Summary
Aeruginosins represent a group of peptide metabolites isolated from various cyanobacterial genera and from marine sponges that potently inhibit different types of serine proteases. Members of this family are characterized by the presence of a 2-carboxy-6-hydroxyoctahydroindole (Choi) moiety. We have identified and fully sequenced a NRPS gene cluster in the genome of the cyanobacterium Planktothrix agardhii CYA126/8. Insertional mutagenesis of a NRPS component led to the discovery and structural elucidation of two new glycopeptides that were designated aeruginoside 126A and aeruginoside 126B. One variant of the aglycone contains a 1-amino-2-(N-amidino-Δ3-pyrrolinyl)ethyl moiety at the C-terminus, the other bears an agmatine residue. In silico analyses of the aeruginoside biosynthetic genes aerA-aerI as well as additional mutagenesis and feeding studies allowed the prediction of enzymatic steps leading to the formation of aeruginosides and the unusual Choi moiety.
Introduction
Cyanobacteria provide a broad range of biologically active compounds, many of which are highly intriguing from a structural point of view [1]. In particular cyanobacterial peptides are often composed of highly unusual amino acids. A good example is the group of aeruginosins, which are characterized by an N-terminal derivative of an aryl lactic acid, a 2-carboxy-6-hydroxyoctahydroindole (Choi) moiety and an arginine derivative at the C-terminal position [2]. Different variants of aeruginosin have been isolated from the planktonic cyanobacterial genera Microcystis, Oscillatoria (Planktothrix), Nodularia, and from marine sponges [2-11]. Aeruginosins are potent inhibitors of serine proteases with different specificities for trypsin and thrombin, respectively. Two specific aeruginosins have also been crystallized bound to their protease targets trypsin and thrombin, respectively, in order to assess the structure-activity relationships [12, 13].
The majority of cyanobacterial peptides is synthesized by modular biosynthetic assembly lines, so-called nonribosomal peptide synthetases (NRPS) [1, 14]. A NRPS module is minimally composed of an amino acid activating adenylation (A) domain, a peptidyl carrier protein (PCP) domain and a condensation (C) domain [15]. In the last decade a number of such NRPS systems have been described from cyanobacteria. Biosynthesis genes for the production of the widespread cyanobacterial hepatotoxin microcystin were detected and characterized in Microcystis, Planktothrix and Anabaena, three distant cyanobacterial genera that form blooms in freshwater habitats [16-19]. The microcystin pathway exemplifies the prevalence of mixed NRPS/polyketide synthase (PKS) pathways, which is a major theme of cyanobacterial biosyntheses. This biosynthetic principle is also seen in several pathways that have been reported from the marine cyanobacterium Lyngbya majuscula, namely the barbamide, jamaicamide, and curacin pathways [20-22]. Natural product biosyntheses from cyanobacteria have further attracted attention because of their unique tailoring enzymes, including diverse forms of halogenases and aromatic prenyl transferases [20, 23, 24]. These fascinating properties of cyanobacterial NRPS/PKS pathways make them particularly interesting for pathway engineering approaches. It was therefore tempting to investigate the natural biosynthetic machinery of the structurally unique aeruginosin and its core moiety Choi.
Here we report the cloning, sequencing and mutational analysis of a gene cluster encoding aeruginosin biosynthesis in Planktothrix agardhii CYA 126/8 as well as the full structural elucidation of two new glycosylated aeruginosin variants, aeruginosides 126A and 126B.
Results and Discussion
Identification of the aeruginoside gene cluster via gene disruption
A candidate gene cluster for the biosynthesis of the aeruginosides was identified in the course of a search for NRPS genes by a degenerate PCR approach in Planktothrix CYA126/8 [16]. Subsequently, individual PCR fragments were cloned and sequenced and used to screen a phagemide library of the strain. As a result, a gene fragment was identified encoding a NRPS module comprising an A domain, a PCP domain and an epimerization (E) domain (Figure 1A). Analysis of the specificity conferring residues of the amino acid binding pocket [25] and comparison with characterized sequences in the database revealed a putative specificity of the A domain for leucine (Figure 1A and Supplementary Table 1). Furthermore, the presence of an epimerization domain suggested that the module could be responsible for the incorporation of a d-leucine moiety into a cyanopeptide structure. Leucine is common in several cyanobacterial peptide families; however, the d stereoisomer is characteristic for some aeruginosin variants that have been described from Microcystis [6]. Although Planktothrix CYA126/8 was known to produce a number of different peptides including putative aeruginosin variants [16], no detailed structural information on aeruginosins from CYA126/8 was available. In order to confirm the proposed involvement of the gene fragment in aeruginosin biosynthesis, an insertional mutagenesis of the gene in Planktothrix CYA126/8 was initiated using a homologous recombination strategy. For this purpose a chloramphenicol resistance cassette was inserted into the NRPS gene fragment and the resulting construct was introduced into the Planktothrix cells by electroporation. Following selection in liquid medium mutant clones were obtained. Subsequent PCR using primers distal to the insertion region revealed the successful insertion of the resistance cassette and the homozygosity of the mutants (Figure 1B).
Figure 1.
Inactivation of NRPS presumably involved in aeruginosin biosynthesis genes by homologous recombination and HPLC analysis of wild type and mutant.
(A) Schematic representation of ΔNRPS-d-Leu knock-out plasmids for the insertional mutagenesis of P. agardhii CYA126/8. The chloramphenicol resistance gene cassette (Cmr) is highlighted in black. (B) PCR amplification with the DNA from wild type (WT) and ΔNRPS-d-Leu mutant (MT) with primers amplifying the region flanking the insertion site. (C) HPLC-DAD profile of extract from Planktothrix agardhii CYA 126/8 wild type. D) HPLC-DAD profile of extract from ΔNRPS-d-Leu mutant. The peaks at min 9.07 (1) and 10.56 (2) represent putative aeruginosins, the peaks at min 26.6 (3) and 33.8 (4) [d-Asp]-microcystin-RR and [d-Asp]-microcystin-LR, respectively.
Identification and structural elucidation of aeruginosides 126A and 126B
The composition of the peptide fraction of wild type cells and of mutant cells cultured in the presence of chloramphenicol was compared by HPLC-DAD analysis. This analysis revealed that in the extract from the mutant three peaks were missing (Figure 1C and D and data not shown). MALDI-TOFMS analysis for two of these indicated molecular masses of 714 and 690, respectively (data not shown). These masses did not match those of any known aeruginosin-type molecules, which suggested that these compounds might be new. Thus, the peaks missing in the mutant were collected from large-scale laboratory cultures of the wild type. Two metabolites, aeruginoside 126A (1) and aeruginoside 126B (2), were obtained in sufficient quantity, 0.8 mg and 0.4 mg, respectively, to allow for a full spectroscopic characterization by NMR.
Positive ion HRESI-TOFMS analysis of 1 yielded a pseudomolecular ion (M+H)+ at m/z 715.4056 suggesting a molecular formula of C36H55N6O9 (calc. 715.4031, Δ= 2.5 mmu). Inspection of the 1H and 13C NMR spectra recorded in DMSO-d6 indicated the presence of two conformers in the ratio of 3:1. Only the data for the major conformer are displayed in Table 1 and will be discussed in the following. The 13C NMR spectrum showed 34 signals two of which were assigned to methyl groups on the basis of the analysis of gHSQC data, 11 to methylenes, 15 to methines and six to quaternary carbon atoms. This suggested that the molecule contained a symmetrical substructure, which was readily assigned to a phenyl group on the basis of the 1H NMR spectrum. This aromatic ring accounted only for four of the thirteen degrees of unsaturation. From the 13C NMR data five of the remaining double bond equivalents were identified in the form of three carbonyl resonances, a guanidine carbon atom plus one C-C double bond. Thus, 1 had to contain four additional rings (Supplementary Figure 1).
Table 1.
NMR Spectral Data for Aeruginoside A (1) in DMSO-d6
| Unit | C/H no. | δH (J in Hz) | δ C | HMBC |
|---|---|---|---|---|
| Plac | 1 | 173.2 | Plac H-2, H-3, Leu NH | |
| 2 | 4.11, dd (8.8, 3.9) | 72.02 | Plac H-3α, H-3β | |
| 3α | 2.95, dd (13.7, 3.9) | 40.4 | Plac H-2, H-5/9 | |
| 3β | 2.65, dd (13.7, 8.8) | |||
| 4 | 138.4 | Plac H-2, H-3α, H-3β, H-6/8 | ||
| 5/9 | 7.21, m | 129.3 | Plac H-3α, H-3β, H-7, H-9/5 | |
| 6/8 | 7.24, m | 127.9 | Plac H-8/6 | |
| 7 | 7.16, m | 125.9 | Plac H-5/9 | |
| Leu | 1 | 169.9 | Leu H-2 | |
| 2 | 4.50, ddd (9.0, 7.9, 4.8) | 48.6 | Leu H-3α | |
| 3α | 1.46, m | 41.0 | Leu H-4, H-5, H-5′ | |
| 3β | 1.34, m | |||
| 4 | 1.48, m | 24.1 | Leu H-3α, H-5, H-5′ | |
| 5 | 0.86, d (6.5) | 21.8 | Leu H-3α, H-3β, H-5′ | |
| 5′ | 0.84, d (6.4) | 23.3 | Leu H-3β, H-5 | |
| NH | 7.76, d (7.9) | |||
| Choi | 1 | 171.4 | Choi H-2, Aeap NH, H-1 | |
| 2 | 4.18, dd (9.0, 7.8) | 59.8 | Choi H-7a | |
| 3α | 2.04, ddd (13.0, 7.8, 6.8) | 30.5 | Choi H-2 | |
| 3β | 1.78, ddd (13.0, 13.0, 9.0) | |||
| 3a | 2.28, m | 36.1 | ||
| 4ax | 2.08, m | 19.5 | ||
| 4eq | 1.47, m | |||
| 5eq | 1.56, m | 24.5 | ||
| 5ax | 1.49, m | |||
| 6 | 3.84, m | 70.05 | Xyl H-1 | |
| 7eq | 2.27, brdd (14.0, 4.8) | 29.2 | ||
| 7ax | 1.57, brdd (14.0, 11.6) | |||
| 7a | 4.15, ddd (11.6, 6.2, 4.8) | 54.11 | Choi H-3α, H-4eq, H-7eq | |
| Aeap | 1α | 3.21, m | 36.6 | Aeap NH, H-2α, H-2β |
| 1β | 3.17, m | |||
| 2α | 2.28, m | 28.2 | Aeap H-1, H-4 | |
| 2β | 2.26, m | |||
| 3 | 136.2 | Aeap H-1, H-2α, H-2β, H-4, H-7 | ||
| 4 | 5.61, brs | 119.1 | Aeap H-2α, H-2β, H-5 | |
| 5 | 4.10, m | 54.08 | ||
| 6 | 154.2 | |||
| 7 | 4.09, m | 55.3 | Aeap H-4 | |
| NH | 7.74, t (5.8) | |||
| Xyl | 1 | 4.81, d (3.4) | 96.5 | Xyl H-3, H-5ax |
| 2 | 3.21, dd (9.4, 3.4) | 71.94 | Xyl H-3 | |
| 3 | 3.40, dd (9.4, 8.8) | 73.4 | Xyl H-1, H-2, H-5ax, H-5eq | |
| 4 | 3.28, m | 70.03 | Xyl H-3, H-5ax | |
| 5eq | 3.39, m | 62.1 | Xyl H-1 | |
| 5ax | 3.34, dd (10.6, 10.6) |
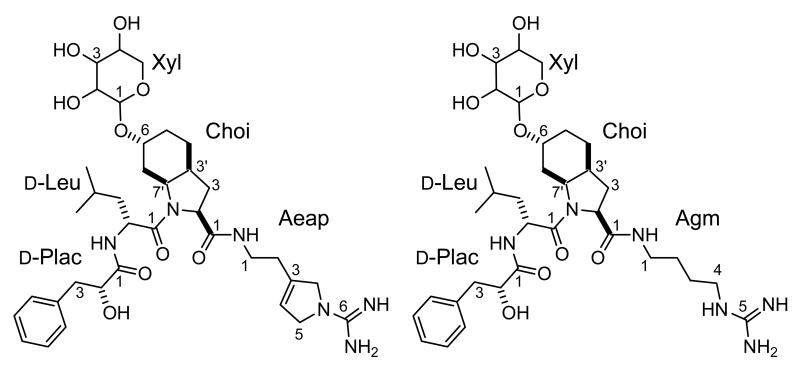
The identification of individual residues was accomplished by means of 1D TOCSY spectra. In this fashion, a leucine residue and a β-substituted lactic acid were readily identified. The large geminal coupling constant for the protons on C-3 of the latter suggested the presence of a π-system in the β-position. Eventually this residue was identified as a phenyl lactic acid (Plac) on the basis of gHMBC data and chemical shift considerations. Thus, the assignment of the latter as an α-hydroxy acid rather than an amino acid is based on the chemical shift for C-2 (72.02 ppm) and H-2 (4.11 ppm). Further inspection of the 13C NMR spectrum of 1 provided evidence for a 2-carboxy-6α-hydroxy-octahydroindole (Choi) subunit with excellent agreement of the chemical shifts observed for 1 and those reported in the literature. The only significant chemical shift difference noted was for the hydroxymethine carbon atom of 1 at δ= 70.05 ppm, which was shifted downfield (Δδ≈ 6 ppm) in comparison to Choi residues in other aeruginosin-type molecules (δ= 63.8 to 64.1 ppm)[4, 6, 8]. Since the correct assignment of the relative stereochemistry of the Choi residue has proven problematic in the past, we paid particular attention to this aspect. The cis stereochemistry of the ring junction follows from the coupling constant 3J= 6.2 Hz observed for H-7a. Confirmation of this stereochemistry by NOE was not possible because of the overlap of the resonances for H-7eq. and H-3a. All expected NOE’s were observed (see Supplementary Figure 2). Finally, the arginine mimetic characteristic of the aeruginosin class of compounds was identified as a 1–amino-2-(N-amidino-Δ3-pyrrolinyl)ethyl (Aeap) residue on the basis of gCOSY and gHMBC data using the NH and the olefinic methine proton resonances as starting points. This assignment was corroborated through chemical shift comparison with literature values [26, 27], which were in excellent agreement.
The subunits of 1 identified thus far account for all but five carbon atoms, five oxygen atoms and ten protons. This elemental composition of the remainder suggested the presence of a carbohydrate moiety in 1, and a proton resonance at 4.81 ppm was tentatively assigned to the anomeric proton of the putative sugar. In the HSQC spectrum the signal for this proton correlates to a resonance at 96.5 ppm, a typical value observed for acetal carbon atoms. Further analysis of gHMBC and gHSQC data led to the identification of three additional hydroxymethine and one hydroxymethylene carbon atom. The relative stereochemistry of the sugar was established on the basis of 1D TOCSY data. The large 3J coupling constants (3J2,3= 9.4 Hz; 3J3,4= 8.8 Hz; 3J4,5ax =10.6 Hz) observed for the hydroxymethine protons on C-2, C-3 and C-4 of the sugar suggested an all-axial arrangement of these protons in a six-membered ring. Thus, the sugar residue in 1 was identified as a xylopyranoside. The small coupling constant between the anomeric proton and H-2 (3J= 3.4 Hz) was indicative of H-1 being equatorial and hence of the presence of an α-glycosidic linkage. The observation of a gHMBC correlation from H-1 of the sugar to C-6 (δ=70.05 ppm) of the Choi residue indicates the site of attachment of the sugar to the aglycone. This connectivity also explains the downfield shift of the signal for this carbon atom, as noted earlier, relative to that found in Choi units bearing a free hydroxyl.
The sequence of the amino acids in the peptide was determined by means of gHMBC correlations from the exchangeable NH proton of the leucine unit to C-1 of Plac and from the NH-1 resonance of Aeap to the carboxyl resonance of Choi. No HMBC correlations were observed in support of the bond between C-1 of leucine and the nitrogen atom of Choi. However, unambiguous evidence for a connection between these units is furnished by very strong NOE correlations between the aminomethine proton of leucine (4.50 ppm) and H-7a as well as H-7eq. of the Choi unit.
Positive ion HRESI-TOFMS analysis of 2 yielded a pseudomolecular ion (M+H)+ at 691.4051, suggesting a molecular formula of C34H54N6O9 (Δ= 2.0 mmu). As in the case of 1, analysis of the NMR spectra of 2 revealed the presence of two conformers. Chemical shift comparison of the NMR spectra of 1 and 2 revealed excellent agreement (Δδ (13C) ≤ 0.1ppm, Δδ (1H) ≤ 0.05ppm) for all but the arginine mimetic portion of the peptides. Analysis of gHMBC and 1D TOCSY data then established that 2 was bearing an agmatine rather than an Aeap moiety. Thus, the structure of 2 closely resembles the recently revised structure of aeruginosin 205 [28]. The absolute stereochemistry of the phenyl lactic acid and the leucine moiety of 1 was determined after acid hydrolysis by chromatography on chiral stationary phase and comparison of retention times to those of authentic standards. The retention times of Plac from 1 matched that of d-Plac and that of leucine matched the retention time of the d-leucine standard. The absolute configuration of the xylose residue could not be established (Table 1, Figure 2). Aeruginoside 126A weakly inhibited porcine pancreas trypsin and bovine plasma thrombin with IC50 values of 67 and 30 μg/ml, respectively, compared to aeruginosin 98-A [5] as a positive control (0.07 and 2.5 μg/ml). Aeruginoside 126A did not inhibit human plasma plasmin at a concentration of 100 μg/ml, compared to aeruginosin 98-A [5] as a positive control with IC50 value of 2.7 μg/ml.
Figure 2.
Structures of aeruginoside 126A (1) and 126B (2). d-Plac; d-phenyl lactic acid, Choi; 2-carboxy-6-hydroxyoctahydroindole, Aeap; 1-amino-2-(N-amidino-Δ3-pyrrolinyl)ethyl, Agm; agmatin, Xyl; xylose.
Analysis of the organisation of the aeruginoside gene cluster and flanking regions
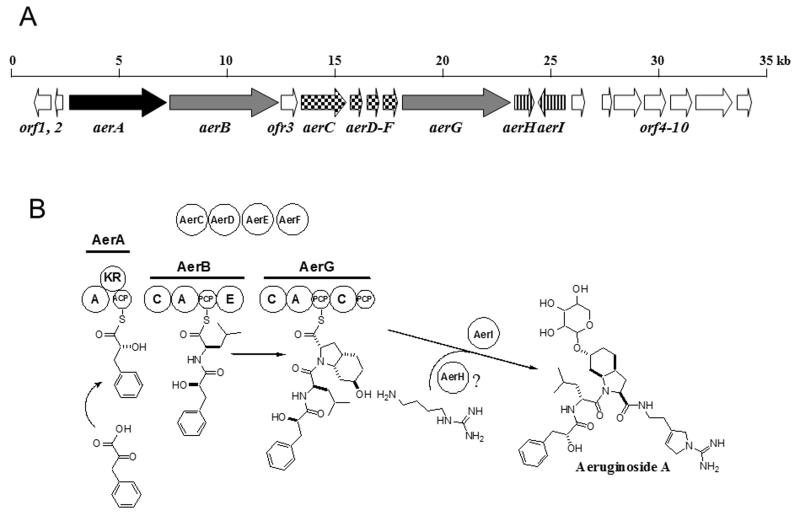
In order to obtain sequence information about the entire gene cluster encoding aeruginosin biosynthesis the original fragment of the aeruginoside gene cluster was used as a probe to screen a genomic phagemid library of Planktothrix CYA126/8. Subsequently 35 kb were sequenced from overlapping phagemide clones spanning the aeruginoside biosynthesis gene cluster and flanking regions. Sequence analysis of the aeruginosin (aer) region revealed the presence of 19 ORFs, nine of which could be assigned to aeruginoside biosynthesis and therefore designated aerA-aerI. The function of ten additional ORFs remained obscure (Table 2, Figure 3A). Nine of the Aer proteins are encoded in the aerA-I cluster. Upstream of the aer region a gene was transcribed in the opposite direction encoding a protein with 51% similarity to the cyanobacterial circadian clock protein KaiA that was not considered to be involved in aeruginoside production (ORF1, Figure 3A). Further upstream a second ORF encoded a protein with no significant similarity to characterized proteins and was also not assigned to the biosynthesis (ORF2, Figure 3A). A third ORF encoded between the aerB and aerC genes did not show any similarity to biosynthetic proteins and was thus not expected to be involved in aeruginoside biosynthesis (ORF3, Figure 3A). Downstream of the aerA-I gene cluster seven additional genes were sequenced. An overview of the deduced functions of these (ORF4-10) is given in Table 2. One of these genes encodes a protein with 46% similarity to a glycosyltransferase and was designated aerI. This tailoring activity is needed for the modification of the aeruginosin base structure and was therefore attributed to aeruginoside biosynthesis. The sequence of the aer gene cluster and flanking regions has been deposited in the EMBL database (accession number AM071396). In the following section the individual Aer proteins are analysed in detail. A schematic representation of the predicted aeruginoside biosynthesis pathway is shown in Figure 3B.
Table 2.
Deduced Functions of ORFs in the Aeruginoside Biosynthetic Gene Cluster
| Protein | Amino Acids | Deduced Function | Sequence Similarity | Identity/SimilarityAccession Number (AA length) |
|---|---|---|---|---|
| AerA | 1416 | PKS (A KR ACP) | BarE (Lyngbya majuscula) | 44%/64% (587)AAN32979 |
| AerB | 1598 | NRPS (C A T E) | NosC (Nostoc sp. GSV224) | 54%/71% (944)AAF17280 |
| AerC | 736 | Oxygenase | Putative diaminopimelate decarboxylase (Burkholderia mallei 10229) |
30%/49% (707)ZP00431845 |
| AerD | 202 | Decarboxylase | BacA (Bacillus amyloliquefaciens AAM90573) | 39%/58% (191)AAM90573 |
| AerE | 213 | Unknown | BacB (Bacillus subtilis) | 26%/45% (208)AAM90569 |
| AerF | 264 | Reductase | BacC (Bacillus subtilis) | 36%/55% (256)AA65204 |
| AerG | 1622 | NRPS (C A T C T) | McyB (Microcystis aeruginosa) | 47%/64% (927)AAE09608 |
| AerH | 308 | Dioxygenase | Thymine dioxygenase (Rhodotorula glutinis) | 30%/50% (517)AAU12179 |
| AerI | 418 | Glycosyltransferase | Spore coat protein SA (Bacillus subtilis) | 26%/46% (336)P46915 |
|
| ||||
| ORF1 | 248 | Circadian clock protein | KaiA (Synechococcus elongatus) | 31%/51% (227)1R8JA |
| ORF2 | 126 | Unknown | no similarity | |
| ORF3 | 288 | Unknown | no similarity | |
| ORF4 | 229 | Unknown | no similarity | |
| ORF5 | 170 | Unknown | Putative reverse transcriptase (Nostoc sp. PCC7120) | 70%/83% (157) BAB77479 |
| ORF6 | 428 | Unknown | Retron-type reverse transcriptase (Anabaena variabilis ATCC29413) |
58%/74% (415)ABA24728 |
| ORF7 | 386 | Oxidoreductase | Putative aldo/keto reductase (Tricodesmium erythraeum IMS101) |
70%/84% (383)ZP00328942 |
| ORF8 | 339 | Sulfotransferase | Putative sulfotransferase (Shewanella sp. PV-4) | 22%/39% (228)EAP04083 |
| ORF9 | 664 | ABC-type transporter | NcpC (Nostoc sp. ATCC 53789) | 57%/75% /663)AAO23332 |
| ORF10 | 257 | Succinate dehydrogenase | Succinate dehydrogenase (Sulfolobus tokodaii) | 33%/53% (238)BAB40685 |
Figure 3.
Biosynthetic pathway of aeruginosides 126A and 126B.A) Schematic representation of the aeruginoside biosynthesis gene cluster of Planktothrix agardhii CYA126/8. Genes encoding NRPS or NRPS/PKS hybrid components are indicated in light and dark grey, respectively. Genes presumably involved in Choi biosynthesis are checkered and tailoring genes are highlighted with stripes. Genes that were not associated with aeruginoside biosynthesis are shown in white.
B) Model for the formation of aeruginoside A and B and predicted domains of AerA-AerI. Each circle represents a NRPS domain or tailoring function. A, adenylation domain; KR, ketoreductase domain; ACP, acyl carrier protein; C, condensation domain; PCP, peptidyl carrier protein; E, epimerization domain.
In silico analysis of AerA-J proteins
The first protein encoded by the aer gene cluster, AerA, represents an unusual PKS-like module comprising a NRPS A-domain, a ketoreductase (KR) domain and an acyl carrier protein (ACP) domain. Analysis of the substrate binding pocket showed closest similarity to the A domain of the microcystin biosynthesis protein McyG that is known to activate diverse phenylpropanoid starter units in vitro ([29], Supplemental Table 1). It is therefore proposed that AerA activates phenylpyruvate, which is reduced by the ketoreductase domain to yield the Plac moiety of aeruginoside. The rare phenylpyruvate starter [30] is likely provided in sufficient amounts by the shikimate pathway.
Further downstream the aerB gene encodes a NRPS module comprising a C, A, PCP and E domain. This gene was initially inactivated resulting in the loss of aeruginoside production. As mentioned above, the substrate activating binding pocket shows close similarity to leucine activating domains (Supplemental Table 1). The AerB module is thus expected to incorporate d-leucine into the peptide structure. AerC contains signature sequences of ring-hydroxylating dioxygenases and a conserved Rieske [2Fe-2S] domain (Table 2).
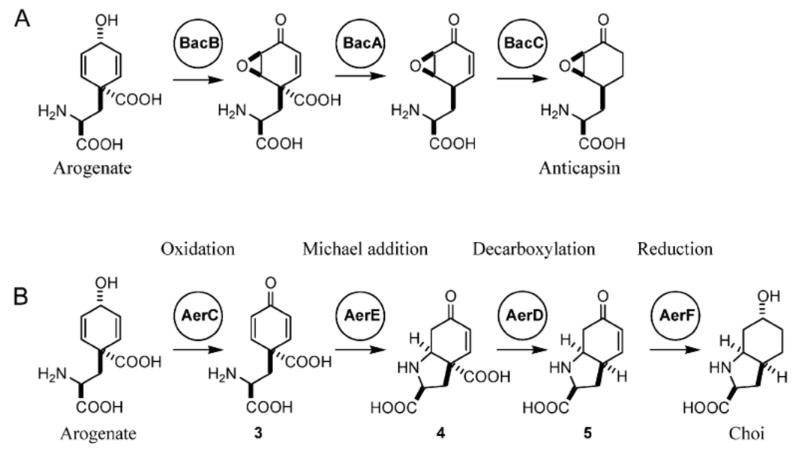
AerD, E and F show 58%, 45% and 55% amino acid sequence similarity to BacA, BacB and BacC of bacilysin biosynthesis. These three enzymes are involved in the biosynthesis of the anticapsin moiety [31] of l-alanyl-(2,3-epoxycylohexan-4-one)l-alanine ( bacilysin). An alignment of the sequences of the Bac enzymes from Bacillus amyloliquefaciens and Bacillus subtilis [31] and of the corresponding Ywf enzymes encoded in the completely sequenced genome of Bacillus subtilis subsp. subtilis str. 168 [32] with AerD, AerE and AerF, respectively, is provided as Supplemental Information (Supplemental Figure 3). Feeding studies and studies of knock out mutants of chorismate mutase have revealed prephenate as precursor of the anticapsin moiety [33]. A motif scan of AerD using protein family analysis software (http://www.sanger.ac.uk/Software/Pfam/search.shtml) showed a conserved prephenate dehydratase signature. Prephenate is a known intermediate of phenylalanine and of tyrosine biosynthesis in many organisms. We conclude that AerD is involved in the biosynthesis of Choi in vivo. BacB and AerE, respectively, contain a conserved cupin domain. Although the role of this domain is not well understood, it is conserved among several dioxygenases [34]. BacB could perform the epoxidation step in anticapsin synthesis. It is not clear, what function could be fulfilled by the related enzyme AerE in Choi biosynthesis. Finally, AerF and BacC, respectively, show close similarity to dehydrogenases and reductases. On the basis of the preceeding analysis we propose that AerC, D, E and F provide the Choi moiety of aeruginoside. A biochemical model for the biosynthetic route to Choi is discussed below.
Downstream of the aerD-F region, the aerG gene encodes a NRPS module with the domain order C, A, PCP, C, and PCP. The analysis of the substrate binding pocket of the A domain revealed closest similarity to enzymes activating proline or methylproline, although part of the specificity conferring residues differ from typical proline-activating domains. This suggests that a proline-like amino acid rather than proline itself is activated (Supplemental Table 1). It is interesting to note that spumigin, a peptide isolated from a Nodularia strain, is similar to aeruginosin but contains a methylproline moiety instead of Choi [35]. In Planktothrix, however, it is expected that AerG directly activates and incorporates the Choi moiety (see also feeding studies below). It is not clear to which extent the second C domain and, in particular, the second PCP domain of AerG are involved in aeruginoside biosynthesis. Amide bond formation principally functions with a free-standing C-domain, such as for the VibH enzyme in vibrobactin biosynthesis [36]. The second C and PCP domains of AerG in Planktothrix may be remnants of a complete NRPS module. It is of note that aeruginosin-type metabolites are known from Microcystis aeruginosa in which the C-terminal carboxyl group of arginine is reduced to the aldehyde and/or alcohol oxidation state [4, 6]. Production of metabolites of the latter type requires a functional, complete module and a reductase in order to effect release of the peptide from the enzyme. The structure of AerG from the Planktothrix aer cluster suggests that in this organism the synthetase may be able to add the agmatine/Aeap moiety to the growing peptide chain with just a C domain. Since agmatine and Aeap lack the carboxyl that would otherwise tether the growing peptide to the synthetase, a thioesterase or domain for a reductive release is not required in the Planktothrix enzyme. We therefore postulate that the release of the peptide from the enzyme complex is triggered by amide bond formation between the PCP-bound carboxyl group of Choi and the primary amine of agmatine or of Aeap.
The AerH protein shows similarity to oxygenases including isopenicillin N synthase and related enzymes from various bacteria. This enzyme may have a function in Aeap biosynthesis from arginine or agmatine. However, since the origin of carbon atoms in the Aeap residue is not presently known, this assignment is speculative at this time. The putative tailoring enzyme AerI shows similarity to glycosyltransferases. A glycosyltransferase is needed for the transfer of the xylose moiety to the hydroxyl group of Choi. Although the specificity of the glycosyltransferase cannot be deduced from the sequence, the close proximity of the gene to aeruginosin biosynthesis genes suggests a role in aeruginoside biosynthesis.
Taken together, the in silico analysis of the enzymes encoded by the aer gene cluster suggests roles for the enzymes in aeruginoside biosynthesis. The majority of enzymatic reactions proposed are in agreement with the synthesis of the tetrapeptide (Figure 3B). However, several of the enzymatic steps remain obscure at this point, particularly as far as Aeap biosynthesis is concerned. In order to obtain further insights into some of the reactions involved in Choi biosynthesis we constructed additional mutants and initiated feeding studies.
Mutagenesis of putative Choi biosynthesis genes and feeding studies
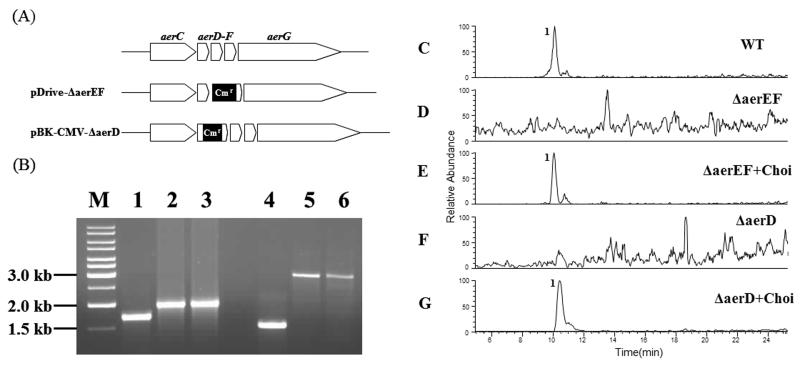
In order to confirm the proposed involvement of AerD, E and F in the synthesis of the Choi moiety, two knock-out constructs were designed. For both constructs a chloramphenicol resistance cassette was inserted into the aerD-F region. The first construct was bearing a deletion in the aerE-F region, whereas in the second experiment the antibiotic cassette was inserted into the aerD gene (Figure 4A). Both constructs were introduced into Planktothrix by electroporation. Mutants obtained after three months were analysed by PCR and revealed the successful homologous recombination of the construct DNA (Figure 4B). Extracts of broths from both mutants, designated ΔaerEF and ΔaerD, were compared to the wild type by LC-ESI-MS (Figure 4C, D and F). While the wild type spectrum revealed the characteristic aeruginoside peak at 10 min (m/z 714.80-715.80), no aeruginoside could be detected in the mutants (Fig. 4C, D and F). Following this observation both mutants were fed with external l-Choi hydrochloride [37] and reanalysed by LC-ESI-MS. Mass spectra of cell mass of the mutant after Choi feeding clearly revealed the production of aeruginoside (Figure 4E and G). These results confirmed the proposed involvement of the aerD-F genes in Choi biosynthesis. Moreover, the results indicate that Choi is directly activated at the AerG enzyme rather than being synthesized as an enzyme-bound intermediate. This conclusion is further supported by the fact that free Choi could be detected in the wild type strain Planktothrix CYA 126/8 and in the ΔaerB mutant strain (data not shown).
Figure 4.
Analysis of putative Choi biosynthesis genes.A-B) Inactivation of putative Choi biosynthesis genes by homologous recombination. (A) Schematic representation of ΔaerEF and ΔaerD knock-out plasmids for the insertional mutagenesis of P. agardhii CYA126/8. Chloramphenicol resistance gene cassettes (Cmr) are highlighted in black.
(B) PCR amplification with the DNA from wild type (lanes 1 and 4), ΔaerEF and ΔaerD mutants (lanes 2 and 5), and ΔaerEF and ΔaerD plasmid constructs (lanes 3 and 6) with primers amplifying the aerD-aerF (lanes 1-3) and the aerD (lanes 4-6) regions, respectively. C-G) LC-ESI MS (Positive SIM, range m/z 714.80-715.80) of extracts from (C) P. agardhii CYA126/8 WT, (D) P. agardhii CYA126/8 ΔAerEF, (E) P. agardhii CYA126/8 ΔAerEF + Choi, (F) P. agardhii CYA126/8 ΔAerD, (G) P. agardhii CYA126/8 ΔAerD + Choi. 1; aeruginoside 126A.
Although AerD-F look strikingly similar to the BacA-C enzymes of Bacillus, the Choi moiety and the anticapsin moiety exhibit only limited structural similarity. An adaptation of the enzymatic reactions of the bacilysin pathway to Choi biosynthesis is not straightforward as the sequence of reactions in Bacillus is not well understood. Both pathways have in common that direct feeding with tyrosine itself did not result in any evidence for its incorporation into either Choi (this work, data not shown) or anticapsin [38]. This suggests that a tyrosine precursor is the obligatory intermediate in these two pathways. In the case of anticapsin biosynthesis it has been suggested that prephenate, an intermediate of phenylalanine biosynthesis, might fulfil this function [33]. This view is supported by the observation that there is strong sequence similarity between BacA and AerD and prephenate decarboxylases. However, although Choi bears an amino group, an enzyme potentially transferring an amino group is not encoded within the aeruginoside gene cluster. Therefore we propose that the amino group of Choi is provided by arogenate (pre-tyrosine), a known intermediate of tyrosine biosynthesis in cyanobacteria [39]. Arogenate could potentially also serve as precursor for anticapsin biosynthesis (Figure 5A). On the basis of these assumptions the hypothetical Choi biosynthetic pathway is proposed as follows (Figure 5B): Arogenate is oxidized by AerC to produce 3, which is cyclized via Michael addition. This step must be enzyme catalyzed in order for the correct diastereomer of Choi to be produced: addition of the α-amino group to the pro-R carbon of the dienone results in the 2S, 3aR, 7aR diastereomer of 4, whereas addition to the pro-S carbon results in the 2S, 3aS, 7aS diastereomer of 4. The resulting vinylogous β-keto acid 4 is decarboxylated by AerD to yield the bicyclic amino acid 5, which may be reduced by AerF (Figure 5B). This model is chemically reasonable but will require additional refinement through future enzymological studies after the complete heterologous expression of the AerD-F region, or through further mutagenesis studies.
Figure 5.
Proposed biosynthetic pathways of the anticapsin moiety of bacilysin and the Choi moiety of aeruginoside. (A) Biosynthetic model for the formation of anticapsin starting from arogenate. B) Biosynthetic model for the formation of Choi starting from arogenate.
Significance
Many cyanobacterial metabolites are infamous as health-threatening toxins that occur in bacterial blooms. However, various natural products from these organisms show important biological activities, such as the aeruginosins, which specifically inhibit serine proteases and are regarded as promising drug candidates. Aeruginosins are also interesting from a structural point of view, as they represent unusual cyanobacterial peptides that contain a unique 2-carboxy-6-hydroxyoctahydroindole (Choi) moiety. We have identified a putative aeruginoside biosynthesis genes in the genome of the freshwater cyanobacterium Planktothrix agardhii CYA126/8 and succeeded in unequivocally proving its identity by directed mutagenesis of a NRPS core component encoded by the cluster. Comparison of the metabolic profiles led to the discovery and structural elucidation of two new glycopeptides, which were named aeruginosides 126A and 126B. The aeruginoside biosynthesis pathway from Planktothrix CYA126/8 provides a novel example for a complex molecular assembly line composed of NRPS featuring an unusual loading module as well as an extender module for the unique Choi moiety and a putative glycosyltransferase. Heterologous expression of aeruginoside pathway genes for sustainable production or engineering of potential aeruginoside-derived drugs would be to be a promising alternative to chemical de novo synthesis in the future.
Experimental section
Bacterial strains and culturing
The axenic strain P. agardhii CYA126/8 was provided by Prof. K. Sivonen (University of Helsinki, Helsinki, Finland). Wild-type and mutant strains were cultured in Z8 medium as described previously [16]. Cultures were maintained under continuous white light (30 μEm−2s−1) on a shaker with 40 or 80 rpm at 23°C. DNA manipulations were performed in Escherichia coli XL1 Blue in LB medium. E. coli XL1 Blue MRF’ Kan strain was used for the excision of lambda phagemids according to the suggestions of the manufacturer (Stratagene, La Jolla, USA).
Construction and screening of the phagemid library
The isolation of chromosomal DNA from P. agardhii CYA126/8 and the construction of a lambda ZAP library were previously reported [16]. Screening of the library was performed by hybridization. Clone gaps were connected by PCR or inverse PCR, respectively, using Pfu polymerase (Stratagene, La Jolla, USA).
Analysis of aeruginosides
Cells of WT strain CYA126/8 and the mutant cultures grown in BG11 medium22 at 20°C (15 μE m−2 s−1) were harvested on preweighed glass fiber filters (GF/C). The cell mass was freeze-dried and the filters were reweighed for the determination of the dry weight (6 mg of DW each for WT and the mutant). Filters were extracted in aqueous MeOH (50/50, v/v) on a shaker for 60 min. The extract was centrifuged and the supernatant cleaned up by solid phase extraction (SPE) using Sep-Pak ® tC18 cartridges (Waters, Vienna, Austria). The tC18 cartridges were conditioned using MeOH followed by equilibration using 25% aq. MeOH (v/v). Samples were applied in 25% aq. MeOH, the cartridge was washed with five bed volumes of the same solvent and aeruginosins were eluted using 80% MeOH (v/v). The cleaned extract was resuspended in 100 μl of 50% MeOH and 100 μl were injected using HPLC-DAD (HP1100) and separated on a LiChrosper® 100, ODS, 5 μm, LiChroCART® 250-4 cartridge system (Merck, Darmstadt, Germany) using a linear gradient of aq. acetonitrile (with 0.05% v/v trifluoracetic acid, TFA) starting with 20% acetonitrile (v/v) and increasing up to 50% acetonitrile within 45 min with a flow of 1 mL min−1. Peptides were detected at 210 nm.
Isolation of aeruginosides
Fourtyfive L of CYA126/8 were grown as described above, harvested by filtration through a sieve with a mesh size of 3 μm using low pressure vacuum and the cell mass was freeze dried. Cells were extracted with 50% MeOH (see above), and the extracts injected after SPE purification as described. HPLC purification was performed using a linear gradient of aqueous acetonitrile (0.05% v/v TFA) starting with 20% acetonitrile (v/v) and increasing up to 30% acetonitrile within 15 min with a flow of 1 mL min−1. The compounds were colorless oils and HPLC analysis of the purified compounds did not show UV absorbing contaminants.
Aeruginoside 126A
Amorphous colorless solid; [α]25d 137 (c 0.0025 H2O); 1H NMR and 13C NMR, see Table 1; HRESITOFMS (positive ion mode) [M+H]+ 715.4056 (715.4031 calculated).
Aeruginoside 126B
Amorphous colorless solid; [α]25d 122 (c 0.0039, H2O); 1H NMR (500 MHz), DMSO-d6 δ Plac: 7.25 (2H, t, J = 7.4 Hz, H-6/8), 7.21 (2H, d, J = 7.4 Hz, H-7), 7.17 (1H, m, H-5/9), 4.11 (1H, dd, J = 8.8, 3.7 Hz, H-2), 2.96 (1H, dd, J = 13.4, 3.7 Hz, H-3α), 2.64 (1H, dd, J = 13.4, 8.8 Hz, H-3β); Leu: 7.77 (1H, d, J = 7.8 Hz, NH), 4.53 (1H, m, H-2), 1.48 (1H, m, H-4), 1.46 (1H, m, H-3α), 1.34 (1H, m, H-3β), 0.87 (3H, d, J = 6.4 Hz, H-5), 0.84 (3H, d, J = 6.8 Hz, H-5′); Choi: 4.18 (1H, dd, J = 8.9, 8.5 Hz, H-2), 4.16 (1H, m, H-7a), 3.85 (1H, m, H-6), 2.30 (1H, m, H-3a), 2.29 (1H, m, H-7eq), 2.09 (1H, m, H-4ax), 2.03 (1H, m, H-3α), 1.83 (1H, m, H-3β), 1.61 (1H, m, H-7ax), 1.57 (1H, brd, J = 12.7 Hz, H-5eq), 1.50 (1H, m, H-5ax) 1.48 (1H, m, H-4eq); Agm: 7.74 (1H, t, J = 5.6 Hz, NH-1), 7.44 (1H, t, J = 5.6 Hz, NH-4), 3.08 (2H, m, H-4), 3.05 (2H, m, H-1), 1.44 (2H, m, H-2), 1.43 (2H, m, H-3); Xyl: 4.82 (1H, d, J = 3.6 Hz, H-1), 3.40 (1H, dd, J = 9.0, 9.0 Hz, H-3), 3.39 (1H, m, H-5eq), 3.34 (1H, dd, J = 11.0, 11.0 Hz, H-5ax), 3.28 (1H, m, H-4), 3.22 (1H, dd, J = 9.0, 3.6 Hz, H-2); 13C NMR (125 MHz, DMSO-d6) δ Plac: 173.2 ((1) C-1), 138.5 ((1) C-4), 129.4 ((2) C-5/9), 128.0 ((2) C-6/8), 126.0 ((2) C-7), 72.06 ((2) C-2), 40.46 ((3) C-3); Leu: 169.8 ((1) C-1), 48.5 ((2) C-2), 41.3 ((3) C-3), 24.2 ((2) C-4), 23.4 ((4) C-5′), 21.9 ((4) C-5); Choi: 171.6 ((1) C-1), 70.12 ((2) C-6), 59.9 ((2) C-2), 54.1 ((2) C-7a), 36.1 ((2) C-3a), 30.5 ((3) C-3), 29.3 ((3) C-7), 24.5 ((3) C-5), 19.5 ((3) C-4); Agm: 156.7 ((1) C-5), 40.56 ((3) C-4), 37.9 ((3) C-1), 26.2 ((3) C-2), 25.8 ((3) C-3); Xyl: 96.6 ((2) C-1), 73.5 ((2) C-3), 72.0 ((2) C-2), 70.07 ((2) C-4), 62.2 ((3) C-5); HRESITOFMS (positive ion mode) [M+H]+ 691.4051 (691.4031 calculated, Δ= 2.0 mmu).
Analysis of stereochemistry of 1
Aeruginoside 126A (0.5 mg) was heated in 0.5 mL 6 M HCl at 110 °C for 16 hrs. After cooling to rt, the solution was extracted with EtOAc (3 × 0.5 mL) and the organic layer was removed under a stream of nitrogen. The residue was dissolved in MeOH and a portion was injected onto a Chiralpak MA(+) column equilibrated in 2 mM aqu. CuSO4/ CH3CN (85:15) and eluted at 0.8 mL min−1. d-Plac eluted at 41 min (retention time l-Plac: 59 min).
The aqueous layer from the hydrolysis was evaporated under a stream of nitrogen and a portion of the residue was injected onto a Chirex D column equilibrated in 2 mM aqu. CuSO4/CH3CN (95: 5, v/v) and eluted at 0.8 mL min−1. d-Leu eluted at 61.7 min (retention time l-Leu: 51.4 min).
Protease inhibition assays
Serine protease inhibitory activities were analysed as described previously [40] using porcine pancreas trypsin (Sigma; St. Louis, USA), bovine plasma thrombin (Sigma; St Louis, USA) and human plasma plasmin (Sigma; St. Louis, USA). Aeruginosin 98A from M. aeruginosa NIES98 was used as a positive control.
Construction of the gene disruption plasmids for aerB, aerD and aerEF
The phagemid pBK-CMV-ΔaerB was digested with XcmI (E1 site in epimerization domain). After blunting of the phagemide ends by Klenow treatment, the 1.4-kb BsaAI fragment from pACYC184 containing the chloramphenicol resistance cassette was inserted. Positive clones containing the antibiotic resistance cassette were identified by restriction analysis. The phagemid pBK-CMV-ΔaerD was digested with EcoRI and XhoI to remove a HindIII recognition site. Subsequently, sticky ends of the plasmid were blunted as described above. The resulting plasmid was purified (Qiaquick, Qiagen, Hilden, Germany) and selfligated. The derived plasmid was digested with HindIII and then blunted as described above. The 1.4-kb BsaAI fragment from pACYC184 containing the chloramphenicol resistance cassette was cloned into the blunted plasmid. Positive clones and the orientation of the antibiotic cassette were identified by restriction analysis.
A 6115 bp gene fragment containing aerEF was amplified by PCR using the primers Pa36_61Arv3 (5′-AAG CAG GAA GCT TCA CTT GG-3′) and Dhg1641rv5 (5′-CCT GCA ATC ACA AAC AAT TAC T-3′) with Pfu polymerase (Stratagene, La Jolla, USA). The resulting PCR product was treated with Taq polymerase (72 °C for 30 min), and purified using a gel purification kit (Qiaquick, Qiagen, Hilden, Germany). The purified DNA was cloned into the pDrive vector (Qiagen, Hilden, Germany). An 1178 bp fragment covering the region between 5′ end of aerE to the mid of aerF was deleted with PacI and blunted by Klenow treatment. The 1.4-kb BsaAI fragment from pACYC184 containing the chloramphenicol resistance cassette was cloned into the blunted plasmid generating the gene disruption plasmid pDrive-ΔaerEF. The antibiotic resistance clones were identified by restriction analysis.
Planktothrix mutagenesis
The three constructs (15 to 20 μg of plasmid DNA) were linearized in the multi-cloning site of the vectors using BamHI. The restricted DNA was column purified (Qiaquick, Qiagen, Hilden, Germany), denaturated at 95 °C for 10 min, and then immediately put onto an ice bath. 50 mL of cultured algal cells were centrifuged at 6000 rpm and washed three times with 1 mM HEPES buffer and centrifuged. Cells were resuspended in 1 mM HEPES and subjected to electroporation with 10 μg of the single strand plasmid DNA (1.0 kV, 25 μF, 200 Ω).
Transformed cyanobacterial cells were inoculated in 100 mL of Z8 medium and cultivated as described above. After three days, 50 μg of chloramphenicol was added to culture medium. After 4 to 6 weeks, a fresh Z8 medium containing 0.5 μg mL−1 chloramphenicol was added to the cultures. Transformants were purified by increasing stepwise the chloramphenicol concentration up to 5 μg mL−1 (8 to 12 weeks).
DNA sequencing and sequence analysis
Sequencing of phagemids was performed by primer walking using the Big Dye terminator cycle sequencing kit (ABI, Foster City, USA). Analyses of DNA sequences and deduced amino acid sequences were performed using the NCBI (National Institute for Biotechnology Information, Bethesda, MD) BLAST server (http://www.ncbi.nlm.nih.gov) and the ExPASy proteomics server (http://www.expasy.ch) of the Swiss Institute of Bioinformatics (Geneve, Switzerland).
HPLC-ESIMS analysis of aeruginosides and feeding experiments
One 200 mL aliquot of wild type and two 200 mL aliquots of the ΔaerD and ΔaerEF mutant cells were cultivated as described above. One aliquot of the ΔaerD and ΔaerEF mutant, respectively, was fed with 2 mg l-Choi•HCl in the early logarithmic phase. All cultures were grown until they reached late logarithmic phase (OD750: 1.0), centrifuged and freeze-dried. Freeze-dried algal cells were extracted twice with 80% MeOH (10 mL / 100 mL cultured cells) and once with MeOH (10 mL / 100 mL cultured cells). Combined 80% MeOH and MeOH extracts were concentrated to an aqueous suspension which was then washed with diethylether. The aqueous layer was extracted with n-BuOH. The n-BuOH layer was evaporated under reduced pressure and dissolved in 50% MeOH (0.5 mL / 100 mL cultured cells). Sample solutions were analysed by LC-ESIMS (Finnigan TSQ Quantum ULTRA ThermoFinnigan Surveyor HPLC system, Thermo Electron Corporation, Waltham, MA, USA) using reversed-phase HPLC (PRONTOSIL 120-5-C18 ace-EPS (4.0 × 250 mm, Bischoff Chromatography, Leonberg, Germany) using a linear gradient of 10 to 80% acetonitrile containing 0.1% AcOH in 35 min at a flow rate of 1.0 mL min−1.
Supplementary Material
Acknowledgement
K. I. was financially supported by JSPS Postdoctoral Fellowships for Research abroad. We thank Dr. O. Scheibner and Mrs. A. Perner for LC-ESI MS analyses, and Dr. M. Ito-Ishida for assistance. The work was supported by a grant of the EU (PEPCY) to T.B and E.D and by the NSF (OCE04-32479) and the NIEHS (P50 ES012740) to T.H.
References
- 1.Welker M, von Döhren H. Cyanobacterial peptides - Nature’s own combinatorial biosynthesis. FEMS Microbiol. Rev. 2006;30:530–563. doi: 10.1111/j.1574-6976.2006.00022.x. [DOI] [PubMed] [Google Scholar]
- 2.Murakami M, Okita Y, Matsuda H, Okino T, Yamaguchi K. Aeruginosin 298-a, a Thrombin and Trypsin-Inhibitor from the Blue-Green-Alga Microcystis-Aeruginosa (NIES-298) Tetrahedron Lett. 1994;35:3129–3132. [Google Scholar]
- 3.Kodani S, Ishida K, Murakami M. Aeruginosin 103-A, a thrombin inhibitor from the cyanobacterium Microcystis viridis. J. Nat. Prod. 1998;61:1046–1048. doi: 10.1021/np980106w. [DOI] [PubMed] [Google Scholar]
- 4.Matsuda H, Okino T, Murakami M, Yamaguchi K. Aeruginosins 102-A and B, new thrombin inhibitors from the cyanobacterium Microcystis viridis (NIES-102) Tetrahedron. 1996;52:14501–14506. [Google Scholar]
- 5.Murakami M, Ishida K, Okino T, Okita Y, Matsuda H, Yamaguchi K. Aeruginosin-98-a and Aeruginosin-98-B, Trypsin-Inhibitors from the Blue-Green-Alga Microcystis-Aeruginosa (NIES-98) Tetrahedron Lett. 1995;36:2785–2788. [Google Scholar]
- 6.Ishida K, Okita Y, Matsuda H, Okino T, Murakami M. Aeruginosins, protease inhibitors from the cyanobacterium Microcystis aeruginosa. Tetrahedron. 1999;55:10971–10988. [Google Scholar]
- 7.Shin HJ, Matsuda H, Murakami M, Yamaguchi K. Aeruginosins 205A and -B, serine protease inhibitory glycopeptides from the cyanobacterium Oscillatoria agardhii (NIES-205) J. Org. Chem. 1997;62:1810–1813. [Google Scholar]
- 8.Banker R, Carmeli S. Inhibitors of serine proteases from a waterbloom of the cyanobacterium Microcystis sp. Tetrahedron. 1999;55:10835–10844. [Google Scholar]
- 9.Ploutno A, Shoshan M, Carmeli S. Three novel protease inhibitors from a natural bloom of the cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 2002;65:973–978. doi: 10.1021/np010597b. [DOI] [PubMed] [Google Scholar]
- 10.Carroll AR, Buchanan MS, Edser A, Hyde E, Simpson M, Quinn RJ. Dysinosins B-D, inhibitors of factor VIIa and thrombin from the Australian sponge Lamellodysidea chlorea. J. Nat. Prod. 2004;67:1291–1294. doi: 10.1021/np049968p. [DOI] [PubMed] [Google Scholar]
- 11.Carroll AR, Pierens GK, Fechner G, de Almeida Leone P, Ngo A, Simpson M, Hyde E, Hooper JNA, Bostrom SL, Musil D, Quinn RJ. Dysinosin A: A novel inhibitor of factor VIIa and thrombin from a new genus and species of Australian sponge of the family dysideidae. J. Am. Chem. Soc. 2002;124:13340–13341. doi: 10.1021/ja020814a. [DOI] [PubMed] [Google Scholar]
- 12.Sandler B, Murakami M, Clardy J. Atomic structure of the trypsin aeruginosin 98-B complex. J. Am. Chem. Soc. 1998;120:595–596. [Google Scholar]
- 13.Steiner JLR, Murakami M, Tulinsky A. Structure of thrombin inhibited by aeruginosin 298-A from a blue-green alga. J. Am. Chem. Soc. 1998;120:597–598. [Google Scholar]
- 14.Dittmann E, Neilan BA, Börner T. Molecular biology of peptide and polyketide biosynthesis in cyanobacteria. Appl. Microbiol. Biotechnol. 2001;57:467–473. doi: 10.1007/s002530100810. [DOI] [PubMed] [Google Scholar]
- 15.Schwarzer D, Finking R, Marahiel MA. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 2003;20:275–287. doi: 10.1039/b111145k. [DOI] [PubMed] [Google Scholar]
- 16.Christiansen G, Fastner J, Erhard M, Börner T, Dittmann E. Microcystin biosynthesis in Planktothrix: Genes, evolution, and manipulation. J. Bacteriol. 2003;185:564–572. doi: 10.1128/JB.185.2.564-572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishizawa T, Ueda A, Asayama M, Fujii K, Harada K, Ochi K, Shirai M. Polyketide synthase gene coupled to the peptide synthetase module involved in the biosynthesis of the cyclic heptapeptide microcystin. J. Biochem. 2000;127:779–789. doi: 10.1093/oxfordjournals.jbchem.a022670. [DOI] [PubMed] [Google Scholar]
- 18.Rouhiainen L, Vakkilainen T, Siemer BL, Buikema W, Haselkorn R, Sivonen K. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl. Environm. Microbiol. 2004;70:686–692. doi: 10.1128/AEM.70.2.686-692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tillett D, Dittmann E, Erhard M, von Döhren H, Börner T, Neilan BA. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chem. Biol. 2000;7:753–764. doi: 10.1016/s1074-5521(00)00021-1. [DOI] [PubMed] [Google Scholar]
- 20.Chang ZX, Flatt P, Gerwick WH, Nguyen VA, Willis CL, Sherman DH. The barbamide biosynthetic gene cluster: a novel marine cyanobacterial system of mixed polyketide synthase (PKS)-non-ribosomal peptide synthetase (NRPS) origin involving an unusual trichloroleucyl starter unit. Gene. 2002;296:235–247. doi: 10.1016/s0378-1119(02)00860-0. [DOI] [PubMed] [Google Scholar]
- 21.Chang ZX, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia JY, Sherman DH, Gerwick WH. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 22.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Structure and biosynthesis of the jamaicamides, new mixed polyketide-peptide neurotoxins from the marine cyanobacterium Lyngbya majuscula. Chem. Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 23.Rouhiainen L, Paulin L, Suomalainen S, Hyytiainen H, Buikema W, Haselkorn R, Sivonen K. Genes encoding synthetases of cyclic depsipeptides, anabaenopeptilides, in Anabaena strain 90. Mol. Microbiol. 2000;37:156–167. doi: 10.1046/j.1365-2958.2000.01982.x. [DOI] [PubMed] [Google Scholar]
- 24.Edwards DJ, Gerwick WH. Lyngbyatoxin biosynthesis: Sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J. Am. Chem. Soc. 2004;126:11432–11433. doi: 10.1021/ja047876g. [DOI] [PubMed] [Google Scholar]
- 25.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 26.Fujii K, Sivonen K, Adachi K, Noguchi K, Shimizu Y, Sano H, Hirayama M, Suzuki M, Harada K. Comparative Study of Toxic and Non-toxic Cyanobacterial Products: A Novel Glycoside, Suomilide, from Non-toxic Nodularia spumigena HKVV. Tetrahedron Lett. 1997;38:5529–5532. [Google Scholar]
- 27.Pluotno A, Carmeli S. Banyasin, and Banyasides A and B, three novel modified peptides from a water bloom of the cyanobacterium Nostoc sp. Tetrahedron. 2005;61:575–583. [Google Scholar]
- 28.Valls N, Vallribera M, Font-Bardia M, Solans X, Bonjoch J. Synthesis of the proposed core of aeruginosins 205 : The new alpha -amino acid (2S,3aS,6R,7aS)-2-carboxy-6-chlorooctahydroindole. Tetrahedron: Asymmetry. 2003;14(10):1241–1244. [Google Scholar]
- 29.Hicks LM, Moffitt MC, Beer LL, Moore BS, Kelleher NL. Structural characterization of in vitro and in vivo intermediates on the loading module of microcystin synthetase. ACS Chem. Biol. 2006;1:93–102. doi: 10.1021/cb500007v. [DOI] [PubMed] [Google Scholar]
- 30.Moore BS, Hertweck C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 31.Steinborn G, Hajirezaei MR, Hofemeister J. bac genes for recombinant bacilysin and anticapsin production in Bacillus host strains. Arch. Microbiol. 2005;183:71–79. doi: 10.1007/s00203-004-0743-8. [DOI] [PubMed] [Google Scholar]
- 32.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Codani JJ, Connerton IF, Cummings NJ, Daniel RA, Denizot F, Devine KM, Dusterhoft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, Haiech J, Harwood CR, Henaut A, Hilbert H, Holsappel S, Hosono S, Hullo MF, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, KlaerrBlanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee SM, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Mellado RP, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, OReilly M, Ogawa K, Ogiwara A, Oudega B, Park SH, Parro V, Pohl TM, Portetelle D, Porwollik S, Prescott AM, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror SJ, Serror P, Shin BS, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Tognoni A, Tosato V, Uchiyama S, Vandenbol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler E, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa HF, Zumstein E, Yoshikawa H, Danchin A. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 33.Hilton MD, Alaeddinoglu NG, Demain AL. Synthesis of Bacilysin by Bacillus subtilis branches from Prephenate of the Aromatic Amino Acid Pathway. J. Bacteriol. 1988;170:482–484. doi: 10.1128/jb.170.1.482-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLuskey K, Cameron S, Hammerschmidt F, Hunter WN. Structure and reactivity of hydroxypropylphosphonic acid epoxidase in fosfomycin biosynthesis by a cation- and flavin-dependent mechanism. Proc. Natl. Acad. Sci. USA. 2005;102:14221–14226. doi: 10.1073/pnas.0504314102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujii K, Sivonen K, Adachi K, Noguchi K, Sano H, Hirayama K, Suzuki M, Harada K. Comparative study of toxic and non-toxic cyanobacterial products: Novel peptides from toxic Nodularia spumigena AV1. Tetrahedron Lett. 1997;38:5525–5528. [Google Scholar]
- 36.Keating TA, Marshall CG, Walsh CT, Keating AE. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nature Struct. Biol. 2002;9:522–526. doi: 10.1038/nsb810. [DOI] [PubMed] [Google Scholar]
- 37.Valls N, Vallribera M, Lopez-Canet M, Bonjoch J. Synthesis of microcin SF608. J. Org. Chem. 2002;67:4945–4950. doi: 10.1021/jo025709y. [DOI] [PubMed] [Google Scholar]
- 38.Roscoe J, Abraham EP. Experiments Relating to Biosynthesis of Bacilysin. Biochem. J. 1966;99:793–800. doi: 10.1042/bj0990793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall GC, Flick MB, Gherna RL, Jensen RA. Biochemical Diversity for Biosynthesis of Aromatic-Amino-Acids among the Cyanobacteria. J. Bacteriol. 1982;149:65–78. doi: 10.1128/jb.149.1.65-78.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin HJ, Murakami M, Matsuda H, Yamaguchi K. Microviridins D-F, serine protease inhibitors from the cyanobacterium Oscillatoria agardhii (NIES-204) Tetrahedron. 1996;52:8159–8168. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.