Abstract
In Trypanosoma cruzi the isoprenoid and sterol biosynthesis pathways are validated targets for chemotherapeutic intervention. In this work we present a study of the genetic diversity observed in genes from these pathways. Using a number of bioinformatic strategies, we first identified genes that were missing and/or were truncated in the T. cruzi genome. Based on this analysis we obtained the complete sequence of the ortholog of the yeast ERG26 gene and identified a non-orthologous homolog of the yeast ERG25 gene (sterol methyl oxidase, SMO), and we propose that the orthologs of ERG25 have been lost in trypanosomes (but not in Leishmanias). Next, starting from a set of 16 T. cruzi strains representative of all extant evolutionary lineages, we amplified and sequenced ∼24 Kbp from 22 genes, identifying a total of 975 SNPs or fixed differences, of which 28% represent non-synonymous changes. We observed genes with a density of substitutions ranging from those close to the average (∼2.5/100 bp) to some showing a high number of changes (11.4/100 bp, for the putative lathosterol oxidase gene). All the genes of the pathway are under apparent purifying selection, but genes coding for the sterol C14-demethylase, the HMG-CoA synthase, and the HMG-CoA reductase have the lowest density of missense SNPs in the panel. Other genes (TcPMK, TcSMO-like) have a relatively high density of non-synonymous SNPs (2.5 and 1.9 every 100 bp, respectively). However, none of the non-synonymous changes identified affect a catalytic or ligand binding site residue. A comparative analysis of the corresponding genes from African trypanosomes and Leishmania shows similar levels of apparent selection for each gene. This information will be essential for future drug development studies focused on this pathway.
Introduction
Trypanosoma cruzi, a protozoan parasite of the order Kinetoplastida, is the causative agent of Chagas Disease, a neglected disease that is endemic in South America, affecting in excess of 8 million people [1]. The currently available drugs used to treat Chagas Disease (Nifurtimox, Benznidazole) have several drawbacks including toxicity, and the fact that they are mostly effective during the acute phase of the infection.
The T. cruzi species has a structured population, with a predominantly clonal mode of reproduction, with infrequent genetic exchange [2], [3]. Through the use of a number of genetic markers the population has been divided into six evolutionary lineages, also called Discrete Typing Units (DTUs) [4], [5], now designated as TcI to TcVI [6]. Lineages TcV and TcVI (this latter lineage includes the strain used for the first genomic sequence of T. cruzi, CL Brener) have a very high degree of heterozygosis. The currently favoured hypothesis suggests that these two lineages originated after one or two ancestral hybridization events [7]–[9]. There are no general agreement regarding the estimated time of divergence of the six T. cruzi lineages; however recent estimations suggest that major lineages (excluding the hybrid lineages TcV and TcVI) diverged 1–4 millions of years ago [9], [10] while the hybrid lineages emerged much more recently (less than 1 million years ago, according to Flores-Lopez and Machado [9], and within the last 60,000 years, according to Lewis MD, et al. [10]). This divergence was accompanied with a diversification of phenotypic and biological properties. Indeed, several investigations suggest that the observed diversity in host preference, cell tropism and drug susceptibility might be properties of different strains and/or lineages [11]–[14].
The parasite ergosterol biosynthesis pathway is one of the major routes for chemotherapeutic intervention against T. cruzi. Humans and trypanosomes share many of the enzymes leading to essential isoprenoid and sterol precursors. However, the mevalonate pathway is more related to that found in bacteria and archaea [15], and the ergosterol pathway is essentially similar to that in fungi, with a number of key steps that differ with the cholesterol biosynthesis pathway of mammals.
Inhibitors that block sterol biosynthesis or the biosynthesis of isoprenoid precursors inhibit growth of the parasite and cause severe morphological defects [16], [17]. Triazole derivatives that inhibit the parasite C14-α sterol demethylase are the most promising compounds, with proved curative activity in murine models of acute and chronic Chagas disease [18]–[22]. And one of them (posaconazole) is undergoing a number of clinical trials. Other ergosterol biosynthesis inhibitors with good potency in vitro or in vivo, or with good activity in an enzymatic assasy include those that target 3-hydroxy-3-methyl-glutaryl-CoA reductase [23]–[25], farnesyl diphosphate syntetase [26], squalene synthase[27], [28], squalene epoxidase/monooxygenase [29], [30], lanosterol synthase/oxidosqualene cyclase [31]–[33], and 24-C sterol methyl transferase [34]–[36], as well as compounds with dual mechanisms of action (ergosterol biosynthesis inhibition and free radical generation, reviewed in [16], [37]).
The azoles, like the triazoles, are used extensively for the treatment of fungal infections with excellent results, though different resistant strains have appeared over time in different species. One of the main resistance mechanisms observed is based on a diminished affinity of the target enzyme for the compound; which is caused by specific mutations in the ERG11/CYP51 gene. Different point mutations were identified in several fungi species as responsible for this azole resistance (reviewed in [38], [39]).
In this paper we analyze the genetic diversity present in the ergosterol biosynthesis pathway of T. cruzi and describe the apparent genetic selective pressure on these genes.
Results
Filling pathway holes: genes involved in sterol biosynthesis in T. cruzi
To analyze the genetic diversity of the T. cruzi sterol biosynthesis pathway (SBP) we decided to sequence all enzymes of the pathway, starting from enzymes that produce the terpenoid backbone precursors, and going down to the last enzyme that produces ergosterol as a product. Therefore, as a first step we looked for T. cruzi genes that were mapped to the corresponding KEGG metabolic pathway maps [40]. SBP genes in KEGG are classified in two maps: the steroid biosynthesis pathway map (TCR00100, www.genome.jp/kegg/pathway/tcr/tcr00100.html, and the terpenoid backbone biosynthesis pathway map (TCR00900, www.genome.jp/kegg/pathway/tcr/tcr00900.html). These maps contain information derived from the T. cruzi CL-Brener reference genome. From this analysis we were able to identify 15 genes mapped to these pathways. However, we also detected a number of holes in the pathway: enzymatic reactions with no enzyme mapped, and cases in which the enzymes available in KEGG were truncated (probably because of genome assembly problems). Therefore, before attempting to amplify and sequence the corresponding genes, we invested some effort in analyzing the existing sequence data to obtain a relevant complement of genes. As mentioned, in one case the corresponding genes from the reference genome were truncated, probably because of genome assembly problems. This was the case of the isopentenyl-diphosphate delta-isomerase gene (TcIDI1), which was cloned and sequenced by Dr. TK Smith (unpublished, GenBank accession number: AJ866772, 1071 bp). The corresponding genes in KEGG, mapped from the CL-Brener genome (Esmeraldo-like and non-Esmeraldo-like alleles) were both shorter, at 537 bp (TcCLB.408799.19), and 540 bp (TcCLB.510431.10). Therefore for this work we used the full-length TcIDI1 sequence obtained from GenBank (AJ866772). This sequence produces an active enzyme when expressed in an heterologous system (Dr. TK Smith, personal communication).
To fill in other identified gaps, we used the Saccharomyces cerevisiae sterol biosynthetic pathway as a reference model (Table 1). The yeast SBP has been studied extensively, and is essentially complete in pathway databases. Using the yeast genes from these pathways, we looked for orthologs in T. cruzi by doing sequence similarity searches (BLAST) against the complete T. cruzi genome or using databases of orthologs compiled from complete genome data, such as the OrthoMCL database [41]. As a result of this strategy, we were able to map five additional genes (see Table 2), which are the orthologs of the S. cerevisiae genes: ERG13 (3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase), ERG7 (lanosterol synthase), ERG26 (C-3 sterol dehydrogenase), ERG3, (C-5 sterol desaturase), and ERG5 (C-22 sterol desaturase). These genes were present in the T. cruzi genome, but were not mapped to the corresponding metabolic maps in KEGG. Some of these enzymes were already characterized biochemically in T. cruzi [25], [32]. In all these cases except for one (ERG26), the identification of the corresponding ortholog did not present further complications. The putative T. cruzi ortholog of the yeast ERG26 gene (TcCLB.510873.10, C3-sterol dehydrogenase) was found in the T. cruzi genome database as a truncated gene of 675 bp (224 aa). This may explain why it was not mapped to the corresponding pathway map in KEGG. Reasoning that this might be a consequence of assembly problems, as in the case of the TcIDI1 gene, we set out to identify the missing portions of this gene by performing BLASTN searches against a database of unassembled genomic reads (GSS or WGS reads) from the CL-Brener genome project (data provided originally by the TIGR-SBRI-KI sequencing consortium). Starting with the truncated TcCLB.510873.10 sequence as query, we identified a number of matching sequence reads (BLASTN, E-value < 10E −40). These sequences were then assembled into a single contiguous sequence, which was used as a query in successive rounds of BLASTN searches, followed by reassembly. This iteration cycle was repeated until we recovered the complete (full-length) sequence of the T. cruzi putative C3-sterol dehydrogenase gene, as judged by its alignment against the yeast ERG26 gene. This reconstructed full-length sequence of the T. cruzi C-3 sterol dehydrogenase gene has 1,221 bp and has been used to design primers for amplification from T. cruzi DNA (see Methods). The final sequence, obtained after amplification and sequencing from CL-Brener DNA, was submitted to GenBank under the accession number JN050853.
Table 1. Genes in the sterol biosynthesis pathway (SBP) of yeast.
| Gene ID/Locus | Molecular Function | EC Number | Pathway Order | Ortholog Group |
| ERG10/YPL028W | Acetoacetyl-CoA thiolase | 2.3.1.9 | 1 | OG4_10214 |
| ERG13/YML126C | 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase | 2.3.3.10 | 2 | OG4_11016 |
| HMG1/2/YML075C, YLR450W | HMG-CoA reductase | 1.1.1.34 | 3 | OG4_11458 |
| ERG12/YMR208W | Mevalonate kinase | 2.7.1.36 | 4 | OG4_11698 |
| ERG8/YMR220W | Phosphomevalonate kinase | 2.7.4.2 | 5 | OG4_15366 |
| ERG19/YNR043W | Mevalonate pyrophosphate decarboxylase | 4.1.1.33 | 6 | OG4_11688 |
| ERG20/YJL167W | Farnesyl pyrophosphate synthetase | 2.5.1.10 | 7 | OG4_11009 |
| IDI1/YPL117C | Isopentenyl diphosphate isomerase (IPP isomerase) | 5.3.3.2 | 7' | OG4_12197 |
| ERG9/YHR190W | Farnesyl-diphosphate farnesyl transferase (squalene synthase) | 2.5.1.21 | 8 | OG4_13084 |
| ERG1/YGR175C | Squalene epoxidase | 1.14.99.7 | 9 | OG4_13490 |
| ERG7/YHR072W | Lanosterol synthase (oxidosqualene cyclase) | 5.4.99.7 | 10 | OG4_11381 |
| ERG11/YHR007C | Lanosterol 14-alpha-demethylase | 1.14.13.70 | 11 | OG4_12975 |
| ERG24/YNL280C | C-14 sterol reductase | 1.3.1.70 | 12 | OG4_12018 |
| ERG25/YGR060W | C-4 methyl sterol oxidase | 1.14.13.72 | 13 | OG4_13007 |
| ERG26/YGL001C | C-3 sterol dehydrogenase | 1.1.1.170 | 14 | OG4_12533 |
| ERG27/YLR100W | 3-keto sterol reductase | 1.1.1.270 | 15 | OG4_16167 |
| ERG6/YML008C | Delta(24)-sterol C-methyltransferase | 2.1.1.41 | 16 | OG4_13307 |
| ERG2/YMR202W | C-8 sterol isomerase | 5.-.-.- | 17 | OG4_14573 |
| ERG3/YLR056W | C-5 sterol desaturase (lathosterol oxidase) | 1.3.3.- | 18 | OG4_12421 |
| ERG5/YMR015C | C-22 sterol desaturase | 1.14.14.- | 19 | OG4_14688 |
| ERG4/YGL012W | C-24(28) sterol reductase | 1.3.1.71 | 20 | OG4_16908 |
The table lists the reference genes used in this work for mapping the corresponding T. cruzi genes. Information for this table was derived from the Saccharomyces Genome Database (SGD, yeastgenome.org). Ortholog group identifiers are from the OrthoMCL Database, version 4 (orthomcl.org).
Table 2. Genes in the Trypanosoma cruzi sterol biosynthesis pathway.
| SBP Ortholog | T. cruzi Locus Identifiers(s) | Current annotation | Ortholog Group | In KEGG? | Gene Name |
| ERG10 | TcCLB.511003.60 | Hypothetical protein | OG4_10214 | Yes | TcACAT |
| ERG13 | TcCLB.511903.40, TcCLB.511071.50 | Hypothetical protein, conserved | OG4_11016 | No | TcHMGS |
| HMG1, HMG2 | TcCLB.509167.20, TcCLB.506831.40 | 3-hydroxy-3-methylglutaryl-CoA reductase, putative | OG4_11458 | Yes | TcHMGR |
| ERG12 | TcCLB.436521.9, TcCLB.509237.10 | Mevalonate kinase, putative | OG4_11698 | Yes | TcMK |
| ERG8 | TcCLB.508277.140, TcCLB.507913.20 | Phosphomevalonate kinase e-like protein, putative | OG4_15366 | Yes | TcPMK |
| ERG19 | TcCLB.507993.330, TcCLB.511281.40 | Diphosphomevalonate decarboxylase, putative | OG4_11688 | Yes | TcMVD |
| ERG20 | TcCLB.511823.70, TcCLB.508323.9 | Farnesyl pyrophosphate synthase, putative | OG4_11009 | Yes | TcFPPS |
| IDI1 | TcCLB.408799.19, TcCLB.510431.10 | Isopentenyl-diphosphate delta-isomerase, putative | OG4_14499 | Yes 6 | TcIDI |
| ERG9 | TcCLB.507897.20, TcCLB.508369.20 | Farnesyl transferase, putative; squalene synthase, putative | OG4_13084 | Yes | TcSQS |
| ERG1 | TcCLB.509589.20, TcCLB.503999.10 | Squalene monooxigenase, putative | OG4_13490 | Yes | TcSQLE |
| ERG7 | TcCLB.508175.70, TcCLB.506825.170 | Lanosterol synthase, putative | OG4_11381 | No | TcOSC |
| ERG11 | TcCLB.506297.260, TcCLB.510101.50 | Lanosterol 14-α-demethylase, putative | OG4_12975 | Yes | Tc14DM, cCYP51 |
| ERG24 | TcCLB.507969.60, TcCLB.507129.30 | C-14 sterol reductase, putative | OG4_12018 | Yes | Tc14SR |
| – | TcCLB.511339.20, TcCLB.511895.69, TcCLB.509235.20 | C-5 sterol desaturase, putative | OG4_20087 | No | TcSMO-like |
| ERG26 | TcCLB.510873.10 | NAD(P)-dependent steroid dehydrogenase protein, putative | OG4_12533 | No | TcNDSDHL |
| ERG27 | – | – | – | No | – |
| ERG6 | TcCLB.504191.10, TcCLB.505683.10, TcCLB.510185.10 | Sterol 24-C-methyltransferase, putative | OG4_13307 | Yes | Tc24SMT |
| ERG2 | TcCLB.510329.90 | C-8 sterol isomerase, putative | OG4_14573 | Yes | Tc8SI |
| ERG3 | TcCLB.473111.10, TcCLB.507853.10 | Lathosterol oxidase, putative | OG4_12421 | No | TcSC5D |
| ERG5 | TcCLB.506945.190 | Cytochrome p450-like protein, putative | OG4_14688 | No | TcSC22D |
| ERG4 | TcCLB.506577.120, TcCLB.507709.90 | Sterol C-24 reductase, putative | OG4_16908 | Yes | TcSC24R |
The table lists T. cruzi genes mapped to either the corresponding KEGG maps, or the yeast SBPs (see Table 1). OrthoMCL identifiers are from the OrthoMCL Database version 4. T. cruzi gene identifiers are those currently available at the TriTrypDB resource. The fifth column shows whether the gene was mapped to the corresponding KEGG metabolic map at the time of this writing. The last column shows the nomenclature used in this work. Some of these gene names were previously used in the literature, and others are used here for the first time, based on the gene names of relevant orthologs. In the case of the TcIDI gene, the two listed alleles are truncated copies of the gene. The full-length gene is deposited in GenBank as AJ866772. While in the case of the TcNSDHL gene, the copy annotated by the genome project is truncated due to genome assembly problems. The full-length gene is deposited in GenBank as JN050853 (this work).
At this point, there were still two gaps present when modeling the T. cruzi SBP on top of the yeast pathway. These correspond to the enzymatic reactions catalyzed by the yeast genes ERG27 (3-keto sterol reductase) and ERG25 (C-4 methyl sterol oxidase). Our failure at identifying the orholog of the yeast ERG27 gene came as no surprise, as the enzymes performing C-3 ketoreduction in land plants and sterol synthesizing bacteria are still unknown [42], [43].
However, a number of putative homologs of ERG25 in T. cruzi were readily identified in sequence similarity searches. In this case, it was difficult to discern which of these were the true orthologs of the yeast ERG25 gene.
Apparent loss of ERG25 homologs in T. cruzi and T. brucei
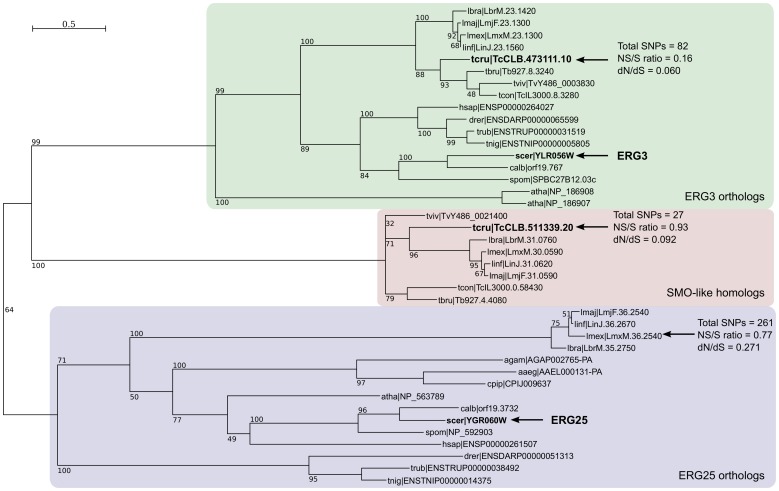
A BLASTP search using the yeast ERG25p as query against kinetoplastid proteomes retrieved 5 T. cruzi significant hits, with 4 of them having a similar length (278-279 aa, See Table S1), sharing a common Pfam Domain (PF04116, Fatty Acid Hydroxylase Superfamily), and a common architecture with 3–4 predicted trans-membrane domains (the fifth gene is a truncated copy). These hits correspond to two pairs of alleles, and are currently annotated as 'C-5 sterol desaturases' in the T. cruzi genome database (or ‘lathosterol oxidases’, both are synonymous terms). As there is detectable similarity between ERG3 and ERG25 genes in yeast as well, a similar search using the yeast ERG3p as query retrieves the same set of T. cruzi loci. Therefore, to identify which loci correspond to the true orthologs of ERG25, we carried out a detailed phylogenetic analysis between these genes, in different organisms. Using the sequences of fungi, plant, insect, human, fish and trypanosomatid enzymes, we obtained a maximum-likelihood tree with a clear segregation of ERG25 and ERG3 orthologs in separate branches (Figure 1). The tree suggests that there is one T. cruzi locus (containing TcCLB.473111.10 and its TcIII-like allele, TcCLB.507853.10) that is the true ortholog of the yeast ERG3 gene. Interestingly, the branch containing ERG25 orthologs does not show genes from African or American trypanosomes (the only kinetoplastid genes are from Leishmanias). The other trypanosomatid genes identified in our BLAST searches are grouped together in a third branch, carrying only trypanosomatid genes. These results are consistent with reciprocal best-hits identified in BLAST searches (see Table S1). Reciprocal or bidirectional best hits provide support for the conjecture that the genes are equivalent orthologs [44], [45]. In these searches, there is a clear drop in the BLAST score when the query sequence is ERG3 or a ERG3 homolog. This score drop is not observed when the query sequence is ERG25 or an ERG25 homolog. Apart from this evidence, there is additional support for this separate branch from the BLAST searches, in the observed reciprocity of best hits. For example when using the L. major gene LmjF.36.2540 as query (grouped with ERG25 orthologs in Fig 1) the best hits are the yeast ERG25 and their orthologs. Also viceversa, when doing the reciprocal BLAST search using the yeast ERG25p as query, the best L. major hit is again LmjF.36.2540, even though in this case the scores are lower than for those obtained for ERG3 orthologs. These bidirectional hits are not observed for the group of trypanosomatid genes grouped in the middle branch in the phylogenetic tree.
Figure 1. Phylogenetic tree of selected ERG3/ERG25 orthologs.
Seleced orthologs from kinetoplastids, plants, vertebrates, invertebrates and fungi were aligned with t_coffee. A phylogenetic reconstruction was calculated using PhyML (LG model, bootstrap resampling with N = 1000). For clarity, highly similar genes/alleles were not included in the final tree. Organism abbreviations are: lbra = L. braziliensis; lmex = L. mexicana; linf = L. infantum; lmaj = L. major; tcru = T. cruzi; tviv = T. vivax; tcon = T. congolense; tbru = T. brucei; agam = Anopheles gambiae; aaeg = Aedes aegypti; cpip = Culex pipiens; hsap = Homo sapiens; calb = Candida albicans; scer = S. cerevisiae; spom = Schizosaccharomyces pombe; atha = A. thaliana; drer = Danio rerio; trub = Takifugu rubripes; tnig = Tetraodon nigroviridis.
Taken together, the most parsimonious interpretation of these results is that the C-4 sterol oxidase gene has been lost in the ancestor of T. cruzi and T. brucei (but not in the ancestor of Leishmanias). The presence of Leishmanial and Trypanosomal proteins grouped in the middle branch of the tree suggests that these group of homologs have a different ancestral origin. However, because C-4 demethylation is essential, we hypothesize that they may have a C-4 sterol oxidase activity (see Discussion). For these reasons we decided to call them Sterol Methyl Oxidase-like (SMO-like) in this work.
Table 2 summarizes the results of our efforts to fill in the missing genes in the pathway, whereas Figures 2 and 3 show the reconstructed isoprenoid and sterol metabolic pathways, respectively. Once the T. cruzi SBP genes were identified, we proceeded to study the genetic diversity by re-sequencing these genes in a panel of T. cruzi strains, representative of all the extant evolutionary lineages of the parasite.
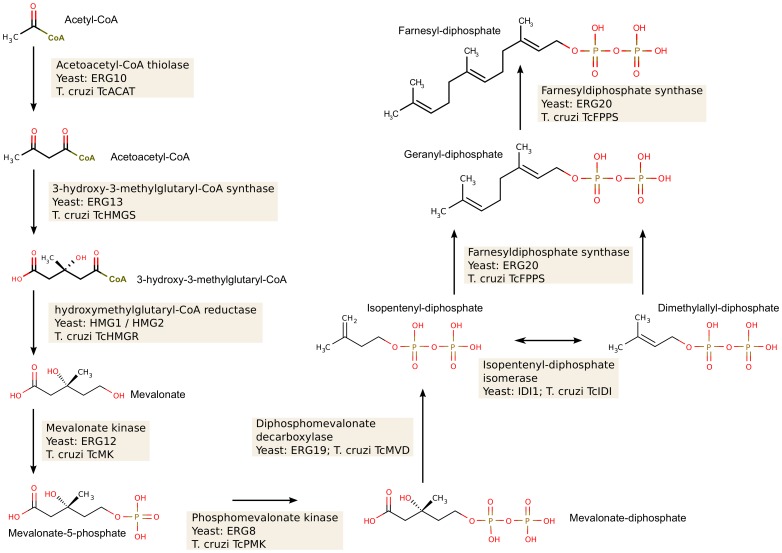
Figure 2. The T. cruzi isoprenoid biosynthesis pathway.
The figure shows the metabolic steps leading to farnesyl-diphosphate, from acetyl-CoA, and the corresponding yeast and T. cruzi enzymes that catalyze these steps. Gene names used are those in Tables 1 and 2.
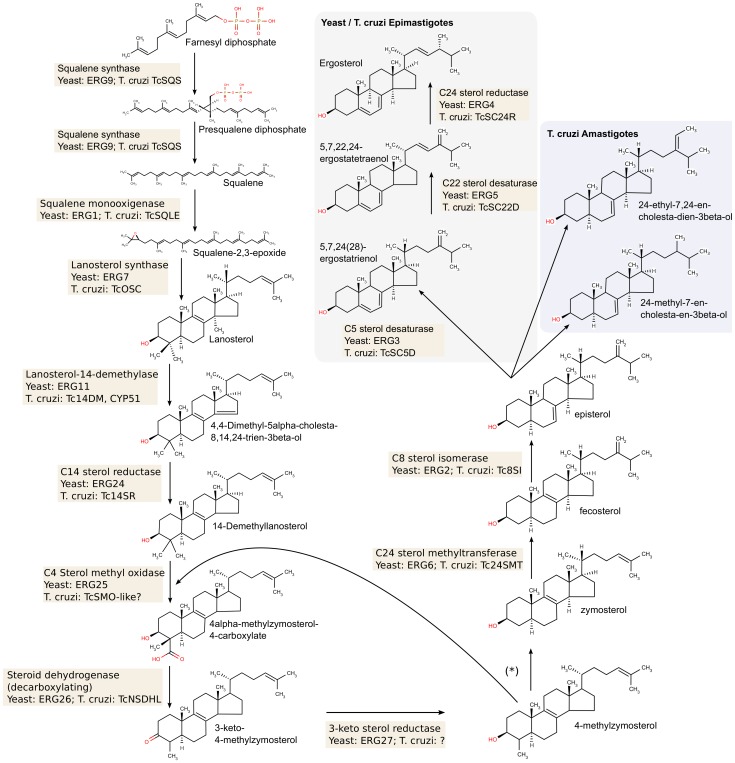
Figure 3. The T. cruzi sterol biosynthesis pathway.
The figure shows the metabolic steps leading from farnesyl-diphosphate to ergosterol (in yeast, and in T. cruzi epimastigotes) or to different 24-alkylsterols (in T. cruzi amastigotes [64]), and the corresponding yeast and T. cruzi enzymes that catalyze these steps. The two methyl groups at C4 are removed in two rounds of successive C4-oxidation, C4-decarboxylation and C3-ketoreduction (*). Gene names used are those in Tables 1 and 2. Unknown/hypothetical assignments are shown with question marks.
Genetic diversity in the sterol biosynthesis pathway
To obtain sequence information from the selected genes we decided to use a methodology based on PCR amplification followed by direct sequencing. Therefore, it was important to reduce the possibility of amplification problems generated by polymorphisms that could prevent the annealing of the primers. A number of aspects were considered to reduce these risks i) we decided to focus our analysis on coding sequences where possible, as they are generally less variable than non-coding sequences; ii) we used the T. cruzi SNP database (TcSNP, http://snps.tcruzi.org) [46], to select the best regions for primer design, avoiding regions with candidate SNPs; and iii) we also limited the size of amplification products, to optimize the quality of the final sequence (see Methods). In this latter case, an optimal size was set to ∼750–800 bp, which would allow us to get complete sequence coverage, with good quality on both strands. Depending on the size of each gene, one or more overlapping amplification products had to be analyzed (the number of amplification products per gene is listed in Table 3).
Table 3. Comparative analysis of the quantity, density and type of SNPs identified in the sterol biosynthesis pathway of Trypanosoma cruzi.
| Polymorphisms (SNPs) | Length (bp) | ||||||||
| Synonymous (S) | Non synonymous (NS) | Ratio | Total | CDS | Resequenced | ||||
| Genes | No. (%) | density | No. (%) | density | NS/S | No. | bp | bp (%) | PCR Fragments |
| TcACAT | 35 (75.00) | 3.27 | 11 (25.00) | 1.09 | 0.33 | 48 | 1188 | 1100 (92.59) | 2 |
| TcHMGS | 37 (86.05) | 2.66 | 6 (13.95) | 0.43 | 0.16 | 43 | 1497 | 1390 (92.85) | 2 |
| TcHMGR | 27 (79.41) | 2.18 | 7 (20.59) | 0.56 | 0.26 | 34 | 1356 | 1240 (91.45) | 2 |
| TcMK | 31 (79.49) | 3.20 | 12 (30.77) | 1.24 | 0.39 | 50* | 1086 | 970 (89.32) | 2 |
| TcPMK | 35 (53.85) | 2.65 | 33 (50.77) | 2.50 | 0.94 | 69* | 1428 | 1320 (92.44) | 2 |
| TcMVD | 32 (62.75) | 3.08 | 19 (37.25) | 1.83 | 0.59 | 51 | 1140 | 1040 (91.23) | 2 |
| TcIDI | 25 (78.13) | 2.50 | 7 (21.88) | 0.70 | 0.28 | 32 | 1071 | 1000 (93.37) | 2 |
| TcFPPS | 22 (66.67) | 2.27 | 11 (33.33) | 1.13 | 0.50 | 36* | 1275 | 970 (76.08) | 2 |
| TcSQS | 32 (69.57) | 2.88 | 14 (30.43) | 1.26 | 0.44 | 46 | 1212 | 1110 (91.58) | 2 |
| TcSQLE | 50 (72.46) | 3.07 | 21 (30.43) | 1.29 | 0.42 | 73* | 1716 | 1630 (94.99) | 3 |
| TcOSC | 61 (66.30) | 2.53 | 31 (33.70) | 1.29 | 0.51 | 92 | 2706 | 2410 (89.06) | 4 |
| Tc14DM | 47 (87.04) | 3.46 | 7 (12.96) | 0.51 | 0.15 | 54 | 1443 | 1360 (94.25) | 2 |
| Tc14SR | 33 (63.46) | 2.58 | 19 (36.54) | 1.48 | 0.58 | 52 | 1371 | 1280 (93.36) | 2 |
| TcSMO-like | 14 (51.85) | 2.09 | 13 (48.15) | 1.94 | 0.93 | 27 | 837 | 670 (80.05) | 1 |
| TcNSDHL | 21 (70.00) | 1.75 | 9 (30.00) | 0.75 | 0.43 | 30 | 1221 | 1200 (98.28) | 2 |
| Tc24SMT | 17 (77.27) | 2.07 | 5 (22.73) | 0.61 | 0.29 | 22 | 1050 | 820 (78.10) | 1 |
| Tc8SI | 18 (78.26) | 3.16 | 5 (21.74) | 0.88 | 0.28 | 23 | 654 | 570 (87.16) | 1 |
| TcSC5D | 70 (89.74) | 9.72 | 11 (14.10) | 1.53 | 0.16 | 82* | 834 | 720 (86.33) | 1 |
| TcSC22D | 29 (69.05) | 2.09 | 13 (30.95) | 0.94 | 0.45 | 42 | 1518 | 1390 (91.57) | 2 |
| TcSC24R | 53 (75.71) | 3.79 | 17 (24.29) | 1.21 | 0.32 | 70 | 1467 | 1400 (95.43) | 2 |
| Total/Avg | 692 (71.71) | 2.93 | 273 (28.29) | 1.16 | 0.39 | 975 | 26070 | 23590 (90.50) | 39 |
SNP counts marked with * indicate totals that do not result from the sum of synonymous + non-synonymous substitutions, in some cases because of the presence of two substitutions in the same codon, and in other cases, because some SNPs fall in the non-coding region of the gene.
All amplification products were analyzed with a software package (PolyPhred, see Methods) that allows the identification of heterozygous peaks in the chromatograms. Therefore, for all selected genes, we identified two types of sequence polymorphisms: i) allelic variation within a strain/clone (heterozygous peaks identified by Polyphred); and ii) variation between strain/clones (identified in a multiple sequence alignment of sequenced products). For every gene, we identified the position of the variant sites, and the type of change introduced (synonymous or non synonymous). The complete information for each gene is available as supplementary material (Table S2), and a table summarizing these results is included herein (Table 3). Using this strategy, we generated ∼24 Kb of sequence, from 16 strains covering the 6 major T. cruzi evolutionary lineages. Overall we attained a coverage of ∼90% of the total coding sequence for the selected genes. From this analysis we identified 975 polymorphic sites, producing 965 codon changes, which generate 692 synonymous changes (72%) and 273 non-synonymous changes (28%, see Table 3). Interestingly, we uncovered heterozygous SNPs in all genes of the pathway, even for those which are currently represented in the reference genome in an haploid state (e.g. TcACAT, and TcSC22D). The genes Tc24SMT, TcHMGR and TcNSDHL had the lowest SNP density (2.68, 2.74 and 2.50 every 100 bp respectively), while TcSC5D had the highest SNP density (11.39 every 100 bp). This density is at least double that which is found in other SBP genes. Nevertheless, this gene is under an apparently high purifying selection, as suggested by its low dN/dS ratio (Table 4). Upon further investigation of this gene, we noticed a number of informative SNPs that could be exploited in a lineage typing assay. Therefore, we performed a separate re-sequencing experiment for this gene, in an expanded panel of strains, and developed two alternative typing assays based on this locus [47].
Table 4. Comparative analysis of the genetic diversity present in the sterol biosynthesis pathway of different kinetoplastids.
| dN/dS (ω) | |||
| Yeast gene, Std symbol | Af Tryps | Leish | Tcr |
| ERG10/ACAT | – | – | 0.028 |
| ERG13/HMGS | 0.057 | 0.048 | 0.054 |
| HMG1/HMGR | 0.065 | 0.052 | 0.057 |
| ERG12/MK | 0.077 | 0.076 | 0.077 |
| ERG8/PMK | 0.110† | 0.122† | 0.122† |
| ERG19/MVD | 0.067 | 0.072 | 0.059 |
| IDI1/IDI | 0.051* | 0.043* | 0.048* |
| ERG20/FPPS | 0.060 | 0.061 | 0.065 |
| ERG9/FDFT, SQS | 0.085 | 0.087 | 0.093 |
| ERG1/SQLE | 0.115† | 0.107† | 0.115† |
| ERG7/LSS, OSC | 0.109† | 0.113† | 0.113† |
| ERG11/14DM, CYP51 | 0.042* | 0.040* | 0.039* |
| ERG24/14SR, TM7SF2 | 0.142† | 0.109† | 0.110† |
| ERG25/SMO/SC4MOL | – | 0.271 | – |
| –/SMO-like | 0.080 | 0.082 | 0.092 |
| ERG26/NSDHL | 0.051* | 0.057 | 0.058 |
| ERG27/3KSR | – | – | – |
| ERG6/24SMT | 0.065 | 0.062 | 0.070 |
| ERG2/8SI | 0.115† | 0.100 | 0.097 |
| ERG3/SC5D | 0.069 | 0.059 | 0.060 |
| ERG5/SC22D | – | 0.028* | 0.107† |
| ERG4/SC24R | – | 0.097 | 0.070 |
| Average | 0.080 | 0.075 | 0.079 |
| Standard deviation | 0.028 | 0.028 | 0.026 |
The table shows the dN/dS (ω) ratio observed in the african trypanosome lineage (Af Tryps) (T. brucei brucei, T. brucei gambiense, T. evansi, T. congolense, T. vivax), in the Leishmania lineage (Leish) (L. major, L. infantum, L. braziliensis, and L. mexicana), and in the T. cruzi lineage (Tcr, sequences from TcI{TcVI DTUs). The values that are above (†) or under (*) a standard deviation from the average of each column are marked.
We next assessed the accumulation of changes at the protein level (missense polymorphisms), and observed that the genes TcSMO-like, TcMVD and TcPMK have the highest non-synonymous SNP density (1.82, 1.94 and 2.50 non-synonymous SNPs every 100 bp respectively). In spite of this, the dN/dS ratio for these genes is still low, ranging from 0.028 (TcACAT) to 0.122 (TcPMK), indicating the relatively high degree of conservation of T. cruzi SBP genes.
Lineage-specific and intra-lineage differences
All the collected data on sequence diversity was also analyzed in the context of the current separation of T. cruzi in discrete evolutionary lineages. For this analysis we considered three types of polymorphisms or fixed differences: i) lineage specific polymorphisms (LSPs) (these are the polymorphic sites that could differentiate one lineage from all the others; ii) intra-lineage polymorphisms (ILP) (differences between strains belonging to the same lineage); and iii) heterozygous sites (those that differ between alleles of a single diploid individual). In this comparative analysis we found that the lineage TcI showed more than one LSP every 200 bp, while in the other lineages, the LSP density was at least four times lower (data not shown). This data agrees with the hypothesis that TcI is one of the ancestral lineages [6], [9]. Analyzing the ILP distribution, we found that the TcIV lineage presents the highest density of ILPs, with almost one ILP every 100 bp. Interestingly more than ∼90% of the ILPs observed in TcIV are restricted to the comparison between the South American strain CanIII with two strains from North America (Dog Theis and 92122102R, data not shown). The observed differences among TcIV strains from South and North America are in agreement with previous observations using different markers which suggested a phylogenetic divergence between TcIV from these two regions [4], [10], [48]–[54]. Indeed, our results show that the number of differences between CanIII and these other TcIV strains is even greater than those observed between lineage TcV and TcVI (the two hybrid lineages). Our results therefore agree with previous publications that describe a considerable phylogenetic distance between the CanIII and Dog Theis strains, even though they are still placed into the same evolutionary lineage based on current typing methods. As expected, the hybrid lineages (TcV and TcVI) show the highest level of heterozygosity, with more than 3 heterozygote sites every 200 bp; while TcII showed an intermediate level of heterozygosity with approximately 3 sites every 2 Kb, and the other lineages showed less than 10 sites in the 24 Kb analyzed.
Potentially important non-synonymous changes
In many cases, variations in susceptibility or resistance to a drug are associated to mutations that occur near the interaction site of a substrate or an inhibitor. To analyze the potential relevance of the non-synonymous SNPs found, we gathered relevant information from the literature on the selected targets, their PFAM domains, PROSITE motifs, and the available crystallographic structures in the Protein Data Bank (PDB) for the T. cruzi enzyme or their orthologs (see Methods). Starting with these crystal structures, we first identified residues near the co-crystallized ligands (substrate, inhibitor or co-factor). For this we established a maximum distance limit of 7Å from each corresponding ligand. This analysis revealed a number of potentially important changes in the Tc14DM (TcCYP51), and TcMK genes.
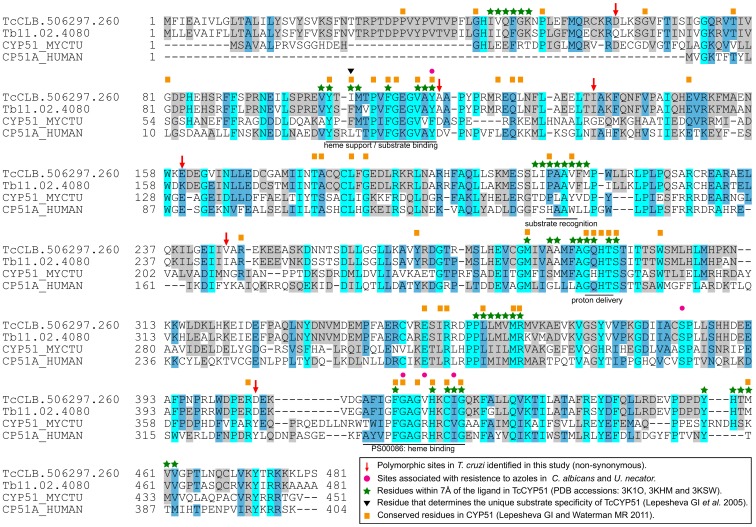
In the azole target gene, Tc14DM (TcCYP51), we identified only 5 non-synonymous substitutions, none of which affect key residues of the enzyme (see Figure 4). However, after mapping these substitutions on top of the available TcCYP51 structure [55], we identified a number of potentially important substitutions that lie just next to important residues, or are within 7Å of the co-crystallized ligand. One of these is a A117S substitution that sits just next to a tyrosine (Y116) that is within 7Å of the ligand, and that has been shown to be involved in the generation of resistance to azoles in C. albicans and U. necator [56], [57]. In trypanosomatids, residue 117 marks the start of a short helix in the structure, that is uniquely found in trypanosomatids [58]. Alanine at residue 117 is present in T. brucei and was found in strains from the TcIII lineage in homozygosis and in heterozygosis in hybrid strains, while Serine at this position was found in homozygosis in strains from lineages TcI, TcII, and TcIV.
Figure 4. Alignment of T. cruzi, T. brucei, M. tuberculosis and human Lanosterol 14-α demethylases, showing the non-synonymous changes identified in this work (red arrows).
Important residues either in Tc14DM or in the CYP51 family are noted [100], [101], as well as residues associated with resistance to azoles in C. albicans and U. necator [56], [57]. PS00086 is the Prosite Cytochrome 450 motif (cysteine heme-iron ligand signature).
In the case of the TcMK gene, we identified 12 non-synonymous changes. One of these is a H29Y change, located in a conserved region of the sequence, close to a group of residues that are involved in substrate binding (at <7Å of the ligand (mevalonate) in the L. major structure (LmjF.31.0560, PDB: 2HFU) [59]. Histidine is encoded by the genes from strains M5631 and X109/2, both from lineage TcIII, while tyrosine is encoded in all other strains, including M6241, also of lineage TcIII. The second possibly important change in TcMK is a G287A substitution, also located in a very conserved motif of the MK gene, that is part of the GHMP kinase family domain (Pfam PF08544). This particular motif, contains a number of highly conserved glycines, some of which are within 7Å of the ligand in the L. major MK. In this polymorphic position strains from lineage TcV (Sc43 and Mn) encode glycine and alanine in heterozygosis, while all the other strains encode glycine in homozygosis.
For other targets, crystallographic structures were not available, and so we resorted to analyze non-synonymous substitutions in the context of functionally important motifs or domains. In the case of the squalene epoxidase gene (TcSQLE), we identified two changes (S306G and I307L) that stand between highly conserved proline residues (at 95% and 94% identity within the squalene epoxidase domain, PF08491). The I307L substitution is apparently conservative, as these are the two most frequent residues in this position in the Pfam domain. In contrast, the S306G substitution introduces a glycine that is completely absent at this position in the 429 sequences currently available for this protein family in the Pfam database. Serine is the second most frequent residue in this position, and is present in 14% of the sequences, all from fungi (glutamine is the most frequent residue, in 50% of the sequences). The Ser residue occurs in lineage TcII in homocygosis, and in lineages TcV and TcVI in heterocygosis, and its conservation in fungi suggests that this is the ancestral character at this position. Finally in the C-14 sterol reductase gene (Tc14SR), we identified two consecutive substitutions (P208H, V209F) in strains 92122102R and Dog Theis (lineage TcIV), that fall within the sterol reductase family signature motif (PS01017). However the substitutions are conservative, because they are both described by the motif pattern.
The phosphomevalonate kinase of T. cruzi is not under a strong purifying selection
The phosphomevalonate kinases (PMKs) of pathogenic bacteria, fungi, and trypanosomes are attractive targets for the design of selective inhibitors, because the same phosphorylation reaction in humans and other animals is catalyzed by a non-orthologous enzyme [60]. In the first group of organisms, phosphorylation of mevalonate is performed by orthologs of the yeast ERG8 gene, while in animals it is performed by a group of orthologs of the human PMK gene (hPMK). These two groups of enzymes differ in a number of kinetic, biophysical properties, and in the ATP-binding motifs (ERG8-like kinases contain a protein kinase motif, while orthologs of the human enzyme have a P-loop or “Walker A” motif)[60], [61]. Analysis of the TcPMK gene in different strains of T. cruzi showed that the gene has accumulated 68 changes (35 synonymous, 33 non-synonymous, see Table 3), and 1 non-sense substitution (see below) in these independently evolving lineages. Based on the dN/dS ratio, the gene is under purifying selection (dN/dS < 1, see Table 4). However, it displays the highest dN/dS ratio of the pathway, the highest density of missense SNPs (2.5 non-synonymous SNPs/100 bp), as well as the highest ratio of synonymous to non-synonymous SNPs (∼1). Taken together, these data show that this is the gene that is apparently under more relaxed selection in the SBP pathway of T. cruzi.
Moreover, an interesting substitution was observed at codon 136 (407 bp) in this gene in the T. cruzi strain IVV (TcII). In this strain, one allele encoded a Serine (TCA), as in all other strains, whereas the second allele encoded a premature STOP codon (TAA). All ERG8-like PMKs are composed of two GHMP kinase domains: an N-terminal domain (Pfam PF00288) located in the middle of the protein (starting at residue 160 in T. cruzi), and a C-terminal domain (PF08544) closer to the C-termini. In T. cruzi, the nonsense mutation in the IVV strain is located upstream of this first domain (PF00288). Considering that the next possible translational start codon is located downstream of this domain, the IVV strain is therefore probably producing a very short non-functional protein from this allele, or two truncated proteins, both devoid of this domain. Thus, the most plausible hypothesis is that the IVV strain carries only one functional PMK allele, and can be considered naturally hemizygous for the PMK gene.
Comparative analysis of genetic diversity in kinetoplastid sterol biosynthesis pathways
Although phylogenetically related, kinetoplastid parasites have evolved different adaptations to their host environments. This is particularly evident in the case of ergosterol dependency: both T. cruzi and Leishmania have an essential requirement for ergosterol and/or other 24-alkylsterols, and are unable to survive by salvaging cholesterol from the host [62]. Whereas sterol biosynthesis in Trypanosoma brucei, is apparently suppressed in bloodstream forms, relying instead on receptor-mediated endocytosis of host low-density lipoproteins carrying cholesterol [63]. Apart from differences in their strict requirement for de novo synthesis of sterols, there are also differences in the exact type and abundance of synthesized sterols, which may be explained by differences in the complement of genes and/or their activities or regulation. As an example, it has been described that T. cruzi amastigotes lack Δ5,7 sterols, suggesting a lack of Δ5 desaturase activity in this life-cycle stage [62], [64]. Based on these premises, we decided to investigate the accumulation of nucleotide changes (fixed differences) in genes from the sterol biosynthesis pathway in African Trypanosomes, and Leishmanias, reasoning that the selection acting on these genes could be different in each case. For this we identified the corresponding orthologs of the yeast and T. cruzi genes used in this work (see Table 2) in currently available kinetoplastid genomes (see Methods). The available information includes that of T. brucei brucei (2 strains), T. brucei gambiense, Trypanosoma evansi, Trypanosoma congolense and Trypanosoma vivax; and that of 4 subspecies of Leishmania (major, infantum, braziliensis and mexicana). Using this information we proceeded to analyze the nucleotide substitutions observed in each group. A summary of this analysis is presented in Table 4 (and in Table S3). Apart from the absence of some genes that were already discussed, other notable missing genes were the orthologs of the ERG10 (ACAT/thiolase) in African Trypanosomes and Leishmanias, and the ERG5 (SC22D) and ERG4 (SC24R) in T. brucei. The reaction catalyzed by the orthologs of ERG10 in other trypanosomatids, is catalyzed in T. brucei by another thiolase [15] (Tb927.8.2540, ortholog of POT1, see Table S4). However, whereas the absence of the sterol reductase has been noticed by others recently [65], we also noticed the absence of a gene encoding the C22 sterol desaturase in this organism.
Because the genetic distances within each phylogenetic group are different, it would be perhaps incorrect to compare nucleotide changes for each gene across these groups of organisms in a direct way. However, it is still possible to analyze and compare the information on sequence diversity within each group (e.g. column-wise in Table 4). In the table, we marked the genes with the highest and lowest dN/dS ratios within the pathway, and those deviating > 1 standard deviation. When looking at data in this way, a number of observations can be made: the less and most diverse genes in each pathway correlate very well: PMK, SQLE, OSC and 14SR belong to the most diverse genes of the pathway, while IDI1 and 14DM are the most conserved genes in all species. The only exceptions are the NSDHL and 8SI, which belong, respectively, to the less and most conserved genes only in African trypanosomes, and SC22D, which belongs to the group of highly conserved genes in Leishmania but to the group of less conserved genes in T. cruzi. Altogether, these data show that although there is a general high apparent purifying selection over the SBP of trypanosomatids (all dN/dS values are <<1), there are differences in the restrictions imposed to diversification in each group of organisms.
Discussion
The main goal of our work was to study the genetic diversity present in the isoprenoid and sterol biosynthesis pathway (SBP) of T. cruzi. These pathways are validated chemotherapeutic targets for Chagas Disease, with a number of compounds currently undergoing clinical trials. Because the T. cruzi SBP pathway appeared to be incomplete in metabolic pathway databases such as KEGG when we started this work, and because the annotation of the SBP genes was also incomplete, we had to perform a small-scale bioinformatics analysis to fill in the gaps in available sequence and annotation. This task was performed primarily based on the well studied S. cerevisiae ergosterol biosynthesis pathway. As a result of this strategy, the majority of the genes of the pathway have now been identified, with the exception of the orthologs of the yeast gene ERG27, which encodes a 3-keto sterol reductase. As mentioned, the gene(s) responsible for this activity in land plants and sterol synthesizing bacteria have not been identified yet [43]. It is therefore highly likely that the trypanosomatid 3-keto sterol reductase is phylogenetically closer to the plant enzymes, and that once this elusive gene is identified it will be readily identified in trypanosomatids.
We selected 21 genes from this pathway to build a genetic diversity profile from representative strains of the six major evolutionary lineages of T. cruzi. For this analysis we used at least 2 strains for each evolutionary lineage therefore effectively sampling a large genetic space. Although it is certainly likely that other SNPs or fixed differences can be discovered when sequencing from new isolates, most probably these new mutations will correspond to changes that are unique to the new isolate (e.g. introduced during the clonal expansion of this particular isolate). In our experience, when expanding our re-sequencing analysis of the TcSC5D gene with 13 additional strains from lineages TcI-TcVI, only 6 new polymorphic sites were identified (see Fig S1 in [47]). Considering that this is the gene with the highest density of SNPs in the panel (82 polymorphic sites found in the present analysis), by doubling the number of strains sampled we only obtained a mild increase of sequence diversity information. Thus, we consider that in the current study we have covered a significant amount of the genetic diversity space of these pathways.
The strategy employed consisted in the generation of overlapping amplification products of approximately 750 bp (with ∼100 bp of overlap) for each gene, followed by direct sequencing of amplification products in both strands. As a result, the majority of the sequenced bases were read at least twice, with coverage in both strands. The primers were designed based on the CL-Brener genome sequence, and the majority of them were designed against the corresponding coding sequence to reduce the possibility of amplification problems (under the hypothesis that coding sequences are much less polymorphic than non-coding sequences). Moreover, when designing primers we avoided SNPs already identified from sequences in the public domain by checking against the TcSNP database. This strategy enabled the amplification and sequencing of all the selected gene fragments in strains from all the lineages, except for the first amplification product of the TcMK gene, that could not be amplified initially from lineage I. These strains carry a SNP that is specific for TcI and that mapped exactly at the 3'-end of the forward primer (see Table S5). We fixed this primer after recent genomic data was made available for a number of Tcruzi I strains (Sylvio X10 [66], JR cl4 (accessed trough TriTrypDB [67]), and TcAdriana (Westergaard G and Vazquez M, unpublished results).
Our results show that the TcSC5D gene is the most polymorphic gene in the pathway, with 11 SNPs per 100 bp (for a total of 82 SNPs in 720 bp analyzed). This SNP density is more than twofold larger than those observed for other re-sequenced genes. Despite this, the gene is under strong purification pressure, as judged by the ratio of missense and sense SNPs (or its dN/dS ratio). The Tc14DM and TcIDI1 genes had the lowest dN/dS ratios in the panel. Interestingly, the orthologs in Leishmanias and African trypanosomes also display the lowest ratios in each case (See Table 4), highlighting the level of apparent selection pressure for these enzymes in trypanosomatids. Furthermore the Tc14DM, TcHMGS and TcHMGR genes have the lowest density of missense SNPs in the panel, with only 0.51, 0.43 and 0.56 non synonymous SNPs every 100 bp respectively, so at least from these genetic evidence alone, these genes would appear to be the best candidates for drug development in T. cruzi.
The SMO-like (or SC5D-like) genes of T. cruzi and Leishmania
As described above, when looking for the T. cruzi orthologs of the ERG25 (C4-methyl oxidase) gene, we identified two homologs (two pairs of allelic variants from the hybrid genome), which are members of the Fatty Acid Hydroxylase superfamily (Pfam Domain PF04116). Based on best-reciprocal BLAST hits and careful examination we concluded that these genes (TcCLB.511339.20, TcCLB.509235.20) are not orthologous to ERG25, and are probably divergent homologs of a different ancestral gene. Our phylogenetic reconstruction suggests that Leishmanias have retained the ancestral ortholog of ERG25, although the apparent selection acting on this gene is more relaxed than that observed for other genes in the pathway (see Table 4). In any case, based on this analysis it is tempting to speculate that the group of genes we call SMO-like, which are present in all trypanosomatid species analyzed in this work, are the ones that are responsible for the essential C4-demethylation step.
Interestingly, when performing BLASTP searches of SMO-like proteins against fungal genomes, we noticed a number of SUR2 (Sphinganine C4-hydroxylase) homologs among the significant hits (see Table S1). In yeast, SUR2 (also a member of the FA hydroxylase superfamily) catalyzes the conversion of sphinganine to phytosphingosine in sphingolipid biosynthesis. Recently, the presence of phytosphingosine in trypanosomes was demonstrated by mass spectrometry [68], and the authors also show that the biosynthesis of phytosphingosine is driven by bi-functional hydroxylase/desaturase enzymes. However, the trypanosomatid genes they identified as responsible for this activity are not orthologs of yeast SUR2 (see Figure 2 in Vacchina et al. [68]). In our reciprocal BLAST searches using the yeast SUR2 protein as query against trypanosomatid genomes, we always retrieve the same set of ERG3 orthologs, and SMO-like genes. Overall these data suggest that in trypanosomes the orthologs of both SUR2 and ERG25 have been lost, and that the SMO-like genes grouped in the middle branch in Figure 1 could represent an ancestral hydroxylase/desaturase that has adjusted (or is still adjusting) to a new functional niche in these organisms (cellular localization or time/stage of expression, etc). Interestingly the expression of this gene is higher in amastigotes and trypomastigotes, the two life cycle stages that occur in the mammalian host (data from Minning et al. [69]).
The protein sequences encoded by these genes show the three canonical conserved histidine boxes (HxxxH, HxxHH, and HxxHH) present in all fatty acid hydroxylase family members. The distribution of the accumulated changes is shown in Figure S1. None of the non-synonymous changes affect these highly conserved motifs, and at least a third of these (depending on the membrane topology prediction) are predicted to be exposed (not embedded in the membrane). This is important because, as reviewed in [70], the evolution of new regioselectivities in these enzymes would not involve the active site, but adjacent sequences. However, the failure to predict a reasonable topology (see Fig S1) points to the need to do an in-depth study of the membrane topology of this protein.
The use of dN/dS ratios
Although appealing because it normalizes substitutions over silent vs non-silent sites, the dN/dS ratio is extremely sensitive to violation of a number of assumptions, such as the need to use data from divergent lineages, and is also affected by differences in the time since divergence and/or effective population sizes of compared species, among others [71]. This has been noticed in many studies, particularly in the case of bacterial species (even those with highly clonal populations) [72]. As shown by these and other authors, the dN/dS ratio is sometimes unstable, displaying a time dependency that makes it difficult to compare genomes at differing levels of divergence [73].
In preliminary studies with genes from these pathways, we came across a number of puzzling observations: some highly conserved genes in T. brucei showed unusually high (>>1) dN/dS values. This was due to the restricted number of genomes analyzed (3), which resulted in a low SNP frequency. Therefore the dN/dS ratio calculation did not provide enough depth for a reliable analysis as most genes had very few (or none) synonymous substitutions. This effect disappeared when adding sequences from other african trypanosomes (more distant lineages), which prompted us to investigate dN/dS trajectories.
As shown in the Figure S2, for pairwise comparisons where the number of polymorphisms is low, the dN/dS ratios are unstable and tend to display large values particularly for comparisons between highly conserved pairs of sequences. This should not be interpreted as an apparent diversifying selection, as these dN/dS values tend to (paradoxically) stabilize when more divergent sequences are compared. These results are in line with previous observations, and point to the use of care when analyzing dN/dS values. As shown by others [71]–[73] the use of dN/dS should be restricted to sequences from divergent lineages (with no sexual exchange among them). However, in the case of trypanosomatids this is difficult to enforce. Both within the T. cruzi and Leishmania lineages there are natural hybrids [7], [8], [49], [74], [75], which suggests that, although infrequent, genetic exchange have occurred in the wild. Therefore, the possibility of recombination between the parental alleles in these hybrids cannot be excluded. This is also the case for T. brucei, which has an extant capacity for both clonal and sexual propagation with varying degrees of inbreeding or out-crossing, with T. brucei gambiense showing a strict clonality [76], whereas different subpopulations of T. b. rhodesiense show both clonality and epidemic or close to panmictic behavior [77].
For all these reasons we believe that extreme care should be used when interpreting the meaning of isolated dN/dS values (e.g. in Table 4), or the meaning of comparisons of dN/dS across taxa in the absence of the expected trajectories of dN/dS over time (because of the critical effect of time since divergence of the lineages). This makes it difficult to answer simple questions such as whether the C-3 sterol dehydrogenase gene (ERG26 ortholog) is under the same apparent selection in T. cruzi, the African trypanosomes and Leishmanias. The Table 4 shows that the dN/dS values for this gene are similar in all cases (0.058 in T. cruzi, 0.057 in Leishmania, and 0.051 in African tryps). However, these genes differ wildly in their SNP densities (see Table S3), which makes it difficult to provide a simple answer.
Conclusions
In this study we have explored the genetic diversity present in an important pathway of T. cruzi, which is the current target of a number of drugs undergoing clinical trials. Our analysis show that the targets of current drugs are highly conserved across all evolutionary lineages. We have also filled a number of holes in the pathway by completing the sequences of a number of genes that were missing or truncated in the current reference genome. Finally by comparing genes across other important trypanosomatids, we show than in spite of differences in diversity, all trypanosomes show a mostly conserved set of enzymes.
Materials and Methods
T. cruzi stocks and strains
Strains used in this study (and the corresponding current lineage classification) were: Sylvio X10 cl1 and Dm28 (TcI); MAS1 cl1, TU18 cl93, IVV cl4 (TcII); M6241 cl6, M5631 cl5 and X109/2 (TcIII); CanIII cl1, Dog Theis and 92122102R (TcIV); Sc43 cl9 and MN cl2 (TcV); Tulahuen cl2, CL Brener and P63 cl1 (TcVI).
Oligonucleotides and gene identifiers
For each selected gene, a number of primers were designed for PCR-amplification. Using information from the TcSNP resource, we selected primers to avoid known polymorphic bases. Taking into account that in the direct Sanger sequencing of PCR products the chromatogram quality is optimal in the range from 50 to 700 bp, a desirable length of the amplification products was set around 750 bp. This length would also maximize our ability to sequence both strands of the amplification product, with good quality. Depending on the size of each selected gene, one or more overlapping amplification products were obtained. The list of the designed primers for each gene and the size of the corresponding amplification product is shown in Table S5. In only one case we had to design a separate primer (Tc-Mev-kinase26-TcI-fw) to amplify a fragment in one lineage (TcI) because there was a SNP at the 3' end of the primer that was absent in the release 1 of the TcSNP database.
Amplification and sequencing
Selected fragments were amplified by PCR using Taq polymerase (Invitrogen) in a Biometra T Professional Gradient 96 cycler. Amplification mixtures contained 10 pmol of each primer, PCR buffer (Invitrogen), 1.5 mM MgCl2, 50 ng of genomic DNA, 200 µM dNTPs, 2.5 U Taq polymerase (Invitrogen), and water to a final volume of 25 µl. After denaturing at 94°C for 2 minutes, thermal cycling was performed for 35 cycles at 94°C for 30 seconds, followed by 30 seconds at a temperature set to 5°C less than the melting temperature of the selected primers, followed by 72°C for 30 seconds. Reactions were finished by a 5 minute incubation at 72°C. Amplification products were checked in 1.2% agarose gels stained with ethidium bromide to verify the presence of a single amplification product. Next, an aliquot (10 µl) of the amplification reaction was treated with 1 U of Exonuclease I (Fermentas) and 10 U of Shrimp Alkaline Phosphatase (Fermentas) for 45 minutes at 37°C and then for 30 minutes at 80°C to inactivate these enzymes. Subsequently two sequencing reactions were prepared, each with one of the primers used for the amplification of the product. Sequencing was carried out in an Applied Biosystems 3130 capillary sequencer using a Big-Dye terminator cycle sequencing kit, according to the instructions of the manufacturer.
SNP identification and scoring
Gene fragments were PCR-amplified from every strain of the panel and sequenced in both strands. Base calling of chromatograms, assembly of sequences, detection of polymorphisms and manual inspection of assembled sequences and polymorphisms was done using a software package composed of Phred (version 0.020425.c) [78], [79], Phrap (version 0.9909329), Polyphred (version 5.04) [80] and Consed (version 15.0) [81]. Basecalling of chromatograms was done by Phred. Sequences were then assembled by Phrap. Polyphred was used to process phrap assemblies to detect polymorphic sites. All candidate SNPs identified by PolyPhred (score >70/99), including heterozygous peaks, were visualized with Consed. A few false positives, and false negatives were removed/added after this manual inspection.
Other sequences used in this study
For the comparative analysis of SBP genes in kinetoplastid genomes, we have obtained the corresponding protein sequences for each of the yeast and/or T. cruzi orthologs used in this study from the TriTrypDB database [67]. A BLAST search using the corresponding SBP gene as query was used to identify the corresponding ortholog. This has been cross-checked by inspection of ortholog clusters at the OrthoMCL database [41]. The sequences used belong to the following species/strains: T. brucei brucei strains TREU927/4 [82] and Lister 427 (George Cross, unpublished), Trypanosoma brucei gambiense [83]; Trypanosoma congolense and Trypanosoma vivax [84]; Trypanosoma evansi (STIB 805) (unpublished); L. major [85], L. infantum [86], L. braziliensis [86], and L. mexicana [87].
Alignment and calculation of dN/dS
For calculation of dN/dS ratios (ω) nucleotide sequences were first trimmed so that they all start and end at the corresponding START and STOP codons, and/or they have a similar length that is also a multiple of 3. Nucleotide alignments for each gene were then obtained using TranslatorX, which produces a codon alignment guided by aminoacid information [88]. Using these alignments as input, together with an unrooted phylogenetic tree with 3 branches containing T. cruzi lineages, Leishmania species and African trypanosome species, we ran the codeml program from the PAML package [89] with parameters “model = 0; NSSites = 0” to obtain optimized lengths for these branches by maximum likelihood. Finally, using these optimized branch lengths, we calculated the dN/dS ratio (ω) for each of the 3 branches by running again codeml with the following parameters:“model = 2; NSSites = 0”.
Mapping substitutions on three dimensional structures
Crystallographic structures of enzymes of the sterol biosynthetic pathway were obtained from the Protein Data Bank (PDB, http://www.rcsb.org). Using the molecular graphics viewer VMD (version 2.8.6) [90] together with the multiple sequence alignment plugin for VMD [91] we mapped sequences from different strains on top of the reference sequence from which the structure was obtained. Atomic distances from each residue to the co-crystallized ligand were measured using standard tools implemented in VMD. Structures used in this work (and their source organism, and T. cruzi homolog) were: 1wl5 (human, TcACAT) [92]; 2fa3 (Brassica juncea, TcHMGS) [93]; 3bgl (human, TcHMGR) [94]; 2hfu (L. major, TcMK) [59]; 2hke (T. brucei, TcMVD)[95]; 1yhm (T. cruzi, TcFPPS) [96]; 1ezf (human, TcSQS) [97]; 1w6k (human, TcOSC) [98]; 3khm, 3k1o, 3ksw (T. cruzi, TcCYP51) [99].
Data deposition
The sequences reported in this paper have been deposited in the GenBank database under the following accession numbers: JN050313-JN050853, HQ586972-HQ586973, and KF290395-KF290460. Heterozygous sequence polymorphisms have been submitted as ambiguities in the sequences using standard IUPAC notation. Sequence polymorphisms identified between different strains/clones are available as supplementary material, and will be also available in a future release of the TcSNP database (http://snps.tcruzi.org, [46]).
Supporting Information
Distribution of observed SNPs in the TcSMO-like genes of T. cruzi . Based on the prediction of trans-membrane spanning domains (see TMHMM probability plot at the bottom), we created two alternative representations, following [70]. The distribution of synonymous and non-synonymous SNPs is shown according to these models. The representations differ in the presence/absence of the second (non-predicted) trans-membrane domain. In these two representations the location of the 3rd histidine box always lies on the opposite side of the membrane. Both topologies may be wrong and an in-depth study may be required to establish the correct topology of these proteins.
(PDF)
Stability of the dN/dS ratio for the trypanosomatid isoprenoid and ergosterol biosynthesis genes. The dN/dS ratio was calculated for all possible pairwise comparisons of each gene within each phylogenetic group/branch (e.g. the genes for all lanosterol demethylases were aligned with each other within the T. cruzi group, the Leishmania group and the T. brucei/African trypanosomes group). The plot therefore summarizes data from all genes in the pathways, and for all species analyzed. The dN/dS values were then sorted, grouped in bins of 10 and plotted. The error bars show the standard deviation within each bin.
(PDF)
Reciprocal BLASTP searches between fungal and kinetoplastid ERG25/ERG3 homologs. The file contains a summary of BLASTP searches against fungal and kinetoplastid protein databases. BLAST searches using the yeast ERG3/ERG25 protein sequences as query, were run at the TriTrypDB BLAST server against a database of Kinetoplastid proteomes from reference and draft genomes. BLAST searches using a number of putative T. cruzi orthologs of these yeast genes were run at the SGD Fungal BLAST Server, against a database containing a selection of fungal genomes. Each BLASTP search is shown in a separate tab in the Excel workbook.
(XLS)
List of nucleotide changes (SNPs, fixed differences) identified for each gene analyzed. The excel file contains one spreadsheet per gene with information on the location of each SNP relative to the start codon, the PolyPhred score for the SNP, and the character state of the SNP in each strain/lineage.
(XLS)
Comparative genetic diversity of kinetoplastid SBP genes.
(XLS)
Presence of two types of thiolases in T. cruzi . The file contains the summary of results from reciprocal BLASTP searches between fungal and kinetoplastid ERG10/POT1 homologs.
(XLS)
List of oligonucleotide primers and amplification products analyzed in this study. The table lists the oligonucleotide primers used to amplify each PCR product, the corresponding product size, and the number of products per gene/locus.
(XLS)
Acknowledgments
We would like to thank Patricio Diosque (Universidad Nacional de Salta, Argentina) for the generous gift of T. cruzi genomic DNA of different reference strains and isolates. We would also like to thank Antonio Uttaro (Universidad Nacional de Rosario, Argentina), Terry Smith (University of St Andrews, Scotland, UK), and Patricio Diosque for critical reading of the manuscript, and Francisco Pisciottano (Universidad de Buenos Aires) for helpful advice with the dN/dS analysis.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All sequences described are available from GenBank under accession numbers JN050313-JN050853, HQ586972-HQ586973, and KF290395-KF290460.
Funding Statement
This work was supported by grants from the National Agency for the Promotion of Science and Technology of Argentina (Grants PICT-2005-38209 and PICT-2010-1479, http://www.agencia.gob.ar), the National Research Council of Argentina (CONICET, Grant PIP-5454, http://www.conicet.gob.ar), and the Special Programme for Research and Training in Tropical Diseases (WHO / UNDP / UNICEF / World Bank, Grant A50830, http://www.who.int/tdr). ROC is now a fellow of CONICET and was also supported during this work by a fellowship from the University of San Martín. FA is a member of the Research Career of CONICET. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rassi AJ, Rassi A, Marin Neto JA (2010) Chagas disease. Lancet 375: 1388–1402. [DOI] [PubMed] [Google Scholar]
- 2. Tibayrenc M, Ayala FJ (2002) The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol 18: 405–410. [DOI] [PubMed] [Google Scholar]
- 3. Tibayrenc M, Ayala FJ (2013) How clonal are Trypanosoma and Leishmania? Trends Parasitol 29: 264–269. [DOI] [PubMed] [Google Scholar]
- 4. Barnabé C, Brisse S, Tibayrenc M (2000) Population structure and genetic typing of Trypanosoma cruzi, the agent of Chagas disease: a multilocus enzyme electrophoresis approach. Parasitology 120 (Pt 5): 513–526. [DOI] [PubMed] [Google Scholar]
- 5. Brisse S, Barnabé C, Tibayrenc M (2000) Identification of six Trypanosoma cruzi phylogenetic lineages by random amplified polymorphic DNA and multilocus enzyme electrophoresis. Int J Parasitol 30: 35–44. [DOI] [PubMed] [Google Scholar]
- 6. Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, et al. (2012) The revised Try-panosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol 12: 240–253. [DOI] [PubMed] [Google Scholar]
- 7. Machado CA, Ayala FJ (2001) Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc Natl Acad Sci U S A 98: 7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Westenberger SJ, Barnabé C, Campbell DA, Sturm NR (2005) Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 171: 527–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flores López CA, Machado CA (2011) Analyses of 32 loci clarify phylogenetic relationships among Trypanosoma cruzi lineages and support a single hybridization prior to human contact. PLoS Negl Trop Dis 5: e1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lewis MD, Llewellyn MS, Yeo M, Acosta N, Gaunt MW, et al. (2011) Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids. PLoS Negl Trop Dis 5: e1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buscaglia CA, Di Noia JM (2003) Trypanosoma cruzi clonal diversity and the epidemiology of Chagas' disease. Microbes Infect 5: 419–427. [DOI] [PubMed] [Google Scholar]
- 12. Vago AR, Andrade LO, Leite AA, d'Avila Reis D, Macedo AM, et al. (2000) Genetic characterization of Trypanosoma cruzi directly from tissues of patients with chronic Chagas disease: differential distribution of genetic types into diverse organs. Am J Pathol 156: 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andrade LO, Machado CR, Chiari E, Pena SD, Macedo AM (1999) Differential tissue distribution of diverse clones of Trypanosoma cruzi in infected mice. Mol Biochem Parasitol 100: 163–172. [DOI] [PubMed] [Google Scholar]
- 14. Andrade LO, Galvão LMC, Meirelles MdNSL, Chiari E, Pena SDJ, et al. (2010) Differential tissue tropism of Trypanosoma cruzi strains: an in vitro study. Mem Inst Oswaldo Cruz 105: 834–837. [DOI] [PubMed] [Google Scholar]
- 15. Smith TK, Bütikofer P (2010) Lipid metabolism in Trypanosoma brucei. Mol Biochem Parasitol 172: 66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Urbina JA (2009) Ergosterol biosynthesis and drug development for Chagas disease. Mem Inst Oswaldo Cruz 104 Suppl 1311–318. [DOI] [PubMed] [Google Scholar]
- 17. Kessler RL, Soares MJ, Probst CM, Krieger MA (2013) Trypanosoma cruzi response to sterol biosynthesis inhibitors: morphophysiological alterations leading to cell death. PLoS One 8: e55497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Urbina JA, Payares G, Molina J, Sanoja C, Liendo A, et al. (1996) Cure of short- and long-term experimental Chagas' disease using D0870. Science 273: 969–971. [DOI] [PubMed] [Google Scholar]
- 19. Buckner FS, Joubert BM, Boyle SM, Eastman RT, Verlinde CLMJ, et al. (2003) Cloning and analysis of Trypanosoma cruzi lanosterol 14α-demethylase. Mol Biochem Parasitol 132: 75–81. [DOI] [PubMed] [Google Scholar]
- 20. Silva DT, de Meirelles MdNSL, Almeida D, Urbina JA, Pereira MCS (2006) Cytoskeleton reassembly in cardiomyocytes infected by Trypanosoma cruzi is triggered by treatment with ergosterol biosynthesis inhibitors. Int J Antimicrob Agents 27: 530–537. [DOI] [PubMed] [Google Scholar]
- 21. Lepesheva GI, Villalta F, Waterman MR (2011) Targeting Trypanosoma cruzi sterol 14α-demethylase (CYP51). Adv Parasitol 75: 65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buckner FS, Urbina JA (2012) Recent Developments in Sterol 14-demethylase Inhibitors for Chagas Disease. Int J Parasitol Drugs Drug Resist 2: 236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Florin Christensen M, Florin Christensen J, Garin C, Isola E, Brenner RR, et al. (1990) Inhibition of Trypanosoma cruzi growth and sterol biosynthesis by lovastatin. Biochem Biophys Res Commun 166: 1441–1445. [DOI] [PubMed] [Google Scholar]
- 24. Urbina JA, Lazardi K, Marchan E, Visbal G, Aguirre T, et al. (1993) Mevinolin (lovastatin) potentiates the antiproliferative effects of ketoconazole and terbinafine against Trypanosoma (Schizotrypanum) cruzi: in vitro and in vivo studies. Antimicrob Agents Chemother 37: 580–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hurtado-Guerrrero R, Peña-Díaz J, Montalvetti A, Ruiz-Pérez LM, González-Pacanowska D (2002) Kinetic properties and inhibition of trypanosoma cruzi 3-hydroxy-3-methylglutaryl coa reductase. FEBS Lett 510: 141–144. [DOI] [PubMed] [Google Scholar]
- 26. Rosso VS, Szajnman SH, Malayil L, Galizzi M, Moreno SNJ, et al. (2011) Synthesis and biological evaluation of new 2-alkylaminoethyl-1,1-bisphosphonic acids against Trypanosoma cruzi and Toxoplasma gondii targeting farnesyl diphosphate synthase. Bioorg Med Chem 19: 2211–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sealey-Cardona M, Cammerer S, Jones S, Ruiz-Pérez LM, Brun R, et al. (2007) Kinetic characterization of squalene synthase from trypanosoma cruzi: selective inhibition by quinuclidine derivatives. Antimicrob Agents Chemother 51: 2123–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodrígues Poveda CA, González Pacanowska D, Szajnman SH, Rodríguez JB (2012) 2-alkylaminoethyl-1,1-bisphosphonic acids are potent inhibitors of the enzymatic activity of Trypanosoma cruzi squalene synthase. Antimicrob Agents Chemother 56: 4483–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gerpe A, Odreman Nuñez I, Draper P, Boiani L, Urbina JA, et al. (2008) Heteroallyl-containing 5-nitrofuranes as new anti-Trypanosoma cruzi agents with a dual mechanism of action. Bioorg Med Chem 16: 569–577. [DOI] [PubMed] [Google Scholar]
- 30. Gerpe A, Alvarez G, Benítez D, Boiani L, Quiroga M, et al. (2009) 5-Nitrofuranes and 5-nitrothiophenes with anti-Trypanosoma cruzi activity and ability to accumulate squalene. Bioorg Med Chem 17: 7500–7509. [DOI] [PubMed] [Google Scholar]
- 31. Buckner FS, Griffin JH, Wilson AJ, Van Voorhis WC (2001) Potent anti-Trypanosoma cruzi activities of oxidosqualene cyclase inhibitors. Antimicrob Agents Chemother 45: 1210–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oliaro-Bosso S, Viola F, Matsuda S, Cravotto G, Tagliapietra S, et al. (2004) Umbelliferone aminoalkyl derivatives as inhibitors of oxidosqualene cyclases from saccharomyces cerevisiae, trypanosoma cruzi, and pneumocystis carinii. Lipids 39: 1007–1012. [DOI] [PubMed] [Google Scholar]
- 33. Oliaro Bosso S, Viola F, Taramino S, Tagliapietra S, Barge A, et al. (2007) Inhibitory effect of umbelliferone aminoalkyl derivatives on oxidosqualene cyclases from S. cerevisiae, T. cruzi, P. carinii, H. sapiens, and A. thaliana: a structure-activity study. ChemMedChem 2: 226–233. [DOI] [PubMed] [Google Scholar]
- 34. Magaraci F, Jimenez Jimenez C, Rodrigues C, Rodrigues JCF, Vianna Braga M, et al. (2003) Azasterols as inhibitors of sterol 24-methyltransferase in Leishmania species and Trypanosoma cruzi. J Med Chem 46: 4714–4727. [DOI] [PubMed] [Google Scholar]
- 35. Lorente SO, Rodrigues JCF, Jiménez Jiménez C, Joyce Menekse M, Rodrigues C, et al. (2004) Novel azasterols as potential agents for treatment of leishmaniasis and trypanosomiasis. Antimicrob Agents Chemother 48: 2937–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Braga MV, Magaraci F, Lorente SO, Gilbert I, de Souza W (2005) Effects of inhibitors of Delta24(25)-sterol methyl transferase on the ultrastructure of epimastigotes of Trypanosoma cruzi. Microsc Microanal 11: 506–515. [DOI] [PubMed] [Google Scholar]
- 37. Urbina JA, Docampo R (2003) Specific chemotherapy of Chagas disease: controversies and advances. Trends Parasitol 19: 495–501. [DOI] [PubMed] [Google Scholar]
- 38. Lupetti A, Danesi R, Campa M, Del Tacca M, Kelly S (2002) Molecular basis of resistance to azole antifungals. Trends Mol Med 8: 76–81. [DOI] [PubMed] [Google Scholar]
- 39. Morio F, Loge C, Besse B, Hennequin C, Le Pape P (2010) Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn Microbiol Infect Dis 66: 373–384. [DOI] [PubMed] [Google Scholar]
- 40. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen F, Mackey AJ, Stoeckert CJ Jr, Roos DS (2006) OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34: D363–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bouvier F, Rahier A, Camara B (2005) Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res 44: 357–429. [DOI] [PubMed] [Google Scholar]
- 43. Desmond E, Gribaldo S (2009) Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol Evol 1: 364–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolf YI, Koonin EV (2012) A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes. Genome Biol Evol 4: 1286–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koonin EV (2005) Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet 39: 309–338. [DOI] [PubMed] [Google Scholar]
- 46. Ackermann AA, Carmona SJ, Agüero F (2009) TcSNP: a database of genetic variation in Trypanosoma cruzi. Nucleic Acids Res 37: D544–D549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cosentino RO, Agüero F (2012) A simple strain typing assay for Trypanosoma cruzi: discrimination of major evolutionary lineages from a single amplification product. PLoS Negl Trop Dis 6: e1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brisse S, Verhoef J, Tibayrenc M (2001) Characterisation of large and small subunit rRNA and mini-exon genes further supports the distinction of six Trypanosoma cruzi lineages. Int J Parasitol 31: 1218–1226. [DOI] [PubMed] [Google Scholar]
- 49. Brisse S, Henriksson J, Barnabé C, Douzery EJP, Berkvens D, et al. (2003) Evidence for genetic exchange and hybridization in Trypanosoma cruzi based on nucleotide sequences and molecular karyotype. Infect Genet Evol 2: 173–183. [DOI] [PubMed] [Google Scholar]
- 50. de Freitas JM, Augusto Pinto L, Pimenta JR, Bastos Rodrigues L, Gonçalves VF, et al. (2006) Ancestral genomes, sex, and the population structure of Trypanosoma cruzi. PLoS Pathog 2: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marcili A, Lima L, Valente VC, Valente SA, Batista JS, et al. (2009) Comparative phylogeography of Trypanosoma cruzi TCIIc: new hosts, association with terrestrial ecotopes, and spatial clustering. Infect Genet Evol 9: 1265–1274. [DOI] [PubMed] [Google Scholar]
- 52. Lewis MD, Ma J, Yeo M, Carrasco HJ, Llewellyn MS, et al. (2009) Genotyping of Trypanosoma cruzi: systematic selection of assays allowing rapid and accurate discrimination of all known lineages. Am J Trop Med Hyg 81: 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yeo M, Mauricio IL, Messenger LA, Lewis MD, Llewellyn MS, et al. (2011) Multilocus sequence typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi. PLoS Negl Trop Dis 5: e1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lauthier JJ, Tomasini N, Barnabé C, Monje Rumi MM, Alberti D'Amato AM, et al. (2012) Candidate targets for Multilocus Sequence Typing of Trypanosoma cruzi: Validation using parasite stocks from the Chaco Region and a set of reference strains. Infect Genet Evol 12: 350–358. [DOI] [PubMed] [Google Scholar]
- 55. Chen CK, Leung SSF, Guilbert C, Jacobson MP, McKerrow JH, et al. (2010) Structural characterization of CYP51 from Trypanosoma cruzi and Trypanosoma brucei bound to the antifungal drugs posaconazole and uconazole. PLoS Negl Trop Dis 4: e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kelly SL, Lamb DC, Kelly DE (1999) Y132H substitution in Candida albicans sterol 14alphademethylase confers uconazole resistance by preventing binding to haem. FEMS Microbiol Lett 180: 171–175. [DOI] [PubMed] [Google Scholar]
- 57. Délye C, Laigret F, Corio Costet MF (1997) A mutation in the 14 alpha-demethylase gene of Uncinula necator that correlates with resistance to a sterol biosynthesis inhibitor. Appl Environ Microbiol 63: 2966–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lepesheva GI, Park HW, Hargrove TY, Vanhollebeke B, Wawrzak Z, et al. (2010) Crystal structures of Trypanosoma brucei sterol 14alpha-demethylase and implications for selective treatment of human infections. J Biol Chem 285: 1773–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sgraja T, Smith TK, Hunter WN (2007) Structure, substrate recognition and reactivity of Leishmania major mevalonate kinase. BMC Struct Biol 7: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Houten SM, Waterham HR (2001) Nonorthologous gene displacement of phosphomevalonate 665 kinase. Mol Genet Metab 72: 273–276. [DOI] [PubMed] [Google Scholar]
- 61. Chang Q, Yan XX, Gu SY, Liu JF, Liang DC (2008) Crystal structure of human phosphomevalonate kinase at 1.8 A resolution. Proteins 73: 254–258. [DOI] [PubMed] [Google Scholar]
- 62. Roberts CW, McLeod R, Rice DW, Ginger M, Chance ML, et al. (2003) Fatty acid and sterol metabolism: potential antimicrobial targets in apicomplexan and trypanosomatid parasitic protozoa. Mol Biochem Parasitol 126: 129–142. [DOI] [PubMed] [Google Scholar]
- 63. Coppens I, Courtoy PJ (2000) The adaptative mechanisms of Trypanosoma brucei for sterol homeostasis in its different life-cycle environments. Annu Rev Microbiol 54: 129–156. [DOI] [PubMed] [Google Scholar]
- 64. Liendo A, Visbal G, Piras MM, Piras R, Urbina JA (1999) Sterol composition and biosynthesis in Trypanosoma cruzi amastigotes. Mol Biochem Parasitol 104: 81–91. [DOI] [PubMed] [Google Scholar]
- 65. Nes CR, Singha UK, Liu J, Ganapathy K, Villalta F, et al. (2012) Novel sterol metabolic network of Trypanosoma brucei procyclic and bloodstream forms. Biochem J 443: 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Franzén O, Ochaya S, Sherwood E, Lewis MD, Llewellyn MS, et al. (2011) Shotgun sequencing analysis of Trypanosoma cruzi I Sylvio X10/1 and comparison with T. cruzi VI CL Brener. PLoS Negl Trop Dis 5: e984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, et al. (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38: D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vacchina P, Tripodi KEJ, Escalante AM, Uttaro AD (2012) Characterization of bifunctional sphingolipid Δ4-desaturases/C4-hydroxylases of trypanosomatids by liquid chromatographyelectrospray tandem mass spectrometry. Mol Biochem Parasitol 184: 29–38. [DOI] [PubMed] [Google Scholar]
- 69. Minning TA, Weatherly DB, Atwood J, Orlando R, Tarleton RL (2009) The steady-state transcriptome of the four major life-cycle stages of Trypanosoma cruzi. BMC Genomics 10: 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sperling P, Ternes P, Zank TK, Heinz E (2003) The evolution of desaturases. Prostaglandins Leukot Essent Fatty Acids 68: 73–95. [DOI] [PubMed] [Google Scholar]
- 71. Kryazhimskiy S, Plotkin JB (2008) The population genetics of dN/dS. PLoS Genet 4: e1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Castillo Ramírez S, Harris SR, Holden MTG, He M, Parkhill J, et al. (2011) The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog 7: e1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rocha EPC, Smith JM, Hurst LD, Holden MTG, Cooper JE, et al. (2006) Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol 239: 226–235. [DOI] [PubMed] [Google Scholar]
- 74. Ravel C, Cortes S, Pratlong F, Morio F, Dedet JP, et al. (2006) First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major. Int J Parasitol 36: 1383–1388. [DOI] [PubMed] [Google Scholar]
- 75. Lukes J, Mauricio IL, Schönian G, Dujardin JC, Soteriadou K, et al. (2007) Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci U S A 104: 9375–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Koffi M, De Meeûs T, Bucheton B, Solano P, Camara M, et al. (2009) Population genetics of Trypanosoma brucei gambiense, the agent of sleeping sickness in Western Africa. Proc Natl Acad Sci U S A 106: 209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Duffy CW, MacLean L, Sweeney L, Cooper A, Turner MRC, et al. (2013) Population genetics of Trypanosoma brucei rhodesiense: clonality and diversity within and between foci. PLoS Negl Trop Dis 7: e2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185. [DOI] [PubMed] [Google Scholar]
- 79. Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194. [PubMed] [Google Scholar]
- 80. Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using uorescence-based resequencing. Nucleic Acids Res 25: 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gordon D (2003) Viewing and editing assembled sequences using Consed. Curr Protoc Bioinformatics Chapter 11: Unit11.2. [DOI] [PubMed] [Google Scholar]
- 82. Berriman M, Ghedin E, Hertz Fowler C, Blandin G, Renauld H, et al. (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309: 416–422. [DOI] [PubMed] [Google Scholar]
- 83. Jackson AP, Sanders M, Berry A, McQuillan J, Aslett MA, et al. (2010) The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human african trypanosomiasis. PLoS Negl Trop Dis 4: e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jackson AP, Berry A, Aslett M, Allison HC, Burton P, et al. (2012) Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. Proc Natl Acad Sci U S A 109: 3416–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, et al. (2005) The genome of the kinetoplastid parasite, Leishmania major. Science 309: 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, et al. (2007) Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, et al. (2011) Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res 21: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Abascal F, Zardoya R, Telford MJ (2010) TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38: W7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Bielawski JP, Yang Z (2004) A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol 59: 121–132. [DOI] [PubMed] [Google Scholar]
- 90.Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14: 33–8, 27–8. [DOI] [PubMed]
- 91. Eargle J, Wright D, Luthey Schulten Z (2006) Multiple Alignment of protein structures and sequences for VMD. Bioinformatics 22: 504–506. [DOI] [PubMed] [Google Scholar]
- 92. Kursula P, Sikkilä H, Fukao T, Kondo N, Wierenga RK (2005) High resolution crystal structures of human cytosolic thiolase (CT): a comparison of the active sites of human CT, bacterial thiolase, and bacterial KAS I. J Mol Biol. 347: 189–201. [DOI] [PubMed] [Google Scholar]
- 93. Pojer F, Ferrer JL, Richard SB, Nagegowda DA, Chye ML, et al. (2006) 743 Structural basis for the design of potent and species-specific inhibitors of 3-hydroxy-3-methylglutaryl CoA synthases. Proc Natl Acad Sci U S A 103: 11491–11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Park WKC, Kennedy RM, Larsen SD, Miller S, Roth BD, et al. (2008) Hepatoselectivity of statins: design and synthesis of 4-sulfamoyl pyrroles as HMG-CoA reductase inhibitors. Bioorg Med Chem Lett 18: 1151–1156. [DOI] [PubMed] [Google Scholar]
- 95. Byres E, Alphey MS, Smith TK, Hunter WN (2007) Crystal structures of Trypanosoma brucei and Staphylococcus aureus mevalonate diphosphate decarboxylase inform on the determinants of specificity and reactivity. J Mol Biol 371: 540–553. [DOI] [PubMed] [Google Scholar]
- 96. Gabelli SB, McLellan JS, Montalvetti A, Oldfield E, Docampo R, et al. (2006) Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: implications for drug design. Proteins 62: 80–88. [DOI] [PubMed] [Google Scholar]
- 97. Pandit J, Danley DE, Schulte GK, Mazzalupo S, Pauly TA, et al. (2000) Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis. J Biol Chem 275: 30610–30617. [DOI] [PubMed] [Google Scholar]
- 98. Thoma R, Schulz Gasch T, D'Arcy B, Benz J, Aebi J, et al. (2004) Insight into steroid scaffold formation from the structure of human oxidosqualene cyclase. Nature 432: 118–122. [DOI] [PubMed] [Google Scholar]
- 99. Lepesheva GI, Hargrove TY, Anderson S, Kleshchenko Y, Furtak V, et al. (2010) Structural insights into inhibition of sterol 14alpha-demethylase in the human pathogen Trypanosoma cruzi. J Biol Chem 285: 25582–25590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lepesheva GI, Zaitseva NG, Nes WD, Zhou W, Arase M, et al. (2006) CYP51 from Trypanosoma cruzi: a phyla-specific residue in the B' helix defines substrate preferences of sterol 14alphademethylase. J Biol Chem 281: 3577–3585. [DOI] [PubMed] [Google Scholar]
- 101. Lepesheva GI, Waterman MR (2011) Structural basis for conservation in the CYP51 family. Biochim Biophys Acta 1814: 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of observed SNPs in the TcSMO-like genes of T. cruzi . Based on the prediction of trans-membrane spanning domains (see TMHMM probability plot at the bottom), we created two alternative representations, following [70]. The distribution of synonymous and non-synonymous SNPs is shown according to these models. The representations differ in the presence/absence of the second (non-predicted) trans-membrane domain. In these two representations the location of the 3rd histidine box always lies on the opposite side of the membrane. Both topologies may be wrong and an in-depth study may be required to establish the correct topology of these proteins.
(PDF)
Stability of the dN/dS ratio for the trypanosomatid isoprenoid and ergosterol biosynthesis genes. The dN/dS ratio was calculated for all possible pairwise comparisons of each gene within each phylogenetic group/branch (e.g. the genes for all lanosterol demethylases were aligned with each other within the T. cruzi group, the Leishmania group and the T. brucei/African trypanosomes group). The plot therefore summarizes data from all genes in the pathways, and for all species analyzed. The dN/dS values were then sorted, grouped in bins of 10 and plotted. The error bars show the standard deviation within each bin.
(PDF)
Reciprocal BLASTP searches between fungal and kinetoplastid ERG25/ERG3 homologs. The file contains a summary of BLASTP searches against fungal and kinetoplastid protein databases. BLAST searches using the yeast ERG3/ERG25 protein sequences as query, were run at the TriTrypDB BLAST server against a database of Kinetoplastid proteomes from reference and draft genomes. BLAST searches using a number of putative T. cruzi orthologs of these yeast genes were run at the SGD Fungal BLAST Server, against a database containing a selection of fungal genomes. Each BLASTP search is shown in a separate tab in the Excel workbook.
(XLS)
List of nucleotide changes (SNPs, fixed differences) identified for each gene analyzed. The excel file contains one spreadsheet per gene with information on the location of each SNP relative to the start codon, the PolyPhred score for the SNP, and the character state of the SNP in each strain/lineage.
(XLS)
Comparative genetic diversity of kinetoplastid SBP genes.
(XLS)
Presence of two types of thiolases in T. cruzi . The file contains the summary of results from reciprocal BLASTP searches between fungal and kinetoplastid ERG10/POT1 homologs.
(XLS)
List of oligonucleotide primers and amplification products analyzed in this study. The table lists the oligonucleotide primers used to amplify each PCR product, the corresponding product size, and the number of products per gene/locus.
(XLS)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All sequences described are available from GenBank under accession numbers JN050313-JN050853, HQ586972-HQ586973, and KF290395-KF290460.