Abstract
Ergothioneine is a small, sulfur-containing metabolite (229 Da) synthesized by various species of bacteria and fungi, which can accumulate to millimolar levels in tissues or cells (e.g. erythrocytes) of higher eukaryotes. It is commonly marketed as a dietary supplement due to its proposed protective and antioxidative functions. In this study we report the genes forming the two-step ergothioneine biosynthetic pathway in the fission yeast, Schizosaccharomyces pombe. We identified the first gene, egt1+ (SPBC1604.01), by sequence homology to previously published genes from Neurospora crassa and Mycobacterium smegmatis. We showed, using metabolomic analysis, that the Δegt1 deletion mutant completely lacked ergothioneine and its precursors (trimethyl histidine/hercynine and hercynylcysteine sulfoxide). Since the second step of ergothioneine biosynthesis has not been characterized in eukaryotes, we examined four putative homologs (Nfs1/SPBC21D10.11c, SPAC11D3.10, SPCC777.03c, and SPBC660.12c) of the corresponding mycobacterial enzyme EgtE. Among deletion mutants of these genes, only one (ΔSPBC660.12c, designated Δegt2) showed a substantial decrease in ergothioneine, accompanied by accumulation of its immediate precursor, hercynylcysteine sulfoxide. Ergothioneine-deficient strains exhibited no phenotypic defects during vegetative growth or quiescence. To effectively study the role of ergothioneine, we constructed an egt1+ overexpression system by replacing its native promoter with the nmt1+ promoter, which is inducible in the absence of thiamine. We employed three versions of the nmt1 promoter with increasing strength of expression and confirmed corresponding accumulations of ergothioneine. We quantified the intracellular concentration of ergothioneine in S. pombe (0.3, 157.4, 41.6, and up to 1606.3 µM in vegetative, nitrogen-starved, glucose-starved, and egt1+-overexpressing cells, respectively) and described its gradual accumulation under long-term quiescence. Finally, we demonstrated that the ergothioneine pathway can also synthesize selenoneine, a selenium-containing derivative of ergothioneine, when the culture medium is supplemented with selenium. We further found that selenoneine biosynthesis involves a novel intermediate compound, hercynylselenocysteine.
Introduction
Ergothioneine (EGT) is a sulfur-containing Nα,Nα,Nα-trimethyl-L-histidine-derived metabolite that is synthesized by various species of bacteria and fungi (recently extensively reviewed by Cheah and Halliwell [1]). Higher organisms obtain EGT in food and accumulate it in certain tissues up to millimolar levels [2], [3] through a specific transporter, ETT/OCTN1 [4]. In mammals, large amounts of EGT are found in erythrocytes, bone marrow, liver, kidney, eye lens, and seminal fluid [2], [5], [6]. Nevertheless, EGT is neither a nutrient (it is virtually unmetabolized in humans) nor a vitamin (it is non-essential). EGT is commonly marketed as a dietary supplement or nutraceutical, due to its anti-oxidant properties in vitro, reported in numerous publications [3], [7]–[10]. Direct scavenging of free radicals and chelation of transition metals are the most widely cited possible functions of EGT [10], [11]. However, so far, no rigorous research has conclusively demonstrated any benefit of EGT in vivo. It is unclear whether EGT consumption contributes to human health, and if it does, what daily intake is optimal. It is thus of considerable interest for biologists and medical scientists to uncover the physiological mechanism of EGT at the molecular level. Recently, biosynthetic pathways for EGT have been characterized in Mycobacterium smegmatis [12] and Neurospora crassa [13], allowing the use of genetic methods.
The fission yeast, Schizosaccharomyces pombe, is a suitable model organism for the study of cell division and quiescence [14]–[16]. We have previously established a method of comprehensive metabolomic analysis in S. pombe using liquid chromatography-mass spectrometry (LC-MS) [17]. Among the several hundred observed metabolites, we also identified EGT and described its accumulation under glucose starvation [18] and nitrogen starvation [19]. In addition, we reported abnormally high accumulations of EGT in the proteasome regulatory subunit mutant mts3-1, which suffers from severe oxidative stress caused by mitochondrial dysfunctions in G0 arrest [20], and in the Krüppel-like zinc-finger transcription factor deletion mutant Δklf1, which exhibits cell wall defects in long-term quiescence, accompanied by up-regulation of mitochondrial transcripts [21]. Interestingly, EGT is not synthesized by the budding yeast, Saccharomyces cerevisiae [22], suggesting that not all fungi require EGT for normal cellular function. Due to its simplicity and advanced genetics, S. pombe might represent an ideal unicellular, eukaryotic system to study the biochemical role of EGT.
Here we report identification of genes forming the two-step biosynthetic pathway of EGT from histidine in S. pombe, through the combined use of genetic and metabolomic approaches. Among other compounds, we were able to identify the direct precursor of EGT, hercynylcysteine sulfoxide. We constructed an EGT and hercynylcysteine sulfoxide overproduction system utilizing three different overexpression strains with the inducible nmt1+ promoter. We quantified intracellular EGT content in wild type (WT) as well as overexpression cells by constructing a calibration curve from pure EGT standard injections into the LC-MS. Further, we show the accumulation of EGT under long-term quiescence. Finally, we demonstrate that the EGT pathway can also synthesize selenoneine, a selenium-containing derivative of EGT, the production of which involves a novel intermediate compound, hercynylselenocysteine.
Results
EGT biosynthesis pathway
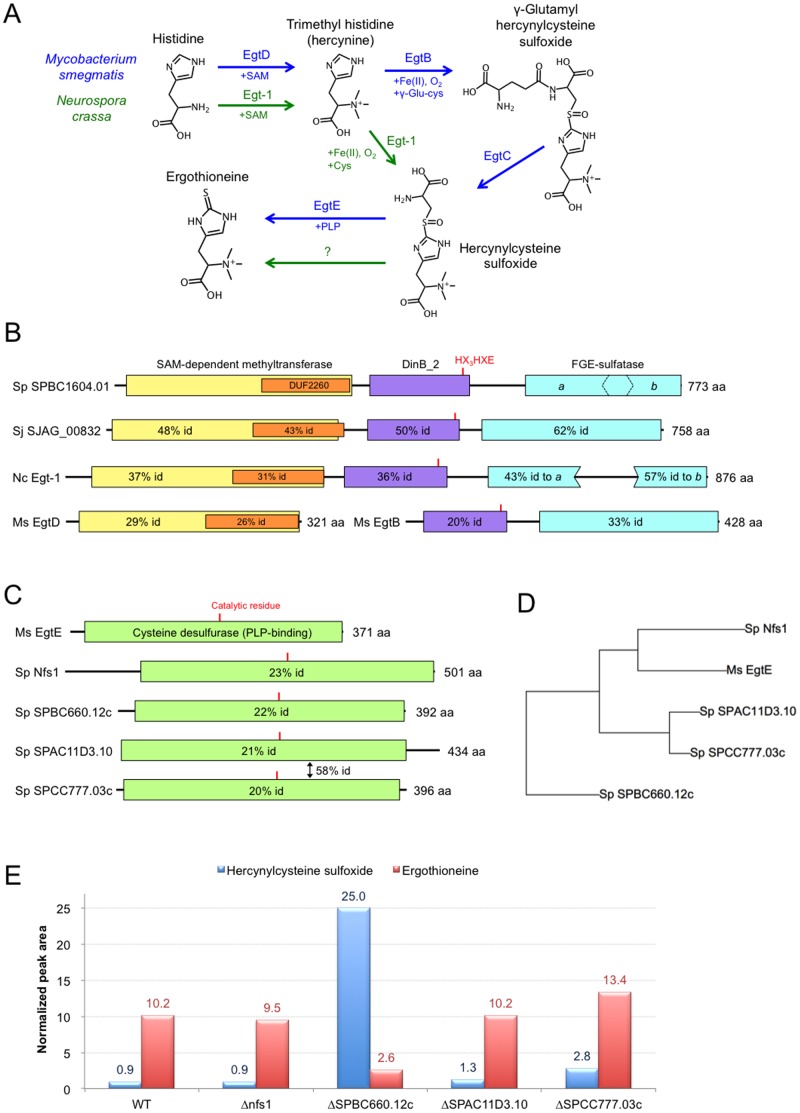
The reported EGT biosynthetic pathways in M. smegmatis and N. crassa are schematized in Figure 1A. On the basis of sequence homology, the S. pombe locus, mug158+/SPBC1604.01, was previously suggested to encode the main EGT biosynthetic enzyme [12], [13]. This enzyme, with 773 amino acids, catalyzes triple methylation of histidine to hercynine (Nα,Nα,Nα-trimethyl histidine) and subsequent conjugation with cysteine and oxygen to form hercynylcysteine sulfoxide. mug158+/SPBC1604.01 is a distant homolog of the mycobacterial EgtD and EgtB genes, encoding a single fusion protein. Figure 1B shows the domain structure of this protein, according to the Conserved Domain Database [23], in comparison with its homologs in Schizosaccharomyces japonicus (a relative of S. pombe), N. crassa, and M. smegmatis. The exact locations and sequences of individual domains are shown in Table S1 and the amino acid sequence alignment is shown in Figure S1. Due to its structure and sequence similarities to NcEgt-1, we presume that the S. pombe homolog also utilizes cysteine as a substrate, rather than using γ-glutamyl-cysteine, as in the case of the bacterial EgtB enzyme. The bacterial pathway requires a subsequent removal of the glutamyl residue by another enzyme, EgtC [12], which has no obvious homolog in the genome of S. pombe. Since the mug158+/SPBC1604.01 gene encodes the first step of EGT synthesis, we hereafter designate it egt1+.
Figure 1. Characterization of EGT biosynthesis in S. pombe.
A. Previously published EGT biosynthesis pathways in M. smegmatis and N. crassa. The Egt-1 protein in N. crassa is a fusion enzyme that catalyzes two different reactions. B. Comparison of the conserved domain structure of S. pombe protein SPBC1604.01 (designated Egt1 in this manuscript) with S. japonicus SJAG_00832 and EGT biosynthesis proteins in N. crassa (Egt-1) and M. smegmatis (EgtD and EgtB). All proteins are composed of: 1. an S-adenosyl-L-methionone (SAM)-dependent methyltransferase domain, including DUF2260, a domain of unknown function; 2. an uncharacterized DinB_2 domain, including an iron-binding motif HX3HXE; and 3. a formylglycine generating enzyme (FGE)-sulfatase domain. Percentage identity (% id) of amino acid sequences is indicated in comparison to the corresponding sequence in S. pombe. C. Comparison of conserved domain structure of M. smegmatis protein EgtE with its four putative homologs in S. pombe. All proteins contain a single pyridoxal phosphate (PLP)-binding cysteine desulfurase domain. The conserved catalytic residues (PLP binding sites) are indicated by red lines. Percentage identity (% id) of the amino acid sequences is indicated in comparison to the corresponding sequence in M. smegmatis. D. Phylogenetic tree visualizing the similarity of amino acid sequences of M. smegmatis EgtE and its putative homologs in S. pombe. E. Normalized peak areas of hercynylcysteine sulfoxide and EGT obtained by metabolomic analysis of WT and deletion mutant S. pombe strains. Cells were nitrogen-starved prior to analysis (24 h in EMM2-N medium) to induce EGT synthesis.
The final step of EGT biosynthesis in bacteria is represented by EgtE, a pyridoxal phosphate (PLP)-binding cysteine desulfurase that forms the end product, EGT, from hercynylcysteine sulfoxide, by cleaving the cysteine residue at the sulfur atom. To date, the EgtE homolog has not been characterized in N. crassa. The S. pombe genome contains four cysteine desulfurases that could be homologous to EgtE: Nfs1 (SPBC21D10.11c), SPBC660.12c, SPAC11D3.10, and SPCC777.03c (Figure 1C; location and sequences of conserved domains in Table S2; amino acid sequence alignment in Figure S2). Among these, SPAC11D3.10, and SPCC777.03c have highly similar sequences, possibly originating from horizontal gene transfer (Figure 1D).
To identify the EgtE homolog in S. pombe, we obtained deletion mutants from the Bioneer haploid deletion library [24], cultivated them under nitrogen starvation (EMM2 medium lacking NH4Cl, hereafter designated EMM2-N) for 24 hours to induce EGT production, and performed a metabolomic analysis. Among the four deletion mutants tested, ΔSPBC660.12c was the only one showing a substantial decrease in EGT and an increase in its precursor, hercynylcysteine sulfoxide (Figure 1E; numerical results of all LC-MS measurements shown in Table S3). We thus designated the SPBC660.12c locus egt2+, as it represents the gene primarily responsible for the second step of EGT biosynthesis in S. pombe.
Verification of egt1+ and egt2+ by metabolomic analysis
To verify correct assignment of the egt1+ and egt2+ genes, we newly constructed full deletion mutants, Δegt1 and Δegt2, by replacing the target loci in the WT h− 972 strain with the kanamycin resistance marker (kanMX). Correct integration of the kanMX modules into the new strains was verified by PCR. No difference from WT in cell size or shape was observed in these strains under any of the analyzed conditions. Table 1 shows the results of metabolome analysis of the constructed strains under starvation (EMM2-N or EMM2 medium with low concentration of glucose, 1.1 mM, hereafter designated EMM2-LG, for 24 hours). Data regarding EGT and its precursors, starting with histidine, are shown. We confirmed the absence of all pathway intermediates in Δegt1, and the accumulation of hercynylcysteine sulfoxide in Δegt2. A small amount of EGT was still found in Δegt2 under starvation. To determine whether any other cysteine desulfurase might contribute to this enzymatic reaction, we constructed multiple deletion mutants of SPBC660.12c (egt2+) with the other candidate EgtE homologs, but even in successfully constructed double and triple mutants, a significant amount of EGT still remained (Figure S3). This, however, is not a surprising result, as Seebeck [12] previously demonstrated that hercynylcysteine sulfoxide could spontaneously convert into EGT in the presence of PLP. Furthermore, as also shown by Seebeck [12], this reaction could be catalyzed by an unrelated PLP-binding β-lyase originating from Erwinia tasmaniensis. Thus, we suspect the residual EGT found in Δegt2 might be a product of a hercynylcysteine sulfoxide reaction with PLP, possibly catalyzed by an unrelated PLP-binding enzyme (S. pombe genome contains at least 26 PLP-binding enzymes, according to PomBase [25]).
Table 1. Normalized peak areas of the four compounds composing the EGT biosynthetic pathway obtained by metabolomic analysis of WT and newly constructed strains.
| Compound, peak m/z and retention time/Strain, | Histidine | Trimethyl-histidine (hercynine) | Hercynyl-cysteine sulfoxide | Ergothio-neine | |
| cultivation condition | 156.077 m/z @12.4 min | 198.124 m/z @10.3 min | 333.123 m/z @12.2 min | 230.096 m/z @12.6 min | |
| EMM2 | 14.4 | 1.7 | 0 | 0.1 | |
| WT 972 | EMM2-N (24 h) | 2.2 | 3.2 | 2.2 | 13.7 |
| EMM2-LG (24 h) | 55.4 | 65.9 | 3.5 | 6.7 | |
| EMM2 | 10.7 | 0.3 | 0 | 0 | |
| Δegt1 | EMM2-N (24 h) | 2.8 | 0.1 | 0 | 0 |
| EMM2-LG (24 h) | 54.1 | 0 | 0 | 0 | |
| EMM2 | 11.4 | 0.9 | 3.9 | 0 | |
| Δegt2 | EMM2-N (24 h) | 1.9 | 1.8 | 58.2 | 3.1 |
| EMM2-LG (24 h) | 61.1 | 64.7 | 44.1 | 1.6 | |
Values were measured from metabolome samples of four different S. pombe strains in three different cultivation conditions, as indicated. Mass values (m/z) and LC retention times (min) of each peak are included for reference.
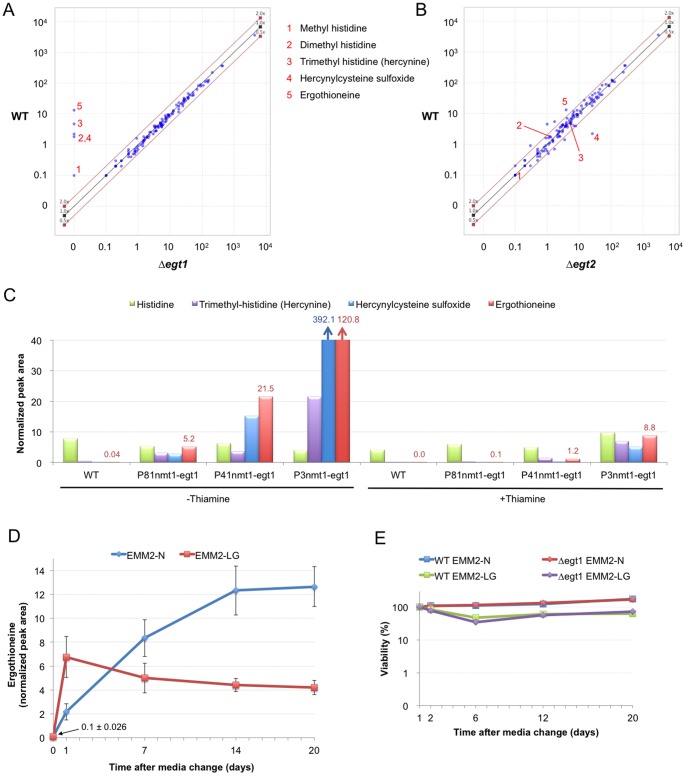
Neither the Δegt1 nor the Δegt2 strain showed any growth defects during cultivation in either rich (YE) or minimal (EMM2) culture media. Furthermore, deletion of egt1+ or egt2+ caused no significant perturbation to the intracellular metabolome of quiescent cells (Figures 2A and 2B). Apart from the disappearance of EGT and its precursors, all other metabolite levels remained within the range of common experimental error.
Figure 2. Characterization of Δegt1 and Δegt2 strains.
A and B. A scatter plot comparing results of metabolomic analysis of WT vs. Δegt1 (A) or Δegt2 (B) strains under nitrogen starvation (24 h in EMM2-N medium). Each dot represents a single identified metabolite. Values on both scales indicate normalized peak areas of metabolite peaks in corresponding strains. Red diagonal lines indicate a 2-fold difference. C. Results of metabolomic analysis of WT and egt1+ overexpression strains. Cells were cultivated at 26°C in the EMM2 medium lacking thiamine for at least 24 h. Cultures indicated +Thiamine were cultivated in the presence of 5 µg/ml thiamine for 24 h. Normalized peak areas of compounds composing the EGT pathway are shown. D. Time course metabolomic analysis of quiescent S. pombe cultures under nitrogen (EMM2-N) and glucose (EMM2-LG) starvation. Values represent means ± standard deviations of normalized peak areas of EGT in three independent cell cultures. E. Time course viability results of WT and Δegt1 deletion mutant cultures under nitrogen (EMM2-N) and glucose (EMM2-LG) starvation.
Conservation of egt1+ and egt2+ in other species
As observed by Seebeck [12], the EGT biosynthestic pathway can be found in a relatively small number of eukaryotes, notably in the phyla Basidiomycota (most species) and Ascomycota (most species in subphyla Pezizomycotina and Schizosaccharomycetes). We summarized homologs of EGT biosynthesis genes in several organisms closely related to S. pombe (Table 2). The exact locations of the conserved domains in these enzymes are shown in Tables S4 and S5. According to sequence alignment (Figures S4 and S5), conserved domains show higher homology among species than inter-domain regions. Methyltransferase and FGE-sulfatase domains contain long non-homologous sequence inserts in U. maydis UM00197. The function of the inserts in these two domains is unknown, however, they are not unusual among various species [13].
Table 2. Closest homologs of S. pombe Egt1 and Egt2 proteins in selected species.
| Organism | Closest homolog of S. pombe Egt1 | Closest homolog of S. pombe Egt2 |
| Schizosaccharomyces japonicus | SJAG_00832 | SJAG_03856 |
| Schizosaccharomyces octosporus | SOCG_01424 | SOCG_02548 |
| Neurospora crassa | NCU04343 (NcEgt-1) | NCU11365 |
| Aspergillus niger | An15g05880 | An02g02030 or An05g02190 |
| Aspergillus oryzae | Ao090012000265 | Ao090026000291 |
| Ustilago maydis | UM00197 | UM04128 |
Candidate homologs were searched using the on-line version of the Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov) for protein sequences (blastp) and candidates with the best similarity scores (lowest blastp E-values) were selected.
Based on sequence homology to S. pombe Egt2 (33% amino acid sequence identity), we propose that the NCU11365 gene may encode the second enzyme for EGT biosynthesis in N. crassa. Bello et al. [13] previously suggested the NCU04636 gene (19% identity to Egt2), which appears to correspond to nfs1 +, the mitochondrial cysteine desulfurase in S. pombe (63% identity). Both S. pombe nfs1 + and N. crassa NCU04636 appear to be the closest homologs of mycobacterial EgtE (as also indicated in Figure 1D). However, we did not detect any change in hercynylcysteine sulfoxide levels in the Δnfs1 deletion mutant (Figure 1E). Furthermore, the nfs1 + gene has apparent homologs in species that do not produce EGT, such as S. cerevisiae or humans (both genes called NFS1).
Overexpression of egt1+
To study the overproduction effect of EGT in S. pombe, we applied the method of Bähler et al. [26] to replace the egt1+ native promoter with the nmt1+ promoter, which is inducible in the absence of thiamine [27]. We employed three versions of the nmt1 promoter plasmid with increasing strength of expression and constructed three strains P81nmt1-egt1+, P41nmt1-egt1+, and P3nmt1-egt1+, respectively. Using metabolomic analysis we confirmed the accumulation of EGT and its precursors in these strains, and this accumulation was effectively suppressed by the addition of 5 µg/ml thiamine to the EMM2 medium (Figure 2C).
Intracellular EGT content
Wild type S. pombe cells contain only trace amounts of EGT under normal vegetative conditions (Table 1). However, EGT increases in quiescent cells under starvation [18], [19]. We measured the areas of EGT peaks under vegetative, quiescent, and egt1+-overexpressing conditions, and converted them to absolute concentrations using a calibration curve based upon pure EGT injections ranging from 1 fmol - 10 nmol (Figure S6). Intracellular volume was assumed to be 148.5 µm3 for vegetative cells [28]. For nitrogen- and glucose-starved cells, intracellular volumes were estimated as 1/3 and 2/3 of the vegetative cell volume, respectively. The resulting intracellular concentrations are shown in Table 3.
Table 3. Absolute intracellular EGT concentrations ( µM) in S. pombe cells.
| Cell condition | Culture medium | Intracellular EGT ( µM) |
| WT vegetative | EMM2 | 0.3 |
| WT nitrogen starvation | EMM2-N (24 h) | 157.4 |
| WT glucose starvation | EMM2-LG (24 h) | 41.6 |
| P81nmt1-egt1+ | EMM2 | 32.4 |
| P41nmt1-egt1+ | EMM2 | 181.2 |
| P3nmt1-egt1+ | EMM2 | 1606.3 |
Intracellular concentrations were derived from measured normalized peak areas using a calibration curve generated by injections of pure EGT in 10-fold dilution steps. The detailed calculation method is described in Figure S6.
To assess long-term variation in EGT content, we measured the level of intracellular EGT using metabolomic analysis during a 20-day time course under quiescence induced by nitrogen (EMM2-N) and glucose (EMM2-LG) starvation, respectively (Figure 2D). Cells showed time-dependent accumulation of EGT, despite being deprived of nutrients, suggesting that EGT might support cellular health under long-term quiescence. However, no loss of viability was observed in Δegt1 mutant during 20 days of starvation (Figure 2E).
Contribution of egt1+ to oxidative stress response
As EGT is generally considered to be a physiological antioxidant, and the ΔNcEgt-1 mutant was reportedly sensitive to tert-butyl hydroperoxide in N. crassa [13], we performed spot test experiments on EMM2 plates containing oxidants hydrogen peroxide and tert-butyl hydroperoxide, using the deletion and overexpression mutants described above (Figure S7). However, we did not observe any sensitivity or resistance of these strains compared to WT 972 strain, suggesting that egt1+ might not be among the primary mechanisms that protect S. pombe from exogenous peroxide.
Biosynthesis of selenoneine
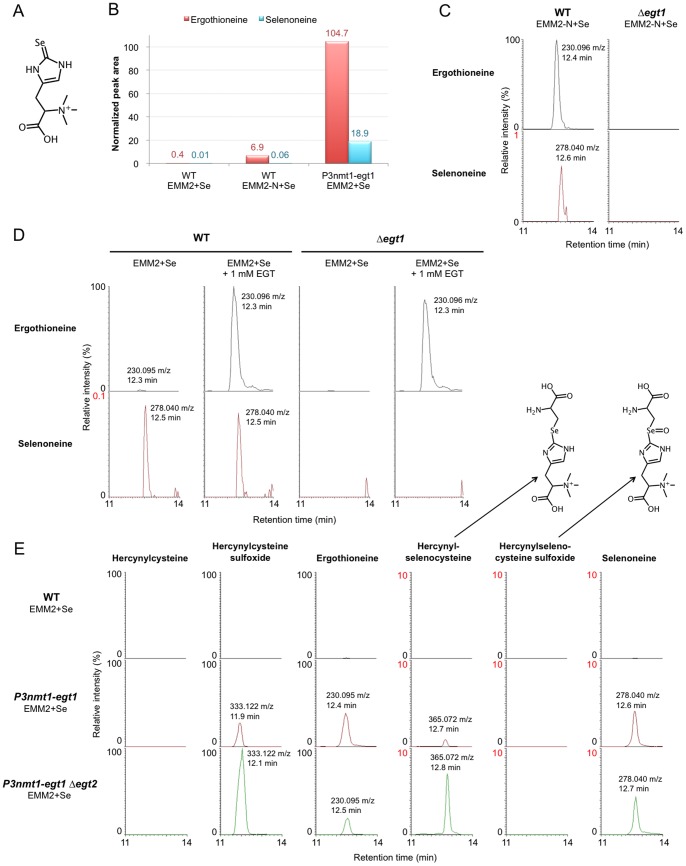
Selenoneine (Figure 3A) is a selenium-containing derivative of EGT found in tuna, and possibly implicated in methylmercury detoxification [29], [30]. As the EMM2 medium does not normally contain selenium, it is not surprising that no selenoneine was detected in previous S. pombe metabolome data sets. To test whether S. pombe can produce selenoneine, we first examined cell cultivation in a liquid EMM2 medium supplemented with various concentrations of Na2SeO4 (Figure S8). The medium with 10 µM Na2SeO4 (hereafter designated EMM2+Se) was selected for further experiments, as S. pombe cells exhibited quite normal (albeit slightly slower) proliferation in this condition. Metabolomic analysis was performed on WT vegetative cells, WT nitrogen-starved cells (24 h in EMM2-N+Se), and P3nmt1-egt1+ overexpression mutant cells. We could clearly observe accumulation of selenoneine in the P3nmt1-egt1+ strain cultivated in EMM2+Se medium (Figure 3B), suggesting that the overexpressed egt1+ gene was also responsible for selenoneine synthesis. A tiny signal of selenoneine (<1% of the EGT signal intensity) could also be detected in WT nitrogen-starved cells, and the signals of both EGT and selenoneine disappeared in the Δegt1 deletion mutant (Figure 3C).
Figure 3. Production of selenoneine in S. pombe.
A. Chemical structure of selenoneine. B. Results of metabolomic analysis of WT and P3nmt1-egt1+ strains in EMM2+Se and EMM2-N+Se media. Normalized peak areas of EGT and selenoneine are shown. C. Extracted ion chromatograms of EGT and selenoneine masses in raw LC-MS data acquired from metabolome samples of nitrogen-starved cells (24 h in EMM2-N+Se medium) of WT and Δegt1 strains. Note that the intensity scale of the selenoneine plot is 1% relative to that of the EGT plot. D. Extracted ion chromatograms of EGT and selenoneine masses in raw LC-MS data acquired from metabolome samples of WT and Δegt1 strains cultivated in EMM2+Se medium with and without supplementation with 1 mM pure EGT. Note that the intensity scale of the selenoneine plot is 0.1% relative to that of the EGT plot. E. Extracted ion chromatograms of six compound masses in WT, P3nmt1-egt1+, and P3nmt1-egt1+ Δegt2 strains. The plots of hercynylcysteine and hercynylselenocysteine sulfoxide show mass values calculated from their predicted chemical formulas (C12H21N4O4S+ = 317.128 m/z for hercynylcysteine, and C12H21N4O5Se+ = 381.067 m/z for hercynylselenocysteine sulfoxide, respectively). Note that the intensity scale of the plots in the right half of the figure is adjusted to 10% relative to the plots in the left half of the figure.
To rule out the possibility that selenoneine was produced by direct conversion from ergothioneine (without requiring any egt1+ activity), we performed two additional experiments. First, pure EGT was mixed with an equal concentration of Na2SeO4 in vitro and incubated at room temperature for 24 h. No selenoneine signal was detected in this mixture (Figure S9). In the second experiment we supplemented the Δegt1 mutant with 1 mM EGT, which was apparently transported into the cells and produced a strong EGT peak. No selenoneine was detected in this case either (Figure 3D). Furthermore, the weak, but clearly detectable selenoneine signal found in WT cells did not increase as a result of EGT supplementation. These results suggest that egt1+ activity is indispensable for selenoneine biosynthesis in S. pombe.
As we did not observe any signal of the presumed intermediate, hercynyl-selenocysteine sulfoxide, we constructed a double mutant of P3nmt1-egt1+ and Δegt2. This mutant should accumulate large amounts of the intermediate, assuming that the Egt1/Egt2 pathway is used for selenoneine synthesis. Surprisingly, we found a strong signal of hercynylselenocysteine (not sulfoxide) in this mutant (Figure 3E). This signal was also found in the P3nmt1-egt1+ single mutant, but further increased in the double mutant with Δegt2. We thus conclude that selenoneine biosynthesis, unlike EGT biosynthesis, does not produce a sulfoxide as its intermediate, but produces hercynylselenocysteine instead.
Since selenium naturally occurs in a very characteristic set of isotopes, we verified the identities of selenoneine and hercynylselenocysteine peaks by checking their isotope distribution patterns (Figure S10). Finally, to check whether the EGT/selenoneine pathway could possibly be involved in selenium detoxification in S. pombe, we performed a spot test experiment on EMM2 plates containing various concentrations of Na2SeO4. However, none of the analyzed mutants showed any growth differences compared with WT cells (Figure S11).
Discussion
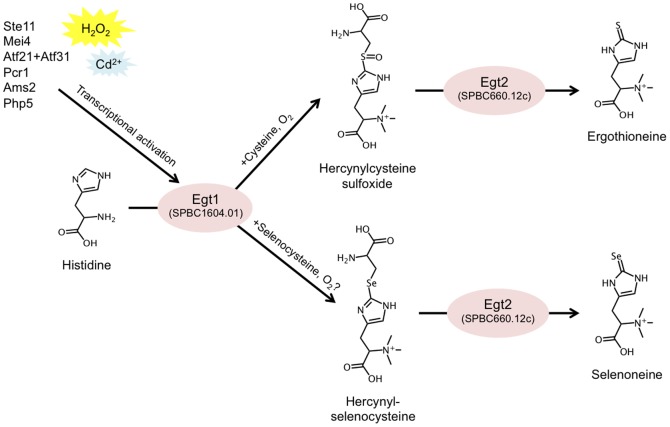
In this study we applied metabolomic analysis to identify the egt1+ and egt2+ genes composing the EGT and selenoneine biosynthetic pathway in S. pombe (Figure 4). These two genes have no homologs in S. cerevisiae, consistent with the fact that budding yeast do not produce EGT. In the future, a comparison between these two yeasts that appear similar, but are genetically rather distant, might provide useful clues regarding the native physiology of EGT or selenoneine. The presence or absence of the EGT pathway could be related to differences in the ecology of these two species.
Figure 4. Summary of the described EGT and selenoneine biosynthetic pathway in S. pombe, and its transcriptional regulation.
In an environmental stress study, transcription of the egt1+ gene was up-regulated ∼10-fold in the presence of 0.5 mM H2O2, and ∼4-fold in the presence of Cd2+ [31]. It thus appears that egt1+ is strongly stress-responsive, providing support for the hypothesized antioxidative role of EGT. In addition, the egt1+ locus was previously named mug158 +, denoting Meiotically Up-regulated Gene 158, due to its up-regulation upon entry into meiosis [32]. The egt1+ promoter contains the TR-box motif 5′-TTCTTTGTTY-3′, recognized by the sexual development transcription factor, Ste11 [33], and a 5′-GTAAAYA-3′-binding motif recognized by the forkhead transcription factor, Mei4 [34]. Indeed, the egt1+ transcript was up-regulated in strains overexpressing ste11 + or mei4+ [35], [36]. Also, egt1+ was up-regulated in a strain overexpressing both basic leucine zipper (bZIP) transcription factors atf21+ and atf31+ [36]. Furthermore, egt1+ was previously reported as a putative regulatory target of the transcription factors, Ams2, Php5, and Pcr1, by analysis of multiple genome-wide microarray datasets [37]. Ams2 is a cell cycle-dependent GATA factor activated during replication [38]. Php5 is a subunit of the CCAAT-binding complex, which regulates respiration, based on glucose and iron availability [39]. Pcr1 is a CREB/ATF protein involved in stress response and sexual development [40], [41]. In addition, the 5′ UTR region of egt1+ contains the CuSE element sequence, 5′-DWDDHGCTGD-3′, which is recognized by the Cuf1 transcription factor and activated by copper deficiency [42]. However, no transcriptional activation of egt1+ was found under varying copper levels [43], suggesting this 5′ UTR DNA fragment might not correspond to an actual Cuf1 binding site. In conclusion, it seems that egt1+ might be under regulatory control of a variety of different transcription factors.
In our study, the Δegt1 deletion mutant showed a complete absence of EGT and all its precursors. In the Δegt2 deletion mutant, some amount of EGT remained, consistent with results previously reported by Seebeck [12], that hercynylcysteine sulfoxide can spontaneously convert to EGT in the presence of PLP. Judging from the wealth of published transcriptome data, transcription of egt2+ does not vary in response to environmental conditions [31], and is only mildly up-regulated (∼2-fold) during meiosis [32], suggesting that egt1+ represents the main regulatory element in this pathway. The Egt2 enzyme could be ubiquitously present in the cells, simply converting any available hercynylcysteine sulfoxide into EGT.
Overexpression strains, P81nmt1-egt1+, P41nmt1-egt1+, and P3nmt1-egt1+, showed gradual accumulation of EGT in standard EMM2 medium without thiamine, providing an additional confirmation of the correct egt1+ assignment. Interestingly, P3nmt1-egt1+ cells contained a large amount of hercynylcysteine sulfoxide. A possible explanation is that the activity of Egt1 in this strain was higher than the native activity of Egt2, resulting in accumulation of the metabolic intermediate between these two enzymes. These overexpression strains will be invaluable for verification of any proposed EGT mechanism. We also demonstrated that the P3nmt1-egt1+ strain could synthesize a considerable amount of selenoneine when selenium was supplemented in the culture medium. Interestingly, selenoneine biosynthesis, unlike EGT biosynthesis, did not involve a formation of a sulfoxide intermediate, but rather involved a simple conjugate compound, hercynylselenocysteine. It is unknown whether egt1 +-mediated synthesis of hercynylselenocysteine requires oxygen, as in the case of EGT synthesis. The P3nmt1-egt1+ strain proved to be very useful to elucidate the complete pathway, and we propose that this strain could also be employed industrially to produce EGT, as well as hercynylcysteine sulfoxide, hercynylselenocysteine, or selenoneine (the latter three compounds are not commercially available at present). The signal intensity of selenoneine found in WT nitrogen-starved cells was rather low; however, selenoneine might potentially have an interesting function in S. pombe. Selenoneine can be found in humans [44], and it was suggested that its radical-scavenging activity is even higher than that of EGT [30]. Importantly, we showed that selenoneine does not seem to be involved in detoxification of selenium in S. pombe.
From a wider perspective, the ubiquitous presence of EGT in living organisms, from bacteria to humans, suggests a crucially important function, yet this function has not been convincingly demonstrated so far. We found only trace amounts (0.3 µM) of EGT in vegetatively growing S. pombe cells; however, under starvation-induced quiescence it increased several hundred-fold. That, together with reported up-regulation of the egt1+ transcript in the presence of H2O2 and in meiosis, implies a supportive or protective role of EGT for long-term hibernation of cells. However, the Δegt1 deletion strain did not show any obvious defects in either sporulation or in quiescence. Furthermore, we did not detect any sensitivity of the Δegt1 mutant to common oxidants H2O2 and tert-butyl hydroperoxide. As cells contain multiple redundant systems to deal with oxidative stress, it is possible that the lack of EGT could be compensated by another mechanism. On the other hand, the observation that the highly overexpressed P3nmt1-egt1+ strain, which accumulated EGT well beyond normal physiological levels, did not acquire any resistance to the tested oxidants, is intriguing. An attractive possibility exists that the true physiological purpose of EGT might lie in a yet unexplored – and possibly unexpected – area. We propose that genetic and metabolomic analyses, together with the collection of S. pombe strains introduced in this manuscript, may provide ideal tools to further investigate the in vivo role of this enigmatic compound.
Materials and Methods
Amino acid sequence alignment and bioinformatic analysis
Amino acid sequence alignment was performed using the on-line version (http://www.ncbi.nlm.nih.gov/tools/cobalt/) of the Constraint-based Multiple Alignment Tool (COBALT) [45] with default parameters. A phylogenetic tree was generated from the on-line COBALT tool (using the Fast Minimum Evolution method with Max Seq Difference set to 0.9 and other parameters set to default values) and visualized using Analyses of Phylogenetics and Evolution (APE) software [46]. Percentage identity of the amino acid sequences was calculated using the Clustal-Omega algorithm [47] with default parameters.
Strains and growth conditions
The S. pombe strains used in this manuscript are listed in Table 4. The synthetic minimal medium (EMM2), rich yeast extract medium (YE) and sporulation-inducing medium (MEA) recipes were used as published previously [48]. The following variants of the liquid EMM2 medium were used: EMM2-N (EMM2 lacking NH4Cl), EMM2-LG (EMM2 containing 1.1 mM – or 0.2 g/l – glucose), EMM2+Se (EMM2 containing additional 10 µM Na2SeO4), and EMM2-N+Se (EMM2-N containing additional 10 µM Na2SeO4). Cell cultures were cultivated at 26°C.
Table 4. S. pombe strains used in this manuscript.
| Strain name | Genotype | Source |
| 972 | h− (WT) | Leupold (1950) [52] |
| 975 | h+ (WT) | |
| KS1366 | h− Δsty1::ura4+ ura4-D18 | Shiozaki and Russell (1995) [53] |
| TP1701 | h− Δnfs1::kanMX4 | Strains from the Bioneer haploid deletion mutant collection [24] were backcrossed with WT 972 to remove auxotrophic markers. |
| TP1704 | h− ΔSPBC660.12c::kanMX4 | |
| TP1705 | h− ΔSPAC11D3.10::kanMX4 | |
| TP1706 | h+ ΔSPCC777.03c::kanMX4 | |
| TP1707 | h− ΔSPAC11D3.10::hphMX6 | Marker switch of TP1705 to hphMX6 |
| TP1732 | h− ΔSPBC660.12c::natMX6 | Marker switch of TP1704 to natMX6 |
| TP1733 | h+ ΔSPBC660.12c::natMX6 | TP1732 crossed with WT 975 |
| TP1736 | h− ΔSPBC660.12c::natMX6 Δnfs1::kanMX4 | TP1701 crossed with TP1733 |
| TP1737 | h− ΔSPBC660.12c::natMX6 ΔSPCC777.03c::kanMX4 | TP1706 crossed with TP1732 |
| TP1739 | h+ ΔSPBC660.12c::natMX6 ΔSPAC11D3.10::hphMX6 | TP1707 crossed with TP1733 |
| TP1740 | h− ΔSPCC777.03c::kanMX4 ΔSPAC11D3.10::hphMX6 | TP1706 crossed with TP1707 |
| TP1743 | h− ΔSPBC660.12c::natMX6 ΔSPAC11D3.10::hphMX6 ΔSPCC777.03c::kanMX4 | TP1739 crossed with TP1740 |
| TP1770 | h− Δegt1::kanMX6 | |
| TP1771 | h− Δegt2::kanMX6 | |
| TP1857 | h− egt1::P81nmt1-egt1+ | Constructed as part of this study. |
| TP1855 | h− egt1::P41nmt1-egt1+ | |
| TP1803 | h− egt1::P3nmt1-egt1+ | |
| TP1813 | h− Δegt2::hphMX6 | Marker switch of TP1771 to hphMX6 |
| TP1814 | h+ Δegt2::hphMX6 | TP1813 crossed with WT 975 |
| TP1879 | h− egt1::P3nmt1-egt1+ Δegt2::hphMX6 | TP1803 crossed with TP1814 |
Construction of mutants
All DNA recombinant strains were constructed using a two-step PCR method. In the first step, two approximately 500-bp regions were amplified using genomic DNA of the WT 972 strain as a template, corresponding to the forward and reverse ends of the recombination cassette. In the second step, both modules were combined with the appropriate plasmid containing the kanamycin resistance marker (kanMX6). Transformants were selected by resistance to geneticin (G418) and correct integrations were verified by PCR. Primer sequences used for all PCR amplifications are included in Table S6.
The gene disruption strains (TP1770 and TP1771) were constructed by replacing the target open reading frames with the kanamycin resistance marker. The pFA6a-kanMX6 plasmid [26] was used as a template for construction of replacement cassettes. The overexpression strains integrating the nmt1+ promoter (TP1857, TP1855, and TP1803) were constructed using the pFA6a-kanMX6-P81nmt1, pFA6a-kanMX6-P41nmt1, and pFA6a-kanMX6-P3nmt1 plasmids [26] as templates.
Hygromycin-resistant (hphMX6) and clonNAT-resistant (natMX6) versions of the deletion strains (TP1707, TP1732, and TP1813) were prepared by PCR amplification of a marker switch cassette and transformation of the cassette into the original strains, as described previously [49]. The pAG32 and pAG25 plasmids [50] were used as templates for the construction of the marker switching cassettes. Crosses were performed by sporulation on an MEA plate followed by tetrad dissection of the formed asci and selection on YE plates containing the appropriate combination of drugs.
Metabolome sample preparation
Metabolomic analysis was performed as previously described [17]. Briefly, cells from cultures (40 ml/sample, 3.3×106 cells/ml for vegetative cells, or 107 cells/ml for nitrogen-starved cells, respectively) were collected by vacuum filtration and immediately quenched in 25 ml of −40°C methanol. Cells were harvested by centrifugation at −20°C and constant amounts of internal standards (10 nmol of HEPES and PIPES) were added to each sample. Cells were disrupted using a Multi-Beads Shocker (Yasui Kikai, Osaka, Japan). Proteins were removed by filtering on an Amicon Ultra 10-kDa cut-off filter (Millipore, Billerica, USA) and samples were concentrated by vacuum evaporation. Finally, each sample was re-suspended in 40 µl of 50% acetonitrile and 1 µl was used for LC-MS analysis.
LC-MS analysis
LC-MS data were obtained using a Paradigm MS4 HPLC system (Michrom Bioresources, Auburn, USA) coupled to an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, USA). LC separation was performed on a ZIC-pHILIC column (Merck SeQuant, Umeå, Sweden; 150×2.1 mm, 5 µm particle size). Acetonitrile (A) and 10 mM ammonium carbonate buffer, pH 9.3 (B) were used as mobile phase, with gradient elution from 80% A to 20% A in 30 min at a flow rate of 100 µl/min. Peak areas of metabolites of interest were measured using the MZmine 2.10 software [51] and normalized by the weighted contribution of the peak areas of the spiked internal standards (HEPES and PIPES). Identification of metabolites reported in this manuscript was based on their theoretical m/z values and MS/MS fragmentation data, with the exception of EGT, the retention time of which was verified by analyzing a pure standard (obtained from Tetrahedron, Vincennes, France). The identity of selenium-containing compounds was verified by their isotope distribution patterns (Figure S10).
Spot test assays
Plate media were prepared using standard EMM2 recipe [48] with 17 g/l agar. Reagents were added to the media after autoclaving to final concentrations as indicated. Cells were cultivated to a concentration of 5×106 cells/ml and serially diluted in 6 steps (5-fold dilution in each step). 5 µl of the diluted cultures was plated in each spot. Plates were incubated at 26°C for 6 days.
Viability measurement
Cell viability was measured by plating approximately 300 cells on a YE agar plate, incubating the plate at 26°C for 4–5 days, and counting the number of colonies formed. Viability was calculated as the percentage of the number of formed colonies against the number of colonies formed at the first time point.
Supporting Information
Amino acid sequence alignment of S. pombe SPBC1604.01, S. japonicus SJAG_00832, N. crassa Egt-1, M. smegmatis EgtD, and M. smegmatis EgtB proteins. Alignment was generated using the COBALT algorithm. Conserved domains are indicated according to their location in S. pombe SPBC1604.01.
(TIF)
Amino acid sequence alignment of M. smegmatis EgtE protein and its four putative homologs in S. pombe . Alignment was generated using the COBALT algorithm. The conserved catalytic residue (PLP binding site) is indicated by a red arrow.
(TIF)
Normalized peak areas of EGT and its precursors obtained by metabolomic analysis of WT, the Δ SPBC660.12c single deletion mutant, and multiple deletion mutants with other putative homologs of mycobacterial EgtE. Cells were nitrogen-starved prior to analysis (24 h in EMM2-N medium) to induce EGT synthesis.
(TIF)
Amino acid sequence alignment of S. pombe SPBC1604.01 (Egt1) protein and its closest homologs in selected species. Alignment was generated using the COBALT algorithm. Conserved domains are indicated according to their location in S. pombe Egt1.
(TIF)
Amino acid sequence alignment of S. pombe SPBC660.12c (Egt2) protein and its closest homologs in other species. Alignment was generated using the COBALT algorithm. Conserved domains are indicated according to their location in S. pombe Egt2. The conserved catalytic residue (PLP binding site) is indicated by a red arrow.
(TIF)
Absolute quantification of EGT content in cells. A calibration curve was constructed by performing LC-MS injections of pure ergothioneine in 10-fold dilution steps, containing a constant amount of HEPES and PIPES standards (250 pmol each) for normalization (upper panel). Normalized peak areas were plotted against injected amounts and a regression curve was generated using Microsoft Excel. The formula to calculate absolute amount (fmol) from normalized peak area was derived from the regression curve formula (middle panel). Normalized peak areas of EGT were converted into absolute intracellular concentrations using estimated average cellular volumes (bottom panel).
(TIF)
Spot test results on hydrogen peroxide and tert -butyl hydroperoxide agar plates. WT, deletion, and overexpression strains described in this manuscript were serially diluted and grown on EMM2 plates supplemented with increasing concentrations of oxidants hydrogen peroxide (H2O2) and tert-butyl hydroperoxide (t-BOOH). The stress-sensitive Δsty1 strain was used as a positive control.
(TIF)
Cell number increase in liquid EMM2 medium supplemented with Na2SeO4. Relative cell number increase in 24 h was measured in liquid EMM2 medium supplemented with increasing concentrations of Na2SeO4. Cell cultures were incubated at 26°C.
(TIF)
Analysis of a mixture of EGT and selenium in vitro . Extracted ion chromatograms of EGT and selenoneine masses are shown for 1 mM EGT, 1 mM Na2SeO4, and mixture of both, incubated at room temperature for 24 h. Note that the intensity scale of the selenoneine plot is 0.1% relative to that of the EGT plot.
(TIF)
Verification of the identity of selenoneine and hercynylselenocysteine by their isotopic patterns. Comparison of detected vs. calculated isotope distribution patterns of selenoneine (A) and hercynylselenocysteine (B). Theoretical isotope patterns were generated from the corresponding chemical formulas using the Xcalibur software (Thermo Fisher Scientific, Waltham, USA).
(TIF)
Spot test results on Na2SeO4 agar plates. WT, deletion, and overexpression strains described in this manuscript were serially diluted and grown on EMM2 plates supplemented with increasing concentrations of Na2SeO4. The stress-sensitive Δsty1 strain was used as a positive control.
(TIF)
Locations and amino acid sequences of conserved protein domains shown in Figure 1B and Figure S1.
(XLSX)
Locations and amino acid sequences of conserved protein domains shown in Figure 1C and Figure S2.
(XLSX)
Results of all LC-MS measurements of the intermediates in the EGT biosynthetic pathway.
(XLSX)
Locations and amino acid sequences of conserved protein domains of Egt1 homologs shown in Table 2 and Figure S4.
(XLSX)
Locations and amino acid sequences of conserved protein domains of Egt2 homologs shown in Table 2 and Figure S5.
(XLSX)
Sequences of oligonucleotide primers used for PCR amplifications.
(XLSX)
Acknowledgments
We are greatly indebted to Eulalia Lopez for providing excellent technical assistance during the initial stage of this work, and to Dr. Kojiro Takeda for numerous fruitful discussions and generous advice regarding oxidative stress physiology. Further, we thank Dr. Sandra Codlin, Dr. Jürg Bähler, and Addgene for providing DNA plasmids, Dr. Kenichi Sajiki for providing the backcrossed versions of the Bioneer library strains, Dr. Norihiko Nakazawa for assistance with PCR experiments, Romanas Chaleckis for suggesting the use of selenium, and Dr. Steven D. Aird for editing the manuscript.
Funding Statement
This study was funded by the Okinawa Institute of Science and Technology Promotion Corporation (until October 2011) and Okinawa Institute of Science and Technology Graduate University (from November 2011). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cheah IK, Halliwell B (2012) Ergothioneine; antioxidant potential, physiological function and role in disease. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1822: 784–793. [DOI] [PubMed] [Google Scholar]
- 2. Shires TK, Brummel MC, Pulido JS, Stegink LD (1997) Ergothioneine distribution in bovine and porcine ocular tissues. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol 117: 117–120. [DOI] [PubMed] [Google Scholar]
- 3. Ey J, Schömig E, Taubert D (2007) Dietary sources and antioxidant effects of ergothioneine. Journal of agricultural and food chemistry 55: 6466–6474. [DOI] [PubMed] [Google Scholar]
- 4. Gründemann D, Harlfinger S, Golz S, Geerts A, Lazar A, et al. (2005) Discovery of the ergothioneine transporter. Proceedings of the National Academy of Sciences of the United States of America 102: 5256–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Melville DB, Horner WH, Lubschez R (1954) Tissue ergothioneine. J Biol Chem 206: 221–228. [PubMed] [Google Scholar]
- 6. Leone E, Mann T (1951) Ergothioneine in the seminal vesicle secretion. Nature 168: 205–206. [DOI] [PubMed] [Google Scholar]
- 7. Paul BD, Snyder SH (2009) The unusual amino acid L-ergothioneine is a physiologic cytoprotectant. Cell death and differentiation 17: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hartman PE (1990) Ergothioneine as antioxidant. Methods in enzymology 186: 310–318. [DOI] [PubMed] [Google Scholar]
- 9. Markova NG, Karaman-Jurukovska N, Dong KK, Damaghi N, Smiles KA, et al. (2009) Skin cells and tissue are capable of using L-ergothioneine as an integral component of their antioxidant defense system. Free Radic Biol Med 46: 1168–1176. [DOI] [PubMed] [Google Scholar]
- 10. Akanmu D, Cecchini R, Aruoma OI, Halliwell B (1991) The antioxidant action of ergothioneine. Archives of biochemistry and biophysics 288: 10–16. [DOI] [PubMed] [Google Scholar]
- 11. Zhu BZ, Mao L, Fan RM, Zhu JG, Zhang YN, et al. (2011) Ergothioneine Prevents Copper-Induced Oxidative Damage to DNA and Protein by Forming a Redox-Inactive Ergothioneine−Copper Complex. Chemical Research in Toxicology 24: 30–34. [DOI] [PubMed] [Google Scholar]
- 12. Seebeck FP (2010) In vitro reconstitution of Mycobacterial ergothioneine biosynthesis. J Am Chem Soc 132: 6632–6633. [DOI] [PubMed] [Google Scholar]
- 13. Bello MH, Barrera-Perez V, Morin D, Epstein L (2012) The Neurospora crassa mutant NcΔEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal genetics and biology 49: 160–172. [DOI] [PubMed] [Google Scholar]
- 14. Yanagida M (2009) Cellular quiescence: are controlling genes conserved? Trends Cell Biol 19: 705–715. [DOI] [PubMed] [Google Scholar]
- 15. Nurse P (1975) Genetic control of cell size at cell division in yeast. Nature 256: 547–551. [DOI] [PubMed] [Google Scholar]
- 16. Sajiki K, Hatanaka M, Nakamura T, Takeda K, Shimanuki M, et al. (2009) Genetic control of cellular quiescence in S. pombe. J Cell Sci 122: 1418–1429. [DOI] [PubMed] [Google Scholar]
- 17. Pluskal T, Nakamura T, Villar-Briones A, Yanagida M (2010) Metabolic profiling of the fission yeast S. pombe: quantification of compounds under different temperatures and genetic perturbation. Molecular BioSystems 6: 172. [DOI] [PubMed] [Google Scholar]
- 18. Pluskal T, Hayashi T, Saitoh S, Fujisawa A, Yanagida M (2011) Specific biomarkers for stochastic division patterns and starvation-induced quiescence under limited glucose levels in fission yeast. FEBS J 278: 1299–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sajiki K, Pluskal T, Shimanuki M, Yanagida M (2013) Metabolomic Analysis of Fission Yeast at the Onset of Nitrogen Starvation. Metabolites 3: 1118–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takeda K, Yoshida T, Kikuchi S, Nagao K, Kokubu A, et al. (2010) Synergistic roles of the proteasome and autophagy for mitochondrial maintenance and chronological lifespan in fission yeast. Proceedings of the National Academy of Sciences 107: 3540–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shimanuki M, Uehara L, Pluskal T, Yoshida T, Kokubu A, et al. (2013) Klf1, a C2H2 Zinc Finger-Transcription Factor, Is Required for Cell Wall Maintenance during Long-Term Quiescence in Differentiated G0 Phase. PLOS ONE 8: e78545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Genghof DS (1970) Biosynthesis of ergothioneine and hercynine by fungi and Actinomycetales. J Bacteriol 103: 475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, et al. (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41: D348–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim DU, Hayles J, Kim D, Wood V, Park HO, et al. (2010) Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 28: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, et al. (2012) PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 40: D695–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, et al. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. [DOI] [PubMed] [Google Scholar]
- 27. Maundrell K (1990) nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem 265: 10857–10864. [PubMed] [Google Scholar]
- 28. Mitchison JM (1957) The growth of single cells. I. Schizosaccharomyces pombe. Exp Cell Res 13: 244–262. [DOI] [PubMed] [Google Scholar]
- 29. Yamashita Y, Yabu T, Yamashita M (2010) Discovery of the strong antioxidant selenoneine in tuna and selenium redox metabolism. World J Biol Chem 1: 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamashita Y, Yamashita M (2010) Identification of a novel selenium-containing compound, selenoneine, as the predominant chemical form of organic selenium in the blood of bluefin tuna. J Biol Chem 285: 18134–18138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen D, Toone WM, Mata J, Lyne R, Burns G, et al. (2003) Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell 14: 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mata J, Lyne R, Burns G, Bahler J (2002) The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet 32: 143–147. [DOI] [PubMed] [Google Scholar]
- 33. Sugimoto A, Iino Y, Maeda T, Watanabe Y, Yamamoto M (1991) Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes & Development 5: 1990–1999. [DOI] [PubMed] [Google Scholar]
- 34. Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, et al. (1998) The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol Cell Biol 18: 2118–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mata J, Bähler J (2006) Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proceedings of the National Academy of Sciences of the United States of America 103: 15517–15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mata J, Wilbrey A, Bahler J (2007) Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol 8: R217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bushel PR, Heard NA, Gutman R, Liu L, Peddada SD, et al. (2009) Dissecting the fission yeast regulatory network reveals phase-specific control elements of its cell cycle. BMC Syst Biol 3: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen ES, Saitoh S, Yanagida M, Takahashi K (2003) A cell cycle-regulated GATA factor promotes centromeric localization of CENP-A in fission yeast. Mol Cell 11: 175–187. [DOI] [PubMed] [Google Scholar]
- 39. Mercier A, Pelletier B, Labbe S (2006) A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell 5: 1866–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sanso M, Gogol M, Ayte J, Seidel C, Hidalgo E (2008) Transcription factors Pcr1 and Atf1 have distinct roles in stress- and Sty1-dependent gene regulation. Eukaryot Cell 7: 826–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Watanabe Y, Yamamoto M (1996) Schizosaccharomyces pombe pcr1+ encodes a CREB/ATF protein involved in regulation of gene expression for sexual development. Mol Cell Biol 16: 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beaudoin J (2001) The Fission Yeast Copper-sensing Transcription Factor Cuf1 Regulates the Copper Transporter Gene Expression through an Ace1/Amt1-like Recognition Sequence. Journal of Biological Chemistry 276: 15472–15480. [DOI] [PubMed] [Google Scholar]
- 43. Rustici G, van Bakel H, Lackner DH, Holstege FC, Wijmenga C, et al. (2007) Global transcriptional responses of fission and budding yeast to changes in copper and iron levels: a comparative study. Genome Biol 8: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamashita M, Yamashita Y, Ando T, Wakamiya J, Akiba S (2013) Identification and determination of selenoneine, 2-selenyl-N alpha, N alpha, N alpha -trimethyl-L-histidine, as the major organic selenium in blood cells in a fish-eating population on remote Japanese Islands. Biol Trace Elem Res 156: 36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Papadopoulos JS, Agarwala R (2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23: 1073–1079. [DOI] [PubMed] [Google Scholar]
- 46. Paradis E, Claude J, Strimmer K (2004) APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- 47. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Forsburg SL, Rhind N (2006) Basic methods for fission yeast. Yeast 23: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM (2005) Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22: 1013–1019. [DOI] [PubMed] [Google Scholar]
- 50. Goldstein AL, McCusker JH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- 51. Pluskal T, Castillo S, Villar-Briones A, Oresic M (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leupold U (1950) Die Vererbung von Homothallie und Heterothallie bei Schizosaccharomyces pombe. C R Lab Carsberg Sér Physiol 24: 381–480. [Google Scholar]
- 53. Shiozaki K, Russell P (1995) Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature 378: 739–743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence alignment of S. pombe SPBC1604.01, S. japonicus SJAG_00832, N. crassa Egt-1, M. smegmatis EgtD, and M. smegmatis EgtB proteins. Alignment was generated using the COBALT algorithm. Conserved domains are indicated according to their location in S. pombe SPBC1604.01.
(TIF)
Amino acid sequence alignment of M. smegmatis EgtE protein and its four putative homologs in S. pombe . Alignment was generated using the COBALT algorithm. The conserved catalytic residue (PLP binding site) is indicated by a red arrow.
(TIF)
Normalized peak areas of EGT and its precursors obtained by metabolomic analysis of WT, the Δ SPBC660.12c single deletion mutant, and multiple deletion mutants with other putative homologs of mycobacterial EgtE. Cells were nitrogen-starved prior to analysis (24 h in EMM2-N medium) to induce EGT synthesis.
(TIF)
Amino acid sequence alignment of S. pombe SPBC1604.01 (Egt1) protein and its closest homologs in selected species. Alignment was generated using the COBALT algorithm. Conserved domains are indicated according to their location in S. pombe Egt1.
(TIF)
Amino acid sequence alignment of S. pombe SPBC660.12c (Egt2) protein and its closest homologs in other species. Alignment was generated using the COBALT algorithm. Conserved domains are indicated according to their location in S. pombe Egt2. The conserved catalytic residue (PLP binding site) is indicated by a red arrow.
(TIF)
Absolute quantification of EGT content in cells. A calibration curve was constructed by performing LC-MS injections of pure ergothioneine in 10-fold dilution steps, containing a constant amount of HEPES and PIPES standards (250 pmol each) for normalization (upper panel). Normalized peak areas were plotted against injected amounts and a regression curve was generated using Microsoft Excel. The formula to calculate absolute amount (fmol) from normalized peak area was derived from the regression curve formula (middle panel). Normalized peak areas of EGT were converted into absolute intracellular concentrations using estimated average cellular volumes (bottom panel).
(TIF)
Spot test results on hydrogen peroxide and tert -butyl hydroperoxide agar plates. WT, deletion, and overexpression strains described in this manuscript were serially diluted and grown on EMM2 plates supplemented with increasing concentrations of oxidants hydrogen peroxide (H2O2) and tert-butyl hydroperoxide (t-BOOH). The stress-sensitive Δsty1 strain was used as a positive control.
(TIF)
Cell number increase in liquid EMM2 medium supplemented with Na2SeO4. Relative cell number increase in 24 h was measured in liquid EMM2 medium supplemented with increasing concentrations of Na2SeO4. Cell cultures were incubated at 26°C.
(TIF)
Analysis of a mixture of EGT and selenium in vitro . Extracted ion chromatograms of EGT and selenoneine masses are shown for 1 mM EGT, 1 mM Na2SeO4, and mixture of both, incubated at room temperature for 24 h. Note that the intensity scale of the selenoneine plot is 0.1% relative to that of the EGT plot.
(TIF)
Verification of the identity of selenoneine and hercynylselenocysteine by their isotopic patterns. Comparison of detected vs. calculated isotope distribution patterns of selenoneine (A) and hercynylselenocysteine (B). Theoretical isotope patterns were generated from the corresponding chemical formulas using the Xcalibur software (Thermo Fisher Scientific, Waltham, USA).
(TIF)
Spot test results on Na2SeO4 agar plates. WT, deletion, and overexpression strains described in this manuscript were serially diluted and grown on EMM2 plates supplemented with increasing concentrations of Na2SeO4. The stress-sensitive Δsty1 strain was used as a positive control.
(TIF)
Locations and amino acid sequences of conserved protein domains shown in Figure 1B and Figure S1.
(XLSX)
Locations and amino acid sequences of conserved protein domains shown in Figure 1C and Figure S2.
(XLSX)
Results of all LC-MS measurements of the intermediates in the EGT biosynthetic pathway.
(XLSX)
Locations and amino acid sequences of conserved protein domains of Egt1 homologs shown in Table 2 and Figure S4.
(XLSX)
Locations and amino acid sequences of conserved protein domains of Egt2 homologs shown in Table 2 and Figure S5.
(XLSX)
Sequences of oligonucleotide primers used for PCR amplifications.
(XLSX)