Summary
Many bacterial pathogens utilize the 2-C-methyl-D-erythritol 4-phosphate pathway for biosynthesizing isoprenoid precursors, a pathway that is vital for bacterial survival and absent from human cells, providing a potential source of drug targets. However, the characterization of 4-diphosphocytidyl-2-C-methyl-D-erythritol (CDP-ME) kinase (IspE) has been hindered due to a lack of enantiopure CDP-ME and difficulty in obtaining pure IspE. Here, enantiopure CDP-ME was chemically synthesized and recombinant IspE from bacterial pathogens were purified and characterized. Although gene disruption was not possible in Mycobacterium tuberculosis, IspE is essential in Mycobacterium smegmatis. The biochemical and kinetic characteristics of IspE provide the basis for development of a high throughput screen and structural characterization.
Introduction
Isoprenoids are one of the largest groups of natural products, including 35,000 primary and secondary metabolites (Chang and Keasling, 2006; Hunter, 2007). Despite functional and structural diversity, all isoprenoids are derived from isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), produced by the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway in most bacterial pathogens, including Mycobacterium tuberculosis (tuberculosis), Burkholderia mallei (glanders), Salmonella enterica serovar typhimurium (S. Typhi; typhoid fever), and Vibrio cholerae (cholera). The early steps in the biosynthesis of bacterial isoprenoids have been demonstrated to be essential in both Gram-positive and Gram-negative bacteria (Brown and Parish, 2008; Buetow et al., 2007; Cornish et al., 2006; Eoh et al., 2007; Freiberg et al., 2001; Sauret-Gueto et al., 2003) and the MEP pathway is not present in mammalian cells. Thus, the MEP pathway is considered to be a good source of potential drug targets (Rohmer, 1998; Eoh et al., 2009; Testa and Brown, 2003).
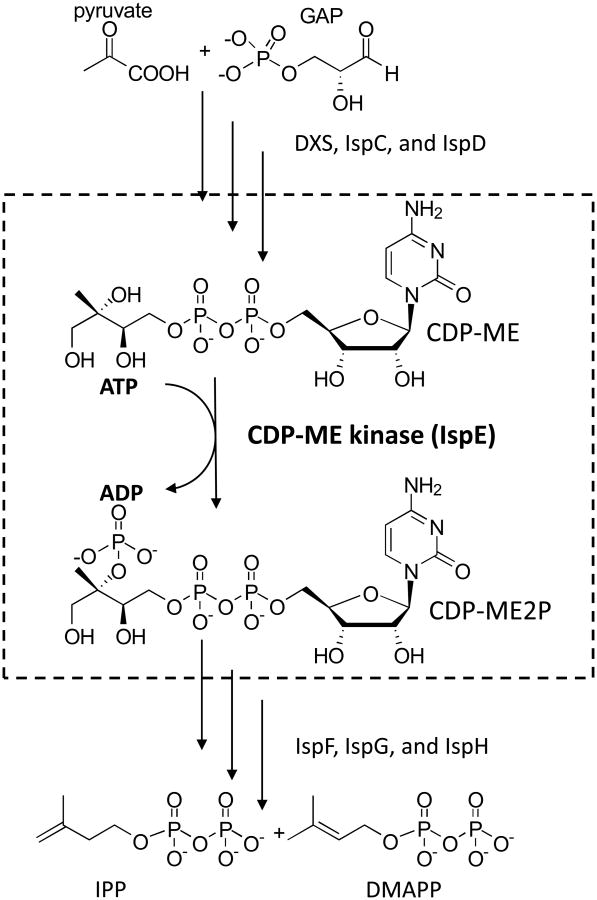
The fourth enzyme in the MEP pathway (4-diphosphocytidyl-2-C-methyl-D-erythritol (CDP-ME) kinase, IspE, EC2.7.1.148) was first identified in Escherichia coli (Luttgen et al., 2000) and tomato (Rohdich et al., 2000). IspE catalyses the conversion of CDP-ME to 4-diphosphocytidyl-2-C-methyl-D-erythritol-2-phosphate (CDP-ME2P) in an ATP dependent manner (Fig. 1) (Eoh et al., 2009; Luttgen et al., 2000). Even though sequence alignments and crystal structures (Miallau et al., 2003) showed that bacterial IspE has a high level of similarity to the ATP-dependent GHMP kinase superfamily (Andreassi and Leyh, 2004; Romanowski et al., 2002; Yang et al., 2002; Zhou et al., 2000), which includes galactose kinase, homoserine kinase, mevalonate kinase and phosphomevalonate kinase, significant differences in the catalytic and substrate binding sites were observed, suggesting that bacterial IspE may be a potential drug target (Miallau et al., 2003; Eoh et al., 2009; Sgraja et al., 2008). However, characterization of IspE from bacteria pathogens to humans has been hampered due to the lack of a source of enantiopure CDP-ME and difficulties encountered in purification of IspE from pathogenic species (Sgraja et al., 2008). Although in silico studies indicate that the putative M. tuberculosis IspE (Rv1011) has no predicted transmembrane domains and is cytosolic, previous attempts to purify the native form of IspE from M. tuberculosis failed (Sgraja et al., 2008). Purification of recombinant M. tuberculosis IspE was made possible by generating genetically truncated proteins. Of eight truncated versions of M. tuberculosis IspE generated, one showed kinetic properties similar to those obtained using native cytosolic protein isolated from wild type M. tuberculosis H37Rv. The truncation strategy also allowed purification of recombinant IspE enzymes from B. mallei, S. Typhi, and V. cholerae. In the present study, we provide direct evidence of IspE essentiality in Mycobacterium smegmatis using allelic disruption (Jackson et al., 2000; Pan et al., 2001). In addition, enantiopure CDP-ME was chemically synthesized, improving the methods we previously reported for chemo-enzymatic synthesis (Narayanasamy et al., 2008), and recombinant IspE from pathogens with potential to be utilized as agents of bioterrorism (Clarke, 2005; Pappas et al., 2006) (M. tuberculosis, B. mallei, S. Typhi, and V. cholerae) was purified.
Fig. 1.
The MEP pathway and the reaction catalyzed by IspE. Starting with pyruvate and glyceraldehyde 3-phosphate (GAP), the sequential activities of DXS, IspC, and IspD produce CDP-ME, the substrate of IspE. IspE phosphorylates the tertiary hydroxyl group of CDP-ME forming CDP-ME2P. The reaction catalyzed by IspE is indicated by the dotted box. Abbreviations; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; IspC, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; IspD, 4-diphosphocytidyl-2-C-methyl-D-erythritol synthase; IspE, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; IspF, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; IspG, 1-hydroxyl-2-methyl-2(E)-butenyl 4-diphosphate synthase; IspH, 1-hydroxy-2-methyl-2(E)-butenyl 4-diphosphate reductase; CDP-ME, 4-diphosphocytidyl-2-C-methyl-D-erythritol; CDP-ME2P, CDP-ME 2 phosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate.
The elucidation of the biochemical and kinetic properties of IspE from human pathogens provides the groundwork for development of high throughput screening (HTS) assays and, potentially, identification of novel antimicrobials.
Results
Synthesis of enantiopure CDP-ME
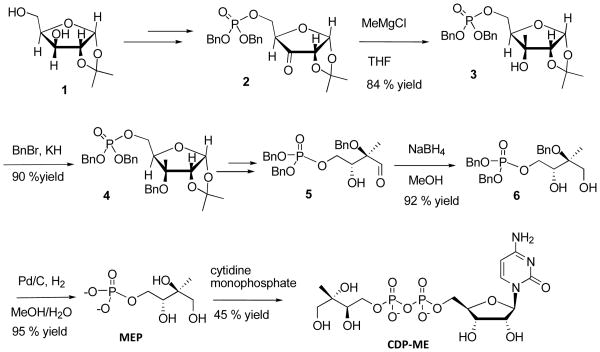
Although the synthetic scheme of 3 from 1 is identical to a previous report [(Narayanasamy et al., 2008), Scheme 1], several steps for synthesizing enantiopure MEP were improved over that the report. At first, the tertiary hydroxyl group 3 was protected by benzyl bromide, after treating 3 with KH to yield 90% of 4. In this step, KH was found to be faster and better than NaH (Narayanasamy et al., 2008). The aldehyde 5 was reduced with sodium borohydride forming benzyl protected MEP precursor 6, which after hydrogenolysis with Pd/C in water/methanol medium and without acid workup led to enantiopure MEP, which was characterized by 1H-NMR, MS and optical rotation. The optical rotations of 3, 4, and 6 are newly identified as +33.0, +13.0, and +10.0, respectively (Experimental Procedures and supplemental data S1). Freshly prepared MEP was coupled with cytidine 5′-monophosphate (CMP), which was titrated with triethyl amine leading to the formation of corresponding triethylammonium CMP. The phosphoester moiety was activated by triflouroacetyl anhydride followed by conversion to phosphoramide by treatment with methylimidazole and then coupled with the tributylammonium salt of MEP. The crude material was purified to obtain CDP-ME. Enantiopure intermediates and CDP-ME were confirmed by 1H NMR and high resolution mass spectrometry (Experimental Procedures and supplemental data, S1 and S2). The methyl group at the 2 position of MEP moiety in CDP-ME was observed as a singlet at 1.10 ppm and the cytidine terminus was observed as a doublet at both 7.94 and 6.09 ppm (Rohdich et al., 2000; Cassera et al., 2004; Richard et al., 2001). Optical rotation of the enantiopure CDP-ME was found to be +13.7 in water. Thus enantiopure MEP was synthesized and the coupling reaction of MEP with CMP was satisfactory, yielding enantiopure CDP-ME.
Scheme 1.
Synthetic scheme of enantiopure CDP-ME. Enantiopure 2-C-methyl-D-erythritol 4-phosphate (MEP) was synthesized from commercially available 1,2-O-isopropylidene-α-D-xylofuranose 3. Subsequently, MEP is coupled with CMP to give a 45% yield of pure CDP-ME.
Determination of mycobacterial IspE essentiality using M. smegmatis
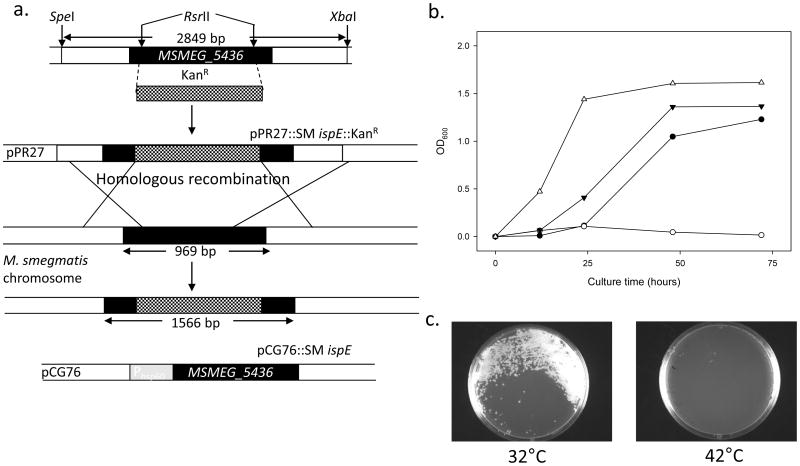
An allelic disruption strategy was employed to show M. smegmatis MSMEG_5436 essentiality as previously described (Jackson et al., 2000). The pPR27∷SM ispE∷KanR plasmid has a temperature sensitive mycobacterial origin of replication that facilitates recombination generating a single homologous-recombination event at the M. smegmatis MSMEG_5436 locus. The plasmid also harbors the sacB counter selectable marker (Pelicic et al., 1997) and the xylE marker (Curcic et al., 1994). Plasmid pPR27∷SM ispE∷KanR was electroporated into M. smegmatis mc2155, and transformants were selected on LB plates containing 30 μg/ml of Kanamycin (Kan) at 32°C. One transformant was then propagated in LB broth containing 30 μg/ml of Kan at 32°C and plated onto LB plates containing 30 μg/ml of Kan at 42°C. The temperature-sensitive origin containing pPR27∷SM ispE∷KanR is not able to replicate at 42°C and the Kan-resistant colonies have necessarily integrated part or all of the plasmid into M. smegmatis mc2155 chromosome, yielding single-crossover strains. The resulting colonies were analyzed for their XylE phenotype with catechol (Pan et al., 2001) and yellow colonies were further used. Double-crossover events were generated by plating a total of 30 yellow, single-crossover strains onto LB plates containing 30 μg/ml Kanamycin and sucrose at 42°C, leading to the disruption of the M. smegmatis MESG_5436 gene; no colonies were observed, suggesting that M. smegmatis MSMEG_5436 is essential for M. smegmatis normal growth. To confirm the essentiality of the gene, a rescue plasmid (pCG76∷SM ispE) was prepared and electroporated into the single-crossover strains prior to generation of the double-crossover. The single-crossover strains containing pCG76∷SM ispE were grown in LB broth containing 30 μg/ml Kan at 32°C and then plated onto LB plates containing 30 μg/ml Kan and sucrose at 32°C. The resulting colonies were analyzed for their XylE phenotype with catechol (Pan et al., 2001), and a total of 30 white colonies were obtained as the double-crossover strains. The double-crossover strains were confirmed by PCR by using primer sets (MutSmispE-F and R, Supplemental Data S3) (data not shown). As a final experiment to confirm that M. smegmatis MSMEG_5436 is essential for growth, M. smegmatis double-crossover strains containing pCG76∷SM ispE were shown to be unable to grow at 42°C (Fig. 2b), due to loss of the plasmid and its insert. On the other hand, M. smegmatis double-crossover strains containing pCG76∷SM ispE were shown to be able to grow at 32°C and M. smegmatis wild-type strains grew normally at both 32°C and 42°C. The similar results were observed on the LB plates containing 30 μg/ml Kan (Fig. 2c).
Fig. 2.
Essentiality of M. smegmatis IspE for the bacterial growth. Panel a: Schematic diagram of showing recombination in M. smegmatis mc2155. Black boxes indicate coding sequence of IspE (MSMEG_5436). Hatched regions represent the intragenic DNA fragment replaced with a Kanamycin (Kan) resistant cassette (KanR) from pUC4K. For generation of MSMEG_5436 disrupted KanR construct, KanR was cloned into RsrII sites of MSMEG_5436 and ligated with pPR27 yielding pPR27∷Sm ispE∷KanR. After homologous recombination, selection for Kan resistance, sucrose resistance and xylE phenotype resulted in colonies with a disrupted MSMEG_5436, only in the presence of rescue vector pCG76∷Sm ispE, which contains temperature sensitive origin of replication and heat shock protein 60 promoter (Phsp60). Panel b: Growth curves of M. smegmatis mc2155 strains at 32 and 42°C. M. smegmatis mc2155 containing a rescue plasmid pCG76∷Sm ispE at 32°C (solid circles) or at 42°C (open circles), M. smegmatis mc2155 wild-type at 32°C (solid triangles) or at 42°C (open triangles). All cultures were grown in LB broth containing appropriate antibiotics (See Experimental Procedures). Panel c: Growth patterns of M. smegmatis mc2155 double-crossover strains containing pCG76∷Sm ispE at 32°C (left) and 42°C (right) on LB plates with appropriate antibiotics.
Identification of putative IspE from M. tuberculosis, B. mallei, S. Typhi, and V. cholerae
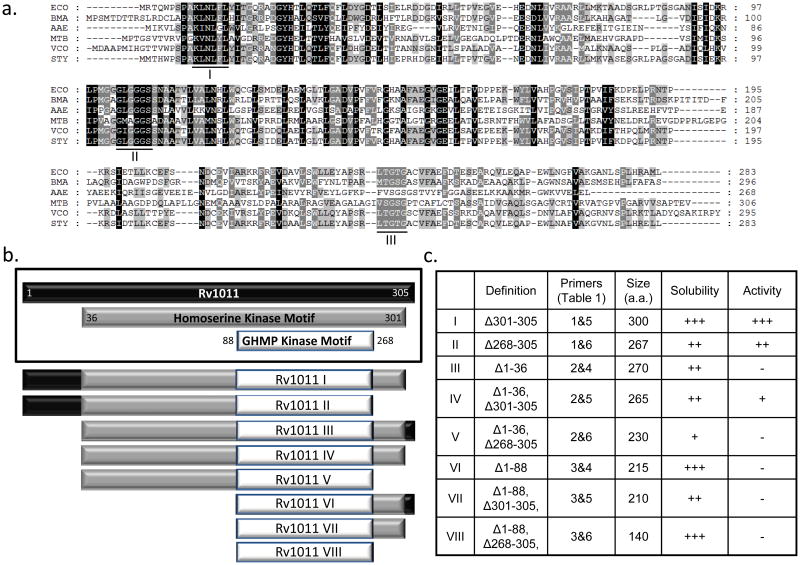
The putative M. tuberculosis, B. mallei, S. Typhi, and V. cholerae IspE proteins have 22%, 44%, 90%, and 59% identity with E. coli IspE (Luttgen et al., 2000), respectively. All reported crucial domains, including the glycine–rich domains and substrate binding motifs of the E. coli ortholog (Luttgen et al., 2000; Miallau et al., 2003) are well conserved in all IspE orthologs (Fig. 3a).
Fig. 3.
Identification of M. tuberculosis IspE and purification strategy. Panel a: Alignment of the amino acid sequence of E. coli IspE (ECO) and the putative IspE amino acids of M. tuberculosis (Rv1011) (MTB), B. mallei (BMA), S. Typhi (STY), V. cholerae (VCH), and previously characterized Aquifex aeolicus (AAE) (Sgraja et al., 2008). Identities are highlighted in black and similarities in gray. The amino acids reported to be involved in CDP-ME binding (I) and ATP-binding (II + III) reported in E. coli IspE (Miallau et al., 2003) are underlined. Panel b: Rv1011 contains two predicted kinase motifs, a homoserine (ThrB) kinase (a.a. 36-299) and a GHMP Kinase (a.a. 88-268)]. A series of eight fragments were engineered as indicated. Panel c: Characteristics of the eight recombinant Rv1011 fragments. The primers used to amplify the fragments are listed in Supplemental Data S3. The solubility was determined by using SDS-PAGE analysis and the activities were determined using the radioisotope based in vitro IspE assay. − no; +, low; ++, intermediate; +++, good.
Purification of active and soluble recombinant IspE
Although M. tuberculosis IspE is predicted to be cytosolic, purification of soluble, active IspE in native form proved difficult. To facilitate the purification of reasonable quantities of M. tuberculosis IspE, eight truncated versions of IspE (Rv1011 I through Rv1011 VIII) were cloned, expressed and purified (Fig. 3b). M. tuberculosis IspE contains two predicted functional motifs; a homoserine kinase (ThrB) motif and a GHMP kinase motif (Fig. 3b); thus, sequences of the genetic constructs were based on the motif distribution, for example, Rv1011 IV corresponds to the ThrB motif and Rv1011 VIII, the smallest fragment, corresponds to the GHMP kinase motif. The solubility of all of the expressed proteins was dramatically improved compared with that of intact Rv1011, giving yields of ~ 1 mg of purified protein/L of culture. Of the expressed proteins, only Rv1011 I (Δ301-305), Rv1011 II (Δ268-365), and Rv1011 IV showed IspE activity; the other 5 Rv1011 fragments were inactive under the conditions tested. Rv1011 I showed the greatest catalytic activity (Fig. 3c). Rv1011 IV, containing only the ThrB and GHMP kinase domains, showed very weak IspE activity and the activity of Rv1011 II, missing C-terminus of ThrB motif was intermediate. Therefore, recombinant Rv1011 I was used for further characterization.
The three IspE orthologs from B. mallei, S. Typhi, and V. cholerae are also predicted to be cytosolic and the native forms also proved difficult to express as soluble, active protein; however, as for the M. tuberculosis IspE, enzymes from B. mallei, S. Typhi, and V. cholerae could be expressed and purified after truncation of the C-terminal five amino acids, again resulting in yields of ~1 mg of purified protein/L of cuture. Three truncated versions of bacterial IspE enzymes of B. mallei, S. Typhi, and V. cholerae were designated mBME, mSTE, and mVCE. The induction and expression of Rv1011 I, mBME, mSTE and mVCE was confirmed by Western blot analysis with the expected molecular weights of 30.8, 31.4, 30.2, and 31.3 kDa, respectively (Supplemental Data S4).
In vitro reaction requirements of IspE orthologs
The activities of purified Rv1011 I, mBME, mSTE and mVCE used in this study were linear with increasing protein up to at least 194.8, 190.9, 198.5, 191.6 pmols, respectively, and all reactions were linear with time up to 30 min. In order to confirm the nature of the product, assay the mixtures were incubated, lyophilized, dissolved in 100 mM aq. ammonium bicarbonate and subjected to size exclusion chromatography on Bio-Gel® P-2 gel fine columns eluted with 100 mM aq. ammonium bicarbonate; fractions containing putative product were then subjected to further chromatography on benzyl DEAE cellulose anion exchange columns, eluted using a gradient of 0- 0.5 M aq. ammonium bicarbonate and subsequent analysis by MS revealed a peak at 633 m/z corresponding to [M+2NH4]+.
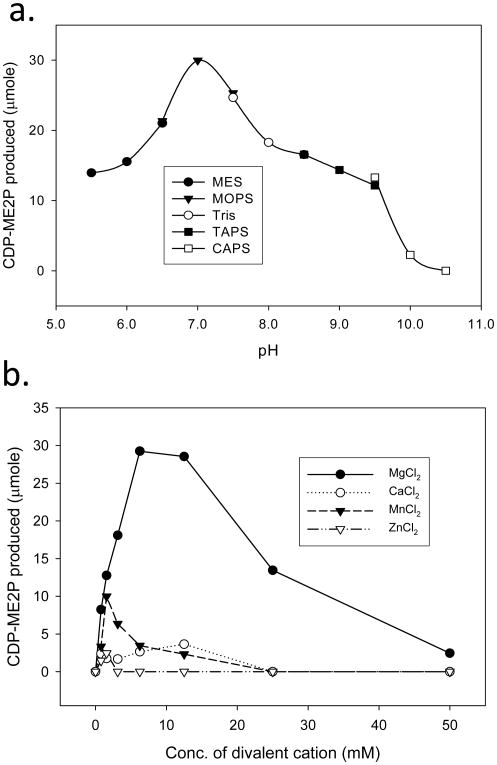
The optimal pH and divalent cation concentrations for all four IspE orthologs were determined. Rv1011 I was active over a broad pH range (6.0 to 9.0), with optimal activity at pH 7.0 (Fig. 4a). The pH optima for mBME, mSTE and mVCE were similar to that of Rv1011 I, although the pH range at which Rv1011 I was active was somewhat narrower than those of the other three IspE orthologs (Supplemental data, S5). Optimal activity for mBME, mSTE and mVCE was observed at pH 8.0, 8.0, and 7.5, respectively.
Fig. 4.
Biochemical properties of M. tuberculosis IspE. a. The optimal pH for catalytic activity was determined using MES (pH 5.5 - pH 7.0), MOPS (pH 7.0 - pH 7.5), Tris (pH 7.5 - pH 8.5), TAPS (pH 8.5 - pH 9.0), and CAPS (pH 9.0 - pH 10.5) using appropriate counter ions. b. Divalent cations (Mg2+, Mn2+, Zn2+, or Ca2+) were added to the reaction mixtures at the indicated concentrations. The reaction mixtures were as described in Experimental Procedures.
The activity of Rv1011 I is absolutely dependent on the presence of divalent cations (Fig. 4b), the addition of 5 mM EDTA to the reaction mixture completely abolished the activity. The activity of Rv1011 I was optimal in the presence of ~ 6 mM Mg2+. Other divalent cations including Mn2+, Ca2+, or Zn2+ supported little or no synthesis of CDP-ME2P at any concentration tested (Fig. 4b). Similar results were observed when mBME, mSTE and mVCE were tested (supplemental data, S5).
Kinetic properties of the IspE orthologs
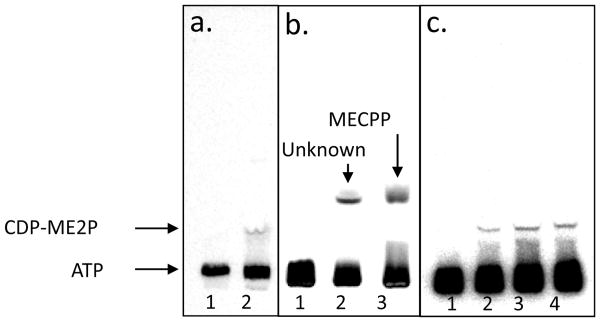
Reaction mixtures containing enantiopure CDP-ME, MgCl2, [γ-32P]ATP, and IspE generated a single radioactive product with an Rf value of 0.13 was subjected to TLC analysis (Fig. 5a); this product corresponds to CDP-ME2P (Kishida et al., 2003; Testa et al., 2006). The apparent KmCDP–ME and KmATP for wild type M. tuberculosis H37Rv (in a crude cytosolic preparation) were calculated to be 206.7 ± 12.5 μM and 20.7 ± 1.2 μM, respectively. The KmCDP–ME and KmATP for purified Rv1011 I (97.2 pmol) were calculated to be 327.8 ± 21.4 μM and 75.2 ± 0.9 μM, respectively (Table 1). The KmCDP–ME values of mBME, mSTE, and mVCE were 31.3 ± 1.5, 25.4 ± 0.8, a nd 52.9 ± 11.7 μM, respectively, in the presence of 100 μM ATP. The KmATP values of mBME, mSTE and mVCE were 76.6 ± 1.1, 9.1 ± 0.2, and 8.8 ±μ 1.3 μM, respectively, in the presence of 100 μM CDP-ME. The calculated kcat and kcat/Km values are summarized in Table 1.
Fig. 5.
TLC analysis of the product generated by Rv1011 I. a. The reaction mixtures containing CDP-ME, MgCl2, and [γ-32P]ATP without Rv1011 I (lane 1) and with Rv1011 I (lane 2). b. Co-migration experiment with a reaction containing [γ-32P]ATP, CDP-ME, and Rv1011 I in the presence of MnCl2 (lane 2) and a reaction containing [γ-32P]ATP, CDP-ME, Rv1011 I, and M. tuberculosis IspF (Eoh et al., 2009) in the presence of MgCl2 (lane 3). c. Reaction mixture containing CDP-ME and different amount of wild type M. tuberculosis IspE. No (lane 1), 20 μg (lane 2), 40 μg (lane 3), and 60 μg (lane 4) of M. tuberculosis IspE were used. The migration positions of ATP, CDP-ME2P, and MECPP are indicated by the arrows.
Table 1. Calculated kinetic parameters for bacterial IspE orthologs.
| Source of IspE | Substrates |
Km (μM) |
Vmax (μmole/min) |
kcat (S-1) |
kcat/Km (S-1μM-1) |
|---|---|---|---|---|---|
| Wild type M. tuberculosis IspE | CDP-ME | 206.7 ± 12.5 | 2.4 | N.A. | N.A. |
| ATP | 20.7 ± 1.2 | 2.0 | N.A. | N.A. | |
| Rv1011 I (Δ301-305) | CDP-ME | 327.8 ± 21.4 | 2.8 | 480.1 | 1.5 |
| ATP | 75.2 ± 0.9 | 2.5 | 428.7 | 5.7 | |
| B. mallei IspE | CDP-ME | 31.3 ± 1.5 | 2.7 | 469.8 | 15.0 |
| ATP | 76.6 ± 1.1 | 2.4 | 417.6 | 5.5 | |
| S. Typhi IspE | CDP-ME | 25.4 ± 0.8 | 2.6 | 454.0 | 17.9 |
| ATP | 9.1 ± 0.2 | 2.1 | 366.7 | 40.2 | |
| V. cholerae IspE | CDP-ME | 52.9 ± 11.7 | 2.9 | 487.0 | 9.2 |
| ATP | 8.8 ± 1.3 | 2.1 | 352.7 | 40.1 |
The Km and Vmax values were calculated from averaged data from three independent experiments, using nonlinear regression analysis (SigmaPlot V.8.02A). In the reaction mixtures, 97.2 pmol of Rv1011 I, 95.5 pmol of B. mallei IspE, 99.2 pmol of S. Typhi IspE, and 95.8 pmol of V. cholerae IspE were used. N.A., not applicable.
Determination of Z′–Factor values
The kinetic data determined above was used to develop a fluorescence based in vitro assay that is amenable to HTS. In this assay, the release of ATP is coupled to pyruvate kinase, pyruvate oxidase, and horseradish peroxidase, to produce a fluorescent dye (resorufin) which can be measured spectrophotometically and is proportional to IspE catalytic activity. The Z′-Factor values for the spectrophotometry based in vitro assay were calculated based on 96 negative controls (without enzyme) and 96 positive controls (with enzyme) in microplates using the equation:
where σ represents the standard deviation and μ is the mean of each set of data points (Zhang et al., 1999). The Z′-Factor values for the 50 μl assays using Rv1011 I was calculated to be 0.82.
Identification of an unknown product
Interestingly, a product other than CDP-ME2P was produced, apparently as the sole product, when the reaction mixtures contained M. tuberculosis IspE or mSTE and Mn2+ or Zn2+ (Fig. 5b). TLC analysis showed that the Rf value of the unknown product is 0.34 and the amount of product generated was proportional to the amount of Mn2+ or Zn2+ added to the reaction mixtures (data not shown). Co-migration experiments of the unknown product with 2C-methyl-D-erythritol 2,4-cyclodiphosphate (MECPP) showed that the Rf values of two products were similar (Fig. 5b), and the unknown product did not co-migrate with authentic 2C-methyl-D-erythritol diphosphate. Thus, the data suggest that the new product could be MECPP. However, reaction mixtures containing cytosolic fractions of wild type M. tuberculosis H37Rv without addition of exogenous divalent cations produced only CDP-ME2P (Fig. 5c), indicating M. tuberculosis IspE only produces CDP-ME2P under physiological conditions; therefore, the unidentified product was not further characterized.
Discussion
M. tuberculosis IspD and IspF have previously been shown to be essential (Buetow et al., 2007; Eoh et al., 2007), indicating that the M. tuberculosis MEP pathway is vital for M. tuberculosis growth (Sassetti et al., 2003). While, M. tuberculosis IspE has been predicted to be essential (Sassetti et al., 2003), this has yet to be formally demonstrated. Attempts were made to demonstrate ispE essentiality in M. tuberculosis using previously reported methods (Eoh et al., 2007; Parish and Stoker, 2000). Of more than 140 single crossover strains tested, none had chromosomal ispE deleted, suggesting M. tuberculosis ispE is essential for the bacillary survival. However, attempts to obtain a chromosomal deletion in the merodiploid background (complemented) were unsuccessful due to unidentified technical difficulties, possibly a lack of site specific recombination in the ispE locus. Therefore, M. smegmatis MSMEG_5436 (an ispE homolog) was utilized to confirm the essentiality of mycobacterial IspE by an allelic disruption method, in that species. These results strongly suggest that ispE is essential in M. tuberculosis as was predicted (Sassetti et al., 2003).
Although the protein is predicted to be cytosolic, purification of full-length native M. tuberculosis IspE was not possible under any conditions tested, as reported previously (Sgraja et al., 2008). Based on sequence alignments with orthologs from E. coli and Aquifex aeolicus, which can be expressed as soluble native enzymes, it is not clear why this should be the case; however, this was also true for native IspE from the other pathogens studied here including B. malli, S. Typhi, and V. cholerae. Truncation of the amino acids at N-terminal or C-terminal end of M. tuberculosis IspE improved the solubility and retained the activity of the expressed proteins. Notably, the truncated version of M. tuberculosis IspE, Rv1011 I (Fig. 3b), obtained by removal of five amino acids (A-P-T-E-V) at C-terminus showed levels of activity comparable to those of wild type IspE found in M. tuberculosis cytosol.
Out of eight truncated versions of M. tuberculosis IspE generated in this study, only three (Rv1011 I, II and IV) showed IspE activity and only Rv1011 I and II, harboring intact ThrB, GHMP kinase domains and N-termini (amino acids 1- 36) showed significant IspE activities, suggesting that the three conserved motifs underlined in Fig. 3a are critical for the activity. The first region (K-L-N-L-F-L, of E. coli IspE) is reported to be involved in CDP-ME binding (Miallau et al., 2003; Sgraja et al., 2008). The second region (G-L-G-G-G-S, of E. coli IspE) and third region (L-T-G-T-G, of E. coli IspE) constitute an ATP binding motif (Sgraja et al., 2008; Miallau et al., 2003). The N-terminal region (amino acids 1 - 36) of M. tuberculosis IspE is also reported to be involved in forming CDP-ME binding domain (Miallau et al., 2003). The five amino acids removed from M. tuberculosis IspE overhang the C-terminus of E. coli IspE in multiple alignments (Fig. 3a) and purified Rv1011 I showed IspE activity similar to wild type IspE. Therefore, the five amino acids at C-terminus of M. tuberculosis IspE are relatively less critical for activity but appear to be a major determinant for solubility of heterologously expressed recombinant proteins. Soluble, active IspE enzymes from B. mallei, S. Typhi, and V. cholerae were also obtained by truncating five amino acids at the C-terminus, even though these amino acids do not align with each other, are not conserved between the species and their removal does not result in generation of a conserved C-terminus.
The pH and divalent cation usage required for optimal activities of the bacterial IspE orthologs tested here are somewhat different from those of previously reported for the A. aeolicus IspE (Sgraja et al., 2008). The activities of four IspE orthologs studied here were retained at acidic pH (5.5) and abolished at basic pH (> 10.0). However, the activity of A. aeolicus IspE was very low at acidic pH (5.5 - 6.0) and retained at basic pH (9.0). In terms of divalent cation usage, Mg2+ at 6 mM supported optimal IspE activity and concentrations higher than 10 mM Mg2+ inhibited the IspE activity for all four species used here. Divalent cations other than Mg2+ used in this study supported significantly reduced IspE activity; Mn2+ allowed production of CDP-ME2P at low concentrations and Zn2+ did not support IspE activity (Fig. 4b and supplemental data, S5). Ca2+ supported mBME and mVCE activities at concentrations below 25 mM. However, Mn2+ supports A. aeolicus IspE activity at levels comparable to Mg2+ (Sgraja et al., 2008). It should be noted that the characterization of A. aeolicus IspE was performed coupled to pyruvate kinase and lactate dehydrogenase; therefore, the reported effects of pH and divalent cations on A. aeolicus IspE may be complicated.
The kinetic parameters obtained using Rv1011 I were comparable to those of wild type IspE in M. tuberculosis cytosolic fraction (Table 1). The KmCDP-ME value of Rv1011 I is similar to that reported for E. coli (150 μM) (Bernal et al., 2005) and A. aeolicus (121 μM) (Sgraja et al., 2008). The KmCDP-ME values of mBME, mSTE, and mVCE are similar each other, but are lower than those reported for the IspE enzymes of E. coli, A. aeolicus and Rv1011 I. The KmATP values for Rv1011 I and mBME are similar but somewhat higher than KmATP values for mSTE and mVCE; however, all four values are lower than those previously reported for E. coli (420 μM) (Bernal et al., 2005) and A. aeolicus (222 μM) (Sgraja et al., 2008). Although the Vmax and kcat values of IspE enzymes from all four species studied here are similar, the kcat/Km values were somewhat different depending upon the origin of enzyme.
Rv1011 I and mSME produced an unknown compound in the presence of Mn2+ or Zn2+, which has Rf value of 0.34 as determined by TLC analysis (Fig. 5b). A previous report indicates that MECPP, the product of IspF, has an Rf value of 0.34 on TLC in the same solvent system (Testa et al., 2006; Kishida et al., 2003). Co-migration experiments showed that new compound has the same Rf value as MECPP (Fig. 5b) although the compound was not rigorously characterized. It is interesting that IspE may be able to generate MECPP from CDP-ME without producing CDP-ME2P under specific in vitro conditions. One report predicted that CDP-ME2P is unstable and naturally hydrolyzed (Kishida et al., 2003); however, the amount of the new compound produced was proportional to the amount of added Mn2+ or Zn2+ in our hands. It should be kept in mind that production of this compound was not observed in reaction mixtures containing wild type IspE in cytosol and endogenous divalent cations, suggesting that CDP-ME2P is the only product of M. tuberculosis IspE in physiological conditions and that the alternate activity is likely an artifact.
To date, characterization and crystallization of IspE from bacterial pathogens have been hampered by a lack of substrate and an inability to isolate a significant quantity of the enzyme. These studies identified a way to obtain the soluble and active IspE enzymes from bacterial pathogens. Moreover, we developed a fluorescence based IspE assay with an excellent Z′-factor value, suggesting that this coupled assay is sufficiently robust to screen for inhibitors. Thus, the results reported here will allow to development of HTS and facilitate elucidation of crystal structures to aid in the identification of IspE specific inhibitors.
Significance
An allelic disruption strategy was employed to confirm the prediction that mycobacterial IspE is essential for bacterial survival; in addition, this study provides first the characterization of IspE from pathogenic bacteria (M. tuberculosis, B. mallei, S. Typhi, and V. cholera). A radiochemical based assay was developed and utilized to directly identify the enzymatic properties of IspE orthologs from the bacterial pathogens. The fluorescence based IspE assay provides a facile and inexpensive HTS assay to screen for IspE specific inhibitors. Since IspE is not targeted by any current anti-microbial drugs, identification of IspE inhibitors could contribute to disease control by providing leads to new drugs for patients with multiple drug resistant or extensively drug resistant tuberculosis or those infected with other pathogenic bacteria. This study lays the groundwork for structural based studies of IspE from pathogenic bacteria and HTS.
Experimental Procedures
Materials
Genomic DNA and a cytosolic fraction isolated from wild type M. tuberculosis H37Rv were provided by TB Vaccine Testing and Research Materials Contract, Colorado State University (NIH NIAID N01-AI-40091). S. Typhi (700931D) and V. cholerae (39315D) genomic DNA were purchased from the American Type Culture Collection (ATCC, Manassas, VA). B. mallei genomic DNA was kindly provided by Dr. Herbert Schweitzer (Colorado State University). All PCR reagents and cloning materials were purchased from Qiagen (Valencia, CA). His-select nickel affinity resin, ATP and reagents were obtained from Sigma-Aldrich (St. Louis, MO). [γ-32P]ATP (6000 Ci/mmol) and PD-10 columns were purchased from Amersham Biosciences (Pittsburgh, PA). The ADP Quest HS Kinase Assay Kit was purchased from GE healthcare Bio-Sciences Corp. (Piscataway, NJ). All other reagents and solvents were of at least analytical grade.
Synthesis of enantiopure CDP-ME
Approximately 0.54 mmol of tertiary alcohol 3 was dissolved in 10 mL of freshly distilled dry tetrahydrofuran (THF) and cooled to 0 °C. To the cooled solution, 1.08 mmol of potassium hydride was added and stirred for 30 min at room temperature. The reaction mixture was cooled to 0 °C again and 1.35 mmol of benzyl bromide was added slowly, warmed to room temperature and stirred for an additional 2 hr. The reaction was quenched by addition of saturated ammonium chloride solution and extracted with ethyl acetate. The crude mixture was purified by silica gel column chromatography using ethyl acetate: hexane (7:3, v/v) as the solvent, yielding 0.486 mmol of benzylated product (90% yield) 4.
Data for 3
1H-NMR (CDCl3, 300 MHz): δ 7.34 (m, 10H), 5.78 (d, 1H, J = 3.6 Hz), 5.02-5.09 (m, 4H), 4.03-4.26 (m, 3H), 3.96 (dd, 1H, J = 2.7 & 7.8 Hz), 1.56 (s, 3H), 1.36 (s, 3H), 1.12 (s, 3H).; 13C-NMR (CDCl3, 75 MHz):135.7, 135.6, 128.6, 128.5, 128.4, 128.0, 127.9, 112.7, 103.4, 83.9, 80.2, 76.1, 69.4, 69.3, 65.9, 26.5, 18.1.; IR (neat, cm−-1): 3588, 2966, 2362, 2336, 1652, 1614. HRMS (ESI) C23H30O8P (M+H+): calculated 465.1673 and found 465.1680; [α]D 33.0 (c 0.5, CHCl3).
Data for 4
1H-NMR (CDCl3, 300 MHz): δ 7.34 (m, 15H), 5.80 (d, 1H, J = 3.6 Hz), 5.02-5.07 (m, 4H), 4.54-4.67 (m, 2H), 4.08-4.35 (m, 4H), 1.60 (s, 3H), 1.37 (s, 3H), 1.20 (s, 3H). HRMS (ESI) C30H36O8P (M+H+): calculated 555.5758 and found 555.5762 ; [α]D 13.0 (c 0.3, CHCl3).
For MEP and cytidine 5′-monophosphate (CMP) coupling, 0.15 mmol of CMP was dissolved in 1 mL of acetonitrile followed by addition of 0.6 mmol of N,N, dimethyl aniline and 0.15 mmol of triethylamine at 0 °C. Then, 0.76 mmol of trifluoroacetic anhydride in acetonitrile was slowly added to the mixture and stirred for 15 min. Excess TFA and anhydride was removed under reduced pressure. To the above reaction mixture, a combination of 0.45 mmol of 1-methyl imidazole and 0.76 mmol of triethylamine in acetonitrile was added slowly and stirred for 30 more min. The activated CMP was added to the mixture of 0.125 mmol of MEP and activated 4 A° molecular sieves in acetonitrile at 0 °C and stirred for 2 hr. After the reaction was completed, the mixture was extracted with chloroform and the aqueous layer was lyophilized. CDP-ME was purified by sequential chromatography on a Bio-Gel® P-2 gel fine column using 100 mM aq. ammonium bicarbonate followed by further purification on a benzyl DEAE cellulose anion exchange column, using a gradient of 0 - 0.5 M aq. ammonium bicarbonate solution. This resulted in a 45 % yield of CDP-ME.
Data for CDP-ME
1H-NMR (D2O, 300 MHz): δ 7.94 (d, 1H, J = 7.5 Hz), 6.09 (d, 1H, J = 7.5 Hz), 5.96 (d, 1H, J = 4.2 Hz), 4.27 (m, 6H), 3.95 (m, 1H), 3.80 (d, 1H, J = 10.5 Hz), 3.57 (d, 1H, J = 11.7 Hz), 3.44 (d, 1H, J = 11.7 Hz), 1.10 (s, 3H). HRMS (ESI) C14H25O14N3P2 (M-H+) calculated 520.0728 and found 520.0724; [α]D 13.7 (c 0.2, H2O).
Construction of vectors for the determination of mycobacterial IspE essentiality
Initially, attempts were made to demonstrate ispE essentiality in M. tuberculosis using previously reported methods (Eoh et al., 2007; Parish and Stoker, 2000); however, attempts to obtain a chromosomal deletion in the merodiploid background (complemented) were unsuccessful, due to unidentified technical difficulties. Thus, we utilized allelic disruption to demonstrate essentiality in the related species, M. smegmatis. The construction of M. smegmatis ispE ortholog (MSMEG_5436) replacement plasmid (pPR27∷Sm ispE∷KanR) and a rescue plasmid (pCG76-Sm ispE) is illustrated in Fig 2a. Briefly, a 2849 bp DNA fragment containing M. smegmatis ispE ortholog (MSMEG_5436), 940 bp upstream, and 940 bp downstream of the gene was amplified by PCR using the MutSmispE-F and R (Supplemental Data S3). The DNA fragment was cloned to pGEM T plasmid and a 1250 bp Kanamycin resistance cassette (KanR) was cut out with HincII from plasmid pUC4K, and inserted into the Klenow fragment (Invitrogen)-filled RsrII site of the MSMEG_5436 gene. A 2849 bp SM ispE∷KanR fragment was cut by SpeI and XbaI and moved to the same enzyme site of pPR27 (temperature sensitive mycobacterial origin of replication; sacB, xylE, Gentamicin resistant) (Pelicic et al., 1997), yielding pPR27∷SM ispE∷KanR, the vector used to achieve homologous recombination event at the SMEG_5436 locus of M. smegmatis. The pPR27 used in this study has the mycobacterial temperature-sensitive origin of replication. Thus, pPR27∷SM ispE∷KanR can replicate at 32°C but is efficiently lost at 42 °C and above (Pelicic et al., 1997; Pan et al., 2001). In addition, the plasmid also contains the counterselectable marker sacB from Bacillus subtilis (Pelicic et al., 1997) for use in selection of the double-crossover event in the presence of sucrose.
The entire M. smegmatis MESG_5436 gene was amplified by PCR with using primer set (SmispE-F and R) (Supplemental Data S3) and cut with NdeI and BamHI and ligated into the same enzyme sites downstream of the heat shock promoter Phsp60 in plasmid pCG76 (E. coli/Mycobacterium shuttle vector carrying a temperature sensitive origin of replication, and streptomycin resistance), yielding pCG76∷SM ispE (Fig. 2a). Plasmid pCG76 carries the same temperature-sensitive mycobacterial replication origin as pPR27 and thus can replicate at the permissive temperature of 32°C but is abrogated at 42°C and above (Jackson et al., 2000).
PCR amplification of putative ispE orthologs of M. tuberculosis, B. mallei, S. Typhi, and V. cholerae
Homologs of E. coli IspE from the genomes of bacterial pathogens were identified by sequence alignment. The M. tuberculosis H37Rv genome is available on the Tuberculist website (http://genolist.pasteur.fr/Tuberculist) and other three bacterial genome sequences are available on the Genbank of National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/Genbank/). The DNA sequences were used to design oligonucleotides for amplification of putative ispE open reading frames. The PCR primers (Supplemental Data S3) were designed to contain NdeI or HindIII restriction enzyme sites in the forward and reverse primers, respectively.
Cloning of eight M. tuberculosis ispE fragments
Purification of the native form of the putative M. tuberculosis IspE (Rv1011) proved difficult as reported previously (Sgraja et al., 2008); however, soluble M. tuberculosis IspE was obtained by removal of amino acid residues at either the N-terminus, C-terminus or both termini. Genetic modifications were designed using conserved domain searches for M. tuberculosis IspE as a query performed by tools available in the NCBI (specialized BLAST) (Fig. 3b). The PCR products of eight Rv1011 fragments were amplified using the primer sets shown in Supplemental Data S3. For amplification of Rv1011 fragments I through VIII (Fig. 3b), primers were designed based on the sequences available in TubercuList (Supplemental Data S3) and were synthesized by Macromolecular Resources (Colorado State University). The eight amplified PCR products were digested with NdeI and HindIII, and ligated into the pET28a(+) vector (EMD Biosciences, Inc., San Diego, CA). The ligation mixtures were used to transform the E. coli DH5α cloning host (Life Technologies, Rockville, MD), generating DH5α[pET28a(+)∷ Rv1011 I] through DH5α[pET28a(+)∷ Rv1011 VIII]), which were subsequently purified using Gene Extraction Kits (Qiagen, Valencia, CA) and sequenced by Macromolecular Resources.
Cloning of truncated ispE orthologs of B. mallei, S. Typhi, and V. cholerae
Forward and reverse primers for amplifying ispE of B. mallei, S. Typhi, and V. cholerae were designed to remove the five C-terminal amino acids (Supplemental Data S3), resulting in generation of three truncated versions of bacterial IspE enzymes. Expression constructs, pET28a(+)∷mBME, pET28a(+)∷mSTE, and pET28a(+)∷mVCE were generated as described above.
Expression and purification of the recombinant proteins
Recombinant Rv1011 I through VIII, mBME, mSTE, and mVCE were expressed and purified as previously described (Eoh et al., 2007; Mao et al., 2008). Briefly, transformation of expression E. coli host, BL21(DE3) (Novagen, Madison, WI), with expression plasmids afforded the recombinant strains, BL21(DE3)[pET28a(+)∷ Rv1011 I] through BL21(DE3)[pET28a(+)∷ Rv1011 VIII], BL21(DE3)[pET28a(+)∷mBME], BL21(DE3)[pET28a(+)∷mSTE], and BL21(DE3)[pET28a(+)∷mVCE]. Cells were grown to an O.D600 of 0.6, treated with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 20 °C for 16 hr, harvested by centrifugation, resuspended in binding buffer [50 mM 4-morpholine propane sulfonic acid (MOPS) (pH7.9), 1 mM MgCl2, 10% glycerol, and 1 mM β-mercaptoethanol], and disrupted by sonication using a Sanyo Soniprep 150 (Integrated Services, TCP Inc, Palisdes Park, NJ) on ice. The recombinant proteins carrying hexa-His tags were purified by immobilized metal affinity chromatography (IMAC) using a Ni-NTA resin (Sigma-Aldrich, St. Luis, MO) and analyzed by SDS-PAGE and Western blot analysis with using an anti-His antibody (Sigma-Aldrich). Fractions containing pure recombinant protein were desalted using PD-10 columns and concentrated using a 5,000 molecular weight cutoff Centricon (Millipore, Bedford, MA).
In vitro radiochemical IspE assay
For enzymatic characterization, IspE assays were performed in reaction mixtures containing 50 mM Tris-Cl (pH 7.0), 100 μM [γ-32P]ATP (100 dpm/pmol), 2 mM DTT, 100 μM CDP-ME, and 5 mM MgCl2 in a 50 μl final reaction volume. Reactions were initiated by addition of recombinant IspE (97.2 pmol of Rv1011 I, 95.5 pmol of mBME, 99.2 pmol of mSTE, and 95.8 pmol of mVCE), incubated at 37 °C for 30 min, and terminated by the addition of 10 mM of EDTA (pH 8.0). TLC analysis was performed by transferring 10 μl of the reaction mixture to a TLC plate (Polygam Sil N-HR, Macherey and Nagel) and developing with n-propanol/ethyl acetate/H2O (6:1:3,v/v/v). Location and quantitation of radiolabeled products on the TLC plates was determined by using a Molecular Dynamics Typhoon 8600 Phosphoimager.
Fluorescence based IspE assay
For high throughput screening purposes, ADP released from the IspE catalyzed reaction was coupled to commercially available the ADP Quest HS Kinase Assay Kit containing pyruvate kinase, pyruvate oxidase, and horseradish peroxidase, to produce a fluorescent dye (resorufin). The reaction mixtures, containing 50 mM Tris-Cl (pH 7.0), 100 μM ATP, 100 μM CDP-ME, 5 mM phosphatase inhibitor (Roche Bioscience, Palo Alto, CA), 5 mM MgCl2 and 97.2 pmols Rv1011 I in a 50μl final reaction volume in 96 well black microplates with clear bottom (Costar, NY), were incubated at 37 °C for 30 min. Then, 25 μl of reagent A and 50 μl of reagent B (ADP Quest HS Kinase Assay Kit) were added, sequentially. After incubating for another 15 min at room temperature, fluorescence was measured using a SynergyTM Multi-Detection Microplate Reader (BioTek instruments) at an excitation wavelength of 530 nm and emission wavelength of 590 nm.
Determination of Z′–Factor value
Z′–factor values for fluorescence based IspE assays were determined as was previously reported (Zhang et al., 1999). All steps of the assay were performed using an automated fluid transfer system (Precision XS microplate sample processor, BioTek instruments) on 96-well microplates. A total of 96 assays were performed for both positive (with enzyme) and negative controls (without enzyme).
Enzymatic properties
The effect of ATP concentration on IspE activity was determined using a constant concentration of CDP-ME (100 μM) and varying concentrations of ATP. The effect of CDP-ME concentration was determined using a constant concentration of ATP (100 μM) and varying concentrations of CDP-ME. The Km and Vmax values of substrates for the enzyme were calculated using non-linear regression analysis (SigmaPlot V.8.02A). To determine the pH optima of Rv1011 and IspE orthologs, appropriate buffers (MES, MOPS, Tris, TAPS, or CAPS) were used. Optimal concentrations for divalent cations were determined in assay mixtures containing MgCl2, CaCl2, ZnCl2, or MnCl2 at the indicated concentrations.
Characterization of an unknown product generated by M. tuberculosis IspE
An unknown product produced by Rv1011 in the presence of Mn2+ or Zn2+ was scraped from the TLC plates and extracted with n-propanol/ethyl acetate/H2O (6:1:3, v/v/v). The residual silica powder was removed by filtering and the solvent was removed under N2 gas. The extracts were analyzed by mass spectrometry and 1H NMR. The mobility of the unknown product and M. tuberculosis 2-C-methyl-D-erythritol 2,4 cyclodiphosphate (MECPP) on TLC plates was also compared. Authentic MECPP was obtained by enzymatic synthesis in reaction mixtures containing Rv1011 I, CDP-ME, [γ-32P]ATP, and 112.6 pmol of recombinant M. tuberculosis IspF (Rv3581c) (Eoh et al., 2009), which had been incubated at 37 °C for 30 min.
Supplementary Material
Acknowledgments
Funding for this research was provided by grants from the National Institutes of Allergy and Infectious Diseases, NIH: AI-065357.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreassi JL, Leyh TS. Molecular functions of conserved aspects of the GHMP kinase family. Biochemistry. 2004;43:14594–14601. doi: 10.1021/bi048963o. [DOI] [PubMed] [Google Scholar]
- Bernal C, Mendez E, Terencio J, Boronat A, Imperial S. A spectrophotometric assay for the determination of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase activity. Anal Biochem. 2005;340:245–251. doi: 10.1016/j.ab.2005.01.055. [DOI] [PubMed] [Google Scholar]
- Brown AC, Parish T. Dxr is essential in Mycobacterium tuberculosis and fosmidomycin resistance is due to a lack of uptake. BMC Microbiol. 2008;8:78. doi: 10.1186/1471-2180-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buetow L, Brown AC, Parish T, Hunter WN. The structure of Mycobacteria 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase, an essential enzyme, provides a platform for drug discovery. BMC Struct Biol. 2007;7:68. doi: 10.1186/1472-6807-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassera MB, Gozzo FC, D'Alexandri FL, Merino EF, del Portillo HA, Peres VJ, Almeida IC, Eberlin MN, Wunderlich G, Wiesner J, Jomaa H, Kimura EA, Katzin AM. The methylerythritol phosphate pathway is functionally active in all intraerythrocytic stages of Plasmodium falciparum. J Biol Chem. 2004;279:51749–51759. doi: 10.1074/jbc.M408360200. [DOI] [PubMed] [Google Scholar]
- Chang MC, Keasling JD. Production of isoprenoid pharmaceuticals by engineered microbes. Nat Chem Biol. 2006;2:674–681. doi: 10.1038/nchembio836. [DOI] [PubMed] [Google Scholar]
- Clarke SC. Bacteria as potential tools in bioterrorism, with an emphasis on bacterial toxins. Br J Biomed Sci. 2005;62:40–46. doi: 10.1080/09674845.2005.11732685. [DOI] [PubMed] [Google Scholar]
- Cornish RM, Roth JR, Poulter CD. Lethal mutations in the isoprenoid pathway of Salmonella enterica. J Bacteriol. 2006;188:1444–1450. doi: 10.1128/JB.188.4.1444-1450.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcic R, Dhandayuthapani S, Deretic V. Gene expression in mycobacteria: transcriptional fusions based on xylE and analysis of the promoter region of the response regulator mtrA from Mycobacterium tuberculosis. Mol Microbiol. 1994;13:1057–1064. doi: 10.1111/j.1365-2958.1994.tb00496.x. [DOI] [PubMed] [Google Scholar]
- Eoh H, Brennan PJ, Crick DC. The Mycobacterium tuberculosis MEP (2C-methyl-d-erythritol 4-phosphate) pathway as a new drug target. Tuberculosis (Edinb) 2009;89:1–11. doi: 10.1016/j.tube.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eoh H, Brown AC, Buetow L, Hunter WN, Parish T, Kaur D, Brennan PJ, Crick DC. Characterization of the Mycobacterium tuberculosis 4-diphosphocytidyl-2-C-methyl-D-erythritol synthase: potential for drug development. J Bacteriol. 2007;189:8922–8927. doi: 10.1128/JB.00925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freiberg C, Wieland B, Spaltmann F, Ehlert K, Brotz H, Labischinski H. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J Mol Microbiol Biotechnol. 2001;3:483–489. [PubMed] [Google Scholar]
- Hunter WN. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J Biol Chem. 2007;282:21573–21577. doi: 10.1074/jbc.R700005200. [DOI] [PubMed] [Google Scholar]
- Jackson M, Crick DC, Brennan PJ. Phosphatidylinositol is an essential phospholipid of mycobacteria. J Biol Chem. 2000;275:30092–30099. doi: 10.1074/jbc.M004658200. [DOI] [PubMed] [Google Scholar]
- Kishida H, Wada T, Unzai S, Kuzuyama T, Takagi M, Terada T, Shirouzu M, Yokoyama S, Tame JR, Park SY. Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis. Acta Crystallogr D Biol Crystallogr. 2003;59:23–31. doi: 10.1107/s0907444902017705. [DOI] [PubMed] [Google Scholar]
- Luttgen H, Rohdich F, Herz S, Wungsintaweekul J, Hecht S, Schuhr CA, Fellermeier M, Sagner S, Zenk MH, Bacher A, Eisenreich W. Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-D-erythritol. Proc Natl Acad Sci U S A. 2000;97:1062–1067. doi: 10.1073/pnas.97.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Eoh H, He R, Wang Y, Wan B, Franzblau SG, Crick DC, Kozikowski AP. Structure-activity relationships of compounds targeting Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate synthase. Bioorg Med Chem Lett. 2008;18:5320–5323. doi: 10.1016/j.bmcl.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miallau L, Alphey MS, Kemp LE, Leonard GA, McSweeney SM, Hecht S, Bacher A, Eisenreich W, Rohdich F, Hunter WN. Biosynthesis of isoprenoids: crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritol kinase. Proc Natl Acad Sci U S A. 2003;100:9173–9178. doi: 10.1073/pnas.1533425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanasamy P, Eoh H, Crick DC. Chemoenzymatic synthesis of 4-diphosphocytidyl-2-C-methyl-D-erythritol: a substrate for IspE. Tetrahedron Lett. 2008;49:4461–4463. doi: 10.1016/j.tetlet.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Jackson M, Ma Y, McNeil M. Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J Bacteriol. 2001;183:3991–3998. doi: 10.1128/JB.183.13.3991-3998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas G, Panagopoulou P, Christou L, Akritidis N. Category B potential bioterrorism agents: bacteria, viruses, toxins, and foodborne and waterborne pathogens. Infect Dis Clin North Am. 2006;20:395–421. x. doi: 10.1016/j.idc.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Parish T, Stoker NG. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000;146(Pt 8):1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- Pelicic V, Jackson M, Reyrat JM, Jacobs WR, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard SB, Bowman ME, Kwiatkowski W, Kang I, Chow C, Lillo AM, Cane DE, Noel JP. Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate-independent isoprenoid biosynthesis. Nat Struct Biol. 2001;8:641–648. doi: 10.1038/89691. [DOI] [PubMed] [Google Scholar]
- Rohdich F, Schuhr CA, Hecht S, Herz S, Wungsintaweekul J, Eisenreich W, Zenk MH, Bacher A. Biosynthesis of isoprenoids. A rapid method for the preparation of isotope-labeled 4-diphosphocytidyl-2C-methyl-D-erythritol. J Am Chem Soc. 2000;122:9571–9574. [Google Scholar]
- Rohmer M. Isoprenoid biosynthesis via the mevalonate-independent route, a novel target for antibacterial drugs? Prog Drug Res. 1998;50:135–154. doi: 10.1007/978-3-0348-8833-2_3. [DOI] [PubMed] [Google Scholar]
- Romanowski MJ, Bonanno JB, Burley SK. Crystal structure of the Streptococcus pneumoniae phosphomevalonate kinase, a member of the GHMP kinase superfamily. Proteins. 2002;47:568–571. doi: 10.1002/prot.10118. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Sauret-Gueto S, Ramos-Valdivia A, Ibanez E, Boronat A, Rodriguez-Concepcion M. Identification of lethal mutations in Escherichia coli genes encoding enzymes of the methylerythritol phosphate pathway. Biochem Biophys Res Commun. 2003;307:408–415. doi: 10.1016/s0006-291x(03)01211-7. [DOI] [PubMed] [Google Scholar]
- Sgraja T, Alphey MS, Ghilagaber S, Marquez R, Robertson MN, Hemmings JL, Lauw S, Rohdich F, Bacher A, Eisenreich W, Illarionova V, Hunter WN. Characterization of Aquifex aeolicus 4-diphosphocytidyl-2C-methyl-d-erythritol kinase - ligand recognition in a template for antimicrobial drug discovery. FEBS J. 2008;275:2779–2794. doi: 10.1111/j.1742-4658.2008.06418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa CA, Brown MJ. The methylerythritol phosphate pathway and its significance as a novel drug target. Curr Pharm Biotechnol. 2003;4:248–259. doi: 10.2174/1389201033489784. [DOI] [PubMed] [Google Scholar]
- Testa CA, Lherbet C, Pojer F, Noel JP, Poulter CD. Cloning and expression of IspDF from Mesorhizobium loti. Characterization of a bifunctional protein that catalyzes non-consecutive steps in the methylerythritol phosphate pathway. Biochim Biophys Acta. 2006;1764:85–96. doi: 10.1016/j.bbapap.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Yang D, Shipman LW, Roessner CA, Scott AI, Sacchettini JC. Structure of the Methanococcus jannaschii mevalonate kinase, a member of the GHMP kinase superfamily. J Biol Chem. 2002;277:9462–9467. doi: 10.1074/jbc.M110787200. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zhou T, Daugherty M, Grishin NV, Osterman AL, Zhang H. Structure and mechanism of homoserine kinase: prototype for the GHMP kinase superfamily. Structure. 2000;8:1247–1257. doi: 10.1016/s0969-2126(00)00533-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.