Abstract
The skeleton affords a framework and structural support for vertebrates, while also facilitating movement, protecting vital organs, and providing a reservoir of minerals and cells for immune system and vascular homeostasis. The mechanical and biological functions of the skeleton are inextricably linked to the size and shape of individual bones, the diversity of which is dependent in part upon differential growth and proliferation. Perturbation of bone development, growth and proliferation, can result in congenital skeletal anomalies, which affect approximately 1 in 3000 live births [1]. Ribosome biogenesis is integral to all cell growth and proliferation through its roles in translating mRNAs and building proteins. Disruption of any steps in the process of ribosome biogenesis can lead to congenital disorders termed ribosomopathies. In this review, we discuss the role of ribosome biogenesis in skeletal development and in the pathogenesis of congenital skeletal anomalies.
Keywords: ribosome biogenesis, skeletal development, Treacher Collins syndrome, POAD syndrome, Diamond Blackfan anemia, Shwachman Diamond syndrome, Robert's syndrome, Cartilage Hair Hypoplasia, Bowen Conradi syndrome
Introduction
Ribosome Biogenesis
The ribosome is a large ribonucloprotein machine that translates mRNA into protein to synthesize all protein within the cell. The eukaryotic ribosome catalyzes protein synthesis through distinct and yet collaborative roles of its two subunits during translation. The small 40S subunit decodes mRNA sequence while the large 60S subunit links amino acids through peptide bonds [2]. As ribosomes are universally responsible for the quality and quantity of proteins in all cells, ribosome production is highly regulated by, and integrated with, many cellular processes including growth, proliferation, differentiation, and hypertrophy.
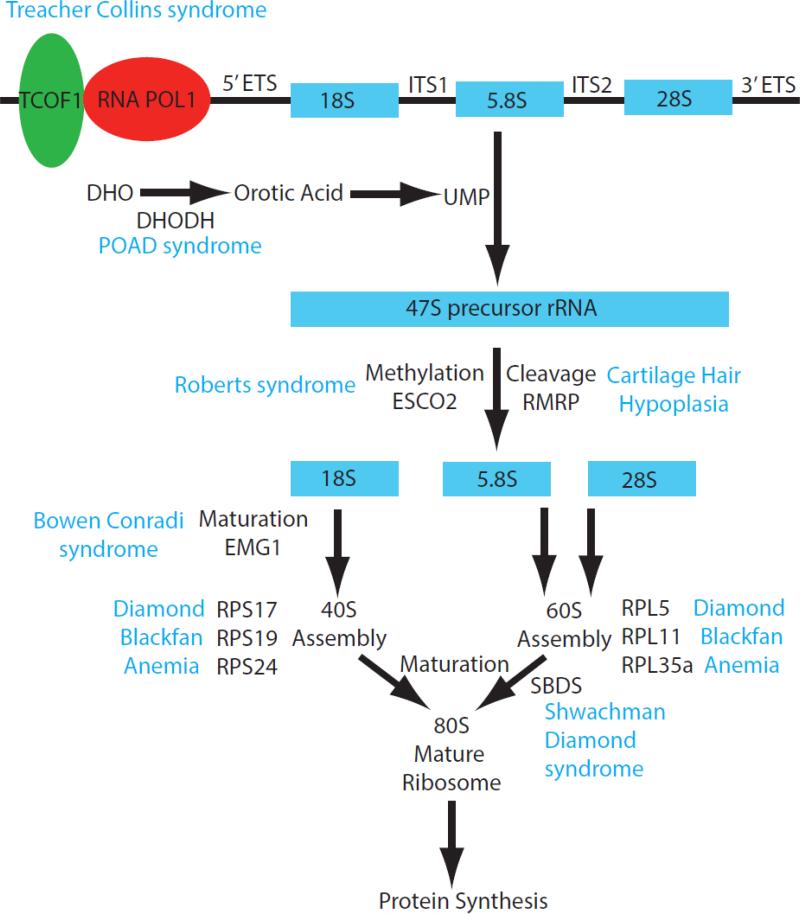
Ribosome biogenesis, the process of making ribosomes, is a complex and metabolically expensive endeavor that involves coordination of all three RNA polymerases. Ribosomes are an assembly of 4 rRNAs transcribed by RNA polymerase I and III, in addition to approximately 80 ribosomal proteins, accessory proteins and about 70 small nucleolar RNAs (snoRNA) all transcribed by RNA polymerase II[2,3]. Ribosome biogenesis begins with transcription of both the 47S precursor ribosomal RNA (rRNA) by RNA polymerase I (RNA Pol I) in the nucleolus and the 5S rRNA by RNA polymerase III in the nucleus. Transcription of rRNA is the rate-limiting step in ribosome production and accounts for 60% of the overall transcription in eukaryotic cells [4]. Additionally, a significant proportion of mRNA transcription by RNA polymerase II in the nucleus is also required for the production of the ribosomal proteins [5]. Following transcription, the 47S rRNA precursor is cleaved into a 45S rRNA which is modified covalently at nearly 200 nucleotides by snoRNPs, bound by ribosomal proteins, and further cleaved into 5.8S, 18S, and 28S rRNAs. Ultimately, the 18S rRNA and 32 small subunit ribosomal proteins (RPSs) are assembled into the 40S subunit and the 5S, 5.8S, and 28S with 47 large subunit ribosomal proteins (RPLs) into the 60S ribosomal subunits. These ribosomal subunits are then exported to the cytoplasm where they unite to form the translationally active mature 80S ribosome [2,6].
As ribosomes determine the capacity for protein production and their synthesis commandeers much of the cell's metabolic efforts, ribosome biogenesis determines growth, cell division rates, and survival [7]. Given the ribosome's universal importance in all cell types, it is remarkable that disruptions in ribosome biogenesis lead to congenital ribosomopathies with very specific clinical phenotypes that include defects in the craniofacial, axial and limb skeleton (Figure 1). Here we discuss how the etiology and pathogenesis of ribosomopathies can reveal new information about the role of ribosome biogenesis in proliferation, growth, and differentiation in skeletal development.
Figure 1.
Schematic diagram summarizing of the major components of ribosome biogenesis and the intersection of specific genes that lead to the pathogenesis of distinct ribosomopathies.
Ribosome Biogenesis in Skeletal Development
Bones, in their many shapes and sizes, underlie form and function of the vertebrate skeleton. The 206 bones in the human skeleton protect our vital organs, provide a reservoir of minerals, facilitate movement, and underlie the basis for physical appearance. These mechanical and biological functions are possible because bones come in a multitude of shapes and sizes—long, short, flat, and irregular. Skeletal diversity arises in the developing embryo by a stepwise series of events that demarcate when, where, and how bone is built.
Skeletal development begins when loose networks of mesenchymal cells coalesce and condense, prefiguring mature cartilage and bone. Different mesenchymal populations give rise to anatomically distinct groups of bones. Neural crest-derived mesenchyme forms bones in the face, jaw and rostral calvarium whereas mesoderm-derived mesenchyme forms bones in the caudal calvarium, vertebral column, rib cage, and limbs. These mesenchymal populations build bone through either endochondral or intramembranous ossification. During endochondral ossification the mesenchyme initially forms a cartilage scaffold that is later replaced by bone. During intramembranous ossification, the mesenchyme differentiates directly into bone. Skeletal elements that develop through endochondral ossification are largely mesoderm-derived, while those that form by intramembranous ossification are almost exclusively neural crest-derived and localized the craniofacial region [8-11].
The commencement of skeletal development is followed by a long period of growth that progresses throughout fetal development and postnatally through adolescence. The directionality of growth, which modifies the size and shape of bones, is differentially regulated by endochondral and intramembranous ossification. Intramembranous ossification leads to appositional growth, adding new bone at the leading edge or surface to increase thickness or length. During this process osteoblasts deposit osteoid matrix against the bone's surfaces or at the osteogenic fronts. Addition of bone at the leading edge, while useful for growing flat bones in the skull, is not compatible with bones whose ends articulate into a moving joint. This is solved by endochondral ossification, which promotes interstitial growth from the bone's center to increase its length [8-11].
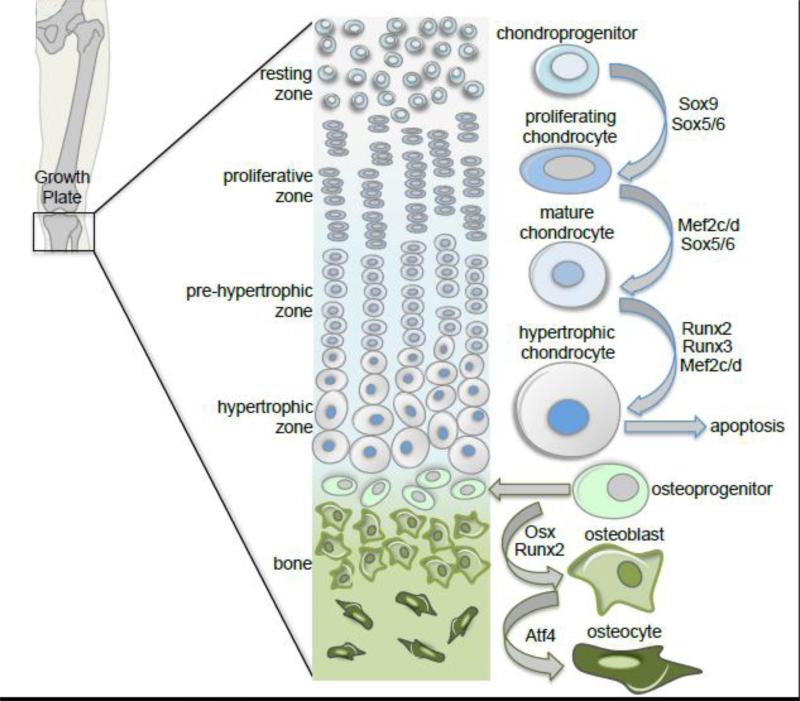
A stratified cartilage structure known as the growth plate is responsible for interstitial growth within endochondral bones. The growth plate, a holdover of the precartilage condensation in the epiphysis, is arranged into four functional zones that represent the developmental progression of endochondral ossification (Figure 2). The resting zone holds a reservoir of chondroprogenitor cells that when activated move into the proliferative zone where they rapidly divide to form columnar stacks parallel to the long axis of the bone. This transition is regulated by the Sox trio: Sox 5, 6 and 9 [12,13]. Proliferating chondrocytes then transition into the zone of hypertrophy were they swell several times their original volume and secrete copious amounts of matrix rich in collagen[14]. This process of chondrocyte maturation is regulated by the transcription factors Mef2c, Mef2d, Runx2, and Runx3 [14-16]. Hypertrophic chondrocytes ultimately undergo cell death in the zone of ossification and osteoblasts invade the void where the calcified cartilage matrix has been eroded by matrix metalloproteinases and osteoclasts. In adulthood, when the long bones have reached their full length, the growth plate cartilage becomes fully replaced by bone.
Figure 2.
Skeletal development proceeds either through endochondral or intramembranous ossification. This schema of the stratified growth plate in a long bone depicts the transition of the chondroprogenitor cell through the developmental progression of endochondral ossification, which includes proliferation, maturation, hypertrophy and ultimately cell death. Osteoprogenitor cells then invade the void left by hypertrophy, differentiate into osteoblasts, and maturate into osteocytes. Differentiation and maturation of osteoprogenitor cells is indistinguishable between endochondral and intramembranous ossification.
Chondrocytes require high translational capacity to meet the demands of proliferation, collagen matrix production, and then cellular hypertrophy within the stratified growth plate. Although a specific role for ribosome biogenesis in chondrocyte proliferation remains to be determined, the skeletal phenotypes of specific ribosomopathies infer such a connection. Moreover, the importance of ribosome biogenesis to general cell growth is well known. The role of ribosome biogenesis in chondrocyte maturation however is clearer due to the molecular link between rRNA transcription and transcriptional regulators of chondrocyte hypotrophy [15,16]. Runx2 and Runx3 associate with rDNA and regulate transcription through interactions with UBF1, an architectural factor that promotes transcription by RNA Pol I [17-19]. However, it is not known whether Runx2 and/or Runx3 regulate rRNA transcription in chondrocytes. Nonetheless, UBF1 is an ideal candidate to control chondrocyte hypertrophy as regulation of UBF1 is the key to the increased ribosome biogenesis and protein accumulation observed in association with cardiomyocyte hypertrophy [20].
While bone can be built through direct or indirect ossification, the role of the osteoblasts in these developmentally distinct processes is indistinguishable. Osteoprogenitor cells, whether located within the periosteum of endochondral bones or the mesenchymal condensation and osteogenic front of intramembranous bones, experience the same process of osteoblast differentiation (Figure 2). Osteoprogenitors are induced to differentiate by growth factors including FGF and BMP (reviewed in [21,22]). Acquisition of osteoblast cell fate requires the activity of the transcription factors Runx2, Osx/SP7, and Atf4 to promote expression of osteoblast specific genes including those that encode for osteoid matrix proteins such as type I collagen, osteocalcin, and osteopontin [23-25]. Once secreted, this collagen-rich osteoid matrix mineralizes upon binding to calcium salts brought to the region through neighboring vasculature. A subpopulation of osteoblasts become entrapped by the mineralized matrix and matures into osteocytes that actively regulate the lifelong process of bone turnover.
Regulators of osteoblast differentiation are unquestionably integrated with ribosome biogenesis. For example NO66, a nucleolar demethylase implicated in ribosome assembly and maturation, interacts with OSX/SP7 and inhibits OSX-mediated promoter activation in preosteoblasts thereby negatively regulating osteoblast differentiation [26,27]. The most well established molecular connector between ribosome biogenesis and osteoblast differentiation is Runx2, the master regulator of bone formation. Runx2 performs two essential functions in osteoprogenitor commitment. Firstly, it promotes osteoblast differentiation by transcriptionally activating bone-specific genes, and secondly it restricts cell division through inhibition of UBF1 and attenuation of rDNA transcription [17,28-31]. Integrating ribosome biogenesis with regulators of bone formation is a means to couple and coordinate osteoprogenitor cell proliferation and differentiation during skeletal development.
Thus ribosome biogenesis plays a critical role in intramembranous ossification of the neural crest-derived craniofacial skeleton and endochondral ossification of the mesoderm-derived limb skeleton. Moreover, mutations in ribosomal proteins and regulators of ribosome biogenesis can cause hypoplasia of the craniofacial bones, shortening of the long bones, and anomalies of the digits. This is exemplified in congenital ribosomopathies with skeletal defects, the etiology and pathogenesis of which are discussed in this review.
Treacher Collins Syndrome
Treacher Collins syndrome (TCS1, MIM 154500; TCS2, MIM 613717) is a congenital disorder of craniofacial development [32] and is also known as mandibulofacial dysostosis and Franschetti-Zwahlen-Klein syndrome [33]. The characteristic cranioskeletal features of Treacher Collins syndrome include hypoplasia of the facial bones, particularly the maxilla, mandible and zygomatic complex [34]. Alterations in the size, shape and position of the external ears are common and usually associated with atresia of the external auditory canals and anomalies of the middle ear ossicles [35]. Other clinical features of Treacher Collins syndrome may include defects in brain development such as microcephaly, mental retardation and psychomotor delay [36-39] however these features are associated with fewer than 5% of affected individuals.
Treacher Collins syndrome occurs with an incidence of about 1 in 50,000 live births and is primarily associated with autosomal dominant mutations in the TCOF1 gene, which is located on chromosome 5 [40]. To date, over 200 largely family-specific mutations have been documented throughout the TCOF1 gene and these include deletions, insertions, splice site, missense and nonsense mutations (http://genoma.ib.usp.br/TCOF1_database/). Deletions ranging in size from 1 to 40 nucleotides are the most common and within that group a reoccurring 5bp deletion in exon 24 accounts for 17% of TCS cases. Recently however, whole exome sequencing revealed causative mutations in POLR1C and POLR1D, which localize to chromosomes 6 and 13 respectively [41]. At least 17 distinct mutations in POLR1D have been described and similar to TCOF1 they elicit their effect in an autosomal dominant manner. In contrast, the 7 distinct mutations in POLR1C associated with Treacher Collins syndrome are all autosomal recessive [41].
Penetrance of the genetic mutations underlying Treacher Collins syndrome is high, yet inter- and intra-familial variation in the severity of the phenotype is a notable feature of the condition [42,43]. No genotype-phenotype correlation has been observed with respect to Treacher Collins syndrome and similarly there is no clear evidence of an association between disease severity and parental origin or type of pathogenic mutation, male or female, sporadic or familial [38,44-47]. The variable severity indicates that genetic background, environmental factors and stochastic events may contribute to the clinical variation observed in patients with Treacher Collins syndrome [48].
TCOF1 encodes a 144 kDa low complexity, serine/alanine-rich, protein known as Treacle [40]. Treacle is a putative nucleolar phosphoprotein that co-localizes with upstream binding factor 1 (UBF1) and RNA Pol I in the nucleolus. Biochemical analyses of siRNA-mediated knockdown of Treacle, have demonstrated that Treacle is essential for the proper transcription of rDNA [49]. Treacle has also been identified as a constituent of human Nop56-associated pre-ribosomal ribonucleoprotein complexes [50] that 2'O-methylate pre-ribosomal RNA during the early stages of pre-RNA processing in the nucleolus [49]. Thus Treacle is contained within an RNP complex in the nucleolus and may specifically regulate multiple steps of the ribosome biogenesis process.
Animal models of Treacher Collins syndrome successfully mimic the characteristic features and variability observed in humans [48] and have been instrumental in deciphering the pathogenesis of this congenital craniofacial disorder. The majority of the cartilage and bone that makes up the craniofacial complex is derived from neural crest cells. Consequently, most craniofacial abnormalities are attributed to problems in neural crest cell development. Tcof1 is broadly expressed during mouse embryogenesis, and between E8.5-10.5, elevated levels of Tcof1 expression are observed in the neuroepithelium and neural crest cell derived facial mesenchyme [51]. This is consistent with a potential role for Tcof1 in the formation and migration of neural crest cells. In support of this idea lineage tracing and gene expression analyses revealed a deficit in the number of migrating neural crest cells by as much as 25% in E8.5 Tcof1+/− mouse embryos compared to wild-type littermates. [51]. This deficiency in neural crest cell number was due to extensive neuroepithelial apoptosis in E8.0-10.5 Tcof1+/− embryos, which diminishes the neural stem cell pool from which neural crest cells are generated. Furthermore, the apoptosis is p53 dependent as nuclear activation and stabilization of p53 was observed in the neuroepithelium of Tcof1+/− embryos [51,52]
Deficient 28S rRNA and defective maturation of the 60S ribosomal subunit has been observed in Tcof1+/− embryos and this correlates with nucleolar stress activation of p53 and p53-dependent neuroepithelial cell death. This leads to decreased numbers of migrating neural crest cells, which underlies the pathogenesis of craniofacial malformations in Treacher Collins syndrome [51,52]. Interestingly, genetic and pharmacological inhibition of p53 in Tcof1+/− embryos can suppress neuroepithelial apoptosis ensuring the normal production of migrating neural crest cells. Remarkably, this can prevent the pathogenesis of craniofacial anomalies characteristic of Treacher Collins syndrome in animal models [52]. In theory this indicates Treacher Collins syndrome may be clinically preventable. Interestingly, the rescue occurred without restoration of ribosome biogenesis, suggesting that merely inhibiting cell death is sufficient to preserve the neural crest cell population required for cranioskeletal development.
Thus, Tcof1+/− mice have provided an important resource to decipher the in vivo cellular basis of Treacher Collins syndrome as well as the biochemical function of Treacle. POLR1D and POLR1C are subunits of RNA Pol I and III, which similar to TCOF1's role as an RNA Pol I binding factor, implicates each of these genes in ribosome biogenesis, which is essential for cell growth and proliferation. It will be interesting in the future, to explore the function of POLR1C and POLR1D and determine whether they share similar or overlapping functions with TCOF1 during embryogenesis confirming Treacher Collins syndrome is a ribosomopathy.
Postaxial Acrofacial Dysostosis
Postaxial Acrofacial dysostosis (POADS, MIM 263750) also known as Miller, Genee–Wiedemann, or Wildervanck–Smith syndromes, is an acrofacial dysostosis syndrome that presents with craniofacial and limb anomalies. The skeletal anomalies includes micrognathia, orofacial clefts, malar hypoplasia, cup-shaped ears combined with postaxial limb deformities, including apparent absence of either the fifth or both the fourth and fifth rays of the hands and feet, with or without ulnar and fibular hypoplasia [53-55]. Many of these characteristics are very similar to Treacher Collins syndrome.
POADS is caused by compound heterozygous mutations in the coding region of dihydroorotate dehydrogenase (DHODH) [56,57]. DHODH is an enzyme associated with the mitochondrial electron transport chain and is required for de novo pyrimidine synthesis. DHODH catalyzes the oxidation of dihydroorotate (DHO) to orotic acid, which is then converted to uracil monophosphate. Uracil is one of the constituent bases of RNA and is integral to ribosome biogenesis. In vivo and in vitro assays have shown that DHODH activity from POADS-associated alleles is diminished. This implies that affected individuals have a deficiency of de novo pyrimidine synthesis, which is consistent with the need for a threshold level of DHODH activity. Furthermore it suggests that a deficiency in ribosome biogenesis may be an underlying feature of POADS..
Similar to the etiology of Treacher Collins syndrome, it is surprising that since uracil and RNA synthesis are global processes,that mutations in DHODH would lead to such tissue specific phenotypes. However, analyses of Dhodh expression in mouse embryos have uncovered spatiotemporally specific activity in the pharyngeal arches, forelimbs, hindlimbs and somites [57]. These are the embryonic precursors of the tissues affected in POADS syndrome. Thus DHODH loss-of-function may result in rate-limiting effects on cell growth and proliferation during specific stages of craniofacial and limb development. Consistent with this idea, treatment of zebrafish with inhibitors of DHODH such as leflunomide, resulted in an almost complete abrogation of neural crest cell development principally by blocking the transcriptional elongation of genes critical for neural crest cell function [58]. This suggests the cellular basis of POADS syndrome may lie in deficient neural crest cell formation and the failure to generate sufficient numbers of migrating neural crest cells, which is analogous to the pathogenesis of Treacher Collins syndrome.
It is important to note however that mutations in Uridine Monophosphate Synthetase (UMPS), which functions immediately downstream of DHODH, result in the pathogenesis of orotic aciduria, Orotic aciduria presents with the classic feature of reduced pyrimidine availability which is megaloblastic anemia. Furthermore, orotic aciduria can be effectively treated with dietary uridine supplementation, demonstrating that the lack of pyrimidines underlies the orotic aciduria disease state. This raises the question as to why individuals with POADS do not have megaloblastic anemia or conversely, why individuals with orotic aciduria do not exhibit skeletal malformations. While is it possible that phenotypic overlap between these disorders has gone unrecognized, it seems more plausible that the underlying basis of POADS may not be restricted to deficient pyrimidine synthesis. Furthermore, there is no clear evidence to date for nuclear localization of DHODH. It will be critical therefore in the future, to better explore the in vivo biochemical function of DHODH and the cellular pathogenesis of POADS syndrome through the generation of Dhodh loss-of-function mammalian animal models.
Diamond Blackfan Anemia
The three genes involved in TCS are all involved in the process of ribosome biogenesis, which leads us to consider the role of neural crest cell formation in other ribosomopathies such as Diamond Blackfan anemia (DBA, MIM 105650). DBA is characterized by anemia, reticulocytopenia, macrocytosis, and a selective decrease or absence of erythroid precursors [59]. However, a range of craniofacial and cardiac defects as well as thumb abnormalities are observed in 40% to 62% of patients [59]. The craniofacial phenotype of DBA has considerable overlap with TCS and can include cleft palate and microtia. Unlike TCS, which exhibits deficient production of rRNA, DBA is caused by mutations in ribosomal proteins including RPS17, RPS19, RPS24, RPL5, RPL11, and RPL35A (reviewed in [59,60]). However, in about half of patients with DBA, the genetic mutation remains unknown.
Mutations in RPL5 are associated with craniofacial and cardiac anomalies more often than mutations in the other RPs linked to DBA [61], which suggests a specific role for RPL5 in neural crest cell development and variable tissue specific roles for individual ribosomal proteins. For example, a zebrafish mutant in rpl11 exhibits metabolic and hematopoietic defects [62], as well as diminished expression of neurogenic markers [63]. The formation of neural crest cells was not investigated in these models, however given the diminished expression of neurogenic markers in rpl11 zebrafish morphants, it seems possible that neural crest cell formation could be impaired by the significant apoptosis observed in these embryos. Interestingly, similar to the phenomenon in Tcof1+/− mice [52], inhibition of p53 prevented the morphant rpl11 induced DBA phenotype [63], and was also successful in preventing the phenotype in mice and zebrafish with mutations in rps19 [64,65].
Recently, L-leucine treatment has been used to improve the phenotype in mouse and zebrafish models of DBA [66,67]. In Rps19-deficient mice, L-leucine administered in the drinking water improved hematopoieis and also led to a down-regulation of p53 [67]. When zebrafish embryos injected with rps14 or rps19 morpholinos (MO) were developed in media supplemented with 100mM L-leucine, this treatment was able to ameliorate craniofacial defects [66]. L-leucine supplementation has also been used to treat anemia in some DBA patients [68,69]. Amino acids such as L-leucine are known to stimulate TORC1 (target of rapamycin complex 1, also known as mTOR), which is an important regulator of ribosome biogenesis, cell growth and cell proliferation. Leucyl-tRNA sythetase (LeuRS), which charges the tRNA with leucine, functions to signal leucine availability to TORC1 [66,70]. TORC1 then acts to stimulate ribosome biogenesis acting on RNA Pol and protein translation. TORC1 will phosphorylate proteins of the RNA Pol I initiation complex to stimulate transcription of rDNA by RNA Pol I [71,72] and also controls phosphorylation of Maf1, which is an important regulator of RNA Pol III [73,74]. Phosphorylation of 4E-BP1 by TORC1 releases it from the translation initiation complex, allowing for translation of ribosomal proteins [75].
Although the impact of L-leucine supplementation on neural crest cell development has not been thoroughly investigated, nutritional supplementation with L-leucine could provide a promising treatment option for other neurocristopathies which have an underlying deficiency in ribosome biogenesis as a part of their pathogenesis. Disruption of ribosomal proteins affects ribosome biogenesis leading to nucleolar stress activation of p53 and cell death. The craniofacial anomalies observed in DBA likely have a similar pathogenesis to TCS of neuroepithelial cell death, diminished NCC formation and proliferation. The investigation of neural crest cell development in specific animal models of DBA is needed in the future to better understand the potential roles of ribosomal proteins and to provide a deeper understanding of the role of neural crest cells in the pathogenesis of DBA.
Roberts Syndrome
Roberts syndrome (RBS, MIM 268300) is characterized by pre- and post-natal growth retardation, bilateral symmetric limb reduction, and craniofacial abnormalities including hypertelorism, cleft lip and palate, and hypoplastic nasal alae [76]. RBS is inherited in an autosomal recessive manner and is caused by mutations in the ESCO2 gene [77], which have been recently shown to influence production of rRNA [78]. ESCO2, Establishment of Cohesion 1 Homolog 2 (Eco1 in yeast), is an acetyl transferase important in assembly of Cohesin. Cohesin is a complex of proteins which binds chromosomes and holds sister chromatids together from DNA replication to cell division [79]. However, Cohesin also has other cellular roles including binding genes with paused RNA Polymerase, facilitating DNA looping to bring together enhancers and promoters, and in DNA repair (reviewed in [80]).
The specific developmental anomalies associated with mutations in ESCO2 have begun to be investigated in animal models. Antisense morpholino oligonucleotides (MOs) knockdown of esco2 in zebrafish, results in embryos with smaller head and eyes, and incorrectly shaped somites at 24 hpf [81]. Later in development additional features such as abnormal pigmentation, reduced craniofacial cartilage and pectoral fin growth, and cardiac defects became apparent. Distorted spindles and disorganized chromosomes were seen in the mitotic cells of esco2 morphants, suggesting that these cells are subsequently unable to proceed through mitosis and undergo apoptosis. Therefore, similar to TCS and DBA, it seems likely that the RBS phenotype may be related, at least in part, to increased cell death and death of neural crest precursors. However, in contrast to TCS and DBA, the cell death in esco2 morphants is p53 independent [81].
Studies in yeast and human cell lines revealed that eco1 mutants exhibited defects in ribosome biogenesis including reduced protein translation [78]. Specifically, production of the methylated 25S and 18S rRNA transcripts was diminished and nucleolar morphology was disrupted in eco1 mutants [78]. Consistent with this, fibroblasts from individuals with RBS showed diminished rRNA production and protein synthesis. This suggests a similar disregulation of ribosome biogenesis underlies the pathogenesis of TCS, DBA and RBS.
Cohesin proteins may therefore normally facilitate production of ribosomal RNA and protein translation. Although RBS is classically considered a cohesinopathy [82], RBS can also be classified as a ribosomopathy given the features of diminished ribosome biogenesis, nucleolar disruption, and apoptosis. It remains to be determined what impact these cohesinopathy mutations have on neural crest cell development, however, future work with RBS models may reveal novel roles for esco2 in neural crest formation, migration, or differentiation and provide new information for the development of therapies to improve the quality of life for RBS patients.
Shwachman Diamond Syndrome
Shwachman Diamond syndrome (SDS, MIM 260400) is characterized primarily by exocrine pancreatic dysfunction, bone marrow failure and skeletal abnormalities [83,84]. The skeletal anomalies include delayed bone age as well as progressive deformities and pathological fractures as well as abnormal development of growth plates and metaphysis, which lead to short stature, [83,85]. Osteopenia is observed in almost all the patients and spinal radiographs of patients often show early signs of osteoporotic vertebral deformities. The skeletal phenotypes and their degree of severity vary considerably between individuals, even among patients with identical mutations and within the same family [86]. However, abnormal endochondral and membranous ossification of the skeleton is a universal feature of SDS.
Shwachman Diamond syndrome is an autosomal recessive disorder arising from mutations in the SBDS gene [87] with an estimated incidence of 1:50,000 live births. About 90% of individuals with SDS carry biallelic mutations in SBDS [87]. Mice homozygous for null alleles of SBDS exhibit early embryonic lethality, indicating that SBDS is essential for normal development [88]. Thus the most common mutations found in SDS patients are thought to be hypomorphic alleles.
The Saccharomyces cerevisiae ortholog of SBDS, Sdo1, functions in ribosome biogenesis [89,90]. However, SBDS in mammalian cells has been implicated in multiple pathways, including ribosome biogenesis [91](Austin et al. 2005), cell motility [92,93], reactive oxygen species, regulation [94] and stabilization of the mitotic spindle [95]. Cells from individuals with SDS exhibit abnormal expression of many genes involved in ribosome biogenesis including rRNA and mRNA processing [96], and display decreased expression of many ribosomal proteins involved in cell growth and survival [97]. More recently, SBDS was shown to function at a late stage in the cytoplasmic maturation of 60S ribosomal subunits.The nascent 60S subunit is typically kept in a functionally inactive state by the trans-acting factor eukaryotic initiation factor 6 (eIF6). SBDS mediates the release of eIF6 from the 60S ribosome which is a pre-requisite for the initiation of 80s ribosome translational activity[98].
The varied clinical phenotypes observed in SDS have been hypothesized to reflect the level of residual SBDS protein expression coupled with critical threshold requirements for SBDS function in meeting translational demands in different tissues at various times during development [99,100]. Although, the expression and role of SBDS in cartilage and bone remains unknown, chondrocytes and osteoblasts of the skeletal system are all characterized by high protein secretory capacity. Impaired SBDS function may therefore mediate its effects through perturbation of global protein translation or more likely through defective translation of critical proteins involved in axial and limb skeletal development. Either way, SDS is a ribosomopathy caused primarily by impaired release of eIF6 and deficient 80S translational activity as a consequence of mutations in the function of the ribosome assembly factor SBDS.
Cartilage Hair Hypoplasia
Cartilage Hair Hypoplasia (CHH, MIM 250250), which is also known as metaphyseal chondrodysplasia McKusick type [101], is characterized by short-limb dwarfism with adults ranging in height from 104-149cm. CHH and other short-limb dwarfism phenotypes are associated with metaphyseal or spondyloepiphyseal dysplasia. The limbs and ribs are most affected, with sparing of the spine and skull. Radiographic studies reveal short and thick tubular bones, with splaying and irregular metaphyseal borders of the growth plates that are widened, scalloped and irregularly sclerotic [102,103]. The costochondral junctions are similarly affected. These metaphyseal changes are present at birth and are usually more severe in the knee region than in the proximal femora helping to distinguish CHH from other metaphyseal chondrodysplasias [104]. CHH is a variant of short-limb dwarfism in which fine, thin and sparse hair is also present, beginning in the newborn period. Other common features include ligamentous laxity, defective T-cell and/or B-cell mediated immunity, hypoplastic anemia, and variable aganglionosis of the intestine, which leads to gastrointestinal anomalies such as Hirschsprung disease in some individuals [103,105-107].
First described in Amish people [101] the incidence of CHH in the Amish is 1 in 1340 births, with a carrier frequency of 1:19. In contrast in Finland, CCH presents with a frequency of 1 in 18 000–23 000 live births with an estimated carrier rate of 1:76 [103]. CHH is an autosomal recessive disorder with equal male-to-female frequency that arises from mutations in the gene for RNAase RMRP, which maps to 9p12 [108]. The founder mutation 70ArG contributes to 92% of Finnish and 48% of non-Finnish patients with CHH [109].
The RMRP gene encodes the untranslated RNA subunit of the ribonucleoprotein endoribonuclease processing complex, RNase MRP. RMRP is a ribonucleoprotein present in the nucleolus and mitochondria and is classified as a snoRNA. SnoRNAs form small nucleolar ribonucleoprotein complexes (snoRNPs) within the nucleolus and are involved in various steps in the synthesis of ribosomal RNA [110]. The yeast ortholog of RMRP is nme1, and it functions in (i) ribosome synthesis, via nucleolar cleavage of preribosomal RNA (pre-rRNA) and rRNA production; (ii), the generation of RNA primers for mitochondrial DNA replication, via cleavage of RNA [110]; and (iii) in the regulation of cell growth via the degradation of cell cycle–regulated mRNA [111]. Mutation of nme1 impacts late-60S ribosomal assembly via defective endonuclease cleavage of the precursor subunit 5.8S rRNA at the ITS-1 A3 site [112,113]. The severity of the 5.8S rRNA–processing defect is directly proportional to the severity of the growth defect in nme1 mutant yeast [114]. Deficiency of a single 60S subunit not only can lead to the breakdown of the entire particle but also can disrupt regulatory signals that feed back on ribosome biogenesis from the secretory machinery[115].
Transfecting human fibroblasts with a wild-type RMRP construct significantly increases the growth rate and production of the cleaved or processed 5.8S rRNA [116]. In contrast the mutated form of RMRP found in individuals with CHH elicits minimal cleavage or processing of the 5.8S rRNA. Furthermore, cells overexpressing the CCH variant of RMRP showed significantly decreased cyclin A2 levels and significantly increased cyclin B2 mRNA levels, indicating a mitotic delay due to decreased mRNA degradation. Thus RMRP gene mutations lead to decreased cell growth by impairing ribosomal assembly and by altering cyclin-dependent cell-cycle regulation, which is consistent with prolonged cell cycle rates and correlates with the reduction in growth rate and clinical phenotype of CHH.
Mutations in RMRP have also been found responsible for two other autosomal recessive skeletal dysplasias. Metaphyseal dysplasia without hypotrichosis (MIM 250460) is an allelic variant of CHH that only exhibit skeletal manifestations [117,118]. The RMRP mutations in Anauxetic dysplasia (MIM 607095) disrupt its endonucleolytic activity, specifically leading to poor processing of rRNA while allowing for normal mRNA processing of B-cyclin [116]. This disorder is characterized by midface hypoplasia and extreme short stature with defects in the growth plate including marked hypocellularity of the resting cartilage, severely reduce numbers of proliferating chondrocytes, and diminished columnization of the hypertrophic zone [119,120].
Bowen Conradi Syndrome
Bowen Conradi syndrome (BWCNS; MIM 211180) is characterized by marked prenatal and postnatal growth retardation, microcephaly, a prominent nose with an absent glabellar angle, micrognathia, joint abnormalities including flexion contractures, camptodactyly, rocker-bottom feet, and severe psychomotor delay [121,122]. These features collectively overlap with those associated with overlap with cerebro-oculo-facial-skeletal syndrome and trisomy 18 disorders. Most individuals with Bowen Conradi syndrome do not survive beyond the first year of life due to complications associated with reduced mobility and failure to thrive.
To date, virtually all Bowen Conradi syndrome affected babies have been born into Hutterite families of the Canadian Prairies with an incidence of about 1 in 355, which suggest a carrier frequency of 1 in 10 in this population [122]. Recently a c.400A/G, p.D86G mutation in the EMG1-encoding gene on chromosome 12p13 was identified in association with Bowen Conradi syndrome making it an autosomal recessive disorder [123].
Very little is known about the expression of EMG1 activity, except that it is broadly expressed in most adult tissues with strongest levels in the heart, liver, and stomach, followed by the kidney and brain. In comparisons between human adult and fetal tissues, semiquantitative PCR revealed EMG1 expression was similar or higher in most adult tissues, except that in the brain, fetal expression appeared higher than that in the adult. The D86G mutation alters the protein structure of EMG1 resulting in educed EMG1 protein levels in fibroblasts of BCS patients [123]. EMG1 can function as a dimer or multidimer and the effect of the D86G mutation is to increase the interaction of EMG1 subunits, 10-fold as compared to the wild-type subunits.
The EMG1 protein is also known as Nep1, which has been identified as being essential in yeast for the biogenesis of 18S ribosomal RNA and the 40S ribosome. The D86 residue in EMG1, which is consistently mutated in association with Bowen Conradi syndrome, is completely conserved across all orthologs analyzed to date [123]. Yeast Emg1 is a component of the small subunit (SSU) processome [124] and plays an essential role in the biogenesis of the ribosomal 40S subunit. In particular, it aids in the maturation of 18S rRNA, where it is thought to participate in methylation [125,126]. In eukaryotes, synthesis and assembly of the ribosomal subunits is a complicated process involving hundreds of factors. The precursor 35S rRNA must undergo many modifications during maturation into 5.8S, 25S, and 18S rRNA [127]. EMG1 is a member of the SPOUT superfamily of methyltransferases, which bind and modify tRNA or rRNA by methylation of the ribose 2’-OH group, guanine N1, or uridine N3 [128]. It has also been shown to play a role in the removal of snR57, the snoRNA component of the snoRNP responsible for the 20-OH ribose methylation of G1570 in 18S rRNA, as well as the recruitment of ribosomal protein RPS19 to the maturing ribosome [129]. Ribosomal biogenesis and the cell cycle are tightly linked, with a checkpoint at the G1/S boundary ensuring sufficient ribosome levels before cell division [130,131]. Notably, depletion of SSU processome components in yeast is known to cause cell-cycle arrest in G1 [132]. In the absence of EMG1, the cell may therefore be unable to produce sufficient 40S ribosomal subunits to sustain normal proliferation.
Conclusions
Despite the widespread expression of ribosomal protein genes and the ubiquitous requirement for protein synthesis, defects in ribosome biogenesis are associated with numerous human skeletal diseases and disorders that vary in phenotype, mode of inheritance, and severity. For example, patients with Treacher Collins syndrome, Postaxial Acrofacial dysostosis, Diamond Blackfan Anemia, Shwachman Diamond syndrome, Cartilage Hair Hypoplasia and Bowen Conradi syndromes, all exhibit skeletal malformations that singularly or combinatorially affect the craniofacial, limb or axial skeleton. However, not all of these conditions present with bone marrow failure. While patients with Diamond Blackfan Anemia, Shwachman Diamond syndrome and Cartilage Hair Hypoplasia display bone marrow failure, this feature has not been observed in Treacher Collins syndrome or Postaxial Acrofacial Dysostosis. This raises the fundamental question of how mutations in ribosome biogenesis associated genes, which might normally have global or widespread roles during organism development, can lead to such selective defects.
The characteristic of broadly expressed genes exhibiting cell-type-specific phenotypes is not unique among ribosomal proteins. However at present there does not appear to be a single unifying factor that links them clinically except for their roles in various aspects of ribosome biogenesis. One possibility however, is that the clinical differences and their variability may be explained by the types of alteration and the magnitude of their effect on ribosome biogenesis. For example, distinct mutations in RMRP give rise to the allelic conditions of Cartilage Hair Hypoplasia and Anauxetic Dysplasia. Mutations in RMRP that reduce ribosomal RNA cleavage are associated with bone dysplasia, whereas mutations that affect mRNA cleavage are associated with hair hypoplasia, immunodeficiency, and dermatologic abnormalities [133]. This also raises a number of issues concerning the spatiotemporal dynamics of ribosome biogenesis and whether different threshold levels of activity are required in one tissue versus another at different times of development. Consistent with this idea, such a cell type specific proliferation requirement has been postulated in the pathogenesis of TCS [51].
While tremendous inroads have been made in understanding the many levels of regulation controlling gene expression, the regulatory control of protein production has garnered much less attention. The historic idea that ribosomes function constitutively to translate the genetic code has recently been challenged. An emerging idea is that ribosome are specialized, built diversely of different rRNAs and combinations of ribosomal proteins and associated factors with variable post-translational modifications. This diversity has a considerable impact on how an mRNA template is translated into functional protein. Even core ribosome components are now thought to exert selective activity by virtue of their interactions with specific cis-acting regulatory elements that are present within subsets of mRNAs [134]. Thus ribosome activity appears to be highly regulated and this may provide an important new mode for governing spatiotemporal gene expression during normal embryonic development and in the pathogenesis of congenital skeletal anomalies.
A common feature shared by TCS, DBA and POADS syndromes is craniofacial skeletal malformations caused by perturbations of neural crest cell development. This phenotypic overlap likely reflects a common molecular function for these proteins in ribosome biogenesis during craniofacial development. Thus, it will be important to understand how TCOF1, POLR1C, POLR1D, ribosomal proteins, and DHODH are functionally integrated in neural crest cell-derived bone. However, TCS, POADS and DBA exhibit distinct abnormalities of the skeleton and this reflects the need to better understand the individual roles of these proteins in skeletal development. Understanding how the mechanisms of disease overlap and diverge in ribosomopathies with skeletal defects will be instrumental in designing realistic avenues for their therapeutic prevention.
Under normal cellular growth conditions, Mdm2 targets p53 for degradation through polyubiquitination. In contrast, under conditions of perturbed ribosome biogenesis and nucleolar stress, unincorporated ribosomal proteins bind to Mdm2 inhibiting its polyubiquitination capacity. This leads to activation and stabilization of p53 and ultimately cell death [135,136]. As a case in point, direct inhibition of p53 dependent apoptosis can successfully preventing the manifestation of ribosomopathy disorders such as TCS (Tcof1) and DBA (Rps19) [52,137]. However, p53 performs a number of important cellular functions, not least of all is its role as a tumor suppressor. Inhibiting p53 therefore carries a substantial risk of cancer or tumorigenesis side-effects, highlighting the need to explore other avenues for ribosomopathy prevention. Interestingly, leucine supplementation has recently been used to successfully treat DBA in humans [68,69] and animal models [66,67]. Embryonic zebrafish and mice which model DBA showed considerably improved craniofacial and hematopoietic development when their diets were supplemented with L-leucine [66,67]. The mechanistic basis for this is in the craniofacial region is that L-leucine stimulates ribosome biogenesis through the mTOR pathway and thus counters the p53 dependent apoptotic loss of neural crest cells. Leucine treatment has been used to successfully treat DBA in humans [68,69]. Thus leucine supplementation may be a possible treatment option for other disorders of ribosome biogenesis. Consistent with this idea, L-leucine was recently shown to ameliorate the development of Roberts syndrome-like abnormalities in zebrafish and patient specific cell based models of the disorder [138].
The disorders described in this review arise due to deficient ribosome biogenesis. However, the converse is also likely to be true, that excessive ribosome biogenesis can also lead to skeletal anomalies. It might be expected that perturbations in signals that lie upstream to regulate levels of ribosome biogenesis in bone may be able to elicit such effects. Consistent with this idea, it has long been known that signaling molecules such as FGF, BMP, Wnt, and Hedgehog spatiotemporally regulate growth, proliferation and differentiation in the craniofacial, limb and axial skeleton. However, how these signals are integrated with ribosome biogenesis as a means to adapt to changing requirements for protein synthesis during bone formation and homeostasis is unclear. Resolving this issue will provide a better understanding of why bone is particularly sensitive to levels of ribosome biogenesis. The convergence of developmentally regulated genes on ribosome specificity provides additional modes for fine tuning translation and influencing fundamental aspects of cell growth and proliferation in the context of embryonic development, evolution and congenital disease.
Highlights.
Ribosome biogenesis is integral to all cell growth and proliferation.
Disruption of ribosome biogenesis leads to congenital disorders termed ribosomopathies.
Congenital skeletal anomalies affect approximately 1 in 3000 live births.
The role of ribosome biogenesis in skeletal development is poorly understood.
Skeletal function is inextricably linked to the size and shape of individual bones
Acknowledgements
A.E.M. is supported by the March of Dimes Basil O'Connor Starter Scholar Award #5-FY12-166, NIH #5P30DE020750-02 to Y. Chai, and start-up funds from the Ostrow School of Dentistry at USC. P.A.T is supported by the Stowers Institute for Medical Research and the National Institute for Dental and Craniofacial Research (DE 016082).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stoll C, Dott B, Roth MP, Alembik Y. Birth prevalence rates of skeletal dysplasias. Clin Genet. 1989;35:88–92. doi: 10.1111/j.1399-0004.1989.tb02912.x. [DOI] [PubMed] [Google Scholar]
- 2.Lafontaine DLJ, Tollervey D. The function and synthesis of ribosomes. Nat Rev Mol Cell Biol. 2001;2:514–520. doi: 10.1038/35080045. [DOI] [PubMed] [Google Scholar]
- 3.Kressler D, Hurt E, Baßler J. Driving ribosome assembly. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Laferte A, Favry E, Sentenac A, Riva M, Carles C, et al. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moss T, Stefanovsky V, Langlois F, Gagnon-Kugler T. A new paradigm for the regulation of the mammalian ribosomal RNA genes. Biochem Soc Trans. 2006;34:1079–1081. doi: 10.1042/BST0341079. [DOI] [PubMed] [Google Scholar]
- 6.Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends in Cell Biology. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 7.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, et al. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 9.Lefebvre V, Bhattaram P. Vertebrate skeletogenesis. Curr Top Dev Biol. 2010;90:291–317. doi: 10.1016/S0070-2153(10)90008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- 11.Long F, Ornitz DM. Development of the endochondral skeleton. Cold Spring Harb Perspect Biol. 2013;5:a008334. doi: 10.1101/cshperspect.a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smits P, Li P, Mandel J, Zhang Z, Deng JM, et al. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 13.Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, et al. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, et al. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, et al. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn. 1999;214:279–290. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 16.Kim IS, Otto F, Zabel B, Mundlos S. Regulation of chondrocyte differentiation by Cbfa1. Mech Dev. 1999;80:159–170. doi: 10.1016/s0925-4773(98)00210-x. [DOI] [PubMed] [Google Scholar]
- 17.Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, et al. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci U S A. 2007;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali SA, Dobson JR, Lian JB, Stein JL, van Wijnen AJ, et al. A RUNX2-HDAC1 co-repressor complex regulates rRNA gene expression by modulating UBF acetylation. J Cell Sci. 2012;125:2732–2739. doi: 10.1242/jcs.100909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pande S, Ali SA, Dowdy C, Zaidi SK, Ito K, et al. Subnuclear targeting of the Runx3 tumor suppressor and its epigenetic association with mitotic chromosomes. J Cell Physiol. 2009;218:473–479. doi: 10.1002/jcp.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannan RD, Stefanovsky V, Taylor L, Moss T, Rothblum LI. Overexpression of the transcription factor UBF1 is sufficient to increase ribosomal DNA transcription in neonatal cardiomyocytes: implications for cardiac hypertrophy. Proc Natl Acad Sci U S A. 1996;93:8750–8755. doi: 10.1073/pnas.93.16.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan M, Cao X. BMP signaling in skeletal development. Biochem Biophys Res Commun. 2005;328:651–657. doi: 10.1016/j.bbrc.2004.11.067. [DOI] [PubMed] [Google Scholar]
- 22.Ornitz DM, Marie PJ. FGF signaling pathways in endochondral and intramembranous bone development and human genetic disease. Genes Dev. 2002;16:1446–1465. doi: 10.1101/gad.990702. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–963. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 26.Eilbracht J, Reichenzeller M, Hergt M, Schnolzer M, Heid H, et al. NO66, a highly conserved dual location protein in the nucleolus and in a special type of synchronously replicating chromatin. Mol Biol Cell. 2004;15:1816–1832. doi: 10.1091/mbc.E03-08-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B. Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 2010;29:68–79. doi: 10.1038/emboj.2009.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pratap J, Galindo M, Zaidi SK, Vradii D, Bhat BM, et al. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 2003;63:5357–5362. [PubMed] [Google Scholar]
- 29.Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, et al. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali SA, Zaidi SK, Dacwag CS, Salma N, Young DW, et al. Phenotypic transcription factors epigenetically mediate cell growth control. Proc Natl Acad Sci U S A. 2008;105:6632–6637. doi: 10.1073/pnas.0800970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaidi SK, Javed A, Choi JY, van Wijnen AJ, Stein JL, et al. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci. 2001;114:3093–3102. doi: 10.1242/jcs.114.17.3093. [DOI] [PubMed] [Google Scholar]
- 32.Treacher Collins E. Case with symmetrical congenital notches in the outer part of each lower lid and defective development of the malar bones. Trans Opthalmol Soc UK. 1900;20:90. [Google Scholar]
- 33.Franceschetti A, Klein D. The mandibulofacial dysostosis; a new hereditary syndrome. Acta Ophthalmol (Copenh) 1949;27:143–224. [PubMed] [Google Scholar]
- 34.Poswillo D. The pathogenesis of the Treacher Collins syndrome (mandibulofacial dysostosis). Br J Oral Surg. 1975;13:1–26. doi: 10.1016/0007-117x(75)90019-0. [DOI] [PubMed] [Google Scholar]
- 35.Stovin JJ, Lyon JA, Jr., Clemmens RL. Mandibulofacial dysostosis. Radiology. 1960;74:225–231. doi: 10.1148/74.2.225. [DOI] [PubMed] [Google Scholar]
- 36.Cohen J, Ghezzi F, Goncalves L, Fuentes JD, Paulyson KJ, et al. Prenatal sonographic diagnosis of Treacher Collins syndrome: a case and review of the literature. Am J Perinatol. 1995;12:416–419. doi: 10.1055/s-2007-994511. [DOI] [PubMed] [Google Scholar]
- 37.Milligan DA, Harlass FE, Duff P, Kopelman JN. Recurrence of Treacher Collins’ syndrome with sonographic findings. Mil Med. 1994;159:250–252. [PubMed] [Google Scholar]
- 38.Teber OA, Gillessen-Kaesbach G, Fischer S, Bohringer S, Albrecht B, et al. Genotyping in 46 patients with tentative diagnosis of Treacher Collins syndrome revealed unexpected phenotypic variation. Eur J Hum Genet. 2004;12:879–890. doi: 10.1038/sj.ejhg.5201260. [DOI] [PubMed] [Google Scholar]
- 39.Sakai D, Dixon J, Dixon MJ, Trainor PA. Mammalian neurogenesis requires Treacle-Plk1 for precise control of spindle orientation, mitotic progression, and maintenance of neural progenitor cells. PLoS Genet. 2012;8:e1002566. doi: 10.1371/journal.pgen.1002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Treacher Collins Syndrome Collaborative Group Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- 41.Dauwerse JG, Dixon J, Seland S, Ruivenkamp CA, van Haeringen A, et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet. 2011;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- 42.Dixon MJ, Marres HA, Edwards SJ, Dixon J, Cremers CW. Treacher Collins syndrome: correlation between clinical and genetic linkage studies. Clin Dysmorphol. 1994;3:96–103. [PubMed] [Google Scholar]
- 43.Marres HA, Cremers CW, Dixon MJ, Huygen PL, Joosten FB. The Treacher Collins syndrome. A clinical, radiological, and genetic linkage study on two pedigrees. Arch Otolaryngol Head Neck Surg. 1995;121:509–514. doi: 10.1001/archotol.1995.01890050009002. [DOI] [PubMed] [Google Scholar]
- 44.Edwards SJ, Gladwin AJ, Dixon MJ. The mutational spectrum in Treacher Collins syndrome reveals a predominance of mutations that create a premature-termination codon. American Journal of Human Genetics. 1997;60:515–524. [PMC free article] [PubMed] [Google Scholar]
- 45.Gladwin AJ, Dixon J, Loftus SK, Edwards S, Wasmuth JJ, et al. Treacher Collins syndrome may result from insertions, deletions or splicing mutations, which introduce a termination codon into the gene. Human Molecular Genetics. 1996;5:1533–1538. doi: 10.1093/hmg/5.10.1533. [DOI] [PubMed] [Google Scholar]
- 46.Isaac C, Marsh KL, Paznekas WA, Dixon J, Dixon MJ, et al. Characterization of the nucleolar gene product, treacle, in Treacher Collins syndrome. Mol Biol Cell. 2000;11:3061–3071. doi: 10.1091/mbc.11.9.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Splendore A, Silva EO, Alonso LG, Richieri-Costa A, Alonso N, et al. High mutation detection rate in TCOF1 among Treacher Collins syndrome patients reveals clustering of mutations and 16 novel pathogenic changes. Hum Mutat. 2000;16:315–322. doi: 10.1002/1098-1004(200010)16:4<315::AID-HUMU4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 48.Dixon J, Dixon MJ. Genetic background has a major effect on the penetrance and severity of craniofacial defects in mice heterozygous for the gene encoding the nucleolar protein Treacle. Dev Dyn. 2004;229:907–914. doi: 10.1002/dvdy.20004. [DOI] [PubMed] [Google Scholar]
- 49.Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci U S A. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayano T, Yanagida M, Yamauchi Y, Shinkawa T, Isobe T, et al. Proteomic analysis of human Nop56p-associated pre-ribosomal ribonucleoprotein complexes. Possible link between Nop56p and the nucleolar protein treacle responsible for Treacher Collins syndrome. J Biol Chem. 2003;278:34309–34319. doi: 10.1074/jbc.M304304200. [DOI] [PubMed] [Google Scholar]
- 51.Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nature Medicine. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Genee E. An extensive form of mandibulo-facial dysostosis. J Genet Hum. 1969;17:45–52. [PubMed] [Google Scholar]
- 54.Miller M, Fineman R, Smith DW. Postaxial acrofacial dysostosis syndrome. J Pediatr. 1979;95:970–975. doi: 10.1016/s0022-3476(79)80285-1. [DOI] [PubMed] [Google Scholar]
- 55.Wiedemann HR. Malformation-retardation syndrome with bilateral absence of the 5th rays in both hands and feets, cleft palate, malformed ears and eyelids, radioulnar synostosis (author's transl). Klin Padiatr. 1973;185:181–186. [PubMed] [Google Scholar]
- 56.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rainger J, Bengani H, Campbell L, Anderson E, Sokhi K, et al. Miller (Genee-Wiedemann) syndrome represents a clinically and biochemically distinct subgroup of postaxial acrofacial dysostosis associated with partial deficiency of DHODH. Hum Mol Genet. 2012;21:3969–3983. doi: 10.1093/hmg/dds218. [DOI] [PubMed] [Google Scholar]
- 58.White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lipton JM, Ellis SR. Diamond-Blackfan Anemia: Diagnosis, Treatment, and Molecular Pathogenesis. Hematol Oncol Clin North Am. 2009;23:261–282. doi: 10.1016/j.hoc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue M-F, et al. Ribosomal Protein L5 and L11 Mutations Are Associated with Cleft Palate and Abnormal Thumbs in Diamond-Blackfan Anemia Patients. American journal of human genetics. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Danilova N, Sakamoto KM, Lin S. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. British Journal of Haematology. 2011;152:217–228. doi: 10.1111/j.1365-2141.2010.08396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. Loss of Ribosomal Protein L11 Affects Zebrafish Embryonic Development through a p53-Dependent Apoptotic Response. PLoS ONE. 2009;4:e4152. doi: 10.1371/journal.pone.0004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaako P, Flygare J, Olsson K, Quere R, Ehinger M, et al. Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Blood. 2011;118:6087–6096. doi: 10.1182/blood-2011-08-371963. [DOI] [PubMed] [Google Scholar]
- 65.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 66.Payne EM, Virgilio M, Narla A, Sun H, Levine M, et al. L-leucine improves the anemia and developmental defects associated with Diamond-Blackfan anemia and del(5q) MDS by activating the mTOR pathway. Blood. 2012;120:2214–2224. doi: 10.1182/blood-2011-10-382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jaako P, Debnath S, Olsson K, Bryder D, Flygare J, et al. Dietary L-leucine improves the anemia in a mouse model for Diamond-Blackfan anemia. Blood. 2012;120:2225–2228. doi: 10.1182/blood-2012-05-431437. [DOI] [PubMed] [Google Scholar]
- 68.Cmejlova J, Dolezalova L, Pospisilova D, Petrtylova K, Petrak J, et al. Translational efficiency in patients with Diamond-Blackfan anemia. Haematologica. 2006;91:1456–1464. [PubMed] [Google Scholar]
- 69.Pospisilova D, Cmejlova J, Hak J, Adam T, Cmejla R. Successful treatment of a Diamond-Blackfan anemia patient with amino acid leucine. Haematologica. 2007;92:e66–e67. doi: 10.3324/haematol.11498. [DOI] [PubMed] [Google Scholar]
- 70.Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, et al. Leucyl-tRNA Synthetase Controls TORC1 via the EGO Complex. Molecular Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 71.Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, et al. mTOR-Dependent Regulation of Ribosomal Gene Transcription Requires S6K1 and Is Mediated by Phosphorylation of the Carboxy-Terminal Activation Domain of the Nucleolar Transcription Factor UBF†. Molecular and Cellular Biology. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes & Development. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wei Y, Tsang CK, Zheng XFS. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 2009;28:2220–2230. doi: 10.1038/emboj.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Michels AA, Robitaille AM, Buczynski-Ruchonnet D, Hodroj W, Reina JH, et al. mTORC1 Directly Phosphorylates and Regulates Human MAF1. Molecular and Cellular Biology. 2010;30:3749–3757. doi: 10.1128/MCB.00319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Powers T, Walter P. Regulation of Ribosome Biogenesis by the Rapamycin-sensitive TOR-signaling Pathway in Saccharomyces cerevisiae. Molecular Biology of the Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Den Berg DJ, Francke U. Roberts syndrome: A review of 100 cases and a new rating system for severity. American Journal of Medical Genetics. 1993;47:1104–1123. doi: 10.1002/ajmg.1320470735. [DOI] [PubMed] [Google Scholar]
- 77.Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 78.Bose T, Lee KK, Lu S, Xu B, Harris B, et al. Cohesin Proteins Promote Ribosomal RNA Production and Protein Translation in Yeast and Human Cells. PLoS Genet. 2012;8:e1002749. doi: 10.1371/journal.pgen.1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nasmyth K, Haering CH. Cohesin: Its Roles and Mechanisms. Annual Review of Genetics. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 80.Dorsett D, Strom L. The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol. 2012;22:R240–250. doi: 10.1016/j.cub.2012.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mönnich M, Kuriger Z, Print CG, Horsfield JA. A Zebrafish Model of Roberts Syndrome Reveals That Esco2 Depletion Interferes with Development by Disrupting the Cell Cycle. PLoS ONE. 2011;6:e20051. doi: 10.1371/journal.pone.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gerton JL. Translational mechanisms at work in the cohesinopathies. Nucleus. 2012;3:520–525. doi: 10.4161/nucl.22800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shwachman H, Diamond LK, Oski FA, Khaw KT. The Syndrome of Pancreatic Insufficiency and Bone Marrow Dysfunction. J Pediatr. 1964;65:645–663. doi: 10.1016/s0022-3476(64)80150-5. [DOI] [PubMed] [Google Scholar]
- 84.Ginzberg H, Shin J, Ellis L, Morrison J, Ip W, et al. Shwachman syndrome: phenotypic manifestations of sibling sets and isolated cases in a large patient cohort are similar. J Pediatr. 1999;135:81–88. doi: 10.1016/s0022-3476(99)70332-x. [DOI] [PubMed] [Google Scholar]
- 85.Aggett PJ, Cavanagh NP, Matthew DJ, Pincott JR, Sutcliffe J, et al. Shwachman's syndrome. A review of 21 cases. Arch Dis Child. 1980;55:331–347. doi: 10.1136/adc.55.5.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Makitie O, Ellis L, Durie PR, Morrison JA, Sochett EB, et al. Skeletal phenotype in patients with Shwachman-Diamond syndrome and mutations in SBDS. Clin Genet. 2004;65:101–112. doi: 10.1111/j.0009-9163.2004.00198.x. [DOI] [PubMed] [Google Scholar]
- 87.Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 88.Zhang S, Shi M, Hui CC, Rommens JM. Loss of the mouse ortholog of the shwachman-diamond syndrome gene (Sbds) results in early embryonic lethality. Mol Cell Biol. 2006;26:6656–6663. doi: 10.1128/MCB.00091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Menne TF, Goyenechea B, Sanchez-Puig N, Wong CC, Tonkin LM, et al. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 90.Moore JBt, Farrar JE, Arceci RJ, Liu JM, Ellis SR. Distinct ribosome maturation defects in yeast models of Diamond-Blackfan anemia and Shwachman-Diamond syndrome. Haematologica. 2010;95:57–64. doi: 10.3324/haematol.2009.012450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Austin KM, Leary RJ, Shimamura A. The Shwachman-Diamond SBDS protein localizes to the nucleolus. Blood. 2005;106:1253–1258. doi: 10.1182/blood-2005-02-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wessels D, Srikantha T, Yi S, Kuhl S, Aravind L, et al. The Shwachman-Bodian-Diamond syndrome gene encodes an RNA-binding protein that localizes to the pseudopod of Dictyostelium amoebae during chemotaxis. J Cell Sci. 2006;119:370–379. doi: 10.1242/jcs.02753. [DOI] [PubMed] [Google Scholar]
- 93.Leung R, Cuddy K, Wang Y, Rommens J, Glogauer M. Sbds is required for Rac2-mediated monocyte migration and signaling downstream of RANK during osteoclastogenesis. Blood. 2011;117:2044–2053. doi: 10.1182/blood-2010-05-282574. [DOI] [PubMed] [Google Scholar]
- 94.Ambekar C, Das B, Yeger H, Dror Y. SBDS-deficiency results in deregulation of reactive oxygen species leading to increased cell death and decreased cell growth. Pediatr Blood Cancer. 2010;55:1138–1144. doi: 10.1002/pbc.22700. [DOI] [PubMed] [Google Scholar]
- 95.Austin KM, Gupta ML, Jr., Coats SA, Tulpule A, Mostoslavsky G, et al. Mitotic spindle destabilization and genomic instability in Shwachman-Diamond syndrome. J Clin Invest. 2008;118:1511–1518. doi: 10.1172/JCI33764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ganapathi KA, Austin KM, Lee CS, Dias A, Malsch MM, et al. The human Shwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA. Blood. 2007;110:1458–1465. doi: 10.1182/blood-2007-02-075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rujkijyanont P, Adams SL, Beyene J, Dror Y. Bone marrow cells from patients with Shwachman-Diamond syndrome abnormally express genes involved in ribosome biogenesis and RNA processing. Br J Haematol. 2009;145:806–815. doi: 10.1111/j.1365-2141.2009.07692.x. [DOI] [PubMed] [Google Scholar]
- 98.Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, et al. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman-Diamond syndrome. Genes Dev. 2011;25:917–929. doi: 10.1101/gad.623011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wong TE, Calicchio ML, Fleming MD, Shimamura A, Harris MH. SBDS protein expression patterns in the bone marrow. Pediatr Blood Cancer. 2010;55:546–549. doi: 10.1002/pbc.22573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wong CC, Traynor D, Basse N, Kay RR, Warren AJ. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011;118:4305–4312. doi: 10.1182/blood-2011-06-353938. [DOI] [PubMed] [Google Scholar]
- 101.McKusick VA, Eldridge R, Hostetler JA, Ruangwit U, Egeland JA. Dwarfism in the Amish. Ii. Cartilage-Hair Hypoplasia. Bull Johns Hopkins Hosp. 1965;116:285–326. [PubMed] [Google Scholar]
- 102.Makitie O, Kaitila I. Cartilage-hair hypoplasia--clinical manifestations in 108 Finnish patients. Eur J Pediatr. 1993;152:211–217. doi: 10.1007/BF01956147. [DOI] [PubMed] [Google Scholar]
- 103.Makitie O, Marttinen E, Kaitila I. Skeletal growth in cartilage-hair hypoplasia. A radiological study of 82 patients. Pediatr Radiol. 1992;22:434–439. doi: 10.1007/BF02013505. [DOI] [PubMed] [Google Scholar]
- 104.Spranger JW, Zabel B, Kennedy J, Jackson G, Briggs M. A disorder resembling pseudoachondroplasia but without COMP mutation. Am J Med Genet A. 2005;132A:20–24. doi: 10.1002/ajmg.a.30350. [DOI] [PubMed] [Google Scholar]
- 105.McKusick VA. Metaphyseal Dysostosis and Thin Hair: A “New” Recessively Inherited Syndrome? Lancet. 1964;1:832–833. doi: 10.1016/s0140-6736(64)93029-6. [DOI] [PubMed] [Google Scholar]
- 106.McKusick VA, Bias WB, Norum RA, Cross HE. Blood groups in two Amish demes. Humangenetik. 1967;5:36–41. doi: 10.1007/BF00286209. [DOI] [PubMed] [Google Scholar]
- 107.Hermanns P, Bertuch AA, Bertin TK, Dawson B, Schmitt ME, et al. Consequences of mutations in the non-coding RMRP RNA in cartilage-hair hypoplasia. Hum Mol Genet. 2005;14:3723–3740. doi: 10.1093/hmg/ddi403. [DOI] [PubMed] [Google Scholar]
- 108.Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 109.Ridanpaa M, Sistonen P, Rockas S, Rimoin DL, Makitie O, et al. Worldwide mutation spectrum in cartilage-hair hypoplasia: ancient founder origin of the major70A-->G mutation of the untranslated RMRP. Eur J Hum Genet. 2002;10:439–447. doi: 10.1038/sj.ejhg.5200824. [DOI] [PubMed] [Google Scholar]
- 110.Welting TJ, van Venrooij WJ, Pruijn GJ. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol Cell Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, et al. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shadel GS, Buckenmeyer GA, Clayton DA, Schmitt ME. Mutational analysis of the RNA component of Saccharomyces cerevisiae RNase MRP reveals distinct nuclear phenotypes. Gene. 2000;245:175–184. doi: 10.1016/s0378-1119(00)00013-5. [DOI] [PubMed] [Google Scholar]
- 115.Zhao Y, Sohn JH, Warner JR. Autoregulation in the biosynthesis of ribosomes. Mol Cell Biol. 2003;23:699–707. doi: 10.1128/MCB.23.2.699-707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thiel CT, Horn D, Zabel B, Ekici AB, Salinas K, et al. Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am J Hum Genet. 2005;77:795–806. doi: 10.1086/497708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Verloes A, Pierard GE, Le Merrer M, Maroteaux P. Recessive metaphyseal dysplasia without hypotrichosis. A syndrome clinically distinct from McKusick cartilage-hair hypoplasia. J Med Genet. 1990;27:693–696. doi: 10.1136/jmg.27.11.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bonafe L, Schmitt K, Eich G, Giedion A, Superti-Furga A. RMRP gene sequence analysis confirms a cartilage-hair hypoplasia variant with only skeletal manifestations and reveals a high density of single-nucleotide polymorphisms. Clin Genet. 2002;61:146–151. doi: 10.1034/j.1399-0004.2002.610210.x. [DOI] [PubMed] [Google Scholar]
- 119.Menger H, Mundlos S, Becker K, Spranger J, Zabel B. An unknown spondylo-meta-epiphyseal dysplasia in sibs with extreme short stature. Am J Med Genet. 1996;63:80–83. doi: 10.1002/(SICI)1096-8628(19960503)63:1<80::AID-AJMG16>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 120.Horn D, Rupprecht E, Kunze J, Spranger J. Anauxetic dysplasia, a spondylometaepiphyseal dysplasia with extreme dwarfism. J Med Genet. 2001;38:262–265. doi: 10.1136/jmg.38.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bowen P, Conradi GJ. Syndrome of skeletal and genitourinary anomalies with unusual facies and failure to thrive in Hutterite sibs. Birth Defects Orig Artic Ser. 1976;12:101–108. [PubMed] [Google Scholar]
- 122.Lowry RB, Innes AM, Bernier FP, McLeod DR, Greenberg CR, et al. Bowen-Conradi syndrome: a clinical and genetic study. Am J Med Genet A. 2003;120A:423–428. doi: 10.1002/ajmg.a.20059. [DOI] [PubMed] [Google Scholar]
- 123.Armistead J, Khatkar S, Meyer B, Mark BL, Patel N, et al. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. Am J Hum Genet. 2009;84:728–739. doi: 10.1016/j.ajhg.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bernstein KA, Gallagher JE, Mitchell BM, Granneman S, Baserga SJ. The small-subunit processome is a ribosome assembly intermediate. Eukaryot Cell. 2004;3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu PC, Thiele DJ. Novel stress-responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol Biol Cell. 2001;12:3644–3657. doi: 10.1091/mbc.12.11.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Eschrich D, Buchhaupt M, Kotter P, Entian KD. Nep1p (Emg1p), a novel protein conserved in eukaryotes and archaea, is involved in ribosome biogenesis. Curr Genet. 2002;40:326–338. doi: 10.1007/s00294-001-0269-4. [DOI] [PubMed] [Google Scholar]
- 127.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 128.Tkaczuk KL, Dunin-Horkawicz S, Purta E, Bujnicki JM. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics. 2007;8:73. doi: 10.1186/1471-2105-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Buchhaupt M, Meyer B, Kotter P, Entian KD. Genetic evidence for 18S rRNA binding and an Rps19p assembly function of yeast nucleolar protein Nep1p. Mol Genet Genomics. 2006;276:273–284. doi: 10.1007/s00438-006-0132-x. [DOI] [PubMed] [Google Scholar]
- 130.Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol Biol Cell. 2007;18:953–964. doi: 10.1091/mbc.E06-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dez C, Tollervey D. Ribosome synthesis meets the cell cycle. Curr Opin Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 132.Bernstein KA, Baserga SJ. The small subunit processome is required for cell cycle progression at G1. Mol Biol Cell. 2004;15:5038–5046. doi: 10.1091/mbc.E04-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thiel CT, Rauch A. The molecular basis of the cartilage-hair hypoplasia-anauxetic dysplasia spectrum. Best Pract Res Clin Endocrinol Metab. 2011;25:131–142. doi: 10.1016/j.beem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 134.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lindstrom MS, Deisenroth C, Zhang Y. Putting a finger on growth surveillance: insight into MDM2 zinc finger-ribosomal protein interactions. Cell Cycle. 2007;6:434–437. doi: 10.4161/cc.6.4.3861. [DOI] [PubMed] [Google Scholar]
- 136.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. Embo Journal. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 138.Xu B, Lee KK, Zhang L, Gerton JL. Stimulation of mTORC1 with L-leucine Rescues Defects Associated with Roberts Syndrome. PLoS Genet. 2013;9:e1003857. doi: 10.1371/journal.pgen.1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]