Summary
Major non primate-primate differences in corticogenesis include the dimensions, precursor lineages and developmental timing of the germinal zones (GZ). microRNAs (miRNAs) of laser dissected GZ compartments and cortical plate (CP) from embryonic E80 macaque visual cortex were deep sequenced. The CP and the GZ including Ventricular Zone (VZ), outer and inner subcompartments of the Outer SubVentricular Zone (OSVZ) in area 17 displayed unique miRNA profiles. miRNAs present in primate, but absent in rodent, contributed disproportionately to the differential expression between GZ sub-regions. Prominent among the validated targets of these miRNAs were cell-cycle and neurogenesis regulators. Co-evolution between the emergent miRNAs and their targets suggested that novel miRNAs became integrated into ancient gene circuitry to exert additional control over proliferation. We conclude that multiple cell-cycle regulatory events contribute to the emergence of primate-specific cortical features, including the OSVZ, generated enlarged supragranular layers, largely responsible for the increased primate cortex computational abilities.
Keywords: OSVZ, VZ, Cell cycle, corticogenesis, macaque monkey, primate, brain development, miRNA, deep sequencing
Introduction
During evolution there is an increase in the size and complexity of the OSVZ, considered to contribute to tangential expansion (Fietz et al., 2010; Lui et al., 2011) and shown to be responsible for the enlarged primate supragranular layers (Lukaszewicz et al., 2005; Smart et al., 2002). Macaque cortical neurons are produced over a 60-day period from E40 to E100 (Rakic, 1974). In contrast to laboratory rodents where the VZ is the prominent GZ throughout corticogenesis, the OSVZ is the major proliferative compartment in the primate from E65 (Betizeau et al., 2013; Martinez-Cerdeno et al., 2012; Smart et al., 2002). The expansion of the OSVZ reaches its climax in the primary visual cortex (area 17), which has augmented supragranular layers as well as increased tangential expansion compared to the adjacent area 18 in the macaque monkey (Lukaszewicz et al., 2006; Smart et al., 2002). The mechanisms responsible for OSVZ expansion are linked to the proliferative capacity of the precursors via cell-cycle regulation (Betizeau et al., 2013; Lukaszewicz et al., 2005). In area 17, the microenvironment of the most basal part of the OSVZ is enriched in embryonic thalamic axons (Smart et al., 2002) that modulate precursor proliferation (Dehay et al., 2001), which raises the possibility of a further radial heterogeneity within area 17 OSVZ. We explored whether these different primate-specific GZ compartments are associated with gene regulatory changes. Because miRNAs, ∼ 22 nucleotide non-coding RNAs, regulate gene expression post-transcriptionally we ascertained miRNA profiles from the macaque embryonic cortical compartments. The readily detectable changes in miRNA expression during differentiation and their frequent de novo appearance at evolutionary divergence points suggest a role in the acquisition of new cell identities and in evolutionary innovation (Hannon, 2002; Heimberg et al., 2008; Kosik, 2009; Nowakowski et al., 2013). To find molecular changes associated with the expansion of the GZ in primates, we laser captured tissue from the compartments of area 17 and 18 in Maccaca fascicularis cerebral cortex at E80 and performed miRseq to obtain a comprehensive non-biased expression pattern of the microRNAs in each compartment.
The present study provides new insight into the molecular distinctions that link anatomical and molecular evolutionary changes in the developing cortex. The data show that the target genes of primate miRNAs that uniquely distinguish the cortical GZ are principally involved in cell-cycle and neurogenesis regulation as well as in human neurodevelopmental disorders.
Results
miRNA profiles distinguish germinal zones of the primate cortex
Seven brain regions were dissected from developing Maccaca fascicularis brains: area 17 and 18 cortical plate (CP), area 17 and 18 ventricular zone (VZ), area 18 outer subventricular zone (OSVZ), and area 17 OSVZ in two compartments: the most apical third (OSVZ17int) and the most basal third OSVZ17ext (Fig S1). miRseq reads from these samples were mapped to well-authenticated miRNAs precursors from miRBase.v16 using the small RNA pipeline from SOLID, and only uniquely mapped reads were counted for miRNA profiling (Table S1; GEO access number: GSE52608). Many hairpins had significant numbers of reads from both strands (Zhou et al., 2012). This dataset contained a total of 766 miRNA precursors or 1532 miRNA arms (5p and 3p), which were loaded to the EdgeR package. After filtering for at least five CPM (counts per million) in at least three libraries, 752 arms remained with 321 of them reported as expressed in primates, but not reported in rodent according to miRBase.v16. A subset of the miRNA reads were validated by digital PCR (Fig S2).
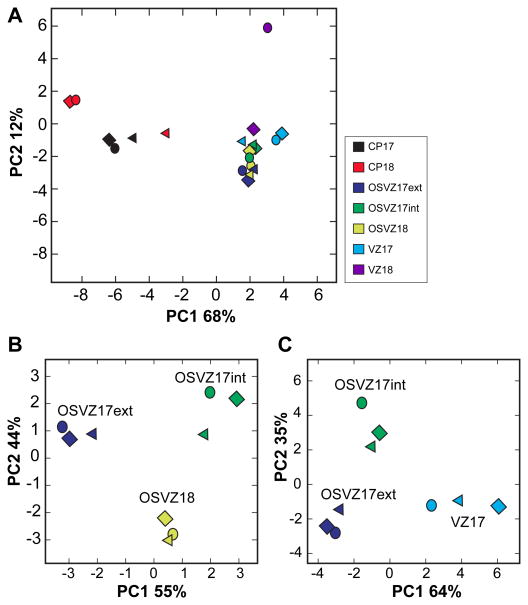
To determine whether the collective variation among the miRNA profiles could distinguish the anatomical regions sampled, principal component analysis (PCA) was applied to the miRNA profiles. PCA can reduce the dimensionality of a data set by finding linear combinations of dimensions (miRNAs in this case) ranked by their importance and projected onto a set of axes. Using all the samples in the analysis, the CP separated from the GZ along PC1 (rank sum test, p<1.0774e-004) with 68% of the total variation (Fig 1A).
Figure 1. miRNA collective variation distinguishes embryonic cortical zones.
(A) PCA was applied to the miRNA profiles of the CP (CP17, CP18) and the GZ (OSVZ17int, OSVZ17ext, OSVZ18, VZ17, VZ18) from E80 macaque embryos. Two clusters stand out— CP and GZ—that are independent of the cortical area. (B) PCA using a subset of the data, the OSVZ of area 17 and 18, separated the OSVZ17 (int and ext) from OSVZ18. (C) Samples from area 17 reveal further distinctions between VZ and OSVZ fractions. See also Figure S1 and Table S1.
These observations prompted a more limited PCA of the GZ to resolve areal differences between OSVZ17int/OSVZ17ext and the neighboring OSVZ18 along PC2 (rank sum test, p<0.0238) (Fig 1B). PC1 evenly spread the three anatomical regions, showing that OSVZ18 is equally dissimilar to OSVZ17int and OSVZ17ext. The PCA also resolved differences within the area 17 GZ (Fig 1C). VZ17 separated from both the OSVZ17 fractions (OSVZ17int and OSVZ17ext) along PC1 (rank sum test, p<0.0238). In addition OSVZ17int separated from the OSVZ17ext and the VZ17 along PC2 (rank sum test, p<0.0238). 17% of the weight contributions to PC2 come from a single miRNA, mir-4271-5p, which is expressed in primates but not in rodents. Thus miRNA profiles can resolve anatomically discrete regions within the developing primate cortex and the dominant difference is between the GZ and the differentiated cells of the CP.
Differentially Expressed (DE) miRNAs point to evolutionary target networks for primate cortex expansion
Having shown that anatomical regions can be distinguished by collective variation of their miRNA profiles, we used the EdgeR package to find the differentially expressed miRNAs among the brain regions. After filtering for a minimum of five CPM in at least three libraries, 752 arms remained for differential expression (DE) analysis (Table S2), and a subset of the DE miRNAs were validated by digital PCR (Table S2, Fig S2). A heat map depicting these data indicates the distinct expression patterns for each miRNA in each compartment with the most prominent distinction between the CP and the GZ (Fig 2A). Among the DE miRNAs, approximately 40% are expressed in primate but not in rodent (Fig 2A; Table S2). A comparison of the OSVZ17 fractions, OSZV17int vs OSVZ17ext shows a higher percentage (54%) of miRNAs that are expressed in primates, but not in rodents (Fig 2C). The molecular distinction within the OSVZ indicated by the miRNA profiles reveals the emergence of a primate developmental compartment below anatomical resolution, the presence of this new compartment remains to be confirmed in the human developing brain. Entering all the known targets for these DE miRNAs into GO using DAVID NCBI web page, the terms cell cycle, cell differentiation, cell migration, and corticogenesis were enriched (FDR<0.05). Among the proven targets of these highly conserved miRNAs are mRNAs that have been associated with area 17 and 18 expansion (Lukaszewicz et al., 2005) such as pax6 (Mi et al., 2013) targeted by mir-450b-5p (Shalom-Feuerstein et al., 2012), p27kip targeted by mir-222-3p (Kim et al., 2009), and cdk4 and ccne2 (cyclin E2) targeted by mir-34c-5p (Toyota et al., 2008). Very few proven targets are known for the primate DE miRNAs not expressed in rodent (Fig 2A-C). Those for which targets are known include cell cycle corticogenesis regulators, i.e. miR-605-5p targets mdm2 (Xiao et al., 2011) and mir-657-3p which targets p21Cip1/Waf1 (Wu et al., 2010).
Figure 2. Heat map for differentially expressed miRNAs.

(A) DE miRNAs were statistically identified by EdgeR, FDR< 0.05. CP (17 and 18) were compared to all GZ (17 and 18). (B) For area 17 two additional comparisons were done: VZ17 to OSVZ17int and to OSVZ17ext. (C) Comparison of OSVZ17: OSVZ17int vs OSVZ17ext. DE miRNAs between OSVZ17int and OSVZ17ext have a higher percentage of miRNAs reported in primates, but absent in mouse (54%) than DE miRNAs between CP and GZ (34%). See also Figure S2 and Table S2.
Trapping targets of Primate miRNAs
RISCtrap (Cambronne et al., 2012) was used to find targets of miRNAs positioned at divergence points within the mammalian radiations. Four were DE miRNAs expressed in primates and Laurasiatheria (a superorder of mammals), but not in rodents (mir-550-3p, mir-3613-5p, mir-1301-3p, mir-1180-3p), and two mir-1260a-5p and mir-1271-5p were expressed in primates and Laurasiatheria, and in a single rodent, the Chinese hamster (miRBase v.16). mir-124-3p, which is an abundant brain miRNA widely present in animal phylogeny including C. elegans and used in previous RISCtrap studies (Cambronne et al., 2012) served as a control. Trapped mRNAs for each miRNA were sequenced and mapped (Table S3; GEO access number: GSE52608). Targets were required to meet these criteria: 1) enriched in a single miRNA pull down as compared to all the other miRNA pull downs at an FDR<0.05; 2) have a binding site for the specific miRNA as predicted by targetscan.org Version 6.2; 3) the presence of an orthologue in macaque (Fig 3A, Table S3). This stringent analysis will miss mRNAs targeted by more than one of the miRNA probes, but will gain in specificity. All of trapped miRNAs were more recently evolved than mir-124-3p, an ancient miRNA, which had an order of magnitude more targets than any of the others (Table S3) that closely matched those targets reported previously (Baek et al., 2008; Cambronne et al., 2012; Chi et al., 2009; Hendrickson et al., 2008; Karginov et al., 2007; Lim et al., 2005) (Table S3). miRNAs that appeared more recently in phylogeny or occupy a more restricted phylogenetic niche, may have fewer mRNA targets than more ancient miRNAs that expanded their targets over longer evolutionary time periods.
Figure 3. RICStrap DE miRNAs and validation.
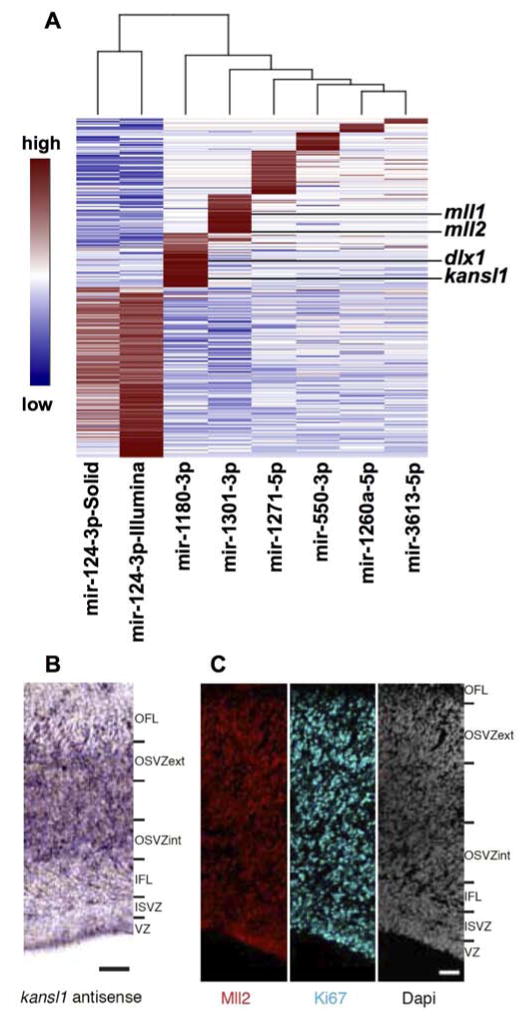
(A) Using the RISCtrap method, DE mRNAs were pulled down for mir-550-3p, mir-3613-5p, mir-1301-3p, mir-1180-3p, mir-1260a-5p, mir-1271-5p. DE mRNAs were determined using the EdgeR package, FDR<0,05. mll1, mll2, kansl1 and dlx1 are targets of miRNAs expressed in primates and laurasiatheria, but not in rodentia. kansl1 mRNA expression was confirmed by ISH (B), and the presence of the MLL2 protein by immunofluorescence (C) on E80 cortex cryosection. Scale bars: 50 μm. See also Figure S3 and Table S3.
The top two enriched functional categories of the RISCtrap targets were brain development (43 mRNAs) and cell cycle (45 mRNAs). Notable among the mir-1301-3p targets was the mRNA for the histone-lysine N-methyltransferase, mll1 (Mixed-Lineage, Leukemia) and mll2, an essential gene for neurogenesis (Lim et al., 2009; Popovic and Zeleznik-Le, 2005). mll1 has eight predicted binding sites, four in the 3′ UTR and four in the CDS; and mll2 has 12 predicted binding sites, two in the 3′ UTR and ten in the CDS. kansl1, a member of the MLL1 complex (http://www.uniprot.org/uniprot/) and dlx1, which is under the control of MLL1 (Lim et al., 2006) and is a key regulator of neurogenesis (Anderson et al., 1997), were both captured in the mir-1180-3p RISCtrap. kansl1 has two predicted binding sites and dlx1 has a single binding site for mir-1180-3p. The presence of these mRNAs in the macaque tissue was confirmed by digital PCR (Fig S3A), and by in situ hybridization (ISH) for kansl1 (Fig 3B). MLL2 protein expression was confirmed by immunofluorescence (Fig 3C). Further validation of these targets beyond their capture in the RISCtrap demonstrated a decrease in KANSL1 and DLX1 proteins in the presence of the mir-1180-3p mimic, and MLL1 protein in the presence of mir-1301-3p mimic (Fig S3B). Luciferase assays supported dlx1 and kansl1 as direct targets of mir-1180-3p and mll1 as direct target of mir-1301-3p (Fig S3C). Also present among the mRNAs trapped by mir-1301-3p were col3a1 and col1a1 (FDR <0.05), two extracellular matrix (ECM) components. Furthermore, mir-29-3p which is proven to down regulate the expression of multiple ECM genes is differentially expressed between the CP and the GZ, in agreement with the report of ECM components being enriched in the human GZ (Fietz et al., 2012).
As we found an over-representation of miRNA that target cell cycle regulators among miRNA both differentially expressed in developing macaque cortex and absent in the rodent, we carried out a more detailed examination of the compartment specific localization of this set of miRNAs. Representing the expression of these miRNAs shows compartment specific miRNA expression patterns within this functional category suggesting that each compartment has evolved its own modifications of the cell cycle regulation (Fig 4A-B). The CP appears to rely on inhibiting cyclin D (2 mirs target it in CP18 and one in CP17) and p21 to inhibit the proliferation in these differentiated cells. Genes involved in neurogenesis such as dlx1 and mll1/2 and kansl1 appear to be released from inhibition in the CP compared to the GZ. Area differences between VZ17 and 18 may be related to the differential cell density between these two compartments. The OSVZ is uniquely identified by a relatively high level of mir-335-5p, which targets rb. The apical and basal portions of the OSVZ17 have distinct profiles of miRNAs cell cycle regulators, and the profile of the external (basal) portion more closely resembles OSVZ18.
Figure 4. DE miRNAs control key points in the cell cycle and in cortical development.
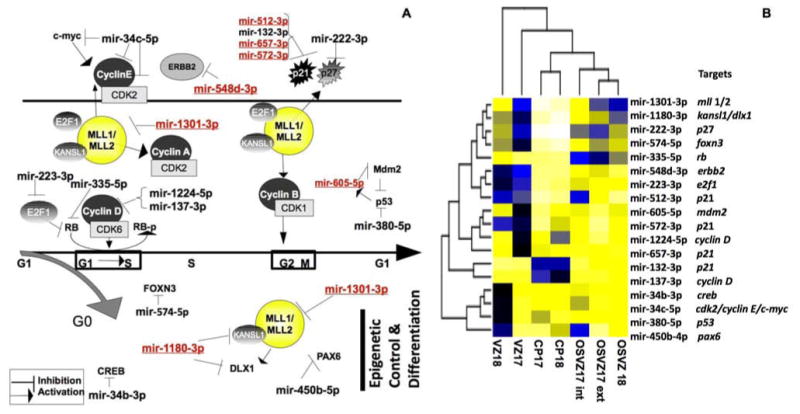
(A) Key proteins which regulate neuronal progenitor cell-cycle and neurogenesis were among the proven targets of the DE miRNAs (Black: miRNAs expressed in primate, Laurasiatheria, and Rodentia; Red, miRNAs expressed in primates and Laurasiatheria, but not in Rodentia). (B) Levels of expression of these DE miRNAs are represented on a heat map.
Co-evolution of miRNA and target site
miRNAs expressed in the developing macaque cortex but not in rodents target genes which are present in both primates and rodents. Hence novel miRNAs often target preexisting mRNAs. Through the phylogenetic history the co-evolution of the target site and the miRNA can be tracked. In addition to the primate and rodent representatives, we selected two additional eutherians, Bos taurus (cow) and Canis familiaris (dog). We assembled the set of validated target mRNAs for the DE miRNAs that are expressed in primates, but not in rodents (included is mir-1271-5p, which is expressed in a single rodent, the Cricetulus griseus (Chinese hamster), miRBase v.16). Of the 27 miRNAs in this category, 34 validated target mRNAs with 48 binding sites were identified and aligned to their 3′ UTR binding sites (Table S4-S5) based upon a seed match (Brennecke et al., 2005; Krek et al., 2005; Lewis et al., 2005). This analysis excluded matches within coding regions because the most effective target sites fall outside the path of the ribosome (Grimson et al., 2007). Of these 27 miRNAs, 21 were found exclusively in primates, and five were found in other eutherians.
38 of the 48 binding sites were completely conserved in human, chimp and rhesus; whereas 10 sites targeted by 10 miRNAs were conserved in human and chimp, but not in rhesus. Five of these had one or more of these features: a single mismatch, another perfectly matched site in the same mRNA targeted by the miRNA, another mRNA with a perfectly matched target. Five of these DE miRNAs in macaque had no validated target site suggesting that the target has not been identified. Among the 38 binding sites conserved across the primates, several categories of evolutionary change occur. In seven cases, the target site is present in the non-primate species suggesting either the newly invented primate miRNA targeted a pre-existing site or the miRNA was lost in the non-primate species and another constraint maintained the target sequence. Most frequently a few nucleotide changes in the MRE (microRNA response element) improved the match with the primate miRNA and strict conservation of the site was observed only in primate suggesting that the target site co-evolved with the miRNA. Cases with partial conservation among species as divergent as dogs, cows, and rodents underscore the difficulties in resolving the basal relationships in the eutherian tree (Cannarozzi et al., 2007; Hou et al., 2009).
Discussion
A key feature of OSVZ primate precursors is their extensive self-renewal and proliferative abilities (Betizeau et al., 2013), properties that lie at the roots of OSVZ expansion and, in turn, of the unique cyto-architectonic features of the primate brain. The present results suggest that a few key miRNAs including some that are specific for the primate lineage and differentially expressed in VZ and OSVZ contributed to the expansion of precursor pools within the developing cortex. To increase the likelihood that miRNAs found only as primate entries in miRBase were primate specific, we blasted the primate entries in miRBase to the rodent genome using the Santa Cruz Browser to see if these sequences exist independently of the tissue analyzed for expression. None of these miRNAs had a perfect match; however some had an imperfect match and others were completely absent. For those that were completely absent from the rodent genome we had high confidence that they are not in the rodent brain. For those that have an imperfect match it is possible that they belong to a family of miRNAs present in rodent and primate. However, even these often had extensive indels relative to the primate miRNA sequences. Combined with their absence from miRBase, it is unlikely they are expressed with the caveat that the exact corresponding tissue in rodent has not been deeply sequenced.
An increase in the noncoding RNA inventory including miRNAs has been noted at evolutionary divergence points (Heimberg et al., 2008). A clearly distinct and prominent OSVZ is a feature of the primate brain, albeit this innovation is foreshadowed in the brains of large rodents and carnivores (Martinez-Cerdeno et al., 2012, Garcia-Moreno et al, 2012). Ongoing expansion of the OSVZ in area 17 resulted in a further subdivision into internal and external compartments distinguished by their miRNA profiles, possibly in relationship with the embryonic thalamocortical projections (Sestan et al., 2001; Smart et al., 2002). Thus, a feature of primate cortex evolution appears to be anatomical segregation of precursor pools into discrete compartments.
DE miRNAs within the developing macaque cortex can uniquely identify mitotic and post-mitotic compartments as well as GZ subregions (Fig 1, 2). Among the RISCtrap identified targets of DE miRNAs present in primates but not in rodents was tcf4 targeted by mir-1301-3p. TCF4 patterns progenitor cells in the developing CNS (Wang et al., 2011) has been associated with schizophrenia (Aberg et al., 2013) and Pitt-Hopkins syndrome, a rare neurodevelopmental genetic disorder characterized by severe intellectual disability and hyperventilation (Zweier et al., 2007). Within the GZ, a set of the DE miRNAs, including those that evolved in the primate lineage, targets the cell-cycle, particularly the G1/S transition, neurogenesis and epigenetic regulation (Fig 2). For example, the primate-specific miR-1180-3p targets kansl1, an evolutionarily conserved regulator of the chromatin modifier KAT8 that functions through histone H4 lysine 16 (H4K16) acetylation and, when haploinsufficient, causes intellectual disability, hypotonia and distinctive facial features associated with the 17q21.31 microdeletion syndrome (Zollino et al., 2012). mir-1180-3p also targets dlx1, a homeodomain transcription factor that controls GZ neurogenesis and has been associated with autism (Liu et al., 2009). Both dlx1 and dlx2 - direct targets of the trxG member mll1 (Lim et al., 2009), an evolutionarily conserved histone H3 lysine 4 methyltransferase that functions by resolving silenced bivalent loci in neural precursors for the induction of neurogenesis (Lim et al., 2009) -bind to E2Fs to promote G1/S progression (Liu et al., 2007; Takeda et al., 2006). Both mll1 and mll2 have multiple target sites for mir-1301-3p present in primates but not in rodents.
The G1/S check point can impact the evolutionary expansion of both the SVZ in rodents (Pilaz et al., 2009) and the OSVZ in primates (Betizeau et al., 2013; Lukaszewicz et al., 2005). Macaque cortical precursors cell-cycle exhibit an unusual temporal regulation with a shortening of G1 and S phase at stage E80 in both VZ and OSVZ (Betizeau et al., 2013; Kornack and Rakic, 1998). The finding that primate-specific miRNAs can target the G1/S transition of OSVZ precursors reveals the potential evolutionary space that existed at the origin of primates within this ancient and extensively regulated cellular process for ongoing brain evolution. Layering novel regulatory elements upon a pre-existing network by bringing pre-existing mRNAs under the control of newly invented miRNAs necessarily involves the co-evolution of these novel miRNAs with target site adaptations. The emergent relationships of a novel miRNA can be depicted as a network with targets as well as competing endogenous RNAs and miRNA sponges (Salmena et al., 2011). A question arises as to how selection can promote the integration of network modifications once an established network is already optimized. Do the preexisting relationships among these RNAs retain their dynamic relationships or does the added layer of control inevitably result in the relaxation of more ancient miRNA controls? Sidestepping these questions is the possibility that the ensemble analysis done here obscures the specific miRNA network relationships in individual cells. Analysis of the total population of cells within a compartment might make the regulatory control system appear more complex than it is and sub-graphs within the network may operate within individual cell types and may even contribute to the emergence of novel cell types (Betizeau et al., 2013).
Experimental Procedures
Animals
Fetuses from timed-pregnant cynomolgus monkeys (Macaca fascicularis, gestation period 165 days) were delivered by caesarian section as previously described (Lukaszewicz et al., 2005). All experiments were in compliance with national and European laws as well as with institutional guidelines concerning animal experimentation. Surgical procedures were in accordance with European requirements 2010/63/UE. The protocol C2EA42-12-11-0402-003 has been reviewed and approved by the Animal Care and Use Committee CELYNE (C2EA #42).
Laser microdissection: see Supplemental methods
Libraries, sequencing, mapping, differential expression
RNA was extracted from each section by microRNeasy kit from Qiagen with DTT to stabilize RNA. Libraries were prepared with SOLiD Total RNA-seq kit (Life Technologies #4452437). Briefly, RNA was ligated with a random adapter overnight at 16°C, first strand cDNA was made using specific primers to the common adapter sequence and run on a 10% urea gel to size select. The resulting cDNA was amplified 15 cycles using primers from the common adapter sequence. Library size was confirmed using an Agilent high sensitivity DNA chip. Small RNA libraries were sequenced on the SOLiD 4 platform. Reads were mapped to well-authenticated primate miRNA precursors from miRBase v.16 using the small RNA pipeline from SOLiD and only uniquely mapped sequences were counted. The inclusion criterion for a miRNA was at least 5 CPM in at least three of the 20 libraries. Data were trimmed mean of M (TMM) normalized using the BioConductor package edgeR v.3.2.1 (Robinson et al., 2010). The same package was used to determine the differential expression of miRNAs across the seven brain regions among all three replicates, i.e. 20 libraries. All comparisons were corrected for multiple testing using a Benjamini-Hochberg procedure. Searches for miRNA conservation utilized mirBase v.16, mirOrtho (Gerlach et al., 2009) and the UCSC Browser (UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly). The data has been deposited to GEO access number: GSE52608).
PCA methods
Principal Component Analysis (PCA) was applied to the mean normalized, mean centered data, for the top 100 more variable miRNA between groups and less variable amongst replicates, using Python 2.6 scripts via SciPy, NumPy. PCA direction was calculated using the average to minimize the effect of the replicates on the vector calculation. The individual replicates were plotted on the new axes. A two-sided Wilcoxon rank-sum test was used to determine the statistical significance of the separation among clusters.
Digital PCR: see Supplemental methods
RISC Trap: see Supplemental methods
Supplementary Material
Acknowledgments
We thank Henry Kennedy for critically reading the manuscript, V. Cortay, A. Bellemin-Ménard for technical assistance, B. Beneyton, M. Valdebenito and M. Séon for excellent animal care. This work was supported by the Sheldon and Miriam Adelson Medical Foundation, NIH grants NS079317 (RHG) and NS076094 (XAC) and LABEX CORTEX (ANR-11-LABX-0042) and LABEX DEVWECAN (ANR-10-LABX-0061) of Université de Lyon (ANR-11-IDEX-0007) operated by the French National Research Agency (ANR). X.A.C. was involved in the commercial licensing of the RISC-trap technology. This did not influence the experimental design, data collection, or analysis of the project data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberg KA, Liu Y, Bukszar J, McClay JL, Khachane AN, Andreassen OA, Blackwood D, Corvin A, Djurovic S, Gurling H, et al. A comprehensive family-based replication study of schizophrenia genes. JAMA Psychiatry. 2013;70:573–581. doi: 10.1001/jamapsychiatry.2013.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein LR. Interneuron migration from basal forebrain to neocortex: Dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Menard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, Dehay C. Precursor diversity and complexity of lineage relationships in the outer subventricular zone (OSVZ) of the primate. Neuron. 2013;80:442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne XA, Shen R, Auer PL, Goodman RH. Capturing microRNA targets using an RNA-induced silencing complex (RISC)-trap approach. Proc Natl Acad Sci U S A. 2012;109:20473–20478. doi: 10.1073/pnas.1218887109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarozzi G, Schneider A, Gonnet G. A phylogenomic study of human, dog, and mouse. PLoS Comput Biol. 2007;3:e2. doi: 10.1371/journal.pcbi.0030002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Savatier P, Cortay V, Kennedy H. Cell-cycle kinetics of neocortical precursors are influenced by embryonic thalamic axons. J Neurosci. 2001;21:201–214. doi: 10.1523/JNEUROSCI.21-01-00201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, Wilsch-Brauninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, Huttner WB. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- Fietz SA, Lachmann R, Brandl H, Kircher M, Samusik N, Schroder R, Lakshmanaperumal N, Henry I, Vogt J, Riehn A, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proc Natl Acad Sci U S A. 2012;109:11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno F, Vasistha NA, Trevia N, Bourne JA, Molnar Z. Compartmentalization of Cerebral Cortical Germinal Zones in a Lissencephalic Primate and Gyrencephalic Rodent. Cereb Cortex. 2012;22:482–492. doi: 10.1093/cercor/bhr312. [DOI] [PubMed] [Google Scholar]
- Gerlach D, Kriventseva EV, Rahman N, Vejnar CE, Zdobnov EM. miROrtho: computational survey of microRNA genes. Nucleic Acids Res. 2009;37:D111–117. doi: 10.1093/nar/gkn707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, Herschlag D, Ferrell JE, Brown PO. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS One. 2008;3:e2126. doi: 10.1371/journal.pone.0002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou ZC, Romero R, Wildman DE. Phylogeny of the Ferungulata (Mammalia: Laurasiatheria) as determined from phylogenomic data. Mol Phylogenet Evol. 2009;52:660–664. doi: 10.1016/j.ympev.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104:19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, Kim VN. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci U S A. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. MicroRNAs tell an evo-devo story. Nat Rev Neurosci. 2009;10:754–759. doi: 10.1038/nrn2713. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Suarez-Farinas M, Naef F, Hacker CR, Menn B, Takebayashi H, Magnasco M, Patil N, Alvarez-Buylla A. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol Cell Neurosci. 2006;31:131–148. doi: 10.1016/j.mcn.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu H, Cheng EH, Hsieh JJ. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 2007;21:2385–2398. doi: 10.1101/gad.1574507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Novosedlik N, Wang A, Hudson ML, Cohen IL, Chudley AE, Forster-Gibson CJ, Lewis SM, Holden JJ. The DLX1 and DLX2 genes and susceptibility to autism spectrum disorders. Eur J Hum Genet. 2009;17:228–235. doi: 10.1038/ejhg.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A, Cortay V, Giroud P, Berland M, Smart I, Kennedy H, Dehay C. The concerted modulation of proliferation and migration contributes to the specification of the cytoarchitecture and dimensions of cortical areas. Cereb Cortex. 2006;16(Suppl 1):i26–34. doi: 10.1093/cercor/bhk011. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz A, Savatier P, Cortay V, Giroud P, Huissoud C, Berland M, Kennedy H, Dehay C. G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron. 2005;47:353–364. doi: 10.1016/j.neuron.2005.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Cerdeno V, Cunningham CL, Camacho J, Antczak JL, Prakash AN, Cziep ME, Walker AI, Noctor SC. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PLoS One. 2012;7:e30178. doi: 10.1371/journal.pone.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi D, Carr CB, Georgala PA, Huang YT, Manuel MN, Jeanes E, Niisato E, Sansom SN, Livesey FJ, Theil T, et al. Pax6 exerts regional control of cortical progenitor proliferation via direct repression of Cdk6 and hypophosphorylation of pRb. Neuron. 2013;78:269–284. doi: 10.1016/j.neuron.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Fotaki V, Pollock A, Sun T, Pratt T, Price DJ. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc Natl Acad Sci U S A. 2013;110:7056–7061. doi: 10.1073/pnas.1219385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz LJ, Patti D, Marcy G, Ollier E, Pfister S, Douglas RJ, Betizeau M, Gautier E, Cortay V, Doerflinger N, et al. Forced G1-phase reduction alters mode of division, neuron number, and laminar phenotype in the cerebral cortex. Proc Natl Acad Sci U S A. 2009;106:21924–21929. doi: 10.1073/pnas.0909894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic R, Zeleznik-Le NJ. MLL: how complex does it get? J Cell Biochem. 2005;95:234–242. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestan N, Rakic P, Donoghue MJ. Independent parcellation of the embryonic visual cortex and thalamus revealed by combinatorial Eph/ephrin gene expression. Curr Biol. 2001;11:39–43. doi: 10.1016/s0960-9822(00)00043-9. [DOI] [PubMed] [Google Scholar]
- Shalom-Feuerstein R, Serror L, De La Forest Divonne S, Petit I, Aberdam E, Camargo L, Damour O, Vigouroux C, Solomon A, Gaggioli C, et al. Pluripotent stem cell model reveals essential roles for miR-450b-5p and miR-184 in embryonic corneal lineage specification. Stem Cells. 2012;30:898–909. doi: 10.1002/stem.1068. [DOI] [PubMed] [Google Scholar]
- Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Chen DY, Westergard TD, Fisher JK, Rubens JA, Sasagawa S, Kan JT, Korsmeyer SJ, Cheng EH, Hsieh JJ. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 2006;20:2397–2409. doi: 10.1101/gad.1449406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- Wang H, Lei Q, Oosterveen T, Ericson J, Matise MP. Tcf/Lef repressors differentially regulate Shh-Gli target gene activation thresholds to generate progenitor patterning in the developing CNS. Development. 2011;138:3711–3721. doi: 10.1242/dev.068270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, Zhan R, He X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- Xiao J, Lin H, Luo X, Wang Z. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 2011;30:524–532. doi: 10.1038/emboj.2010.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Arcila ML, Li Z, Lee EJ, Henzler C, Liu J, Rana TM, Kosik KS. Deep annotation of mouse iso-miR and iso-moR variation. Nucleic Acids Res. 2012;40:5864–5875. doi: 10.1093/nar/gks247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollino M, Orteschi D, Murdolo M, Lattante S, Battaglia D, Stefanini C, Mercuri E, Chiurazzi P, Neri G, Marangi G. Mutations in KANSL1 cause the 17q21.31 microdeletion syndrome phenotype. Nat Genet. 2012;44:636–638. doi: 10.1038/ng.2257. [DOI] [PubMed] [Google Scholar]
- Zweier C, Peippo MM, Hoyer J, Sousa S, Bottani A, Clayton-Smith J, Reardon W, Saraiva J, Cabral A, Gohring I, et al. Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome) Am J Hum Genet. 2007;80:994–1001. doi: 10.1086/515583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.