Abstract
In this paper we describe the nearly complete mitochondrial genome of the leaf-cutter ant Atta laevigata, assembled using transcriptomic libraries from Sanger and Illumina next generation sequencing (NGS), and PCR products. This mitogenome was found to be very large (18,729 bp), given the presence of 30 non-coding intergenic spacers (IGS) spanning 3,808 bp. A portion of the putative control region remained unsequenced. The gene content and organization correspond to that inferred for the ancestral pancrustacea, except for two tRNA gene rearrangements that have been described previously in other ants. The IGS were highly variable in length and dispersed through the mitogenome. This pattern was also found for the other hymenopterans in particular for the monophyletic Apocrita. These spacers with unknown function may be valuable for characterizing genome evolution and distinguishing closely related species and individuals. NGS provided better coverage than Sanger sequencing, especially for tRNA and ribosomal subunit genes, thus facilitating efforts to fill in sequence gaps. The results obtained showed that data from transcriptomic libraries contain valuable information for assembling mitogenomes. The present data also provide a source of molecular markers that will be very important for improving our understanding of genomic evolutionary processes and phylogenetic relationships among hymenopterans.
Introduction
Atta laevigata Smith, 1858 (Hymenoptera: Formicidae: Attini) is a crop pest that is found throughout South America and is widely distributed in Brazil [1], [2]. The prevalence of this agricultural pest is related to its high population density [3] and long life span of the queens [4], resulting in the requirement for a large amount of fresh plant material to maintain the nest. The species cuts leaves from both monocotyledons and dicotyledons plants, including many plantations [5]–[7], as well as a wide variety of native plants from different biomes such as the Cerrado or the rainforest [8], [9]. It is easily recognized based on the very large, shiny head of the soldiers, a characteristic that has led to the popular name “cabeça de vidro” (meaning glass head) in Brazil.
In an aim to better understand the molecular bases of A. laevigata biology, physiology, behavior, and social life, and to find more specific strategies to control the pest, we recently published a partial transcriptome of this species using Sanger sequencing technology [10]. A more complete transcriptome using the Illumina platform is currently being annotated (unpublished data). Characterization of the transcriptome resulted in the retrieval of a large number of mitochondrial sequences. Although ants are highly diverse and represent an ecologically dominant group in terrestrial ecosystems [11], mitogenomes have been described and annotated for only Pristomyrmex punctatus [12] and three species of Solenopsis [13]. The mitogenome of Atta cephalotes [14] is available in GenBank (HQ415764) but annotation is missing, and the mitochondrial genome of Camponotus chromaiodes is not complete in GenBank (JX966368).
Animal mitochondrial DNA (mtDNA) has been used extensively to investigate population structures and in evolutionary and phylogenetic studies at various taxonomic levels, validating its utility as a molecular marker for systematics [15]–[17]. A growing interest in the reconstruction of phylogenetic relationships in Hymenoptera using mitochondrial genomes together with technological improvements and reduced DNA sequencing costs has led to a rapid increase in the number of sequenced mitogenomes [18]–[20].
For many years, mitogenomes were obtained by isolating mitochondria followed by DNA extraction, a procedure that is effective for large organisms but not for small organisms and some tissues [21]. To overcome this and other obstacles, long-range PCR combined with primer walking sequencing has become an alternative approach [21], [22]. More recently, next-generation sequencing (NGS) has been used to generate mtDNA data [20], [21], [23], [24], and expressed sequence tags have been useful for annotating and validating mitochondrial genomes [25].
Here, we describe the mitochondrial genome of a species from the Attini tribe, the leaf-cutter ant A. laevigata, using sequences obtained from transcriptomic libraries followed by PCR procedure to fill in sequence gaps and confirm intergenic regions.
Methods and Materials
Obtaining mitochondrial sequences from transcriptomic libraries
We retrieved mitochondrial sequences from two transcriptomic libraries of A. laevigata, each generated using a pool of soldiers from a single monogynic nest: a Sanger sequencing library (SL) [10] from ants collected in Rio Claro, SP, Brazil (W 22°23.716' and S 47°32.533'); and an Illumina platform library (IL) from ants collected in Botucatu, SP, Brazil (W 48°26.156′ and S 22°50.250′). Despite the fact the ants were collected in different locations, they belong to the same regional group (unpublished data), which is different from those groups previously described [26] based on mitochondrial haplotypes. The ants were collect with IBAMA permit SISBIO 33487-2 and do not involve endangered or protected species and protected area.
The SL data were pre-processed and assembled using the automated pipeline generation system EGene [27]. Sequences of vector (pDONR222) and primer (M13F) were trimmed and high quality sequences (base quality with phred ≥ 20) were selected and assembled into contigs and singlets using the CAP3 software [28], with an overlap percent identity cutoff “p” of 90 and a minimum overlap length cutoff “o” of 50. Functional annotation was based on BLASTX search of contig nucleotide sequences against the non-redundant protein database (nr) of NCBI, performed under the default settings of BLAST2GO [29] and the BLAST E-value of 1.0e−5 and maximum of 20 hits.
For IL, total RNA was extracted using Trizol protocol (Invitrogen). The library was constructed and sequenced at Fasteris SA, in Swiss. The total RNA quality, concentration, and integrity were determined using Qubit Analyzer (Invitrogen) and Bioanalyzer (Agilent). The paired-end library was sequenced in HiSeq 2000 in a single lane of 50 base reads. IL data were submitted to de novo assembly using VELVET [30] with the parameter kmer 43 and the contigs were filtered using BLAST search against ant mitochondrial genes.
For both libraries, contigs were manually verified to exclusion of homopolymer regions to avoid error in the inference of the genomic sequence. All mitochondrial sequences were then mapped onto the mitogenomes of Hymenoptera to generate a first draft of A. laevigata mitogenome (i.e., a mitogenome with gaps), which was used to design new primers for protein coding genes completion and amplification of intergenic regions (described below).
All sequences obtained by transcriptomic libraries and PCR were mapped into the final mitogenome sequence to access the relative cover of each technique (SL, IL, and PCR; Figure 1). For this, we used Bowtie2 [31] and SAMTools [32] and the results were visualized using IGV version 2.3.18 [33].
Figure 1. Contribution of transcriptomic libraries and PCR technique for the assembling of A. laevigata mitochondrial genome.

The figure displays the relative position of the protein coding-genes and ribosomal subunits and the contribution of Sanger library (SL – in blue), Illumina library (IL - black), and PCR fragments (PCR - green) for the final mitogenome assembling. The grey picks represent number of sequences for each codon position in different scale (values between square brackets). The figure is an adaptation of the files generated by Bowtie2 and SAMTools and visualized using IGV program.
Filling the gaps: amplifying and sequencing intergenic regions
Universal and new primers used to fill in the mitochondrial sequence gaps are shown in Table S1 and Figure S1. New primers were designed based on the obtained SL and IL sequences and mapped onto the Hymenoptera mitogenomes. Template DNA was extracted from a single soldier from the Botucatu nest (see below) according to Martins et al. [34]. The PureTaq Ready To Go kit (GE Healthcare) was used for PCR reactions, in total volume of 25 µL, containing 5 pmol of each primer, and ∼100 ng of template and included an initial denaturation of 3 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 45–58°C, and 90 s at 60°C. Amplicons were visualized in a 1% agarose gel, purified using GFX PCR DNA and Gel Band Purification Kit (GE Healthcare), quantified using a NanoDrop 2000 (Thermo Scientific), and sequenced. Amplicons that could not be directly sequenced were cloned into Escherichia coli DH10B using the CloneJET PCR Cloning Kit (Fermentas), and the clones were sequenced. Bidirectional sequences were generated with ABI 3500 (Applied Biosystems), trimmed with EGene system [27], and filtered by length (>100 bp) and quality (phred >20 and 90% minimum identity of window).
All intergenic regions, as well as tRNA and rRNA were obtained or confirmed by sequenced PCR products.
Genome assembly, annotation and analysis
Final mitogenome assembly was based only on IL sequences and PCR fragments obtained from individuals from Botucatu to avoid population polymorphisms. IL and PCR data were aligned using CAP3 [28] and annotated with the program DOGMA [35] and the web server MITOS [36]. The coding regions and ribosomal subunits were manually verified by comparison with two ant mitochondrial genomes (Solenopsis invicta, NC_014672 and Pristomyrmex punctatus, NC_015075) using MEGA version 5 [37]. The sequence data for all coding genes were translated into amino acids to confirm the absence of premature stop codons, i.e., to preclude the sequencing of nuclear mtDNA pseudogenes (numts). Validation of tRNA sequences was performed using the programs tRNAScan-SE [38] and ARWEN [39]. Codon usage, aminoacid translation, A+T content, and base composition for each codon position were obtained using MEGA version 5 [37].
Phylogenetic analysis and comparison of intergenic spacers
We used a Bayesian analysis, as implemented in BEAST software v1.7.5 [40], to infer species relationships following Mao et al. [20]. Mitogenomic sequences for 24 hymenopteran species and two non-hymenopteran were obtained from GenBank (Table 1). Only hymenopteran mitogenomes that were complete for protein-coding and rRNA genes were included in the analyses (24 out of 36 available in Genbank in September 20, 2013).
Table 1. Taxonomy, GenBank accession numbers, and mitogenome sizes of Hymenoptera mitochondrial genomes used for the phylogenetic analysis.
| Order | Family | Species | GenBank N° | Genome size (bp) | IGS bp (N)* | Reference |
| Diptera | Calliphoridae | Cochliomyia hominivorax | NC_002660 | 16,022 | 120 (14) | [41] |
| Lepidoptera | Bombycidae | Bombyx mandarina | NC_003395 | 15,928 | 361 (13) | [42] |
| Hymenoptera | ||||||
| Symphyta | Cephidae | Cephus cinctus | NC_012688 | 19,339 | 311 (20) | [19] |
| Orussidae | Orussus occidentalis | NC_012689 | 15,947 | 127 (12) | [19] | |
| Tenthredinidae | Monocellicampa pruni | JX566509 | 15,169 | 427 (18) | [43] | |
| Apocrita | Apidae | Apis cerana | NC_014295 | 15,895 | 767 (23) | [44] |
| Apidae | Apis florea | NC_021401 | 17,694 | 939 (28) | [45] | |
| Apidae | Apis mellifera ligustica | NC_001566 | 16,343 | 813 (24) | [46] | |
| Apidae | Bombus hypocrita sapporensis | NC_011923 | 15,468 | 1,214 (21) | [47] | |
| Apidae | Bombus ignitus | NC_010967 | 16,434 | 1,063 (24) | [48] | |
| Apidae | Melipona bicolor | NC_004529 | 14,422 | 477 (16) | [49] | |
| Braconidae | Cotesia vestalis | NC_014272 | 15,543 | 252 (24) | [50] | |
| Braconidae | Spathius agrili | NC_014278 | 15,425 | 155 (15) | [50] | |
| Crabronidae | Philanthus triangulum | NC_017007 | 16,029 | 217 (11) | [51] | |
| Evaniidae | Evania appendigaster | NC_013238 | 17,817 | 948 (15) | [52] | |
| Formicidae | Pristomyrmex punctatus | NC_015075 | 16,180 | 779 (28) | [12] | |
| Formicidae | Solenopsis geminata | NC_014669 | 15,552 | 523 (24) | [13] | |
| Formicidae | Solenopsis invicta | NC_014672 | 15,549 | 519 (25) | [13] | |
| Formicidae | Solenopsis richteri | NC_014677 | 15,560 | 523 (25) | [13] | |
| Formicidae | Atta laevigata | KC_346251 | 18,729 | 3,808 (30) | Present study | |
| Ichneumonidae | Diadegma semiclausum | NC_012708 | 18,728 | 1,846 (13) | [53] | |
| Ichneumonidae | Enicospilus sp. | FJ478177 | 15,300 | 281 (14) | [19] | |
| Mutillidae | Radoszkowskius oculata | NC_014485 | 18,442 | 652 (13) | [53] | |
| Scelionidae | Trissolcus basalis | JN903532 | 15,768 | 276 (19) | [20] | |
| Vanhorniidae | Vanhornia eucnemidarum | NC_008323 | 16,574 | 2,626 (23) | [54] | |
| Vespidae | Abispa ephippium | NC_011520 | 16,953 | 1,428 (26) | [55] | |
| Vespidae | Polistes sp. | EU024653 | 14,741 | 660 (20) | [55] | |
*IGS bp: sum of intergenic spacers. N: number of intergenic regions in complete mitogenome (excluding A+T rich region).
Each protein-coding and ribosomal RNA gene was aligned in MEGA version 5 [37] using Muscle [56]. Small portions of clearly missed homologous regions were corrected manually. Data were divided into four partitions: the first, second, and third codon positions and the rRNA genes. The best-fit model GTR+I+G was chosen for all of the partitions and was estimated with MEGA version 5 using a likelihood ratio test according to the Bayesian information criterion. We performed two analyses: one using all partitions and the other excluding the third codon position. The Yule model, starting with a randomly generated tree, was used as a baseline model. The chains were run for 50 million generations, and the tree parameters were sampled every 5,000 generations; 25% of the initial values were discarded as burn-in. Convergence of the runs was confirmed using Tracer v1.4 [57], and the tree was summarized in TreeAnotator v1.6.2 [58] using the maximum clade credibility option as target tree type and mean heights for the node heights.
For all mitogenomes included in the analyses we compared size and number of all available intergenic spacers (IGS), excluding the putative control region after the srRNA gene.
Results and Discussion
Comparison between transcriptomic libraries
Sanger or Illumina libraries were good sources of mitochondrial sequences, providing 45% and 78% of the A. laevigata mitogenome, respectively (Table 2 and Figure 1).
Table 2. Comparison of the transcriptomic libraries for the assembling of A. laevigata mitochondrial genome.
| Gene | Illumina Library | Sanger Library | ||
| Reads | bp* | Reads | bp | |
| trn VMIQ | 15,573 | 667 | 0 | 0 |
| NAD2 | 19,993 | 555 | 0 | 0 |
| trn WCY | 675 | 164 | 0 | 0 |
| COI | 692,055 | 657–117–150–368 | 123 | 1,436 |
| COII | 179,406 | 447 | 30 | 643 |
| COII-trn KD | 68,731 | 693 | 0 | 0 |
| ATP8-6 | 121,623 | 239–155 | 47 | 966 |
| ATP8-6-COIII | 162,863 | 409 | 0 | 0 |
| COIII | 236,772 | 185–114–315 | 43 | 722 |
| NAD3 | 12,569 | 162 | 0 | 0 |
| NAD3-trn ARNSEF | 7,614 | 321 | 0 | 0 |
| trn ARNSEF | 617 | 159 | 0 | 0 |
| NAD5 | 225,379 | 1,552 | 9 | 1,449 |
| NAD4 | 371,624 | 1,302 | 11 | 826 |
| NAD4L | 2,603 | 368 | 0 | 0 |
| NAD6 | 18,327 | 415 | 0 | 0 |
| NAD6-Cytb | 47,000 | 439 | 0 | 0 |
| Cytb | 97,794 | 289–108 | 21 | 970 |
| Cytb-trnS | 136,312 | 935 | 0 | 0 |
| NAD1 | 290,019 | 999 | 6 | 1,365 |
| trnL-lrRNA | 8,217 | 329 | 0 | 0 |
| lrRNA | 292,532 | 861 | 0 | 0 |
| lrRNA-srRNA | 18,127 | 1,036 | 0 | 0 |
| srRNA | 2,955 | 274 | 0 | 0 |
| Total | 3,029,380 | 14,784 | 290 | 8,377 |
*Number of base pairs for each contig. Sizes of non-overlapping contigs for a given gene are separated by a dash.
However, the two sequencing technologies employed herein were very different with respect to sample preparation, time of work with hands on, cost and amount of data generated. SL consumes many work hours (cloning and sequencing) and yields few sequences compared with IL, which can generate millions of reads in a few days with lower costs [59], [60]. Consequently, IL provided greater coverage (14,784 bp) than SL (8,377 bp), resulting in less effort to fill in the remaining sequence gaps. In contrast, SL had the advantage of generating longer reads (average of 931 bp) than IL (average of 462 bp), which facilitated the bioinformatics assembly process. For the COI and COIII genes, IL generated many short and non-overlapping contigs, whereas SL resulted in a single large contig (Table 2). However, IL provided a better indication of gene expression because it generated hundreds or thousands of reads for each gene compared to SL (Figure 1). Table 2 shows that SL recovered 8,377 reads from eight protein-coding genes, whereas IL recovered 2.21 million reads from the same genes. In addition, IL recovered tRNA and ribosomal subunit genes with reduced expression levels that were not sampled using SL.
Sequence composition
A single 18,729 bp sequence was obtained for the A. laevigata mitogenome and submitted to GenBank (KC346251). This sequence is incomplete in the AT-rich control region, which has an estimated size about 150–300 bp based on the length of amplicons. We were unable to sequence this region, which has been shown to be difficult to amplify and sequence in Hymenoptera [19], [54], [55]. We identified the same 37 genes present in other animals: 13 protein-coding genes, two rRNAs, and 22 tRNA genes (Table 3) [61], [62]. Twenty-three genes were encoded by the majority strand (J strand, [63]); 14 were encoded by the opposite (N) strand (Table 3).
Table 3. Mitochondrial genome annotation and A+T content of A. laevigata.
| Gene | Position* | Size (bp) | IGS (bp)# | AT (%) | Start | Stop |
| trnV | (21–89) | 69 | 101 | 88.4 | - | - |
| trnM | 191–261 | 71 | 166 | 72.5 | - | - |
| trnI | 428–499 | 72 | 93 | 82.6 | - | - |
| trnQ | (593–662) | 70 | 189 | 79.7 | - | - |
| ND2 | 852–1832 | 981 | 8 | 87.0 | ATT | TAA |
| trnW | 1841–1910 | 70 | 11 | 85.5 | - | - |
| trnC | (1922–1991) | 70 | 118 | 97.1 | - | - |
| trnY | (2110–2175) | 66 | 202 | 84.8 | - | - |
| COI | 2378–3910 | 1,533 | 160 | 70.2 | ATG | TAA |
| trnL2 | 4071–4141 | 71 | 0 | 78.3 | - | - |
| COII | 4142–4825 | 684 | 196 | 73.7 | ATT | TAA |
| trnK | 5022–5091 | 70 | 236 | 82.6 | - | - |
| trnD | 5328–5396 | 69 | 167 | 88.4 | - | - |
| ATP8 | 5564–5747 | 184 | 1 | 84.2 | ATA | T |
| ATP6 | 5749–6414 | 666 | 91 | 76.4 | ATA | TAG |
| COIII | 6506–7297 | 792 | 215 | 70.0 | ATG | TAA |
| trnG | 7513–7577 | 65 | 0 | 93.8 | - | - |
| ND3 | 7578–7931 | 354 | 57 | 78.8 | ATT | TAA |
| trnA | 7989–8054 | 66 | 85 | 87.9 | - | - |
| trnR | 8140–8213 | 74 | 207 | 87.0 | - | - |
| trnN | 8421–8490 | 70 | −3 | 82.6 | - | - |
| trnS1 | 8488–8548 | 61 | −1 | 83.9 | - | - |
| trnE | 8548–8615 | 68 | −8 | 95.6 | - | - |
| trnF | (8608–8676) | 69 | 13 | 91.3 | - | - |
| ND5 | (8690–10354) | 1,665 | 0 | 79.7 | ATT | TAA |
| trnH | (10355–10427) | 73 | 8 | 82.6 | - | - |
| ND4 | (10436–11782) | 1,347 | 247 | 80.8 | ATA | TAG |
| ND4L | (12030–12305) | 276 | 11 | 86.9 | ATT | TAG |
| trnT | 12317–12386 | 70 | 1 | 89.9 | - | - |
| trnP | (12388–12460) | 73 | 84 | 87.0 | - | - |
| ND6 | 12545–13105 | 561 | 70 | 84.0 | ATG | TAA |
| Cytb | 13176–14294 | 1,119 | 257 | 73.8 | ATG | TAA |
| trnS2 | 14552–14621 | 70 | 322 | 87.0 | - | - |
| ND1 | (14944–15891) | 948 | 176 | 78.6 | ATA | TAA |
| trnL1 | (16068–16138) | 71 | 221 | 81.2 | - | - |
| lrRNA | (16360–17785) | 1,426 | 95 | 83.1 | - | - |
| srRNA | (17881–18675) | 795 | 74+ | 85.5 | - | - |
| Total | 18,729 | 3,882 | 80.8 |
*The J strand is used as reference for position numbers. Parentheses indicate genes encoded by the N strand.
Non-coding intergenic spacer between two adjacent genes. Negative numbers indicate the overlap size in base pairs.
Incomplete sequence.
The A+T content of mitogenome, missing the unsequenced region, was 80.8% (Table 3), which is higher than that found in Solenopsis (77%) and in Pristomyrmex (79.6%) and is consistent with the pattern described for Hymenoptera [55], [13]. Distinct parts of the mitogenome displayed an A+T content that varied from 70% (COIII) to 97.1% (trnC).
Protein-coding genes had an A+T content of 78.8%, which is less than that characterizing the entire genome sequence, as previously shown in Apis mellifera [46] and in Solenopsis [13]. At the third codon position, the A+T content (86.4%) was higher than that of the whole mitogenome; the A+T content of the first and second positions was lower (76.3% and 73.6%, respectively), as reported for other insects [20], [25], [54], [64].
This AT-bias was reflected by the codon usage, as the mitogenome was found to be highly skewed towards codons that are high in A+T content. The four most represented codons were ATT for isoleucine, TTA for leucine, TTT for phenylalanine and ATA for methionine, while codons rich in C and G, such as CTG for leucine, AGC for serine, CGC for arginine and TGC for cysteine, were rarely or never used.
In agreement with Solenopsis mtDNA [13], T-bias was high in all protein-coding regions, especially in the second codon position. There was a discrepancy between these two genomes with respect to G content, which was lower in A. laevigata at all positions.
The A+T content of srRNA and lrRNA was 85.5% and 83.1%, respectively (Table 3), and although we lack some information regarding the A+T content of the control region, these values are consistent with that found in other Hymenoptera that commonly display an elevated A+T content for ribosomal subunits compared with total mtDNA [54], [64]. The srRNA and lrRNA genes of A. laevigata (795 bp and 1,426 bp, respectively) were slightly longer than those of S. invicta and P. punctatus. The precise ends of these rRNAs were difficult to determine because they are usually defined based on the surrounding coding genes or tRNAs (see [19]). In addition, in A. laevigata, there were non-coding sequences surrounding both genes (IGS, see below).
Mitogenome organization
Protein-coding genes and rRNA genes in A. laevigata displayed the same order and orientation as those present in the hypothesized ancestral pancrustacean mitogenome [16], [64], [65] (Figure 2). However, the locations of trnV and trnM indicated distinct rearrangements, as previously reported for P. punctatus and Solenopsis [12], [13]. The position occupied by trnV is uncommon in other Hymenoptera mitogenomes but was recently reported in the wasp Trissolcus basalis [20]. Although these three ants belong to Myrmicinae, Solenopsis and P. punctatus display other rearrangements that are not detected in A. laevigata (Figure 2). Rearrangements of tRNAs are a typical feature of the hymenopteran mitogenome architecture [19], [55].
Figure 2. Organization of the A. laevigata mitogenome compared with those of the ancestor and other ants.
All protein and rRNA-coding genes are in the same direction and position found in other Hymenoptera and hypothetical pancrustacean ancestral sequences. Genes encoded by the N strand are underlined; the remaining genes are encoded by the J strand. The control region of A. laevigata (gray) is incomplete. Shaded genes in pancrustacean ancestral sequence indicate rearrangements and arrows indicate position shifts of tRNA genes compared to it. Black arrow: trnV translocation from the lrRNA-srRNA junction to the srRNA-ND2 junction; grey arrow: trnI-trnQ-trnM became trnM-trnI-trnQ; blue arrow: trnK and trnD swapped positions; red arrow: trnN translocation from the trnA-trnR-trnN-trnS1-trnE-trnF cluster to a position upstream of srRNA, with an inversion. This figure was adapted from Gotzek et al. [13].
All of the predicted tRNA molecules had the typical cloverleaf structure excluding trnS1 (data not shown). In that case, the dihydrouridine arm formed a simple loop, as observed in several species including insects [54], [66]. The tRNA molecules varied between 61 (trnS1) and 74 bp (trnR), and the anticodons were identical to those described for Solenopsis [13] excluding trnN, which consisted of GTT rather than the ATT anticodon found in Solenopsis.
We found only three overlapping regions in the A. laevigata mtDNA (Table 2), and all of them were positioned between tRNA genes: a three-nucleotide overlap between trnN and trnS1, one between trnS1 and trnE, and eight between trnE and trnF (these last two genes occupied different strands). Although it is common to see overlaps between tRNAs and protein-coding genes or between proteins and protein-coding genes (e.g., [25], [54], [64]), overlaps were detected only between tRNAs in A. laevigata.
The start codons ATG, ATA or ATT are common initiation sites in invertebrate mitochondrial genomes [20], [54], [64] and can be assigned to all protein-coding genes (Table 2). The majority of protein-coding genes were predicted to end in TAA, and only three genes (ATP6, ND4, ND4L) terminated with the stop codon TAG. ATP8 lacks a complete stop codon and appears to terminate with a single T from which a stop codon could be created by post-transcriptional polyadenylation, as observed in other animals [67]–[70].
Phylogenetic analyses and intergenic spacers
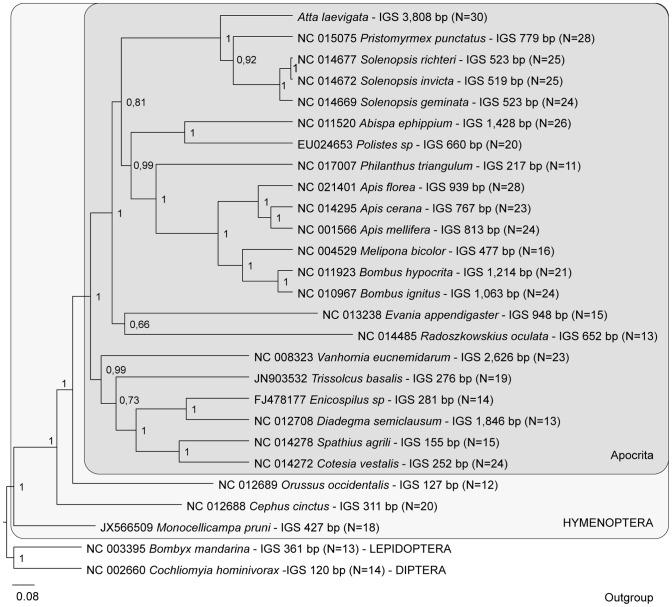
The tree derived from Bayesian inference analyses of the mitochondrial protein-coding gene and rRNAs is shown in Figure 3. The topologies obtained with and without third codon positions were broadly congruent. But the analysis excluding the third codon positions recovered the Apocrita as a monophyletic group, while the analysis with all codon positions recovered a controversial clade, with Vanhornia eucnemidarum out of the Apocrita (Figure S2). This is consistent with previous studies that suggest that the exclusion of the third codon position improves phylogenetic analyses using hymenopteran mitogenomes [71], [51], [20]. The analyses recovered most of the expected relationships on Hymenoptera (according [72]). However, the results obtained here do not support the monophyly of Aculeata (see [72]) because of the position of Radoszkowskius aculata (Aculeata: Mutillidae). Similar result was obtained previously by Kaltenpoth and colleagues [51], and it can be due to a long-branch attraction phenomenon [73] or the inclusion in the analysis of a small number of taxa containing complete genome data.
Figure 3. Bayesian tree derived from mitogenomic analyses.
Dataset included first and second codon positions from protein-coding genes and the rRNA genes. Posterior probabilities are indicated at each node. IGS: sum of intergenic spacers in base pairs. N = number of intergenic spacers.
A remarkable feature of the A. laevigata mitogenome was the presence of IGS spanning 3,808 bp and comprising an average A+T content of 86.1% (Table 3). IGS occurred between almost all of the genes, i.e., in 30 out of the 37 possibilities. Fourteen of them consisted of more than 160 bp, and the longest one contained 322 bp and was located between the trnS2 and ND1 genes. The sizes of these IGS were considerably greater than those commonly found in other insect mtDNAs, which display non-coding nucleotides outside the control (AT-rich) region that are smaller than 50 bp [54].
Unique or few large non-coding intergenic sequences, which are commonly repeated sequences, have been reported to mollusks, nematodes and arthropods, causing their mitogenomes to reach sizes of up to 40 kb [61], [74], [75]. In contrast, the IGS in A. laevigata were relatively short, variable in length, lacked repeats, and were abundantly dispersed through the 19 kb mitogenome. This same pattern was found for the other hymenopteran mitogenomes analyzed here, in particular for the monophyletic Apocrita (Table 1, Figure 3). Despite the fact that the mitochondrial genome of A. cephalotes is not annotated, the data available shows a genome with similar size and containing a large number of IGS.
Although we do not know the function of this IGS in Hymenoptera, it is interesting to note that a range of studies have reported an accelerated rate of gene rearrangement in mitogenomes of Apocrita, when compared with non-apocritans [19], [20], [43], [54]. Together, these data might suggest an association between IGS and number of rearrangements. Further studies characterizing the mitochondrial genomes of additional Hymenoptera species is needed to better understand the role and evolution of these non-coding sequences and the possible association with gene rearrangements.
In Formicidae, the mitogenome of A. laevigata was found to be 2,549 and 3,180 bp longer than that of P. punctatus and of S. invicta, respectively (Table 1, Figure 2). This difference was due primarily to the presence of IGS rather than differences in gene length. It has been noted that the size of the IGS between COI and COII genes increases from lower to higher Attini ants, honey ants, and bees [76], [77], [46]. Thus, variation in the size of the IGS is recognized as an evolutionary marker of social insects. Our data suggest that determination of the IGS position on the mitochondrial genome of Attini ants also may be valuable for phylogenetic studies. Because the IGS is highly variable [78] and informative for studies at subspecies level [79], it may be useful for distinguishing sibling species of Attini ants.
Conclusions
We observed exponential growth in the number of published articles using NGS in the previous few years [80], [81], resulting in the availability of abundant NGS transcriptomic data containing valuable information regarding mitochondrial genes. As demonstrated in the present study, this information is important for initiating the assembly of whole genome sequences. Consequently, these data should be explored to generate more mitogenomes for different species, thus contributing to a better understanding of the phylogenetic relationships and evolutionary history of many groups of organisms.
Ants are a promising group for the application of this mitochondrial genome sequencing strategy, if we consider that A. laevigata mtDNA was only the fifth mitogenome annotated within over 12,000 described species with a dominant ecological role [11]. The mitochondrial genome of A. laevigata is the first one sequenced and annotated for the Attini tribe and can provide basic data for studies investigating population history, molecular systematics, and phylogeography, and also contribute to a better understanding of the mitochondrial rearrangements that occurred during Hymenoptera evolution.
Supporting Information
Primers used to amplify A. laevigata mitogenome. Green: primers designed in this study; blue: primers obtained from the literature.
(TIF)
Bayesian tree for all codon position and rRNA genes. Posterior probabilities are shown at each node.
(TIF)
Primers and annealing temperatures (Ta) for the Atta laevigata mitochondrial regions amplified.
(DOCX)
Acknowledgments
We thank Cintia M. S. Bezerra for collecting and providing ants from Botucatu and the two anonymous reviewers that provided helpful suggestions and comments in the first version of this manuscript.
Data deposition
The Illumina mitochondrial reads of A. laevigata have been deposited to the NCBI Sequence Read Archive (SRR1226545).
Funding Statement
This work was funded by FAPESP 2011/06367-8, FAPESP 11/50226-0, CNPq 311562/2012-4, and CNPq 487639/2012-0 grants. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Borgmeier T (1950) Estudos sôbre Atta (Hym. Formicidae). Rio de Janeiro: Memórias do Instituto Oswaldo Cruz, Tomo 48. [DOI] [PubMed] [Google Scholar]
- 2.Anjos N, Della Lúcia Tm, Mayhé-Nunes AJ (1998) Guia prático sobre formigas cortadeiras em reflorestamentos. Ponte Nova: Editora GarffCor. 97p. [Google Scholar]
- 3.Fowler HG, Silva VP, Saes NB (1986) Population dynamics of leaf-cutting ants: a brief review. In: Lofgren CS, Vander Meer RK, editors. Fire ants and leaf-cutting ants: biology and management. Boulder, Colorado: West-View Press. pp. 123–145. [Google Scholar]
- 4. Keller L (1998) Queen lifespan and colony characteristics in ants and termites. Insectes Soc 45: 235–246. [Google Scholar]
- 5. Delabie JHC, Nascimento IC, Fonseca E, Sgrillo RB, Soares PAO, et al. (1997) Biogeografia das formigas cortadeiras (Hymenoptera; Formicidae; Myrmicinae; Attini) de importância econômica no leste da Bahia e nas regiões periféricas dos estados vizinhos. Agrotrópica 9: 49–58. [Google Scholar]
- 6. Zanetti R, Zanuncio JC, Souza-Silva A, Mendonça LA, Mattos JOS, et al. (2008) Efficiency of products for thermonebulization on the control of Atta laevigata (Hymenoptera: Formicidae) in eucalypus plantations. Cienc Agrotec 32: 1313–1316. [Google Scholar]
- 7. Hernández JV, Jaffé K (1995) Economic damage caused by leaf-cutting ant populations of Atta laevigata (F. Smith) on pine plantations (Pinus caribaeae Mor.) and elements for managing of the pest. An Soc Entomol 24: 287–298. [Google Scholar]
- 8. Vasconcelos HL, Cherret JM (1977) Leaf-cutting ants and early forest regeneration in central Amazonia: effects of herbivory on the seedling establishment. J Trop Ecol 13: 357–370. [Google Scholar]
- 9. Viana LR, Santos JC, Arruda LJ, Santos GP, Fernandes GW (2004) Foraging patterns of the leaf-cutter ant Atta laevigata (Smith) (Myrmicinae: Attini) in an area of cerrado vegetation. Neotrop Entomol 33: 391–393. [Google Scholar]
- 10. Rodovalho CM, Ferro M, Fonseca FPP, Antonio EA, Guilherme IR, et al. (2011) Expressed sequence tags from Atta laevigata and identification of candidate genes for the control of pest leaf-cutting ants. BMC Res Notes 4: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hölldobler B, Wilson EO (1990) The Ants. Massachusetts: Belknap Press of Harvard University. 732p. [Google Scholar]
- 12. Hasegawa E, Kobayashi K, Yagi N, Tsuji K (2011) Complete mitochondrial genomes of normal and cheater morphs in the parthenogenetic ant Pristomyrmex punctatus (Hymenoptera: Formicidae). Myrmecol News 15: 85–90. [Google Scholar]
- 13. Gotzek D, Clarke J, Shoemaker D (2010) Mitochondrial genome evolution in fire ants (Hymenoptera: Formicidae). BMC Evol Biol 10: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suen G, Teiling C, Li L, Holt C, Abouheif E, et al. (2011) The Genome Sequence of the Leaf-Cutter Ant Atta cephalotes Reveals Insights into its Obligate Symbiotic Lifestyle. Plos Genet 7: e1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Avise JC (1995) Mitochondrial DNA polymorphism and a connection between genetics and demography of relevance to conservation. Conser Biol 9: 686–690. [Google Scholar]
- 16. Boore JL, Lavrov DV, Brown WM (1998) Gene translocation links insects and crustaceans. Nature 392: 667–668. [DOI] [PubMed] [Google Scholar]
- 17. Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT (2006) Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst 37: 545–79. [Google Scholar]
- 18. Chiotis M, Jermiin LS, Crozier RH (2000) A molecular framework for the phylogeny of the ant subfamily Dolichoderinae. Mol Phylogenet Evol 17: 108–116. [DOI] [PubMed] [Google Scholar]
- 19. Dowton M, Cameron SL, Dowavic JI, Austin AD, Whiting MF (2009a) Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol Biol Evol 26: 1607–1617. [DOI] [PubMed] [Google Scholar]
- 20. Mao M, Valerio A, Austin AD, Dowton M, Johnson NF (2012) The first mitochondrial genome for the wasp superfamily Platygastroidea: the egg parasitoid Trissolcus basalis . Genome 55: 194–204. [DOI] [PubMed] [Google Scholar]
- 21. Jex AR, Hall RS, Littlewood DTJ, Gasser RB (2010) An integrated pipeline for next-generation sequencing and annotation of mitochondrial genomes. Nucleic Acids Res 38: 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang C, Huang Y (2008) Complete mitochondrial genome of Oxya chinensis (Orthoptera, Acridoidea). Acta Biochim Biophys Sin 40: 7–18. [DOI] [PubMed] [Google Scholar]
- 23. Emblem A, Karlsen BO, Evertsen J, Miller DJ, Moum T, et al. (2012) Mitogenome polymorphism in a single branch sample revealed by SOLiD deep sequencing of the Lophelia pertusa coral genome. Gene 506: 344–349. [DOI] [PubMed] [Google Scholar]
- 24.Hahn C, Bachmann L, Chevreux B (2013) Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res 1–9. [DOI] [PMC free article] [PubMed]
- 25. Margam VM, Coates BS, Hellmich RL, Agunbiade T, Seufferheld MJ, et al. (2011) Mitochondrial genome sequence and expression profiling for the legume pod borer Maruca vitrata (Lepidoptera: Crambidae). Plos One 6: e16444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Solomon SE, Bacci M Jr, Martins J Jr, Vinha GG, Mueller UG (2008) Paleodistributions and Comparative Molecular Phylogeography of Leafcutter Ants (Atta spp.) Provide New Insight into the Origins of Amazonian Diversity. Plos One 3: e2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Durham AM, Kashiwabara AY, Matsunaga FT, Ahagon PH, Rainone F, et al. (2005) EGene: a configurable pipeline generation system for automated sequence analysis. Bioinformatics 21: 2812–2813. [DOI] [PubMed] [Google Scholar]
- 28. Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 30. Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Langmead B, Salzberg S (2012) Fast gapped-read alignment with Bowtie2. Nature Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) 1000 Genome Project Data Processing Subgroup (2009) The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martins J, Solomon SE, Mikheyev AS, Mueller UG, Ortiz A, et al. (2007) Nuclear mitochondrial-like sequences in ants: evidence from Atta cephalotes (Formicidae: Attini). Insect Mol Biol 16: 777–784. [DOI] [PubMed] [Google Scholar]
- 35. Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255. [DOI] [PubMed] [Google Scholar]
- 36. Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, et al. (2013) MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation. Mol Phyl Evol 69: 313–319. [DOI] [PubMed] [Google Scholar]
- 37. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laslett D, Canbäck B (2008) ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24: 172–175. [DOI] [PubMed] [Google Scholar]
- 40. Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lessinger AC, Martins Junqueira AC, Lemos TA, Kemper EL, da Silva FR, et al. (2000) The mitochondrial genome of the primary screwworm fly Cochliomyia hominivorax (Diptera: Calliphoridae). Insect Mol Biol 9: 521–529. [DOI] [PubMed] [Google Scholar]
- 42. Yukuhiro K, Sezutsu H, Itoh M, Shimizu K, Banno Y (2002) Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth, Bombyx mandarina, and its close relative, the domesticated silkmoth, Bombyx mori . Mol Biol Evol 19: 1385–1389. [DOI] [PubMed] [Google Scholar]
- 43.Wei S-J, Wu Q-L, Liu W (2013) Sequencing and characterization of the Monocellicampa pruni (Hymenoptera: Tenthredinidae) mitochondrial genome. Mitochondrial DNA doi:10.3109/19401736.2013.819501. [DOI] [PubMed]
- 44. Tan HW, Liu GH, Dong X, Lin RQ, Song HQ, et al. (2011) The complete mitochondrial genome of the Asiatic cavity-nesting honeybee Apis cerana (Hymenoptera: Apidae). Plos One 6: e23008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang AR, Kim MJ, Park JS, Choi YS, Thapa R, et al. (2013) Complete mitochondrial genome of the dwarf honeybee, Apis florea (Hymenoptera: Apidae). Mitochondrial DNA 24: 208–210. [DOI] [PubMed] [Google Scholar]
- 46. Crozier RH, Crozier YC (1993) The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics 133: 97–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hong MY, Cha SY, Kim DY, Yoon HJ, Kim SR, et al. (2008) Presence of several tRNA-like sequences in the mitochondrial genome of the bumblebee, Bombus hypocrita sapporoensis (Hymenoptera: Apidae). Genes Genomics 30: 307–318. [Google Scholar]
- 48. Cha SY, Yoon HJ, Lee EM, Yoon MH, Hwang JS, et al. (2007) The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae). Gene 392: 206–220. [DOI] [PubMed] [Google Scholar]
- 49. Silvestre D, Dowton M, Arias MA (2008) The mitochondrial genome of the stingless bee Melipona bicolor (Hymenoptera, Apidae, Meliponini): Sequence, gene organization and a unique tRNA translocation event conserved across the tribe Meliponini. Genet Mol Biol 31: 451–460. [Google Scholar]
- 50. Wei SJ, Shi M, Sharkey MJ, van Achterberg C, Chen XX (2010a) Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genomics 11: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kaltenpoth M, Showers Corneli P, Dunn DM, Weiss RB, Strohm E, et al. (2012) Accelerated evolution of mitochondrial but not nuclear genomes of Hymenoptera: new evidence from crabronid wasps. Plos One 7: e32826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wei SJ, Tang P, Zheng LH, Shi M, Chen XX (2010b) The complete mitochondrial genome of Evania appendigaster (Hymenoptera: Evaniidae) has low A+T content and a long intergenic spacer between atp8 and atp6. Mol Biol Rep 37: 1931–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wei SJ, Shi M, He JH, Sharkey M, Chen XX (2009) The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome 52: 308–319. [DOI] [PubMed] [Google Scholar]
- 54. Castro LR, Ruberu K, Dowton M (2006) Mitochondrial genomes of Vanhornia eucnemidarum (Apocrita: Vanhorniidae) and Primeuchroeus spp. (Aculeata: Chrysididae): evidence of rearranged mitochondrial genomes within the Apocrita (Insecta: Hymenoptera). Genome 49: 752–766. [DOI] [PubMed] [Google Scholar]
- 55. Cameron SL, Dowton M, Castro LR, Ruberu K, Whiting MF, et al. (2008) Mitochondrial genome organization and phylogeny of two vespid wasps. Genome 51: 800–808. [DOI] [PubMed] [Google Scholar]
- 56. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rambaut A, Drummond AJ (2007) Tracer v1.4. Available from http://beast.bio.ed.ac.uk/Tracer. Accessed 17 December 2012.
- 58. Drummond A, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wall PK, Leebens-Mack J, Chanderbali AS, Barakat A, Wolcott E, et al. (2009) Comparison of next generation sequencing technologies for transcriptome characterization. BMC Genomics 10: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, et al. (2012) Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol 30: 434–439. [DOI] [PubMed] [Google Scholar]
- 61. Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wallace DC (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39: 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Simon C, Frati F, Beckenbach A, Crespi B, Liu H, et al. (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87: 651–701. [Google Scholar]
- 64. Castro LR, Dowton M (2005) The position of the Hymenoptera within the Holometabola as inferred from the mitochondrial genome of Perga condei (Hymenoptera: Symphyta: Pergidae). Mol Phylogenet Evol 34: 469–479. [DOI] [PubMed] [Google Scholar]
- 65. Flook PK, Rowell CHF, Gellissen G (1995) The sequence, organization, and evolution of the Locusta migratoria mitochondrial genome. J Mol Evol 41: 928–941. [DOI] [PubMed] [Google Scholar]
- 66. Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cyt 141: 173–216. [DOI] [PubMed] [Google Scholar]
- 67. Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474. [DOI] [PubMed] [Google Scholar]
- 68. Nardi F, Carapelli A, Fanciulli PP, Dallai R, Frati F (2001) The complete mitochondrial DNA sequence of the basal hexapod Tetrodontophora bielanensis: evidence for heteroplasmy and tRNA translocations. Mol Biol Evol 18: 1293–1304. [DOI] [PubMed] [Google Scholar]
- 69. Cook CE (2005) The complete mitochondrial genome of the stomatopod crustacean Squilla mantis . BMC Genomics 6: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scheffler IE (2008) Mitochondria. New York: J. Wiley and Sons. 462 p. [Google Scholar]
- 71. Dowton M, Cameron SL, Austin AD, Whiting MF (2009b) Phylogenetic approaches for the analysis of mitochondrial genome sequence data in the Hymenoptera — a lineage with both rapidly and slowly evolving mitochondrial genomes. J Mol Evol 52: 512–519. [DOI] [PubMed] [Google Scholar]
- 72. Klopfstein S, Vilhelmsen L, Heraty JM, Sharkey M, Ronquist R (2013) The Hymenopteran Tree of Life: Evidence from Protein-Coding Genes and Objectively Aligned Ribosomal Data. Plos One 8: e69344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bergsten J (2005) A review of long-branch attraction. Cladistics 21: 163–193. [DOI] [PubMed] [Google Scholar]
- 74. Snyder M, Fraser AR, Laroche J, Gartner-Kepkay KE, Zouros E (1987) Atypical mitochondrial DNA from the deep-sea scallop Placopecten magellanicus . Proc Natl Acad Sci USA 84: 7595–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hyman BC, Beck JL, Weiss KC (1988) Sequence Amplification and Gene Rearrangemenitn Parasitic Nematode Mitochondrial DNA. Genetics 140: 707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wetterer JK, Schultz TR, Meier R (1998) Phylogeny of fungus-growing ants (Tribe Attini) based on mtDNA sequence and morphology. Mol Phylogenet Evol 9: 42–47. [DOI] [PubMed] [Google Scholar]
- 77. Kronauer DJC, Hölldobler B, Gadau J (2004) Phylogenetics of the new world honey ants (genus Myrmecocystus) estimated from mitochondrial DNA sequences. Mol Phylogenet Evol 32: 416–421. [DOI] [PubMed] [Google Scholar]
- 78. Bacci M, Solomon SE, Mueller UG, Martins VG, Carvalho AOR, et al. (2009) Phylogeny of leafcutter ants in the genus Atta Fabricius (Formicidae: Attini) based on mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol 51: 427–437. [DOI] [PubMed] [Google Scholar]
- 79. Franck P, Garnery L, Solignac M, Cornuet JM (1998) The origin of West European subspecies of honeybees (Apis mellifera): new insights from microsatellite and mitochondrial data. Evolution 52: 1119–1134. [DOI] [PubMed] [Google Scholar]
- 80. Kahvejian A, Quackenbush J, Thompson JF (2008) What would you do if you could sequence everything? Nat Biotechnol 26: 1125–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Su Z, Ning B, Fang H, Hong H, Perkins R, et al. (2011) Next-generation Sequencing and its Applications in Molecular Diagnostics. Expert Rev Mol Diagn 11: 333–343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used to amplify A. laevigata mitogenome. Green: primers designed in this study; blue: primers obtained from the literature.
(TIF)
Bayesian tree for all codon position and rRNA genes. Posterior probabilities are shown at each node.
(TIF)
Primers and annealing temperatures (Ta) for the Atta laevigata mitochondrial regions amplified.
(DOCX)