One classic example where polarization plays an important role is in potassium–benzene (π-cation) interactions. The highly polarizable benzene molecule strongly interacts with the ion and such interactions cannot be reproduced without including polarization effects (Caldwell and Kollman 1995, Ponder and Case 2003). In another example, we showed that including polarization energies and additional interacting points (mimicking lone pairs) substantially improved the interaction energy order and hydrogen bond distances for N-methyl-acetamide–water dimers (Cieplak et al 2001), compared to standard AMBER force fields (Cornell et al 1995).
In the following sections, we describe how polarization effects are modeled and applied in molecular simulations.
4. Polarizable force fields
The development of polarizable force fields is a vivid area of current research. The large number of papers published annually and a full issue dedicated to the subject in the Journal of Chemical Theory and Computation (Jorgensen et al 2007) validates its importance. Almost all key Class I force fields have a polarizable companion. Some, such as AMBER ff02, ff02EP (extra points) (Cieplak et al 2001, Wang et al 2006), CHARMM (Lamoureux et al 2003, Patel and Brooks 2004, Patel et al 2004), PIPF-CHARMM (Xie et al 2007), OPLS/PFF (Friesner 2005, Kaminski et al 2004, 2002, Maple et al 2005), OPLS-AAP/OPLS-CM1AP (Jorgensen et al 2007) and GROMOS (Geerke and van Gunsteren 2007a) have been developed as extensions of existing parameterizations. Other force fields, for example, AMOEBA (Ren and Ponder 2003, 2004), SIBFA (Gresh et al 2007), SDFF (Palmo et al 2003) and NEMO (Hermida-Ramon et al 2003), have incorporated a polarization term since their inception. Large portions of polarizable force fields are devoted to a water model for liquid-phase simulations. Reviews of these efforts can be found in articles by Halgren and Damm (2001), Rick and Stuart (2002), Ponder and Case (2003), Mackerell (2004) and Friesner (2005).
The value of developing polarizable force fields includes being able to model a molecular response to varying dielectrics of the environment. This effect is crucial for, say, modeling protein folding events where part of the amino acids form a hydrophobic core and so must be transferred from its water environment to the interior of a protein (Dill et al 1995) that is characterized by an entirely different dielectric environment (Fitch et al 2002, Garcia-Moreno et al 1997). Other examples include RNA folding in an environment full of divalent ions or the folding of membrane proteins in a lipid environment. In all cases, the energy surface needs to be represented as accurately as possible. Unfortunately, we have no unconditional proof that polarizable force fields are important and useful because all are still under development and their parameterization requires much more computing time than additive versions. Expanding force fields for additional polarization terms increases the computation time from 3 to 10 times, depending on implementation. This challenge is partially alleviated by current developments in programming. These include implementation of the particle-mesh Ewald (PME) method (Darden et al 1993) for fast and accurate treatment of electrostatic energy and other progress made in computer technologies.
At present, five groups of methods include polarization effects in force fields: fluctuating charge, Drude oscillator, induced point dipole, electronic polarization via quantum mechanical treatment (QM) (Dehez et al 2007) or mixed QM/MM (Murphy et al 2000, Senn and Thiel 2009), and polarization treatment using continuum solvent (and solute) (Gilson and Honig 1988, Honig and Nicholls 1995, Tan and Luo 2007). Hybrid methods, such as combining fluctuating charge and induced point dipole, have also been developed and tested (Stern et al 1999) but have not been pursued to date.
4.1. Fluctuating charge model
The fluctuating charge (FQ) model is based on the principle of electronegativity equalization (EE): a charge flows between atoms until electronegativities of the atoms become equalized. By adding this effect, a molecule’s charge-state distribution can be coupled to its environment, providing a way to incorporate the effects of polarization. Note that, with this approach, fluctuating charge and electronegativity equalization are used interchangeably. This method has been used in the universal force field (Rappe et al 1992), in a force field developed by Friesner’s group (Banks et al 1999) and in the CHARMM force field (Patel and Brooks 2004, Patel et al 2004).
Rick and Stuart (Xu et al 2002) designed a special polarizable water model, the TIP4P/FQ, which employs fluctuating charge methodology. Exhaustive reviews on this subject, up to 2002, can be found in the paper by Rick and Stuart (2002). Here we provide basic information about the fluctuating charge method. The formulae derived begin by expressing the energy required to create a charge, Q, on an atom as a Taylor expansion, which has been truncated after second-order terms:
| (16) |
Assuming a neutral atomic state as reference point, the energies required to create +1 and −1 charges on an atom, E(+1) and E(−1), can be calculated from equation (16). Combining those results leads to the following:
| (17) |
and
| (18) |
where IP and EA are the ionization potential and electron affinity, and χ0 is electronegativity, as defined by Mulliken (Mulliken 1934). The second-order coefficient (equation (18)) is the Coulomb repulsion between two electrons in the valence orbital (the self-Coulomb integral). It is used as 1/2J 0 to define the atomic ‘hardness’, xmlpi η (Parr and Pearson 1983), which represents the resistance to electron flow to or from an atom. Thus, equation (16) is rewritten:
| (19) |
Equation (19) can be generalized for a set of molecules (M) each containing Ni atoms. The total energy for such a system is taken as
| (20) |
The last term in equation (20), V(riα jβ), represents the system’s nonelectrostatic interactions, including bond stretching, angle bending, dihedral terms and other terms specific for given force fields as well as intra- and intermolecular van der Waals interactions. Optimum charge distribution is achieved by minimizing energy with respect to the charges on each atom:
| (21) |
Since derivatives are equal to electronegativities (equation (17)), such energy minimization is equivalent to equalizing electronegativities. Equation (21) adds to the conditions necessary for maintaining a molecule’s total charge and leads to a set of simultaneous equations that yield a set of self-consistent charges for the given configuration of atoms. Since charges depend on interactions with other charges located on the same molecule or other molecules, their values change with every time step or sampled configuration during a simulation. Consequently, the process has been aptly called the fluctuating charge method (Rick et al 1994). During the charge equalization process, charges may be constrained to movement within a molecule or movement between any atom pairs. Application of the latter option leads to charge-transfer effects. Unfortunately, the model can predict nonphysical, large charge transfers at great distances and generally overestimates the effect (Rick and Stuart 2002). For this same reason, the method may predict too large dipole moments of single molecules, especially in extended systems like polymers. To alleviate the problem, modifications of the fluctuating charge approach were proposed. These include bond charge increment (BCI) (Banks et al 1999), atom–atom charge transfer (AACT) (Chelli et al 1999) and atom-bond electronegativity equalization ABEEM/MM/(Yang and Zhang 2006) methods.
The electronegativities and hardnesses used in the fluctuating charge method are experimental, are parameters derived using quantum mechanics (Rappe et al 1992) or are adjustable parameters, as implemented by Friesner et al (Banks et al 1999, Stern et al 1999) and Patel et al (Patel and Brooks 2004, Patel et al 2004). According to equation (20), the hardnesses are used as atomic and interatomic parameters. The heterogeneous parameters can be derived from atom-type values, as proposed by Patel et al (2004), by employing the combining rule introduced by Nalewajski et al (1988):
| (22) |
where Rαβ is the distance between atoms α and β. This formula is effectively applied to calculate 1–2, 1–3 and 1–4 interaction types. At sufficiently large atom separation (greater than 2.5 Å), the locally screened Coulomb term reverses to proper 1/Rαβ limiting behavior. It has been shown (Patel and Brooks 2004) that the dipole polarizability tensor is related to hardness matrix elements by
| (23) |
where Δr̄ represents atomic coordinates relative to the center of geometry. This relation can be used either to determine a set of hardness parameters, by fitting them to quantum mechanically derived or experimental polarizabilities, or to determine the quality of assumed hardnesses in reproducing molecular polarizability.
Solving the coupled set of equations (21) for the charges can be performed by matrix inversion, iteration or extended Lagrangian methods. The iteration method, the most efficient, is usually used for propagating the charges during molecular dynamics simulations (Patel and Brooks 2004, Rick et al 1994, 1995). In the extended Lagrangian method, the equation of motion for calculating electronic degrees of freedom is given by
| (24) |
where fluctuating charges are assigned fictitious masses mQ,iα and λi is the Lagrange multiplier for molecule i containing atom α, which is related to the total molecular charge neutrality constraint:
| (25) |
The chosen magnitude of the fictitious mass in equation (24) should be small enough to achieve a prompt response to changes in electronic potential but large enough to be able to use reasonable time steps in molecular dynamics simulations.
The fluctuating charge method has already been applied in several cases, such as simple amino acids, dipeptides, tetrapeptides, gas-phase minimization and isolated protein dynamics (Banks et al 1999, Patel et al 2004), as well as in simulations of methanol, ethanol, other organic liquids (Patel et al 2004, 2005a, 2005b), bulk and liquid–vapor interfaces, and a hexane–water interface (Patel and Brooks 2006). This method predicts liquid vaporization enthalpy within 2% and bulk density within 1%, compared to experimental values, while the additive CHARMM C27r force field underestimates vaporization enthalpy by roughly 20%. The fluctuating charge hexane model realistically captures bulk dielectric properties. This model predicts a value 1.94 for the bulk dielectric constant for liquid hexane. This is in good agreement with experimental values, which range from 1.9 to 2.02. Patel and Brooks also proposed a parameterization to model proteins in solvents (Patel et al 2004). Most recently, Patel et al (Davis et al 2009) reoptimized this method together with some of the CHARMM force field parameters and applied it to simulate a DMPC lipid bilayer and to predict dielectric permittivity inside and outside such a bilayer.
One problem with the fluctuating charge method is that it does not reproduce out-of-plane polarization for planar or linear chemical moieties. This is understandable because electronegativity equalization proceeds along the bonds between atoms. This effect could be fixed by using additional out-of-plane sites. Stern et al (1999) demonstrated that fluctuating charges alone are not always sufficient, for example, to reproduce the energy order of alanine tetrapeptide conformers. In this situation, inducible point dipoles perform better (Friesner 2005, Kaminski et al 2002, Stern et al 1999). Thus, the authors initially introduced a hybrid model involving fluctuating charge and inducible dipoles (Stern et al 1999) but later focused on only pure inducible dipole models (Friesner 2005, Kaminski et al 2004, 2002).
Stenhammar et al (2009) generalized the fluctuating charge method by providing a general expression for the distribution of the fluctuating 2l-pole moment Ml that interacts with a continuum dielectric medium. They demonstrated that the solvation free energy of fluctuating electric moments diverges with increasing order of the moment l, but did not suggest a way to correct this problem. Note that the fluctuating charge method has not been reported for nucleic acid applications.
4.2. Drude oscillator model
Drude oscillator methods are also known as shell models. These models incorporate electronic polarizability by representing an atom or ion as a two-particle system: a charged core with charge qi,0 and a charged shell with charge qi,D. The core and shell, also called a Drude particle (Drude 1902), are linked by a harmonic spring. The magnitude of both charges is fixed. Thus electronic polarization is mimicked by relative displacement of both charges due to an external electrostatic field. Atomic polarizability, αi, is related to force constant k of the harmonic spring connecting the core and shell and is determined by (Rick and Stuart 2002). By fitting molecular polarizability data and experimental intermolecular interaction energies and other properties, charge magnitudes and harmonic force constants may be obtained.
The electrostatic energy between atoms is calculated as the sum of all charge–charge Coulombic interactions, the number of interactions needed to compute quadruples. However, there is no necessity to calculate more expensive dipole field tensors as is so for explicitly induced dipole interactions. Since nonelectrostatic interactions, such as short-range repulsion and van der Waals interactions, are purely electronic in nature, they are considered to act only between shells rather than cores (Rick and Stuart 2002). Since repulsion in molecular mechanical force fields is usually modeled as a fairly steep function (e.g. exp(−α R) or R−12) and is associated with shell interaction, then in a natural way, the shell model avoids a polarization catastrophe observed in inducible dipole polarization approaches (Lindan 1995).
As with other approaches polarizable degrees of freedom may be solved iteratively, analytically or by propagating them in molecular dynamics simulations via extended Lagrangian methods (Rick et al 1995), as initially proposed by Mitchell and Fincham (1993). Jacucci et al (1974) report one of the first implementations and applications of this method in their molecular dynamics simulations of optical phonons in solid sodium chloride. Here, the Drude particles were massless. This initial work was later extended by Mitchell and Fincham (1993) who assigned small masses to the shells and determined their motion in the same way as heavy cores by numerically integrating the classical equations of motion. The work was also directed at simulating ions in molten salt states.
Rick and Stuart (2002) reviewed pre-2002 developments and applications of the Drude oscillator method. Recent vigorous implementation and parameterization by van Gunsteren, MacKerell and Roux have generated greater momentum for this method. The Drude particle approach has been incorporated in a general way into CHARMM (Lamoureux et al 2003, 2006) and GROMOS (Geerke and van Gunsteren 2007a) molecular modeling packages, followed by developing force fields for specific groups of molecules. This progress has allowed molecular dynamics simulations to be conducted for biomolecular systems in vacuo and in condensed phases.
Recently, new polarizable water models become available for the CHARMM force field. In the original model, called SWM4-DP (simple water model with four sites and Drude polarizability), the Drude particle, which was attached to an oxygen atom, was assigned a positive charge (Lamoureux et al 2003). It was not intuitive, since the particle was meant to represent the system’s electronic degrees of freedom. Subsequently, this model evolved into the SWM4-NDP (negative Drude particle), with reversed charges on the oxygen atoms and Drude particle (Lamoureux et al 2006). Unlike the standard shell approach, the Lennard-Jones parameters have been associated with the oxygen atom instead of the Drude particle. The SWM4-NDP model does a good job of reproducing bulk water properties at room temperature and pressure, including vaporization enthalpy, the static dielectric constant (79 ± 3 versus 78.4 exp.) and the self-diffusion constant (2.33±0.02×10−5 cm2 s−1 versus 2.3×10−5 cm2 s−1 exp.). However, the first minimum and second maximum of the gO O radial distribution function is flatter (e.g. has less structure compared to experimental curves) and the molecular polarizability is smaller (0.978 Å3) compared with experimental values (1.44 Å3).
The next step in developing polarizable CHARMM force fields was devoted to alkanes, the main goal being to devise a set of transferable electrostatic parameters for CH3, CH2 and CH groups that could be used for lipids or other biomolecules. This force field is quite good at reproducing quantum mechanical molecular polarizabilities and experimental dielectric constants, including their relative ordering among alkenes. Yet, the self-diffusion constants are 5%–11% less than experimental values. Later on, Drude particle parameterization was extended to other types of molecules, such as ethanol (Noskov et al 2005) and other alcohols (including lone pairs on oxygen atoms) (Anisimov et al 2007), aromatic (Lopes et al 2007) and heteroaromatic compounds (Lopes et al 2009), and liquid amides (Harder et al 2008). That parameterization has now been applied to calculate the membrane dipole potential (Harder et al 2009).
In all the above parameterizations for CHARMM force fields, Drude particles are attached to nonhydrogen atoms, and Lennard-Jones parameters are associated with real atoms instead of Drude particles. The 1–2 and 1–3 electrostatic interactions between atoms separated by one and two bonds, respectively, are included, which can cause a polarization catastrophe. To avoid this outcome, screened dipole–dipole interactions have been incorporated for short-range distances, as suggested by Thole (1981) Screening has been implemented by smearing the charge on Drude particles and real atoms. Screened dipole–dipole interactions were then calculated as interactions between a smeared charge represented by the Slater function and a point charge (Noskov et al 2005). In polarizable molecular dynamics simulations, the extended Lagrangian double-thermostat formalism (Lamoureux and Roux 2003) has been used.
In another development, van Gunsteren et al demonstrated a charge-on-spring method to explicitly treat electronic polarization in the GROMOS force field (Yu and van Gunsteren 2005). Also a Drude particle method, it was initially parameterized to reproduce the properties of liquid, gas and crystal phases of water (Geerke and van Gunsteren 2007a, Yu et al 2003, Yu and van Gunsteren 2004, 2005), ethylene glycol (Geerke and van Gunsteren 2007c) and dimethyl ether (Geerke and van Gunsteren 2007b), and ions in aqueous solution (Geerke and van Gunsteren 2007a). In the latter work, however, it is not clear why the authors chose to use such large negative values for the charges-on-spring (−8e). Geerke and van Gunsteren (2007b) carried out the first work that employed a thermodynamics integration method in Drude oscillator polarizable force fields as a way to calculate ΔGpola, which corresponds to the free-energy difference between identical systems described by the polarizable and nonpolarizable models. The authors performed calculations for dimethyl ether as a pure liquid, as a solute in cyclohexane and in aqueous solution, and as a solvent for a chloride ion. It was found that, for the cyclohexane solution and the pure dimethyl ether liquid, ΔGpola is relatively small. The free energy of hydration, ΔGhyd, for the nonpolarizable model of dimethyl ether solute in water was found to be significantly too positive compared to experiment. ΔGpola for the dimethyl ether solute in water was found to be of the same order as this discrepancy, leading to a ΔGhydr value for the polarizable model of dimethyl ether close to experiment. Both those results, from nonpolar and polar solvent simulations, demonstrate that including polarization effects significantly improves transferability of the dimethyl ether parameters. A single set of parameters can be used for a proper description of the dimethyl ether solvation in nonpolar and aqueous environments, in contrast to the nonpolarizable model.
Worth noting is the recent application of the Drude model in modeling thermal conductivity and other transport coefficients for ionic materials (e.g. melted LiCl, NaCl and KCl) (Ohtori et al 2009). The authors demonstrated that agreement with experimental values is almost quantitative. Here again, this confirms that polarization effects are crucial to correctly model thermal conductivity.
So far, there is no consistent and complete Drude model parameterization available for proteins and nucleic acids.
4.3. Induced point dipoles
The induced point dipole model is the most studied approach for molecular polarization. It is incorporated into several current force fields, such as OPLS/PFF (Kaminski et al 2002, Friesner 2005), AMOEBA (Ponder and Case 2003, Ren and Ponder 2003, 2002) and AMBER ff02, ff02EP (extra points) (Cieplak et al 2001) and ff02r1 (Wang et al 2006).
In the classical point dipole approach, polarization energy is described as the interaction between static point charges and the dipole moments they induce. Ponder et al (Ren and Ponder 2003, 2002) expanded on this model by including the interaction between induced dipoles and higher permanent electric moments, up to quadrupoles, that were derived from a distributed multipole analysis (DMA) (Stone 1981). Only the NEMO force field (Holt and Karlstrom 2008) explores the possibility of including interactions between permanent multipoles and higher-order induced multipoles involving higher-order hyperpolarizabilities. Other force fields do not include such contributions, mainly due to computation cost, convergence issues and difficulties in parameterizing such interactions.
Most polarizable water models developed now employ the point dipole approach (Caldwell and Kollman 1995, Cieplak et al 1990, Dang 1998, Ponder and Case 2003, Ren and Ponder 2003, Walsh and Liang 2009).
In a polarizable AMBER version (Cieplak et al 2001, Wang et al 2006), and in other force fields that employ the point dipole model, an additional energy term, Epol, is added to the total energy:
| (26) |
The Epol describes explicit polarization arising from dipolar interaction between permanent partial charges and induced dipoles. It is calculated from the following formula:
| (27) |
Summation runs over all interacting sites i, αi denotes isotropic point polarizability of atom i, is the electrostatic field on atom i due to partial charges (or higher-order multipoles) and Ei is the electrostatic field on atom i due to charges (or higher-order multipoles) and induced dipoles.
| (28) |
is an induced dipole moment at the ith site. Ti j is the dipole–dipole interaction tensor:
| (29) |
where I is the identity matrix and x, y and z are Cartesian components along the vector between atoms i and j at distance ri j.
Most point dipole force field implementations, including AMBER force fields (Case et al 2005, Cieplak et al 2001), use interactions between static charges and induced dipoles to model polarization effects. The AMOEBA (Ren and Ponder 2003) force field, which is incorporated into TINKER (Ren and Ponder 2003) and AMBER (Case et al 2005, 2008) programs, uses interactions between charges, higher-order static atomic multipoles and induced point dipoles. Therefore, in equation (27) represents the electrostatic field on atom i from partial charges and higher permanent moments. Ei represents the sum of and the electrostatic field on atom i due to other induced dipoles. According to this formalism, multipoles at each ith site are represented as a vector: Mi = [qi, μi,x, μi,y, μi,z, Qi,xx, Qi,xy, Qi,xz, …, Qi,zz, …]T, where q, μ and Q are the monopole, dipole and quadrupole moments, respectively. The multipole expansion is limited to quadrupoles and multipole moments are defined in local frames with atomic sites as origins. A molecule’s static multipole moments are determined by Stone’s distributed multipole analysis (Stone 1981). Using the above definition of Mi, equations (28) and (29) can be generalized. In this formalism, the atomic multipole moments are now composed of both permanent and induced contributions (Rasmussen et al 2007, Ren and Ponder 2003, 2004, 2002):
| (30) |
To determine induced multipole moments, equation (28) becomes
| (31) |
where induced multipoles are truncated at dipoles and αi is atomic polarizability. is the interaction tensor between sites i and j. The matrix elements of the multipole interaction T matrix are derived by differentiating inverse distances between sites:
| (32) |
The interaction energy between sites in this generalized approach is
| (33) |
Equations (28) and (31) can be solved for induced dipole moments either iteratively or by matrix inversion. For molecular dynamics simulations, induced dipoles are treated by extended Lagrangian formalism, similarly to that used for fluctuating charge or Drude oscillator polarization models. To date, no higher-order induced multipoles have been used in molecular dynamics.
Computational results critically depend on the quality of molecular polarizability. In most approaches, total molecular polarizability is derived from a distribution of point polarizabilities usually associated with atoms. In nonadditive, also called interactive, polarization models (Applequist et al 1972, Birge 1980, Thole 1981, van Duijnen and Swart 1998), each of a molecule’s polarizable sites is allowed to respond to an external electric field not only from other molecules but from other sites within the same molecule. Consequently, all interacting sites polarize themselves. Under certain conditions, two inducible dipoles at short distances can cause a polarization catastrophe. One reason might be the current method of using point polarizabilities instead of more accurate approaches whereby interactions between atoms are modeled by diffuse charge distribution. To avoid this problem, the 1–2 and 1–3 bonded polarization interactions can be turned off, as is done in AMBER ff02/ff02EP (Cieplak et al 2001) or PFF (Kaminski et al 2004, 2002) force fields. Alternatively, one can apply distance-dependent damping for interactions on short distances or use both procedures. In the AMOEBA force field, a mixed approach is proposed, where both the distance-dependent damping function and exclusion of some of the 1–2 interactions (Ren and Ponder 2002) are applied. In PIPF-CHARMM (Xie et al 2007), both the damping function and exclusion of 1–2, 1–3 and 1–4 interactions are applied. As shown below, Thole’s damping procedure has practically no effect on intramolecular induced dipole charge interactions at distances longer than 1–2 and 1–3 bonds. Thus, it is neither necessary nor consistent to exclude 1–4 interactions and to apply Thole’s damping together.
In 1981, Thole (1981) proposed several schemes for damping interactions between inducible dipoles at short distances. He demonstrated, after Silberstein (1917), that for a diatomic molecule with bond length r, the values of the polarizabilities parallel and perpendicular to the bond axis are as follows:
| (34) |
| (35) |
When r approaches (4αAαB)1/6, α|| goes to infinity, which is a source of polarization catastrophe. Thole proposed methods to avoid it by using the interaction between smeared charge distributions instead of point charges at short distances. This led to changing the interaction tensor T in such a way that it does not behave as r−3 for small interatomic distances. To represent smeared dipole moments, several types of charge densities have been tested (Thole 1981, van Duijnen and Swart 1998). Among them, linear and exponential schemes have garnered the most interest (van Duijnen and Swart 1998):
| (36) |
and
| (37) |
where u = rij/(αiαj)1/6 is the effective distance as a function of atomic polarizabilities α, rij is the distance between atoms i and j, and a is the screening length that controls the width of the smeared charge distribution. Ren and Ponder (2003) employed another Thole’s charge density in the AMOEBA force field, which has been implemented in Tinker and AMBER programs. It has the following form:
| (38) |
Several other force fields adopted this charge density form (Masella et al 2008, Xie et al 2007). However, Thole (1981) demonstrated that the r.m.s. value of relative errors from optimizing atomic polarizabilities for a set of test molecules was the lowest for linear density (equation (41)). Recently, Masia et al (2005) suggested another charge density form, a normalized three-dimensional Gaussian function, which can be used to damp polarization at short range:
| (39) |
Applying the above densities leads to the following compact form of a damped dipole–dipole interaction tensor (Masia et al 2005, Thole 1981, van Duijnen and Swart 1998):
| (40) |
where fe and ft are distance-dependent screening functions. In the linear Thole model (van Duijnen and Swart 1998) fe and ft are defined:
| (41) |
In the exponential Thole model, fe and ft are defined:
| (42) |
For density, as defined by equation (38) (Tinker-exponential model), fe and ft are defined:
| (43) |
For Gaussian density (equation (39)) screening coefficients (Masia et al 2005) are given:
| (44) |
| (45) |
For completeness in the Applequist et al (1972) polarization model, where screening is not present, fe and ft are constant:
| (46) |
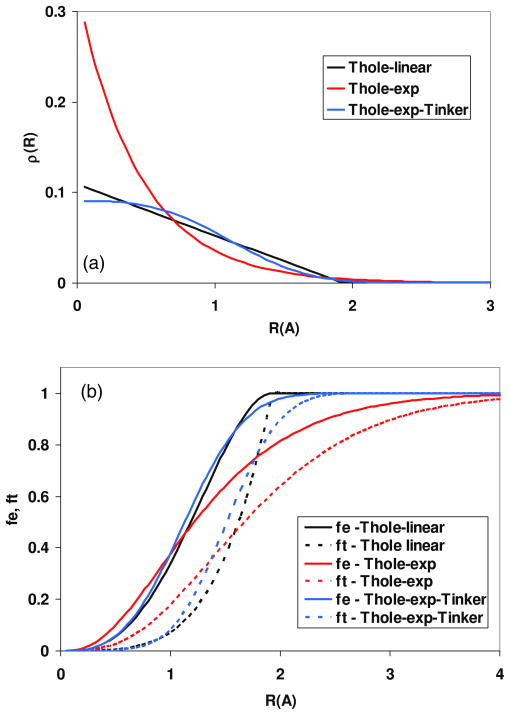
Higher-order modified interaction tensors are presented in original papers by Ponder et al (Ponder and Case 2003, Ren and Ponder 2003, 2002). Theory and implementation of Thole’s polarization scheme into molecular dynamics has also been presented by Burnham et al (1999) in a study of an all-atom polarizable water model derived from first principles. Masia et al (2005) provide an interesting discussion of damping functions and a more detailed derivation of screening functions. They explore the applicability of various screening functions and introduce an additional screening form in which charge distribution is described by a Gaussian function (equation (39)). However, when applying screened point dipole models to ion–CCl4 and ion–H2O systems (ion: Li+, Na+, Mg2+, Ca2+), the linear screening function (equation (36)) yields the best results, followed closely by the exponential function (Tinker-exp, equation (38)), with the Gaussian function taking last place. This agrees with the results of Thole (1981) and with the behavior of density distributions and screening coefficients as a function of distance (see figure 1). The plots presented in figure 1 are computed for a hydrogen atom interacting with a tetrahedral carbon atom using the recently determined set of polarizabilities developed by Wang et al (2009) (see table 2(a)). The method’s parameters specific for C and H were used. The plots show that both Thole-linear and Thole-exp-Tinker behave similarly at short distances, and they screen induced dipoles more effectively, compared to the Thole-exponential model. It is worth noting that damping functions are effective within a 0 to 2 Å range (e.g. distances involving 1–2 and 1–3 interactions).
Figure 1.
(a) Charge densities as a function of distance R (equations (36)–(38)). (b) Screening functions fe and ft (equations (41)–(43)) as a function of distance R between C-sp3 and H atoms (equations (36)–(38)). In each case specific for a given method, atomic polarizabilities were used (see table 2(a)) to calculate effective distance u.
(This figure is in colour only in the electronic version)
Table 2.
Atomic polarizabilities (au) determined by GA (Wang et al 2009) optimization to reproduce experimental molecular polarizabilities for a set of 420 molecules (Bosque and Sales 2002). Four different polarizability models: Applequist, Thole-linear, Thole-exponential and Thole-exponential used in the Tinker program, have been employed. Optimization was performed for the fully interactive model and the model in which induced dipole interactions 1–2 and 1–3 were screened; 1–4 interactions were scaled down by 1.2, as with regular 1–4 electrostatic interactions in the AMBER force field. AUE—average unsigned error, RMSE—root mean square error, APE—average per cent error.
| (a)
| |||||
|---|---|---|---|---|---|
| Applequist | Thole-lin. | Thole-exp | Thole–Tinker | ||
| C1 | 2.83 | 11.32 | 9.22 | 12.03 | sp1 |
| C2 | 4.31 | 11.49 | 10.55 | 12.04 | sp2 |
| C3 | 4.77 | 7.74 | 8.25 | 6.39 | sp3 |
| H | 1.49 | 3.97 | 3.24 | 4.47 | |
| NO | 5.07 | 14.95 | 13.16 | 16.54 | N in nitro |
| N | 3.38 | 8.56 | 7.83 | 8.32 | Other N |
| O2 | 2.26 | 4.33 | 3.81 | 3.44 | sp2 |
| O3 | 2.60 | 4.60 | 4.28 | 4.17 | sp3 |
| F | 3.07 | 4.11 | 3.22 | 4.11 | |
| Cl | 13.78 | 16.78 | 16.89 | 16.86 | |
| Br | 21.01 | 24.07 | 24.58 | 23.75 | |
| I | 33.03 | 37.39 | 38.67 | 37.18 | |
| S4 | 11.33 | 20.64 | 19.40 | 20.10 | S—sulfones |
| S | 14.78 | 24.49 | 23.73 | 23.77 | Other S |
| P | 6.26 | 15.31 | 13.39 | 14.42 | |
| a | — | 2.06 | 2.01 | 0.38 | Screen. length |
| AUE | 3.01 | 1.31 | 1.12 | 1.32 | |
| RMSE | 4.55 | 1.89 | 1.69 | 1.92 | |
| APE (%) | 3.82 | 1.55 | 1.30 | 1.58 | |
| (b)
| |||||
|---|---|---|---|---|---|
| No 1–2, 1–3 dipole interactions 1–4 interactions scaled down by 1.2 |
|||||
| Applequist | Thole-lin | Thole-exp | Thole–Tinker | ||
| C1 | 9.65 | 9.65 | 9.55 | 9.66 | sp1 |
| C2 | 8.76 | 8.72 | 8.61 | 8.71 | sp2 |
| C3 | 6.21 | 6.12 | 5.77 | 6.06 | sp3 |
| H | 2.99 | 3.03 | 3.19 | 3.06 | |
| NO | 10.11 | 10.06 | 9.94 | 10.12 | N in nitro |
| N | 6.30 | 6.26 | 6.09 | 6.24 | Other N |
| O2 | 4.00 | 4.01 | 4.04 | 3.99 | sp2 |
| O3 | 4.09 | 4.07 | 3.97 | 4.08 | sp3 |
| F | 3.36 | 3.40 | 3.55 | 3.43 | |
| Cl | 16.03 | 16.04 | 16.11 | 16.05 | |
| Br | 23.65 | 23.66 | 23.72 | 23.67 | |
| I | 37.65 | 37.63 | 37.57 | 37.64 | |
| S4 | 15.89 | 15.87 | 15.99 | 15.97 | S—sulfones |
| S | 21.48 | 21.46 | 21.36 | 21.44 | Other S |
| P | 12.33 | 12.27 | 12.23 | 12.25 | |
| a | — | 2.39 | 2.11 | 0.79 | Screen. length |
| AUE | 0.98 | 0.98 | 1.02 | 0.99 | |
| RMSE | 1.53 | 1.54 | 1.57 | 1.54 | |
| APE (%) | 1.17 | 1.17 | 1.22 | 1.18 | |
Masia et al (2005) questioned the validity of extending the same intramolecular screening approach into intermolecular interactions.
In the AMBER ff02 force field, induced dipoles are calculated using the Applequist model (e.g. no damping) with atomic polarizabilities, as determined in Applequist’s original paper (Applequist et al 1972). In ff02 and ff02EP (Cieplak et al 2001), as well as ff02r1 (Wang et al 2006) force fields all short-range 1–2 and 1–3 interactions (e.g. between atoms separated by one or two chemical bonds) were excluded. Once those interactions are removed, overall polarizability of the molecule is smaller than that determined experimentally, so this approach leads to a slight underestimation of the polarization effects in those parameterizations.
AMBER polarizable force fields provided the first complete set of parameters for all amino acids and nucleic acids. The AMOEBA force field, as incorporated in Tinker and AMBER programs, also facilitates protein simulations. The AMBER polarizable force field ff02 employs Applequist’s interactive polarization model and almost unchanged van der Waals and internal parameters from an additive version of the AMBER ff99 force field (Cheatham et al 1999, Wang et al 2000). In this force field, a special procedure (described below) has been applied to derive atomic charges consistent with the polarization model. At present, only a limited number of test simulations for proteins have been conducted with AMBER or AMOEBA polarizable force fields (Cieplak et al 2001, Jiao et al 2008, Ponder and Case 2003). Recently, ff02 and ff02EP were used by Sagui et al (Babin et al 2006, Baucom et al 2004) to perform extensive simulations for short DNA oligomers and to compare results with the force field’s additive version. Also, Vladimirov et al (2009) conducted simulations for A-DNA and B-DNA and for the rhodamine 6G-DNA complex using polarizable ff02 and additive ff99 AMBER force fields. Polarization plays an essential role in nucleic acids, which are highly charged polymers. Indeed, in multiple nanosecond-long crystal and liquid DNA simulations, Segui et al demonstrated that a polarizable ff02 force field represents better the crystal structure and sequence-dependent effects observed in an experiment, as measured by helical parameters than the additive ff99 and polarizable ff02EP containing extra interacting points.
Simulations have also been performed using the particle-mesh Ewald method (Toukmaji et al 2000) to calculate interactions between atomic charges and induced dipolar interactions. Polarizable ff02 force field simulations were stable on a nanosecond timescale, and the structures were close to the experimental ones. Helical parameters were reproduced well. For example, the positive opening for A–T base pairs and the negative opening for C–G base pairs were observed; the roll and helical twist reproduced sequence-dependent experimental behaviors. On average, the minor groove width in those simulations reproduced the experimental groove fairly well. On the other hand, the ff02EP, which attempts to better represent interactions, did not behave better or more stably in simulations, possibly because of a lack of van der Waals parameters on extra points carrying charges. Results obtained with ff02EP are slightly inferior to those from ff02, but they are still an improvement over those with a nonpolarizable ff99 force field. In an interesting study, Vladimirov et al (2008, 2009) applied additive ff99 and nonadditive ff02 force fields to study solvent reorganization energies in DNA electron transfers. It has been demonstrated that solvent reorganization energies associated with electron transfer are about 30% lower than those corresponding to an additive force field. However, the effective optical dielectric constant was equal to , which agrees satisfactorily with experimental data and the Marcus theory (Marcus 1993).
The AMOEBA force field has been successfully applied to various molecular systems, including liquid water (Ren and Ponder 2003, 2004), simple ion solvation (Grossfield et al 2003), divalent cation solvation (Piquemal et al 2006b), N-methyl-acetamide dimers, alanine dipeptide conformational study (Ren and Ponder 2002) and several types of organic molecules containing biologically important functional groups (Rasmussen et al 2007). The AMOEBA force field has recently been applied to trypsin with benzamidine and diazamidine complexes (Jiao et al 2008), though extensive testing of this force field for proteins has yet to be done.
Other force fields that employ the inducible point dipole model are PIPF-CHARMM (Xie et al 2007); OPLS-AAP and OPLS-CM1AP, as reported by Jorgensen et al (2007); NEMO (Hermida-Ramon et al 2003, Holt and Karlstrom 2009) and SIBFA (Gresh et al 2007). All have been applied to several cases involving organic molecules, ions, water, dipeptides and others.
PIPF-CHARMM (Xie et al 2007) marks the first attempt to combine the standard CHARMM22 (MacKerell et al 1998b) force field with polarizable point dipoles from one of the Thole (1981) screening functions, the same as that used in the AMOEBA (Ren and Ponder 2002) force field (equation (38)). To reiterate, the screening acts between atoms located at short-range distances but the authors applied a procedure to automatically exclude 1–2, 1–3 and 1–4 interactions within the same molecule. For this approach, parameters from Lennard-Jones nonbonding interactions were maintained as was for the original additive force field. Further, atomic charges were scaled down to compensate for a self-polarization effect as a way to correct dipole moments in the gas phase. As of now, the PIPF has been applied first to simulate liquid amides and alkanes before attempts are made to construct a complete force field for proteins. Results show that PIPF works reasonably well at reproducing experimental heats of vaporization and liquid densities, as well as enhancing dipole moments in liquids compared to the gas phase.
Jorgensen et al (2007) introduced the OPLS-AAP/CM1AP force field. It is used in Monte Carlo simulations, with Epol (equation (27)) computations simplified by neglecting the contribution made by induced dipoles to the electric field. Because the converged iterative solution for induced dipoles is not obtained, the polarization effect is less accurate. Necessary atomic polarizabilities required for this force field have been derived from quantum mechanical calculations on interactions between cation-π aromatic molecules. This force field’s main application focuses on interactions between chloride ions and phenols. Adding inducible dipoles or point polarizabilities on nonhydrogen atoms as a way to include explicit polarization effects has been shown to improve the magnitudes and interaction trends associated with substituent effects in phenol–chloride ion complexes.
Madurga and Vilaseca (2004) published a paper that applies double free-energy perturbation and polarizable force fields to Monte Carlo simulations. They studied how solute polarization influences the conformational equilibrium of 1,2-dichloroethane in aqueous solution. Applequist’s atomic polarizabilities were used together with an iterative procedure to solve for inducible dipoles. Reaction field correction has also been used for both electrostatic interactions and polarization energy. The population of gauche conformations was shown to be greater with polarizable force fields compared to experimental values. The accuracy of this force field still needs to be determined.
The NEMO force field (Hermida-Ramon et al 2003) is based on partitioning the quantum mechanical interaction energy at the Hartree–Fock level into first- and second-order perturbation theory terms. The system’s total energy is the sum of electrostatic, induction, exchange and dispersion energy components. Each is calculated from properties obtained from self-consistent field (SCF) wavefunctions of interacting monomers. Thus, each energy component could be improved separately. Dispersion is represented by the London formula. Electrostatic energy in the NEMO model is calculated between molecular charge distributions represented by a multicenter multipole expansion truncated at the quadrupole moment (Stone 1981). Polarization energy is calculated as the interaction between the same permanent multipoles, up to quadrupoles, and induced atomic dipole moments obtained via isotropic atomic polarizabilities. Thole’s damping procedure (Thole 1981), as defined by equation (38) (Tinker-exponential), is used to screen short-distance interactions. The NEMO method has limited applications today but its usefulness and improvement over empirical force fields has already been demonstrated in calculated glycine dipeptide conformation profiles compared to other empirical force fields (Hermida-Ramon et al 2003), in interactions of ion–water droplets in Monte Carlo simulations (Hagberg et al 2005) and in urea transition from nonplanar to planar conformation in the 99 water droplet complex (Hermida-Ramon et al 2007). Published papers tout new developments in the NEMO model. These include higher-order multipole moments that improved the response of a molecule to an external electrostatic field, which may lead to a more accurate description of intermolecular interactions (Holt and Karlstrom 2009, 2008). However, deriving the general force field for proteins and nucleic acids is difficult and has not yet been attempted.
SIBFA (Gresh et al 2007) is another force field derived from quantum mechanical intermolecular interaction theories. Its history dates back to the 1960s’ pioneering work of Claverie, Rein and Pullman (Claverie and Rein 1969, Pullman et al 1967). They describe polarization energy as the interaction between permanent multipoles, up to quadrupoles, distributed on atoms and bond barycenters and anisotropic point dipole polarizabilities, which are distributed on bond barycenters and heteroatom lone pairs. Both distributed multipoles and polarizabilities are obtained from molecular orbital calculations performed for an entire molecule or molecular fragment. One difficulty with this force field is in determining parameters for a new molecule, and this force field cannot be used for minimization or molecular dynamics simulations because it does not derive analytic gradients from energy terms. Geometry optimization is performed by changing dihedral angles without relaxing valence angles and bond lengths. Piquemal et al (2006a) introduced a promising extension of this force field called GEM-0 (Gaussian Electrostatic Model). This force field uses s-type Gaussian functions to fit atomic charge densities. Total interaction energy is computed in the spirit of SIBFA as the sum of interactions between fragments described on the ab initio level. These include electrostatic, exchange repulsion, polarization and charge-transfer intermolecular interaction energies. In this implementation the polarization energy is calculated between permanent electric fields generated by the density fitting procedure and distributed dipolar polarizabilities, which are computed ab initio with the Garmer and Stevens (1989) approach using B3LYP/aug-cc-pVTZ theory level. The polarization equation is solved iteratively. This force field has been tested on water dimers and larger clusters.
Masella et al offers promise for simulating large protein systems in solvents with the newly developed TCPEp force field (Masella et al 2008), which combines a polarizable solute with a coarse-grained polarizable solvent, where the solvent is treated by polarizable pseudoparticles. This method has been applied to BPTI protein simulations for nanosecond-range molecular dynamics. Results showed that the protein structure along the trajectories was well conserved and solvation thermodynamics was about as accurate as the standard Poisson–Boltzmann continuum method.
The CTPOL (Sakharov and Lim 2009) force field was especially developed to model polarized heavy metal ions from group IIB found in metal binding proteins. It is worth noting here because results showed that, if an ion’s polarizability is not included, ion binding sites in proteins are not correctly represented. Polarizability parameters have been optimized for the CHARMM protein force field.
4.4. Other methods: continuum dielectric and QM/MM
Additional methods that include electronic polarization effects in molecular mechanics are those implicitly treating part of or the entire molecular system as a polarizable continuum dielectric and those explicitly treating electrons.
The continuum dielectric approach includes methods that either explicitly treat a molecule at the atomic level, and the solvent or environment as a continuum medium, or those that treat both solute and solvent as continuum media characterized by different dielectric constants. For both situations, a molecule and continuum medium interact electrostatically by polarizing each other. The (free) energy effect of this interaction can be quantified by numerically solving the Poisson–Boltzmann equation (Gilson and Honig 1988, Honig and Nicholls 1995) or by applying analytical methods based on the generalized Born formalism (Onufriev et al 2002, Still et al 1990). Currently, several ongoing efforts are underway to incorporate explicit terms responsible for polarization effects in continuum dielectric approaches. For example, Maple et al (2005) added explicit induced dipole terms to their PFF polarizable version of the force field by applying self-consistent reaction field methodology. In another development, Schnieders et al (2007) combined previously used explicit solvent and polarizable atomic multipoles of an AMOEBA force field and the Poisson–Boltzmann continuum electrostatic model. Simultaneously, Schnieders and Ponder (2007) incorporated their polarizable force field into the generalized Kirkwood formalism, which is a multipole moment extension of the continuum solvent generalized Born method.
Recently, Tan and Luo (2007), Tan et al (2008) extended the Poisson–Boltzmann-based solvent continuum approach by including solute, which is also treated as a continuum medium. They argue that it is reasonable to treat the electronic polarization of solute and solvent in the same way because the only difference between them is a dielectric constant value. If so, the electrostatic field E can be calculated for the solute by first solving the Poisson–Boltzmann equation and then deriving the polarization P, which is treated as a density of induced dipole moments:
| (47) |
Polarization energy, Vpol, can then be derived as an interaction between induced charge distribution from polarization and the potential due to solute point charges. The method depends on properly deriving atomic charges suitable for this application and accurately determining atomic cavity radii, which have been optimized to reproduce experimental solvation free energies. The authors intend to develop a consistent set of force field parameters for proteins and nucleic acids within the framework of continuum polarization. One parameter of this method includes a solute’s dielectric constant. Tan et al assumed that a universal dielectric constant of 4 is optimal for all molecules tested; however, Truchon et al (2008) found that this value may depend on the class of molecules.
Truchon et al (2008) employed the continuum dielectric to develop a new approach for computing gas-phase molecular polarizabilities based on a finite-difference solution to the Poisson equation. A molecule is treated as an entity with a high dielectric, which produces electronic polarization. A molecule is surrounded by a vacuum dielectric and is immersed in a uniform electric field. By solving the Poisson equation on the grid, the induced dipole and then molecular polarizability can be determined. The method of electronic polarization from internal continuum (EPIC) produces polarizabilities consistent with the quantum mechanical B3LYP/aug-cc-pVTZ approach and experimental values. Calculations require small numbers of parameters, such as those for atomic radii and the inner dielectric. However, their optimum values seem to depend on the class of molecules. In addition, the method is not suitable for determining atomic polarizabilities. The advantages of this approach are its relative simplicity and computational speed. Furthermore, the method avoids polarization catastrophe.
Another methodology combines quantum mechanics with molecular mechanics (QM/MM) simulations. It is typically applied to model large molecular systems in which most of the atoms or degrees of freedom are treated classically with molecular mechanics, while the system’s small fragments are treated quantum mechanically. In the quantum mechanical region, electrons are explicitly treated and, so, polarization effects are included in a natural way. This approach is usually applied to model enzymatic reactions inside the binding site of proteins. More rarely, it is used to model other phenomena, such as protein–ligand docking as described by Friesner et al (Cho et al 2005, Friesner 2005). They developed an algorithm in which fixed charges of ligands obtained from molecular mechanical parameterization are replaced by QM/MM calculations in the protein environment, treating only the ligands as the quantum region. The algorithm was tested on a set of 40 cocrystallized structures taken from the Protein Data Bank. The algorithm does not assume any prior knowledge of native structures of the complexes. It was demonstrated that this algorithm is able in many cases to converge to a native-like structure, which is not possible when performing docking using a fixed charge model. Computational efforts are significantly increased for QM/MM approaches, compared to those for standard molecular mechanics.
Recently, Murphy et al (2000), Friesner and Guallar (2005) and Senn and Thiel (2009) published reviews about implementation and applications of QM/MM methods. Case et al (Seabra et al 2007, Walker et al 2008) detailed quantum mechanical methods in the AMBER molecular mechanics simulation package. The AMBER program supports semiempirical Hamiltonians: PM3, PDDG/PM3, PM3CARB1, AM1, MNDO and PDDG/MNDO, as well as the self-consistent charge density functional tight binding (SCC-DFTB) method.
A new application includes analyzing point-polarizable water models using mixed density functional approaches with molecular mechanics (DFT/MM) (Schropp and Tavan 2008). The authors argue that, to properly calculate induced dipoles on a water molecule, the average electric field 〈E〉 within the volume occupied by the given molecule need to be used together with reduced molecular polarizability: αeff = 0.68αexp. Biswas and Gogonea (2008) demonstrated that polarizable classical atoms can be represented by expansion of point charges into orbitals, then combined with QM/MM calculations. Jensen et al (2005) used QM/MM to identify the difference between the macroscopic electric field and the actual electric field felt by the solute molecule in the solvent. Using only quantum mechanical methods for polarization, Masia (2008) published the Car–Parrinello-type simulation of an ion–water cluster that employs an ab initio polarizable force field.
5. Derivation of polarizability and charge models
The quality of a polarizable force field that employs a point dipole model crucially depends on the polarizability model. Experimental molecular polarizabilities are derived from the Lorentz–Lorenz equation (Born and Wolf 1999):
| (48) |
The equation relates the refractive index n, usually measured at a 5893 Å sodium D-line wavelength, with mean molecular polarizability ᾱ. R is the molar refraction, N0 is Avogadro’s number, and M and d are molecular weight and density, respectively. Information about mean molecular polarizabilities and their anisotropy can also be obtained from the Kerr effect and light scattering depolarization data. Another source of molecular polarizabilities is quantum mechanics. Here, molecular polarizabilities can be determined from a molecule’s response to an applied electric field. The coupled perturbative Hartree–Fock (CPHF) method (Ahlberg and Goscinski 1973) is most popularly used to calculate molecular polarizabilities.
Polarizable force fields most often use distributed atomic polarizabilities. Several methods have been proposed to derive these polarizabilities. All methods are based on fitting atomic polarizabilities to reproduce either experimental or quantum mechanical molecular polarizabilities or quantum mechanical electrostatic potentials. The methods can also be divided into two groups: additive and interactive models, depending on the level of interactions permitted between induced dipoles. In the additive approach (Miller 1990a, 1990b, Stout and Dykstra 1998), polarizable sites are allowed to respond to an external electric field but not to permanent and induced multipoles on other sites within a molecule. In Miller’s approach, atomic polarizabilities depend only on hybridization states and the number of electrons on atoms. With this model, experimental polarizabilities can be reproduced to an average error of 2.2%–2.8% (Miller 1990a) (depending on the formulation) for 400 organic compounds having small molecular weights. However, this approach is not suitable for large molecules like peptides, proteins and nucleic acids.
Interactive models (Applequist et al 1972, Birge 1980, Thole 1981, van Duijnen and Swart 1998) allow polarizable sites in the molecule to interact via either equation (28) or (31), depending on the multipolar expansion level (Ren and Ponder 2003). Applequist derived 16 types of atomic polarizabilities after optimizing them to reproduce molecular polarizabilities for 41 different molecules. Atomic polarizabilities were derived by minimizing the deviation between calculated and experimental molecular polarizabilities. The atomic polarizabilities in this model were much smaller than the values from additive models but, due to intramolecular mutual polarization, they reproduce experimental molecular polarizabilities usually within 1%–5%, depending on the molecule type. Applequist’s model was used in deriving AMBER ff02, ff02EP and ff02r1 force fields (Cieplak et al 2001, Wang et al 2006). As mentioned, for larger molecular systems or condensed-phase simulation, the Applequist model can lead to polarization catastrophe from a failure of converging atomic-induced dipoles located at close range. Thole (1981) in 1981 and later van Duijnen and Swart (1998) used damping functions for the dipole–dipole interaction tensor to derive their own set of atomic polarizabilities. Only nine types of effective isotropic atomic polarizabilities were sufficient to reproduce experimental values for 52 molecules with an overall error of 3.7%. The largest error was observed for a group of substituted aromatic compounds (9.9%). Birge (1980) expanded on Applequist’s work by including electronic repulsion integrals to improve anisotropy when calculating molecular polarizabilities.
Another way to determine atomic polarizabilities for molecular mechanics is using quantum mechanical calculations. Soteras et al (2007) and Dehez et al (2007) applied two distinct procedures. The first approach is based on the second-order perturbation theory. As such, they applied a special distance-dependent scaling factor to reproduce exact molecular polarizabilities in a single Hartree–Fock calculation. The second method requires mapping grids of induction energies using a single high-level quantum mechanical calculation, followed by topological partitioning of the electron density (TPED) response into atomic regions. Induction energies obtained from perturbation theory and TPED approaches have been reported to closely agree. Atomic polarizabilities were obtained by fit-ting them to reproduce QM-PT induction energies via equation (27). This approach led to a model of explicitly interacting distributed (atomic) polarizabilities. Since responses between different molecular subunits (atomic regions) are omitted in the fitting procedure, only a set of effective atomic polarizabilities could be derived.
Ponder et al (Grossfield et al 2003, Jiao et al 2006, Ren and Ponder 2003, 2002) employed quantum mechanical calculations to determine distributed multipole moments (Stone 1981) used in an AMOEBA force field. The multipole moments were subsequently corrected to generate a permanent electrostatic model consistent with the assumed polarization scheme (described in section 4). To calculate the polarization energy, modified Thole’s atomic polarizabilities were used in an AMOEBA force field.
Kaminski et al have adopted a different approach to derive the PFF-OPLS force field (Friesner 2005, Kaminski et al 2004, 2002). To determine atomic polarizabilities, they calculated the molecule’s response to the dipolar probe located in a number of positions around the molecule. Perturbation of the electrostatic potential, defined at a set of grid points outside a molecule’s van der Waals surfaces due to the probe, was then used to fit isotropic atomic polarizabilities. Quantum mechanical calculations of electrostatic potentials were conducted using density functional theory combined with the B3LYP (Becke 1993) method and cc-pVTZ(-f) basis set (Dunning 1989).
Elking et al (2007) have proposed an attractive way to handle induced dipoles and potentially higher multipoles and to derive atom-type polarizabilities. The model is based on interacting Gaussian charge densities. Charge distributions described by Gaussian functions can interact at short distances, thus avoiding polarization catastrophe. Atomic polarizabilities are generated by probing a molecule with point charges or external fields and calculating the response electrostatic potential. The response potential is the difference between quantum mechanical potentials in the presence and absence (vacuum) of the charged probe defined on a grid surrounding the molecule. The model’s parameters (e.g. atomic polarizabilities αi and Gaussian exponents) were fitted to the response of the electrostatic potential. During computations, the Gaussian inducible dipoles are allowed to interact with one another and the probe charge but only the contribution arising from the response potential was used in the fitting procedure, assuming that intramolecular polarization is constant. In this way, computed polarizabilities do not depend on the choice of the permanent multipole model or on intramolecular polarization. Unlike many other approaches, a permanent electrostatic model can be selected after deriving polarizabilities. Actual computations were performed for a number of small molecules and dipeptides, allowing for deriving the set of ‘probed’ (e.g. using a charged probe to define a molecule’s polarization) atomic polarizabilities. The Gaussian polarization model performed slightly better than the Thole-exponential (Tinker-exponential, equation (38)) model for reproducing molecular polarizability tensors and has therefore been incorporated into the AMBER (Case et al 2008) molecular dynamics simulation package and parameterized for AMBER (Wang et al 2000) and GLYCAM (Woods et al 1995) force fields.
Induced dipoles, permanent charges and higher multipole moments determine a molecule’s total electrostatic properties. For these reasons, permanent and distributed atomic multipole moments need to be derived based on fitting to an electrostatic potential and the need to be consistently parameterized depending on the assumed polarization model. In AMBER ff02, ff02EP (Cieplak et al 2001) and ff02r1 (Wang et al 2006) force fields, Applequist’s atomic isotropic polarizabilities are assumed. Cieplak et al (2001) describe a method to determine partial atomic charges for standard residues, such as amino acids and nucleotides. According to their method, atomic charges are iteratively fitted to the differences between the quantum mechanical electrostatic potential and the potential generated by induced dipoles. Iterations stop when an induced molecular dipole moment does not change within some accuracy level (10−3 D). Electrostatic potentials are calculated at a number of points (of the order of 103–104) defined in several shells, beyond the van der Waals envelope, around the molecule. For polarizable AMBER force fields, quantum mechanical ESPQM potentials have been determined at the B3LYP (Becke 1993)-type exchange and correlation functional and the cc-PVTZ (Dunning 1989) basis set.
For the purpose of developing a new AMBER polarizable force field Wang et al (Case et al 2008) derived several sets of atomic polarizabilities for four nonadditive (e.g. dipole interactive models) polarizability models using Applequist, Thole-linear, Thole-exponential and Thole–Tinker-exponential damping approaches (equations (36)–(38) and (46)). The genetic algorithm method (GA) (Wang and Kollman 2001) has been used to determine atomic polarizabilities that reproduce high-quality experimental static molecular polarizabilities. Wang et al performed calculations for a set of 420 molecules previously studied by Bosque and Sales (2002) who performed measurements of their molecular refraction. Results are presented in table 2.
Among the four models, Thole’s exponential scheme comprising 15 atomic polarizability parameters performed best at reproducing experimental data and boasted the lowest average unsigned, root mean square and average per cent errors (table 2(a)). In AMBER polarizable force fields (Cieplak et al 2001, Wang et al 2006) atomic charges are iteratively fitted to the difference (ΔESP) between the quantum mechanically derived electrostatic potential (ESPQM) and the potential generated by induced dipoles (ESPind). Since this procedure quite often became unstable, another set of atomic polarizabilities was derived in which 1–2 and 1–3 dipole interactions were turned off, and 1–4 interactions were scaled down by 1.2, which is consistent with a treatment of the electrostatic interactions between permanent charges. Derived polarizabilities are presented in table 2(b). When atomic polarizabilities are similar to each other in a damping model, with slightly larger errors in an exponential model (equation (37)) as seen here, it confirms that most of the differences between the various polarizability models are related to 1–2 and 1–3 interactions (see figure 1). Removing 1–2 and 1–3 interactions similarly aligns all the models. The polarizabilities that are damped, or are 1–2 and 1–3 excluded, are larger because they need to compensate for lost short–range, dipole–dipole interactions to reproduce total molecular polarizability. The four types of models are now being tested using solvation free-energy calculations and a thermodynamic integration method on a set of small molecules that include amino-acid side-chain analogs. These polarizabilities will be used to construct a newer version of the polarizable AMBER force field for amino acids, nucleic acids and other important biomolecules. The i_RESP (Cieplak 2009), a stand-alone computer program, has been developed to facilitate iterative charge fittings to an effective electrostatic potential, ΔESP, using atomic polarizabilities as an input parameter. The program performs charge fittings in several stages: in the first stage, ESP charges are iteratively fitted to ΔESP (ΔESP = ESPQM − ESPind) and separately fitted for each input molecule. For each individual, iterative step over ΔESP, induced dipoles are determined from the iterative solution of equations (28) and (40). Once each ΔESP converges for each molecule, the standard two-stage RESP-type charge fit is performed using the final values of the iterated ΔESP electrostatic potential. The program is capable of performing a multimolecular, multiconformational restrained fit, as is its predecessor, the RESP program (Bayly et al 1993).
A stand-alone R.E.D. (RESP ESP charge derive) program has been developed to further facilitate charge derivations. Written in the Perl language, it is available online at http://q4md-forcefieldtools.org/RED/ (Dupradeau et al 2008, Pigache et al 2004). The program interfaces quantum mechanical programs with the RESP program and, in the near future, will interface with the i_RESP program to be able to derive atomic charges for polarizable force fields. The R.E.D. program automatically performs all necessary calculations involved in RESP charge fitting without the user needing to handle intermediate files or data. PDB or mol2 files are required for input. Currently, the R.E.D. program can be used together with Gamess (Gordon and Schmidt 2005) and Gaussian03 (Frisch et al 2003) quantum mechanical programs, which perform geometry optimization and generate electrostatic potentials around a molecule. The website also has an online server, ‘R.E.D.Server’, which is available for automatically deriving partial charges. It provides convenient access to the R.E.D and quantum mechanical programs through a web interface.
6. Conclusion
Modeling the effects of polarization in chemical computations is a rapidly growing and exciting area of research. Significant developments published after 2000 have been reviewed here. Incorporating the effects of polarization into classical force fields is a positive step towards improving the accuracy of simulations; however, many other terms and including nonadditive effects still need to be addressed. Most developments in the polarizable force fields for biomolecules relate to explicitly induced dipoles and somewhat less to fluctuating charge and Drude oscillator methods. New methods have begun to emerge, such as those that go beyond point charge models and describe charge densities via spatial functions. Advances have also been made by combining classical mechanics with methods that explicitly treat electrons. Yet most polarizable force fields are currently in the developmental stage. Their parameterization, which usually requires iterative simulations, is a much slower process compared with additive counterparts. This is due to a higher computational burden. However, as we show here, several key comparisons between nonadditive and additive molecular simulations have already been reported, and these support a better agreement of the former compared with experimental data (Babin et al 2006, Baucom et al 2004, Harder et al 2009, Jiao et al 2008). Applying polarizable force fields to large biomolecular simulations is still far from routine and is computationally expensive. We hope that this situation will change given encouraging data and dedicated efforts by those in the field. Overall, the force fields are overdue for much-needed advances.
Acknowledgments
PC, JW and YD are supported by NIH grant GM079383. F-YD thanks CINES, IDRIS and Plateforme de Modélisation et Calcul Scientifique (MεCS)—Université de Picardie Jules Verne for providing computing resources.
References
- Ababou A, van der Vaart A, Gogonea V, Merz KM., Jr Interaction energy decomposition in protein–protein association: a quantum mechanical study of barnase–barstar complex. Biophys Chem. 2007;125:221–36. doi: 10.1016/j.bpc.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Ahlberg R, Goscinski O. Atomic polarizabilities and shielding factors with a local linear response method. J Phys B: At Mol Phys. 1973;6:2254–67. [Google Scholar]
- Allinger N, Young HY, Lii JH. Molecular mechanics. The MM3 force field for hydrocarbons. 1. J Am Chem Soc. 1989;111:8551–66. [Google Scholar]
- Allinger NL, Chen KH, Lii JH, Durkinm KA. Alcohols, ethers, carbohydrates, and related compounds. I. The MM4 force field for simple compounds. J Comput Chem. 2003;24:1447–72. doi: 10.1002/jcc.10268. [DOI] [PubMed] [Google Scholar]
- Anisimov VM, Vorobyov IV, Roux B, Mackerell AD. Polarizable empirical force field for the primary and secondary alcohol series based on the classical Drude model. J Chem Theory Comput. 2007;3:1927–46. doi: 10.1021/ct700100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applequist J, Carl JR, Fung K-K. An atom dipole interaction model for molecular polarizability. Application to polyatomic molecules and determination of atom polarizabilities. J Am Chem Soc. 1972;94:2952–60. [Google Scholar]
- Babin V, Baucom J, Darden TA, Sagui C. Molecular dynamics simulations of DNA with polarizable force fields: convergence of an ideal B-DNA structure to the crystallographic structure. J Phys Chem B. 2006;110:11571–81. doi: 10.1021/jp061421r. [DOI] [PubMed] [Google Scholar]
- Banks J, Kaminski GA, Zhou R, Mainz DT, Derne BJ, Friesner RA. Parameterizing a polarizable force field from ab initio data. I. The fluctuating point charge model. J Chem Phys. 1999;110:741–54. [Google Scholar]
- Baucom J, Transue T, Fuentes-Cabrera M, Krahn JM, Darden TA, Sagui C. Molecular dynamics simulations of the d(CCAACGTTGG)(2) decamer in crystal environment: comparison of atomic point-charge, extra-point, and polarizable force fields. J Chem Phys. 2004;121:6998–7008. doi: 10.1063/1.1788631. [DOI] [PubMed] [Google Scholar]
- Bayly C, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem. 1993;97:10269–80. [Google Scholar]
- Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–52. [Google Scholar]
- Birge R. Calculation of molecular polarizabilities using an anisotropic atom point dipole interaction model which includes the effect of electron repulsion. J Chem Phys. 1980;72:5312–9. [Google Scholar]
- Biswas PK, Gogonea V. A polarizable force-field model for quantum-mechanical–molecular-mechanical Hamiltonian using expansion of point charges into orbitals. J Chem Phys. 2008;129:154108. doi: 10.1063/1.2992527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born M, Wolf E. Principles of Optics: Electromagnetic Theory of Propagation, Interference and Diffraction of Light. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- Bosque R, Sales J. Polarizabilities of solvents from the chemical composition. J Chem Inf Comput Sci. 2002;42:1154–63. doi: 10.1021/ci025528x. [DOI] [PubMed] [Google Scholar]
- Boys S, Bernardi F. The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol Phys. 1970;19:553–66. [Google Scholar]
- Bukowski R, Szalewicz K, Groenenboom GC, van der Avoird A. Predictions of the properties of water from first principles. Science. 2007;315:1249–52. doi: 10.1126/science.1136371. [DOI] [PubMed] [Google Scholar]
- Burnham CJ, Li J, Xantheas SS, Leslie M. The parametrization of a Thole-type all-atom polarizable water model from first principles and its application to the study of water clusters (n = 2–21) and the phonon spectrum of ice Ih. J Chem Phys. 1999;110:4566–81. [Google Scholar]
- Caldwell JW, Kollman PA. Structure and properties of neat liquids using nonadditive molecular dynamics: water, methanol, and N-methylacetamide. J Phys Chem. 1995;99:6208–19. [Google Scholar]
- Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods R. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. AMBER 10. San Francisco, CA: University of California; 2008. [Google Scholar]
- Chalasinski G, Szczesniak MM. Origins of structure and energetics of van der Waals clusters from ab initio calculations. Chem Rev. 1994;94:1723–65. [Google Scholar]
- Cheatham TE, 3rd, Cieplak P, Kollman PA. A modified version of the Cornell et al force field with improved sugar pucker phases and helical repeat. J Biomol Struct Dyn. 1999;16:845–62. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- Chelli RP, Righini R, Califano S. Electrical response in chemical potential equalization schemes. J Phys Chem. 1999;111:8569–75. [Google Scholar]
- Cho AE, Guallar V, Berne BJ, Friesner R. Importance of accurate charges in molecular docking: quantum mechanical/molecular mechanical (QM/MM) approach. J Comput Chem. 2005;26:915–31. doi: 10.1002/jcc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieplak P. i_RESP. La Jolla: Burnham Institute for Medical Research; 2009. [Google Scholar]
- Cieplak P, Caldwell J, Kollman PA. Molecular mechanical models for organic and biological systems. going beyond the atom centered two body additive approximation: aqueous solution free energies of methanol and n-methyl acetamide, nucleic acid bases and amide hydrogen bonding and chloroform/water partition coefficients of the nucleic acid bases. J Comput Chem. 2001;22:1048–57. [Google Scholar]
- Cieplak P, Kollman PA, Lybrand T. A new water potential including polarization: application to gas, phase, liquid and crystal properties of water. J Chem Phys. 1990;92:6755–60. [Google Scholar]
- Cieplak P, Lybrand TP, Kollman PA. Calculation of free energy changes in ion–water clusters using non-additive potentials and Monte Carlo methods. J Chem Phys. 1987;86:6393–404. [Google Scholar]
- Claverie P, Rein R. Theory of intermolecular interactions: the long range terms in the dipole–dipole, monopoles–dipole, and monopoles–bond polarizabilities approximations. Int J Quantum Chem. 1969;3:537–51. [Google Scholar]
- Clementi E, Kistenmacher H, Kolos W, Romano S. Non-additivity in water–ion–water interactions. Theor Chim Acta. 1980;55:257–66. [Google Scholar]
- Cornell W, Cieplak P, Bayly CI, Gould I, Merz KM, Jr, Ferguson D, Spellmeyer D, Fox Th, Caldwell JW, Kollman PA. A second generation force field for simulation proteins, nucleic acids and related organic molecules. J Am Chem Soc. 1995;117:5179–97. [Google Scholar]
- Dang LX. Importance of polarization effects in modeling the hydrogen bond in water using classical molecular dynamics techniques. J Phys Chem B. 1998;102:620–4. [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald: an M * log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–92. [Google Scholar]
- Dauber-Osguthorpe P, Roberts VA, Osguthorpe DJ, Wolff J, Genest M, Hagler AT. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins: Struct Funct Genet. 1988;4:31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- Davis JE, Rahaman O, Patel S. Molecular dynamics simulations of a DMPC bilayer using nonadditive interaction models. Biophys J. 2009;96:385–402. doi: 10.1016/j.bpj.2008.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehez F, Angyan JG, Gutierrez IS, Luque FJ, Schulten K, Chipot C. Modeling induction phenomena in intermolecular interactions with ab initio force field. J Chem Theory Comput. 2007;3:1914–26. doi: 10.1021/ct700156a. [DOI] [PubMed] [Google Scholar]
- Dill K, Bromberg S, Yue K, Fiebig KM, Yee DP, Thomas PD, Chan HS. Principles of protein folding—a perspective from simple exact models. Prot Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R, Kollman PA. Advancing beyond the atom-centered model in additive and nonadditive molecular mechanics. J Comput Chem. 1997;18:1632–46. [Google Scholar]
- Drude P. Theory of Optics. New York: Longmans Green; 1902. [Google Scholar]
- Duan Y, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Dunning T., Jr Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. J Chem Phys. 1989;90:1007–23. [Google Scholar]
- Dupradeau FY, et al. R.E.DD.B.: a database for RESP and ESP atomic charges, and force field libraries. Nucleic Acids Res. 2008;36:D360–7. doi: 10.1093/nar/gkm887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elking D, Darden T, Woods RJ. Gaussian induced dipole polarization model. J Comput Chem. 2007;28:1261–74. doi: 10.1002/jcc.20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch C, Karp DA, Lee KK, Stites WE, Lattman EE, Garcia-Moreno B. Experimental pK (a) values of buried residues: analysis with continuum methods and role of water penetration. Biophys J. 2002;82:3289–304. doi: 10.1016/s0006-3495(02)75670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R, Guallar V. Ab initio quantum chemical and mixed quantum mechanics/molecular mechanics (QM/MM) methods for studying enzymatic catalysis. Annu Rev Phys Chem. 2005;56:389–427. doi: 10.1146/annurev.physchem.55.091602.094410. [DOI] [PubMed] [Google Scholar]
- Friesner RA. Modeling polarization in proteins and protein–ligand complexes: methods and preliminary results. Adv Protein Chem. 2005;72:79–104. doi: 10.1016/S0065-3233(05)72003-9. [DOI] [PubMed] [Google Scholar]
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03. Wallingford CT: Gaussian; 2003. [Google Scholar]
- Garcia-Moreno B, Dwyer JJ, Gittis AG, Lattman EE, Spencer DS, Stites WE. Experimental measurement of the effective dielectric in the hydrophobic core of a protein. Biophys Chem. 1997;64:211–24. doi: 10.1016/s0301-4622(96)02238-7. [DOI] [PubMed] [Google Scholar]
- Garmer DR, Stevens WJ. Transferability of molecular distributed polarizabilities from a simple localized orbital based method. J Phys Chem. 1989;93:8263–70. [Google Scholar]
- Geerke DP, van Gunsteren WF. On the calculation of atomic forces in classical simulation using the charge-on-spring method to explicitly treat electronic polarization. J Chem Theory Comput. 2007a;3:2128–37. doi: 10.1021/ct700164k. [DOI] [PubMed] [Google Scholar]
- Geerke DP, van Gunsteren WF. Calculation of the free energy of polarization: quantifying the effect of explicitly treating electronic polarization on the transferability of force-field parameters. J Phys Chem B. 2007b;111:6425–36. doi: 10.1021/jp0706477. [DOI] [PubMed] [Google Scholar]
- Geerke DP, van Gunsteren WF. The performance of non-polarizable and polarizable force-field parameter sets for ethylene glycol in molecular dynamics simulations of the pure liquid and its aqueous mixtures. Mol Phys. 2007c;105:1861–81. [Google Scholar]
- Gilson MK, Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4:7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- Gordon MS, Schmidt MW. Advances in electronic structure theory: GAMESS a decade later. In: Dykstra CE, Frenking G, Kim KS, Scuseria GE, editors. Theory and Applications of Computational Chemistry: the First Forty Years. Amsterdam: Elsevier; 2005. pp. 1167–89. [Google Scholar]
- Gresh N, Cisneros GA, Darden TA, Piquemal JP. Anisotropic, polarizable molecular mechanics studies of inter- and intramolecular interactions and ligand-macromolecule complexes. a bottom-up strategy. J Chem Theory Comput. 2007;3:1960–86. doi: 10.1021/ct700134r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfield A, Ren P, Ponder JW. Ion solvation thermodynamics from simulation with a polarizable force field. J Am Chem Soc. 2003;125:15671–82. doi: 10.1021/ja037005r. [DOI] [PubMed] [Google Scholar]
- Hagberg D, Brdarski S, Karlstrom G. On the solvation of ions in small water droplets. J Phys Chem B. 2005;109:4111–7. doi: 10.1021/jp045081q. [DOI] [PubMed] [Google Scholar]
- Halgren TA. MMFF VII. Characterization of MMFF94, MMFF94s, and other widely available force fields for conformational energies and for intermolecular-interaction energies and geometries. J Comput Chem. 1999;20:730–48. doi: 10.1002/(SICI)1096-987X(199905)20:7<730::AID-JCC8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Halgren TA, Damm W. Polarizable force fields. Curr Opin Struct Biol. 2001;11:236–42. doi: 10.1016/s0959-440x(00)00196-2. [DOI] [PubMed] [Google Scholar]
- Harder E, Anisimov VM, Whitfield T, Mackerell AD, Roux B. Understanding the dielectric properties of liquid amides from a polarizable force field. J Phys Chem B. 2008;112:3509–21. doi: 10.1021/jp709729d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder E, Mackerell AD, Roux B. Many-body polarization effects and the membrane dipole potential. J Am Chem Soc. 2009;131:2760–1. doi: 10.1021/ja806825g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermida-Ramon JM, Brdarski S, Karlstrom G, Berg U. Inter- and intramolecular potential for the N-formylglycinamide–water system. A comparison between theoretical modeling and empirical force fields. J Comput Chem. 2003;24:161–76. doi: 10.1002/jcc.10159. [DOI] [PubMed] [Google Scholar]
- Hermida-Ramon JM, Ohrn A, Karlstrom G. Planar or nonplanar: what is the structure of urea in aqueous solution? J Phys Chem B. 2007;111:11511–5. doi: 10.1021/jp073579x. [DOI] [PubMed] [Google Scholar]
- Holt A, Karlstrom G. Inclusion of the quadrupole moment when describing polarization. The effect of the dipole–quadrupole polarizability. J Comput Chem. 2008;29:2033–8. doi: 10.1002/jcc.20976. [DOI] [PubMed] [Google Scholar]
- Holt A, Karlstrom G. Improvement of the NEMO potential by inclusion of intramolecular polarization. Int J Quantum Chem. 2009;109:1255–66. [Google Scholar]
- Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–9. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Jacucci G, McDonald IR, Singer K. Introduction of the shell model of ionic polarizability into molecular dynamics calculations. Phys Lett A. 1974;50:141–3. [Google Scholar]
- Jensen L, Swart M, van Duijnen PTh. Microscopic and macroscopic polarization within a combined quantum mechanics and molecular mechanics model. J Chem Phys. 2005;122:034103–14. doi: 10.1063/1.1831271. [DOI] [PubMed] [Google Scholar]
- Jeziorski B, Moszynski R, Szalewicz K. Perturbation theory approach to intermolecular potential energy surfaces of van der Waals complexes. Chem Rev. 1994;94:1887–930. [Google Scholar]
- Jiao D, Golubkov PA, Darden TA, Ren P. Calculation of protein–ligand binding free energy by using a polarizable potential. Proc Natl Acad Sci USA. 2008;105:6290–5. doi: 10.1073/pnas.0711686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao D, King C, Grossfield A, Darden TA, Ren P. Simulation of Ca2+ and Mg2+ solvation using polarizable atomic multipole potential. J Phys Chem B. 2006;110:18553–9. doi: 10.1021/jp062230r. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Jensen KP, Alexandrova AN. Polarization effects for hydrogen-bonded complexes of substituted phenols with water and chloride ion. J Chem Theory Comput. 2007;3:1987–92. doi: 10.1021/ct7001754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–36. [Google Scholar]
- Kaminski GA, Stern HA, Berne BJ, Friesner RA. Development of an accurate and robust polarizable molecular mechanics force field from quantum chemistry. J Phys Chem A. 2004;108:621–7. [Google Scholar]
- Kaminski GA, et al. Development of a polarizable force field for proteins via ab initio quantum chemistry: first generation model and gas phase tests. J Comput Chem. 2002;23:1515–31. doi: 10.1002/jcc.10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux G, Harder E, Vorobyov IV, Roux B, MacKerell AD., Jr A polarizable model of water for molecular dynamics simulations of biomolecules. Chem Phys Lett. 2006;418:245–9. [Google Scholar]
- Lamoureux G, MacKerell AD, Jr, Roux B. A simple polarizable model of water based on classical Drude oscillators. J Chem Phys. 2003;119:5185–97. doi: 10.1063/1.4774577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux G, Roux B. Modeling induced polarization with classical Drude oscillators: theory and molecular dynamics simulation algorithm. J Chem Phys. 2003;119:3025–39. [Google Scholar]
- Lindan PJD. Dynamics with the shell model. Mol Simul. 1995;14:303–12. [Google Scholar]
- London F. Faraday Soc. 1937;33:8. [Google Scholar]
- Lopes PE, Lamoureux G, Mackerell AD., Jr Polarizable empirical force field for nitrogen-containing heteroaromatic compounds based on the classical Drude oscillator. J Comput Chem. 2009;30:1821–38. doi: 10.1002/jcc.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes PE, Lamoureux G, Roux B, Mackerell AD., Jr Polarizable empirical force field for aromatic compounds based on the classical drude oscillator. J Phys Chem B. 2007;111:2873–85. doi: 10.1021/jp0663614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998a;102:3586–616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998b;102:3586–616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Mackerell AD., Jr Empirical force fields for biological macromolecules: overview and issues. J Comput Chem. 2004;25:1584–604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- Madurga S, Vilaseca E. Solvent effect on the conformational equilibrium of 1,2-dichloroethane in water. The role of solute polarization. J Phys Chem A. 2004;108:8439–47. [Google Scholar]
- Maple JR, Cao Y, Damm W, Halgren AT, Kaminski GA, Zhang LY, Friesner RA. A polarizable force field and continuum solvation methodology for modeling of protein–ligand interactions. J Chem Theory Comput. 2005;1:694–715. doi: 10.1021/ct049855i. [DOI] [PubMed] [Google Scholar]
- Marcus RA. Electron transfer reactions in chemistry. Theory and experiment. Rev Mod Phys. 1993;65:599–610. [Google Scholar]
- Margenau H, Kestner NR. Theory of Intermolecular Forces. Oxford; Pergamon: 1971. [Google Scholar]
- Masella M, Borgis D, Cuniasse P. Combining a polarizable force-field and a coarse-grained polarizable solvent model: application to long dynamics simulations of bovine pancreatic trypsin inhibitor. J Comput Chem. 2008;29:1707–24. doi: 10.1002/jcc.20932. [DOI] [PubMed] [Google Scholar]
- Masia M. Ab initio based polarizable force field parametrization. J Chem Phys. 2008;128:184107–4. doi: 10.1063/1.2919161. [DOI] [PubMed] [Google Scholar]
- Masia M, Probst M, Rey R. On the performance of molecular polarization methods. II. Water and carbon tetrachloride close to a cation. J Chem Phys. 2005;123:164505–13. doi: 10.1063/1.2075107. [DOI] [PubMed] [Google Scholar]
- Miller KJ. Additivity methods in molecular polarizability. J Am Chem Soc. 1990a;112:8533–42. [Google Scholar]
- Miller KJ. Calculation of the molecular polarizability tensor. J Am Chem Soc. 1990b;112:8543–51. [Google Scholar]
- Mitchell PJ, Fincham D. Shell model simulations by adiabatic dynamics. J Phys: Condens Matter. 1993;5:1031–8. [Google Scholar]
- Mulliken RS. A new electronegativity scale: together with data on valence states and an ionization potential and electron affinities. J Chem Phys. 1934;2:782–93. [Google Scholar]
- Murphy RB, Philipp DM, Friesner AR. A mixed quantum mechanics/molecular mechanics (QM/MM) method for large-scale modeling of chemistry in protein environments. J Comput Chem. 2000;21:1442–57. [Google Scholar]
- Nalewajski RF, Korchowiec J, Zhou Z. Molecular hardness and softness parameters and their use in chemistry. Int J Quantum Chem Quantum Chem Symp. 1988;22:349–66. [Google Scholar]
- Niketic SR, Rasmussen K. The Consistent Force Field: a Documentation. Berlin: Springer; 1977. p. 212. [Google Scholar]
- Noskov SY, Lamoureux G, Roux B. Molecular dynamics study of hydration in ethanol–water mixtures using a polarizable force field. J Phys Chem B. 2005;109:6705–13. doi: 10.1021/jp045438q. [DOI] [PubMed] [Google Scholar]
- Ohtori N, Salanne M, Madden PA. Calculations of the thermal conductivities of ionic materials by simulation with polarizable interaction potentials. J Chem Phys. 2009;130:104507. doi: 10.1063/1.3086856. [DOI] [PubMed] [Google Scholar]
- Onufriev A, Case DA, Bashford D. Effective Born radii in the generalized Born approximation: the importance of being perfect. J Comput Chem. 2002;23:1297–304. doi: 10.1002/jcc.10126. [DOI] [PubMed] [Google Scholar]
- Palmo K, Mannfors B, Mirkin NG, Krimm S. Potential energy functions: from consistent force fields to spectroscopically determined polarizable force fields. Biopolymers. 2003;68:383–94. doi: 10.1002/bip.10254. [DOI] [PubMed] [Google Scholar]
- Parr RG, Pearson RG. Absolute hardness: companion parameter to absolute electronegativity. J Am Chem Soc. 1983;105:7512–6. [Google Scholar]
- Patel S, Brooks CL., III CHARMM fluctuating charge force field for proteins: I. Parameterization and application to bulk organic liquid simulations. J Comput Chem. 2004;25:1–15. doi: 10.1002/jcc.10355. [DOI] [PubMed] [Google Scholar]
- Patel S, Brooks CL., III Structure, thermodynamics, and liquid–vapor equilibrium of ethanol from molecular-dynamics simulations using nonadditive interactions. J Chem Phys. 2005a;123:164502. doi: 10.1063/1.2009730. [DOI] [PubMed] [Google Scholar]
- Patel S, Brooks CL., III A nonadditive methanol force field: bulk liquid and liquid–vapor interfacial properties via molecular dynamics simulations using a fluctuating charge model. J Chem Phys. 2005b;122:024508–18. doi: 10.1063/1.1827604. [DOI] [PubMed] [Google Scholar]
- Patel S, Mackerell AD, Jr, Brooks CL., III CHARMM fluctuating charge force field for proteins: II protein/solvent properties from molecular dynamics simulations using a nonadditive electrostatic model. J Comput Chem. 2004;25:1504–14. doi: 10.1002/jcc.20077. [DOI] [PubMed] [Google Scholar]
- Patel SA, Brooks CL., III Revisiting the hexane–water interface via molecular dynamics simulations using nonadditive alkane–water potentials. J Chem Phys. 2006;124:204706. doi: 10.1063/1.2198538. [DOI] [PubMed] [Google Scholar]
- Pigache A, Cieplak P, Dupradeau F-Y. Automatic and highly reproducible RESP and ESP charge derivation: application to the development of programs RED and X RED. 227th ACS National Mtg; Anaheim, CA. 2004. [Google Scholar]
- Piquemal JP, Cisneros GA, Reinhardt P, Gresh N, Darden TA. Towards a force field based on density fitting. J Chem Phys. 2006a;124:104101–27. doi: 10.1063/1.2173256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquemal JP, Perera L, Cisneros GA, Ren P, Pedersen LG, Darden TA. Towards accurate solvation dynamics of divalent cations in water using the polarizable amoeba force field: from energetics to structure. J Chem Phys. 2006b;125:054511. doi: 10.1063/1.2234774. [DOI] [PubMed] [Google Scholar]
- Ponder JW, Case DA. Force fields for protein simulations. Adv Protein Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- Pullman B, Claverie P, Caillet J. Interaction energies in hydrogen-bonded purine–pyrimidine triplets. Proc Natl Acad Sci USA. 1967;57:1663–9. doi: 10.1073/pnas.57.6.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappe A, Casewit CJ, Colwell KS, Goddard WA, Skiff WM. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J Am Chem Soc. 1992;114:10024–35. [Google Scholar]
- Rasmussen TD, Ren P, Ponder JW, Jensen F. Force field modeling of conformational energies: importance of multipole moments and intramolecular polarization. Int J Quantum Chem. 2007;107:1390–5. [Google Scholar]
- Ren P, Ponder JW. Consistent treatment of inter- and intramolecular polarization in molecular mechanics calculations. J Comput Chem. 2002;23:1497–506. doi: 10.1002/jcc.10127. [DOI] [PubMed] [Google Scholar]
- Ren P, Ponder JW. Polarizable atomic multipole water model for molecular mechanics simulation. J Phys Chem B. 2003;107:5933–47. [Google Scholar]
- Ren P, Ponder JW. Temperature and pressure dependence of the AMOEBA water model. J Phys Chem B. 2004;108:13427–37. [Google Scholar]
- Rick SW, Stuart SJ. Potentials and algorithms for incorporating polarizability in computer simulations. Rev Comput Chem. 2002;18:89–146. [Google Scholar]
- Rick SW, Stuart SJ, Bader JS, Berne B. Fluctuating charge force fields for aqueous solutions. J Mol Liq. 1995;65/66:31–40. [Google Scholar]
- Rick SW, Stuart SJ, Berne BJ. Dynamical fluctuating charge force fields: application to liquid water. J Chem Phys. 1994;101:6141–56. [Google Scholar]
- Sakharov DV, Lim C. Force fields including charge transfer and local polarization effects: application to proteins containing multi/heavy metal ions. J Comput Chem. 2009;30:191–202. doi: 10.1002/jcc.21048. [DOI] [PubMed] [Google Scholar]
- Schnieders MJ, Baker NA, Ren P, Ponder JW. Polarizable atomic multipole solutes in a Poisson–Boltzmann continuum. J Chem Phys. 2007;126:124114. doi: 10.1063/1.2714528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnieders MJ, Ponder JW. Polarizable atomic multipole solutes in a generalized Kirkwood continuum. J Chem Theory Comput. 2007;3:2083–97. doi: 10.1021/ct7001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schropp B, Tavan P. The polarizability of point-polarizable water models: density functional theory/molecular mechanics results. J Phys Chem B. 2008;112:6233–40. doi: 10.1021/jp0757356. [DOI] [PubMed] [Google Scholar]
- Schuler LD, Daura X, van Gunsteren WF. An improved GROMOS96 force field for aliphatic hydrocarbons in the condensed phase. J Comput Chem. 2001;22:1205–18. [Google Scholar]
- Seabra MGd, Walker RC, Elstner M, Case DA, Roitberg AE. Implementation of the SCC-DFTB method for hybrid QM/MM simulations within the amber molecular dynamics package. J Phys Chem A. 2007;111:5655–64. doi: 10.1021/jp070071l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn HM, Thiel W. QM/MM methods for biomolecular systems. Angew Chem Int Edn Engl. 2009;48:1198–229. doi: 10.1002/anie.200802019. [DOI] [PubMed] [Google Scholar]
- Silberstein L. Phil Mag. 1917;33:92–128. [Google Scholar]
- Sokalski WA, Poirier RA. Cumulative atomic multipole representation of the molecular charge distribution and its basis set dependence. Chem Phys Lett. 1983;98:86–92. [Google Scholar]
- Soteras I, Curutchet C, Bidon-Chanal A, Dehez F, Ángyán JG, Orozco M, Chipot C, Luque FJ. Derivation of distributed models of atomic polarizability for molecular simulations. J Chem Theory Comput. 2007;3:1901–13. doi: 10.1021/ct7001122. [DOI] [PubMed] [Google Scholar]
- Stenhammar J, Linse P, Malmqvist P-A, Karlstrom G. Electric multipole moment fluctuations in polar liquids. J Chem Phys. 2009;130:124521. doi: 10.1063/1.3100312. [DOI] [PubMed] [Google Scholar]
- Stern HA, Kaminski GA, Banks JL, Zhou R, Berne BJ, Friesner RA. Fluctuating charge, polarizable dipole, and combined models: parameterization from ab initio quantum chemistry. J Phys Chem B. 1999;103:4730–7. [Google Scholar]
- Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 1990;112:6127–9. [Google Scholar]
- Stone AJ. Distributed multipole analysis, how to describe a molecular charge distribution. Chem Phys Lett. 1981;83:233–9. [Google Scholar]
- Stone AJ. The Theory of Intermolecular Forces. Oxford: Clarendon; 1996. [Google Scholar]
- Stone AJ, Misquitta AJ. Charge-transfer in symmetry-adapted perturbation theory. Chem Phys Lett. 2009;473:201–5. [Google Scholar]
- Stout JM, Dykstra CE. A distributed model of the electrical response of organic molecules. J Phys Chem. 1998;102:1576–82. [Google Scholar]
- Tan T-H, Luo R. Continuum treatment of electronic polarization effect. J Chem Phys. 2007;126:094103. doi: 10.1063/1.2436871. [DOI] [PubMed] [Google Scholar]
- Tan T-H, Tan C, Luo R. Continuum polarizable force field within the Poisson–Boltzmann framework. J Phys Chem B. 2008;112:7675–88. doi: 10.1021/jp7110988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole B. Molecular polarizabilities calculated with a modified dipole interaction. Chem Phys. 1981;59:341–50. [Google Scholar]
- Toukmaji A, Sagui C, Board JA, Darden T. Efficient particle-mesh Ewald based approach to fixed and induced dipolar interactions. J Chem Phys. 2000:10913–27. [Google Scholar]
- Truchon J-F, Nicholls A, Iftimie RI, Roux B, Bayly CI. Accurate molecular polarizabilities based on continuum electrostatics. J Chem Theory Comput. 2008;4:1480–93. doi: 10.1021/ct800123c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart A, Merz KM., Jr Divide and conquer interaction energy decomposition. J Phys Chem A. 1999;103:3321–9. [Google Scholar]
- van Duijnen PT, Swart M. Molecular and atomic polarizabilities: Thole’s model revisited. J Phys Chem A. 1998;102:2399–407. [Google Scholar]
- Vladimirov E, Ivanova A, Rosch N. Effect of solvent polarization on the reorganization energy of electron transfer from molecular dynamics simulations. J Chem Phys. 2008;129:194515. doi: 10.1063/1.3013456. [DOI] [PubMed] [Google Scholar]
- Vladimirov E, Ivanova A, Rosch N. Solvent reorganization energies in A-DNA, B-DNA, and rhodamine 6G-DNA complexes from molecular dynamics simulations with a polarizable force field. J Phys Chem B. 2009;113:4425–34. doi: 10.1021/jp809774q. [DOI] [PubMed] [Google Scholar]
- Walker RC, Crowley MF, Case DA. The implementation of a fast and accurate QM/MM potential method in Amber. J Comput Chem. 2008;29:1019–31. doi: 10.1002/jcc.20857. [DOI] [PubMed] [Google Scholar]
- Walsh TR, Liang T. A multipole-based water potential with implicit polarization for biomolecular simulations. J Comput Chem. 2009;30:893–9. doi: 10.1002/jcc.21111. [DOI] [PubMed] [Google Scholar]
- Wang J, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules. J Comput Chem. 2000;21:1049–74. [Google Scholar]
- Wang J, Hou T, Duan Y. Molecular polarizability calculations with dipole interactions models. J Comput Chem. 2009 at press. [Google Scholar]
- Wang J, Kollman PA. Automatic parameterization of force field by systematic search and genetic algorithms. J Comput Chem. 2001;22:1219–28. [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–74. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Zhang W, Wu C, Lei H, Cieplak P, Duan Y. Strike a balance: optimization of backbone torsion parameters of AMBER polarizable force field for simulations of proteins and peptides. J Comput Chem. 2006;27:781–90. doi: 10.1002/jcc.20386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshel A, Levitt M. Theoretical studies of enzymic reaction—dielectric, electrostatic and steric stabilization of carbonium-ion in reaction of lysozyme. J Mol Biol. 1976;103:227–49. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- Woods RJ, Dwek RA, Edge CJ, Fraser-Reid B. Molecular mechanics and molecular dynamics simulations of glycoproteins and oligosaccharides. 1. Glycam 93 parameter development. J Phys Chem. 1995;99:3832–46. [Google Scholar]
- Xie W, Pu J, Mackerell AD, Gao J. Development of a polarizable intermolecular potential function (PIPF) for liquid amides and alkanes. J Chem Theory Comput. 2007;3:1878–89. doi: 10.1021/ct700146x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Stern HA, Berne BJ. Can water polarizability be ignored in hydrogen bonds kinetics? J Phys Chem B. 2002;106:2054–60. [Google Scholar]
- Yang Z, Zhang Q. Study of peptide conformation in terms of the ABEEM/MM method. J Comput Chem. 2006;27:1–10. doi: 10.1002/jcc.20317. [DOI] [PubMed] [Google Scholar]
- Yu H, Hansson T, van Gunsteren WF. Development of a simple, self-consistent polarizable model for liquid water. J Chem Phys. 2003;118:221–34. [Google Scholar]
- Yu H, van Gunsteren WF. Charge-on-spring polarizable water models revisited: from water clusters to liquid water to ice. J Chem Phys. 2004;121:9549–64. doi: 10.1063/1.1805516. [DOI] [PubMed] [Google Scholar]
- Yu H, van Gunsteren WF. Accounting for polarization in molecular simulation. Comput Phys Commun. 2005;172:69–85. [Google Scholar]
- Zimmerman SS, Pottle MS, Némethy G, Scheraga HA. Conformational analysis of the 20 naturally occurring amino acid residues using ECEPP. Macromolecules. 1977;10:1–9. doi: 10.1021/ma60055a001. [DOI] [PubMed] [Google Scholar]