Abstract
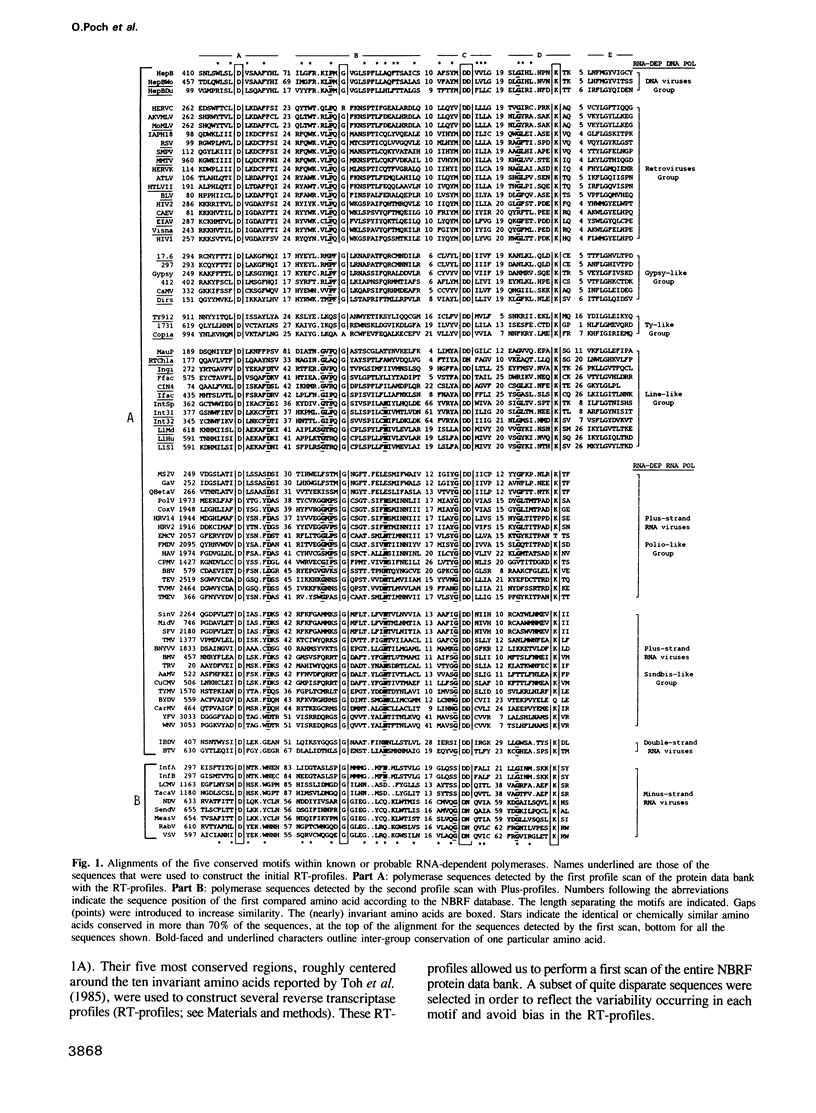
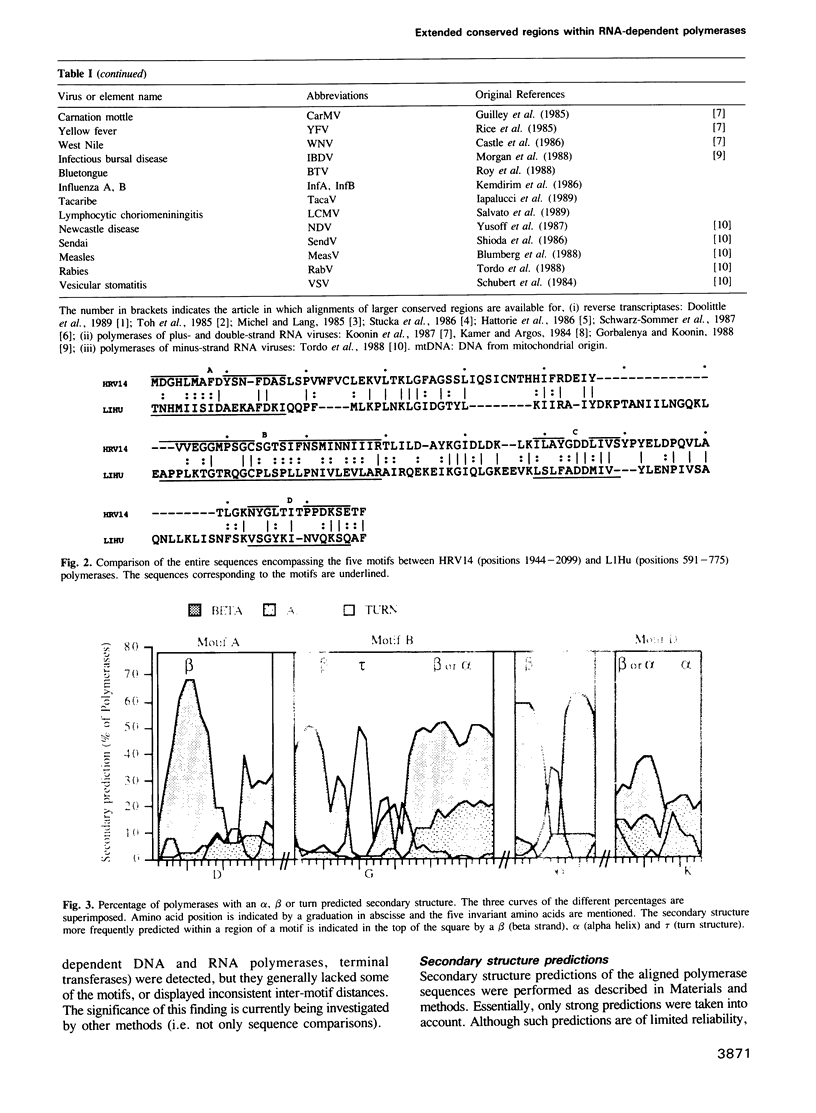
Four consensus sequences are conserved with the same linear arrangement in RNA-dependent DNA polymerases encoded by retroid elements and in RNA-dependent RNA polymerases encoded by plus-, minus- and double-strand RNA viruses. One of these motifs corresponds to the YGDD span previously described by Kamer and Argos (1984). These consensus sequences altogether lead to 4 strictly and 18 conservatively maintained amino acids embedded in a large domain of 120 to 210 amino acids. As judged from secondary structure predictions, each of the 4 motifs, which may cooperate to form a well-ordered domain, places one invariant amino acid in or proximal to turn structures that may be crucial for their correct positioning in a catalytic process. We suggest that this domain may constitute a prerequisite 'polymerase module' implicated in template seating and polymerase activity. At the evolutionary level, the sequence similarities, gap distribution and distances between each motif strongly suggest that the ancestral polymerase module was encoded by an individual genetic element which was most closely related to the plus-strand RNA viruses and the non-viral retroposons. This polymerase module gene may have subsequently propagated in the viral kingdom by distinct gene set recombination events leading to the wide viral variety observed today.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., Dasgupta R., Kaesberg P. Nucleotide sequence of the brome mosaic virus genome and its implications for viral replication. J Mol Biol. 1984 Feb 5;172(4):369–383. doi: 10.1016/s0022-2836(84)80012-1. [DOI] [PubMed] [Google Scholar]
- Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988 Nov 11;16(21):9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A., Zaballos A., Salas M., Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987 Dec 20;6(13):4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B. M., Crowley J. C., Silverman J. I., Menonna J., Cook S. D., Dowling P. C. Measles virus L protein evidences elements of ancestral RNA polymerase. Virology. 1988 Jun;164(2):487–497. doi: 10.1016/0042-6822(88)90563-6. [DOI] [PubMed] [Google Scholar]
- Boccara M., Hamilton W. D., Baulcombe D. C. The organisation and interviral homologies of genes at the 3' end of tobacco rattle virus RNA1. EMBO J. 1986 Feb;5(2):223–229. doi: 10.1002/j.1460-2075.1986.tb04202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer P. H., Gray M. W. Genes encoding a subunit of respiratory NADH dehydrogenase (ND1) and a reverse transcriptase-like protein (RTL) are linked to ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. EMBO J. 1988 Nov;7(11):3501–3508. doi: 10.1002/j.1460-2075.1988.tb03226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonitz S. G., Coruzzi G., Thalenfeld B. E., Tzagoloff A., Macino G. Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit 1 of yeast cytochrme oxidase. J Biol Chem. 1980 Dec 25;255(24):11927–11941. [PubMed] [Google Scholar]
- Callahan P. L., Mizutani S., Colonno R. J. Molecular cloning and complete sequence determination of RNA genome of human rhinovirus type 14. Proc Natl Acad Sci U S A. 1985 Feb;82(3):732–736. doi: 10.1073/pnas.82.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello J., Handelsman K., Lodish H. F. Sequence of Dictyostelium DIRS-1: an apparent retrotransposon with inverted terminal repeats and an internal circle junction sequence. Cell. 1985 Nov;43(1):105–115. doi: 10.1016/0092-8674(85)90016-9. [DOI] [PubMed] [Google Scholar]
- Carroll A. R., Rowlands D. J., Clarke B. E. The complete nucleotide sequence of the RNA coding for the primary translation product of foot and mouth disease virus. Nucleic Acids Res. 1984 Mar 12;12(5):2461–2472. doi: 10.1093/nar/12.5.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle E., Leidner U., Nowak T., Wengler G., Wengler G. Primary structure of the West Nile flavivirus genome region coding for all nonstructural proteins. Virology. 1986 Feb;149(1):10–26. doi: 10.1016/0042-6822(86)90082-6. [DOI] [PubMed] [Google Scholar]
- Chiu I. M., Yaniv A., Dahlberg J. E., Gazit A., Skuntz S. F., Tronick S. R., Aaronson S. A. Nucleotide sequence evidence for relationship of AIDS retrovirus to lentiviruses. 1985 Sep 26-Oct 2Nature. 317(6035):366–368. doi: 10.1038/317366a0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Clare J., Farabaugh P. Nucleotide sequence of a yeast Ty element: evidence for an unusual mechanism of gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2829–2833. doi: 10.1073/pnas.82.9.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J. M., Bricault L. PseqIP: a nonredundant and exhaustive protein sequence data bank generated from 4 major existing collections. Proteins. 1986 Sep;1(1):60–65. doi: 10.1002/prot.340010110. [DOI] [PubMed] [Google Scholar]
- Cornelissen B. J., Brederode F. T., Veeneman G. H., van Boom J. H., Bol J. F. Complete nucleotide sequence of alfalfa mosaic virus RNA 2. Nucleic Acids Res. 1983 May 25;11(10):3019–3025. doi: 10.1093/nar/11.10.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra B., Dasgupta R., Ghosh A., Kaesberg P. Structure of the black beetle virus genome and its functional implications. J Mol Biol. 1985 Mar 20;182(2):183–189. doi: 10.1016/0022-2836(85)90337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nocera P. P., Casari G. Related polypeptides are encoded by Drosophila F elements, I factors, and mammalian L1 sequences. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5843–5847. doi: 10.1073/pnas.84.16.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domier L. L., Franklin K. M., Shahabuddin M., Hellmann G. M., Overmeyer J. H., Hiremath S. T., Siaw M. F., Lomonossoff G. P., Shaw J. G., Rhoads R. E. The nucleotide sequence of tobacco vein mottling virus RNA. Nucleic Acids Res. 1986 Jul 11;14(13):5417–5430. doi: 10.1093/nar/14.13.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Fawcett D. H., Lister C. K., Kellett E., Finnegan D. J. Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell. 1986 Dec 26;47(6):1007–1015. doi: 10.1016/0092-8674(86)90815-9. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D., Merregaert J., Min Jou W., Molemans F., Raeymaekers A., Van den Berghe A. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976 Apr 8;260(5551):500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Fourcade-Peronnet F., d'Auriol L., Becker J., Galibert F., Best-Belpomme M. Primary structure and functional organization of Drosophila 1731 retrotransposon. Nucleic Acids Res. 1988 Jul 11;16(13):6113–6125. doi: 10.1093/nar/16.13.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck A., Guilley H., Jonard G., Richards K., Hirth L. Nucleotide sequence of cauliflower mosaic virus DNA. Cell. 1980 Aug;21(1):285–294. doi: 10.1016/0092-8674(80)90136-1. [DOI] [PubMed] [Google Scholar]
- Franssen H., Leunissen J., Goldbach R., Lomonossoff G., Zimmern D. Homologous sequences in non-structural proteins from cowpea mosaic virus and picornaviruses. EMBO J. 1984 Apr;3(4):855–861. doi: 10.1002/j.1460-2075.1984.tb01896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F., Chen T. N., Mandart E. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: comparison with the hepatitis B virus sequence. J Virol. 1982 Jan;41(1):51–65. doi: 10.1128/jvi.41.1.51-65.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert F., Mandart E., Fitoussi F., Tiollais P., Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979 Oct 25;281(5733):646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Goelet P., Lomonossoff G. P., Butler P. J., Akam M. E., Gait M. J., Karn J. Nucleotide sequence of tobacco mosaic virus RNA. Proc Natl Acad Sci U S A. 1982 Oct;79(19):5818–5822. doi: 10.1073/pnas.79.19.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbach R., Wellink J. Evolution of plus-strand RNA viruses. Intervirology. 1988;29(5):260–267. doi: 10.1159/000150054. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V. Birnavirus RNA polymerase is related to polymerases of positive strand RNA viruses. Nucleic Acids Res. 1988 Aug 11;16(15):7735–7735. doi: 10.1093/nar/16.15.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Homyak M., Edenfield J., Eisenberg D. Profile scanning for three-dimensional structural patterns in protein sequences. Comput Appl Biosci. 1988 Mar;4(1):61–66. doi: 10.1093/bioinformatics/4.1.61. [DOI] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilley H., Carrington J. C., Balàzs E., Jonard G., Richards K., Morris T. J. Nucleotide sequence and genome organization of carnation mottle virus RNA. Nucleic Acids Res. 1985 Sep 25;13(18):6663–6677. doi: 10.1093/nar/13.18.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyader M., Emerman M., Sonigo P., Clavel F., Montagnier L., Alizon M. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature. 1987 Apr 16;326(6114):662–669. doi: 10.1038/326662a0. [DOI] [PubMed] [Google Scholar]
- Hattori M., Kuhara S., Takenaka O., Sakaki Y. L1 family of repetitive DNA sequences in primates may be derived from a sequence encoding a reverse transcriptase-related protein. Nature. 1986 Jun 5;321(6070):625–628. doi: 10.1038/321625a0. [DOI] [PubMed] [Google Scholar]
- Herr W. Nucleotide sequence of AKV murine leukemia virus. J Virol. 1984 Feb;49(2):471–478. doi: 10.1128/jvi.49.2.471-478.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizi A., McGill C., Hughes S. H. Expression of soluble, enzymatically active, human immunodeficiency virus reverse transcriptase in Escherichia coli and analysis of mutants. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1218–1222. doi: 10.1073/pnas.85.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iapalucci S., Lopez R., Rey O., Lopez N., Franze-Fernandez M. T., Cohen G. N., Lucero M., Ochoa A., Zakin M. M. Tacaribe virus L gene encodes a protein of 2210 amino acid residues. Virology. 1989 May;170(1):40–47. doi: 10.1016/0042-6822(89)90349-8. [DOI] [PubMed] [Google Scholar]
- Inokuchi Y., Hirashima A. Interference with viral infection by defective RNA replicase. J Virol. 1987 Dec;61(12):3946–3949. doi: 10.1128/jvi.61.12.3946-3949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi Y., Jacobson A. B., Hirose T., Inayama S., Hirashima A. Analysis of the complete nucleotide sequence of the group IV RNA coliphage SP. Nucleic Acids Res. 1988 Jul 11;16(13):6205–6221. doi: 10.1093/nar/16.13.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi Y., Takahashi R., Hirose T., Inayama S., Jacobson A. B., Hirashima A. The complete nucleotide sequence of the group II RNA coliphage GA. J Biochem. 1986 Apr;99(4):1169–1180. doi: 10.1093/oxfordjournals.jbchem.a135580. [DOI] [PubMed] [Google Scholar]
- Ishihama A., Nagata K. Viral RNA polymerases. CRC Crit Rev Biochem. 1988;23(1):27–76. doi: 10.3109/10409238809103119. [DOI] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemdirim S., Palefsky J., Briedis D. J. Influenza B virus PB1 protein; nucleotide sequence of the genome RNA segment predicts a high degree of structural homology with the corresponding influenza A virus polymerase protein. Virology. 1986 Jul 15;152(1):126–135. doi: 10.1016/0042-6822(86)90378-8. [DOI] [PubMed] [Google Scholar]
- Kimmel B. E., ole-MoiYoi O. K., Young J. R. Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol Cell Biol. 1987 Apr;7(4):1465–1475. doi: 10.1128/mcb.7.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin E. V., Gorbalenia A. E., Chumakov K. M., Donchenko A. P., Blinov V. M. Evoliutsiia RNK-zavisimykh RNK-polimeraz pozitivnykh ribovirusov. Mol Gen Mikrobiol Virusol. 1987 Jul;(7):27–39. [PubMed] [Google Scholar]
- Lang B. F., Ahne F., Bonen L. The mitochondrial genome of the fission yeast Schizosaccharomyces pombe. The cytochrome b gene has an intron closely related to the first two introns in the Saccharomyces cerevisiae cox1 gene. J Mol Biol. 1985 Aug 5;184(3):353–366. doi: 10.1016/0022-2836(85)90286-4. [DOI] [PubMed] [Google Scholar]
- Larder B. A., Purifoy D. J., Powell K. L., Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. 1987 Jun 25-Jul 1Nature. 327(6124):716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- Lazcano A., Fastag J., Gariglio P., Ramírez C., Oró J. On the early evolution of RNA polymerase. J Mol Evol. 1988;27(4):365–376. doi: 10.1007/BF02101199. [DOI] [PubMed] [Google Scholar]
- Loeb D. D., Padgett R. W., Hardies S. C., Shehee W. R., Comer M. B., Edgell M. H., Hutchison C. A., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons. Mol Cell Biol. 1986 Jan;6(1):168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff G. P., Shanks M. The nucleotide sequence of cowpea mosaic virus B RNA. EMBO J. 1983;2(12):2253–2258. doi: 10.1002/j.1460-2075.1983.tb01731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandart E., Kay A., Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984 Mar;49(3):782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlor R. L., Parkhurst S. M., Corces V. G. The Drosophila melanogaster gypsy transposable element encodes putative gene products homologous to retroviral proteins. Mol Cell Biol. 1986 Apr;6(4):1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Lang B. F. Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature. 1985 Aug 15;316(6029):641–643. doi: 10.1038/316641a0. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Waterhouse P. M., Gerlach W. L. Sequence and organization of barley yellow dwarf virus genomic RNA. Nucleic Acids Res. 1988 Jul 11;16(13):6097–6111. doi: 10.1093/nar/16.13.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R., Dixon M., Smith R., Peters G., Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987 Feb;61(2):480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morch M. D., Boyer J. C., Haenni A. L. Overlapping open reading frames revealed by complete nucleotide sequencing of turnip yellow mosaic virus genomic RNA. Nucleic Acids Res. 1988 Jul 11;16(13):6157–6173. doi: 10.1093/nar/16.13.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. M., Macreadie I. G., Harley V. R., Hudson P. J., Azad A. A. Sequence of the small double-stranded RNA genomic segment of infectious bursal disease virus and its deduced 90-kDa product. Virology. 1988 Mar;163(1):240–242. doi: 10.1016/0042-6822(88)90258-9. [DOI] [PubMed] [Google Scholar]
- Mount S. M., Rubin G. M. Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol. 1985 Jul;5(7):1630–1638. doi: 10.1128/mcb.5.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najarian R., Caput D., Gee W., Potter S. J., Renard A., Merryweather J., Van Nest G., Dina D. Primary structure and gene organization of human hepatitis A virus. Proc Natl Acad Sci U S A. 1985 May;82(9):2627–2631. doi: 10.1073/pnas.82.9.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargang F. E., Bell J. B., Stohl L. L., Lambowitz A. M. The DNA sequence and genetic organization of a Neurospora mitochondrial plasmid suggest a relationship to introns and mobile elements. Cell. 1984 Sep;38(2):441–453. doi: 10.1016/0092-8674(84)90499-9. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Ono M., Toh H., Miyata T., Awaya T. Nucleotide sequence of the Syrian hamster intracisternal A-particle gene: close evolutionary relationship of type A particle gene to types B and D oncovirus genes. J Virol. 1985 Aug;55(2):387–394. doi: 10.1128/jvi.55.2.387-394.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M., Yasunaga T., Miyata T., Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986 Nov;60(2):589–598. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden S., Tangy F., Chamorro M., Brahic M. Theiler's virus genome is closely related to that of encephalomyocarditis virus, the prototype cardiovirus. J Virol. 1986 Dec;60(3):1163–1165. doi: 10.1128/jvi.60.3.1163-1165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg A. C., Kirby E. M., Janda M. R., Drake N. L., Duke G. M., Potratz K. F., Collett M. S. The nucleotide and deduced amino acid sequences of the encephalomyocarditis viral polyprotein coding region. Nucleic Acids Res. 1984 Mar 26;12(6):2969–2985. doi: 10.1093/nar/12.6.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repaske R., Steele P. E., O'Neill R. R., Rabson A. B., Martin M. A. Nucleotide sequence of a full-length human endogenous retroviral segment. J Virol. 1985 Jun;54(3):764–772. doi: 10.1128/jvi.54.3.764-772.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaian M. A., Williams R. H., Gordon K. H., Gould A. R., Symons R. H. Nucleotide sequence of cucumber-mosaic-virus RNA 2 reveals a translation product significantly homologous to corresponding proteins of other viruses. Eur J Biochem. 1984 Sep 3;143(2):277–284. doi: 10.1111/j.1432-1033.1984.tb08370.x. [DOI] [PubMed] [Google Scholar]
- Rice C. M., Lenches E. M., Eddy S. R., Shin S. J., Sheets R. L., Strauss J. H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985 Aug 23;229(4715):726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Roy P., Fukusho A., Ritter G. D., Lyon D. Evidence for genetic relationship between RNA and DNA viruses from the sequence homology of a putative polymerase gene of bluetongue virus with that of vaccinia virus: conservation of RNA polymerase genes from diverse species. Nucleic Acids Res. 1988 Dec 23;16(24):11759–11767. doi: 10.1093/nar/16.24.11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagata N., Yasunaga T., Tsuzuku-Kawamura J., Ohishi K., Ogawa Y., Ikawa Y. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc Natl Acad Sci U S A. 1985 Feb;82(3):677–681. doi: 10.1073/pnas.82.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saigo K., Kugimiya W., Matsuo Y., Inouye S., Yoshioka K., Yuki S. Identification of the coding sequence for a reverse transcriptase-like enzyme in a transposable genetic element in Drosophila melanogaster. Nature. 1984 Dec 13;312(5995):659–661. doi: 10.1038/312659a0. [DOI] [PubMed] [Google Scholar]
- Salvato M., Shimomaye E., Oldstone M. B. The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology. 1989 Apr;169(2):377–384. doi: 10.1016/0042-6822(89)90163-3. [DOI] [PubMed] [Google Scholar]
- Schubert M., Harmison G. G., Meier E. Primary structure of the vesicular stomatitis virus polymerase (L) gene: evidence for a high frequency of mutations. J Virol. 1984 Aug;51(2):505–514. doi: 10.1128/jvi.51.2.505-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Leclercq L., Göbel E., Saedler H. Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J. 1987 Dec 20;6(13):3873–3880. doi: 10.1002/j.1460-2075.1987.tb02727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiki M., Hattori S., Hirayama Y., Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K., Takahashi Y., Shimizu N., Gojobori T., Golde D. W., Chen I. S., Miwa M., Sugimura T. Complete nucleotide sequence of an infectious clone of human T-cell leukemia virus type II: an open reading frame for the protease gene. Proc Natl Acad Sci U S A. 1985 May;82(10):3101–3105. doi: 10.1073/pnas.82.10.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Shioda T., Iwasaki K., Shibuta H. Determination of the complete nucleotide sequence of the Sendai virus genome RNA and the predicted amino acid sequences of the F, HN and L proteins. Nucleic Acids Res. 1986 Feb 25;14(4):1545–1563. doi: 10.1093/nar/14.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skern T., Sommergruber W., Blaas D., Pieler C., Kuechler E. Relationship of human rhinovirus strain 2 and poliovirus as indicated by comparison of the polymerase gene regions. Virology. 1984 Jul 15;136(1):125–132. doi: 10.1016/0042-6822(84)90253-8. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Alizon M., Staskus K., Klatzmann D., Cole S., Danos O., Retzel E., Tiollais P., Haase A., Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985 Aug;42(1):369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Barker C., Hunter E., Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986 May 9;45(3):375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- Stephens R. M., Casey J. W., Rice N. R. Equine infectious anemia virus gag and pol genes: relatedness to visna and AIDS virus. Science. 1986 Feb 7;231(4738):589–594. doi: 10.1126/science.3003905. [DOI] [PubMed] [Google Scholar]
- Strauss E. G., Rice C. M., Strauss J. H. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucka R., Hauber J., Feldmann H. Conserved and non-conserved features among the yeast Ty elements. Curr Genet. 1986;11(3):193–200. doi: 10.1007/BF00420606. [DOI] [PubMed] [Google Scholar]
- Stålhandske P. O., Lindberg M., Pettersson U. Replicase gene of coxsackievirus B3. J Virol. 1984 Sep;51(3):742–746. doi: 10.1128/jvi.51.3.742-746.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takkinen K. Complete nucleotide sequence of the nonstructural protein genes of Semliki Forest virus. Nucleic Acids Res. 1986 Jul 25;14(14):5667–5682. doi: 10.1093/nar/14.14.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese N., Roth M., Epstein H., Goff S. P. An insertion mutation in the pol gene of Moloney murine leukemia virus results in temperature-sensitive pol maturation and viral replication. Virology. 1989 Jun;170(2):378–384. doi: 10.1016/0042-6822(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Reverse transcriptases. Retrons in bacteria. Nature. 1989 May 25;339(6222):254–255. doi: 10.1038/339254a0. [DOI] [PubMed] [Google Scholar]
- Toh H., Kikuno R., Hayashida H., Miyata T., Kugimiya W., Inouye S., Yuki S., Saigo K. Close structural resemblance between putative polymerase of a Drosophila transposable genetic element 17.6 and pol gene product of Moloney murine leukaemia virus. EMBO J. 1985 May;4(5):1267–1272. doi: 10.1002/j.1460-2075.1985.tb03771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordo N., Poch O., Ermine A., Keith G., Rougeon F. Completion of the rabies virus genome sequence determination: highly conserved domains among the L (polymerase) proteins of unsegmented negative-strand RNA viruses. Virology. 1988 Aug;165(2):565–576. doi: 10.1016/0042-6822(88)90600-9. [DOI] [PubMed] [Google Scholar]
- Wain-Hobson S., Sonigo P., Danos O., Cole S., Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985 Jan;40(1):9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Yuki S., Inouye S., Ishimaru S., Saigo K. Nucleotide sequence characterization of a Drosophila retrotransposon, 412. Eur J Biochem. 1986 Jul 15;158(2):403–410. doi: 10.1111/j.1432-1033.1986.tb09767.x. [DOI] [PubMed] [Google Scholar]
- Yusoff K., Millar N. S., Chambers P., Emmerson P. T. Nucleotide sequence analysis of the L gene of Newcastle disease virus: homologies with Sendai and vesicular stomatitis viruses. Nucleic Acids Res. 1987 May 26;15(10):3961–3976. doi: 10.1093/nar/15.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]




