Abstract
Changes in metabolic state alter foraging behavior and food preference in animals. Here, I show that normally attractive food becomes repulsive to Caenorhabditis elegans if animals are chronically undernourished as a result of alimentary tract defects. This behavioral plasticity is achieved in two ways: increased food leaving and induction of aversive behavior towards food. A particularly strong food avoider is defective in the chitin synthase that makes the pharyngeal lining. Food avoidance induced by underfeeding is mediated by cGMP signaling in the olfactory neurons AWC and AWB, and the gustatory neurons ASJ and ASK. Food avoidance is enhanced by increased population density and is reduced if the animals are unable to correctly interpret their nutritional state as a result of defects in the AMP kinase or TOR/S6kinase pathways. The TGF-β/DBL-1 pathway suppresses food avoidance and the cellular basis for this is distinct from its role in aversive olfactory learning of harmful food. This study suggests that nutritional state feedback via nutrient sensors, population size and olfactory neurons guides food preference in C. elegans.
KEY WORDS: Aversion, Foraging, Nutritional state, Sensory neurons, TFG-β/DBL-1 pathway
INTRODUCTION
Animals need to integrate sensory information on food quality with post-ingestive feedback to allow them to select food that best supports growth and reproduction. If food is nutrient deficient or harmful, animals develop aversion to that particular food (Provenza, 1996). How this aversion develops mechanistically is unclear. In Caenorhabditis elegans, rejection of pathogenic food is a learnt behavior (Zhang et al., 2005a). It takes about 4 h for animals to learn to avoid pathogenic bacteria. Similarly, preference for good quality food over hard to eat food develops over time in C. elegans (Shtonda and Avery, 2006), suggesting that the animals make their decisions based on their internal metabolic state as well as external information. How the internal nutritional state is used to assign values to sensory information on food quality and abundance is poorly understood. Here, I show that feeding-defective mutants both leave and avoid good food. I use this food-avoidance paradigm to dissect the neuronal basis for this behavior at a sensory level. Food avoidance in feeding-defective animals is primarily mediated by cGMP signaling in the odor-sensing neurons AWC and AWB. A second set of neurons, ASJ and ASK, function together to promote food avoidance. Avoidance behavior is delayed if the animals are unable to monitor their nutritional state correctly, or if they are defective in hen-1, a secreted protein with an LDL motif previously implicated in sensory integration and learning. In addition, I demonstrate that the TGF-β/DBL-1 pathway, which promotes olfactory learning of harmful bacteria (Zhang and Zhang, 2012), suppresses food avoidance in well-fed animals by mechanism that is at least partly distinct from its role in olfactory learning.
RESULTS
Feeding-defective mutants leave and avoid good food
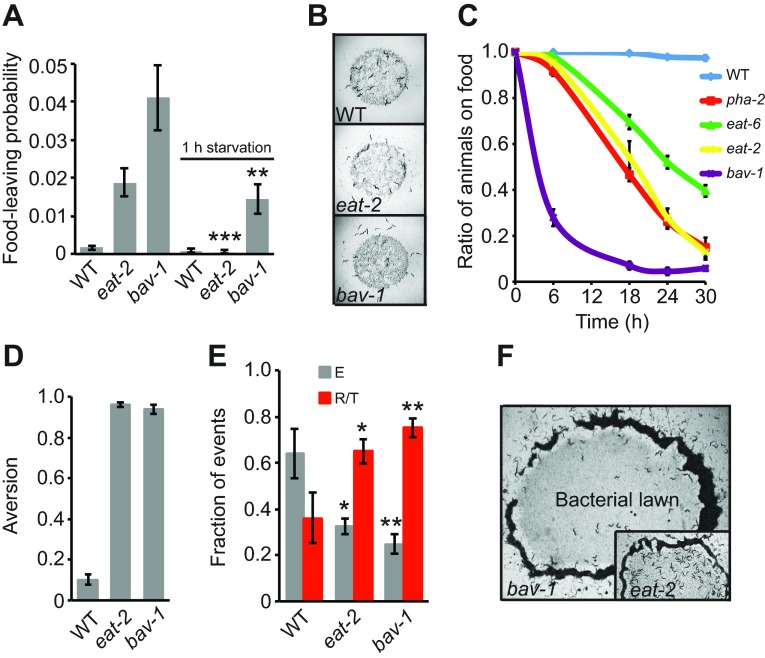
How do animals adjust their behavior to long-term changes in nutritional state? Wild-type hermaphrodite C. elegans accumulate strongly on abundant food and do not leave until food starts to deplete (Milward et al., 2011). Chronic reduction of food intake and hence reduced nutritional state (or dietary restriction) found in feeding-defective mutants such as eat-2, which lack an acetylcholine receptor subunit specifically expressed in the pharyngeal muscle (McKay et al., 2004), and bav-1 (see below) induced a strong food-leaving response even in plentiful food conditions (Fig. 1A,B). Other feeding-defective mutants such as pha-2 (Avery, 1993; Mörck et al., 2004) and eat-6 (Avery, 1993; Davis et al., 1995) behaved similarly, and left good food (Fig. 1C). These chronically food-deprived animals accumulate outside normally palatable food, suggesting that as well as leaving food, they actively avoid it (Fig. 1C,F, supplementary material Movie 1). The strain GE337 shows particularly strong accumulation outside food. I outcrossed this strain and found this phenotype was associated with an allele of a gene I called bav-1, for bacterial avoidance-1. Like feeding-defective mutants, bav-1 animals accumulated in a ring just outside food at higher population densities (Fig. 1F). Importantly, these chronically underfed animals such as eat-2 and bav-1 could still respond to immediate hunger: 1 h of food deprivation drastically reduced their feeding-defective behavior (Fig. 1A). Thus, chronically underfed animals can still respond to the presence or absence of food. However, chronic undernourishment appears to change C. elegans' response to the food from attraction to avoidance.
Fig. 1.
Chronic dietary restriction induces Caenorhabditis elegans to leave and avoid palatable food. (A) eat-2 and bav-1 animals exhibit increased food leaving on abundant food. Withdrawing food for 1 h reduced subsequent food leaving in eat-2 and bav-1 animals. Each genotype was assayed at least six times. WT, wild-type. Error bars represent s.e.m. Student's t-test compares each genotype under the two different conditions: *P<0.05, **P<0.01, ***P<0.005. (B) Distribution of animals on and off food at 10 h. (C) Feeding-defective mutants (pha-2, eat-6, eat-2 and bav-1) accumulate off food. (D) Aversion score of eat-2 and bav-1. (E) Accumulation outside food is associated with increased avoidance behavior when attempting to re-enter the food lawn. Reversals and turns (R/T) were scored manually from recordings at 24 h. At this time point, food depletion leads to high rates of food leaving in WT animals. Number of attempts to re-enter food lawn scored: WT (N2), N=63; eat-2, N=138; bav-1, N=179. E, entry into food. (F) At high population density, eat-2 and bav-1 animals form an avoidance ring outside the food lawn.
To explore food-avoidance behavior more closely, I monitored how animals behaved as they approached the bacterial food lawn. Wild-type animals usually entered the food lawn, and only occasionally reversed and turned away from it. By contrast, chronically underfed animals often reversed and turned when approaching the bacterial lawn, or ‘hesitated’ close to the food edge (hesitation occurs when animals stall close to the food edge and lift their noses and swing their heads) (Fig. 1E). These data suggest that chronic malnourishment alters animals' perception of food quality, leading to food aversion.
To distinguish food-leaving behavior from food avoidance, we asked whether mutants exhibiting high food leaving also invariably avoided food. osm-9 mutants show high food-leaving behavior, both on a depleting food source (Milward et al., 2011) and on a thick lawn (supplementary material Fig. S1A). Although 40% of osm-9 animals were found off food after 24 h on a bacterial lawn, these animals did not show increased reversals and turns upon food approach (supplementary material Fig. S1B). By contrast, in chronically underfed animals there was an increase in both food leaving and aversive behavior to food. eat-2 and bav-1 have an aversion score [Noff/Ntotal, as described elsewhere (Melo and Ruvkun, 2012)] (Fig. 1D) of close to 1, meaning that at the end of the assay almost 100% of the animals are off food.
The food avoider bav-1 is a mutant in the pharynx-specific chitin synthase, chs-2
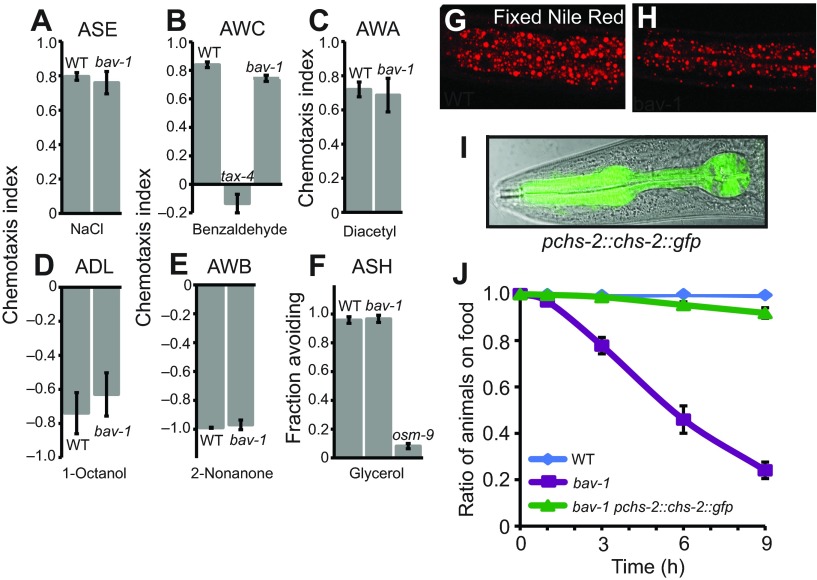
In order to understand why bav-1 animals left and avoided food strongly, I characterized their behavior in more detail. One simple explanation for the avoidance phenotype could be that bav-1 mutants failed to respond the food-associated cues. However, amphid neurons of bav-1 mutants appear to be normal as judged by dye-filling (supplementary material Fig. S2A–C), and the mutants responded normally to a wide range of sensory stimuli (Fig. 2A–F). Thus, bav-1 animals can sense and respond effectively to individual chemosensory cues. However, chs-2 mutants appeared pale, suggesting that they have less fat deposited, and behaved like starved animals (e.g. exhibiting increased nose lifting). Nile Red staining of fixed animals (Fig. 2G,H) as well as Sudan Black staining (data not shown) confirmed that bav-1 mutants have very low fat deposits, much like feeding-defective animals.
Fig. 2.
bav-1 disrupts chitin synthase-2, chs-2. (A–F) bav-1 animals exhibit wild-type-like responses to different chemical cues: NaCl, benzaldehyde, diacetyl, 1-octanol, 2-nonanone and glycerol. All assays were carried out in the absence of food. (G,H) Fat staining of wild-type and bav-1 mutants. Fixed Nile Red staining of L4 wild-type (G) and bav-1 (H) larvae. Images show the intestine posterior to the grinder. (I) chs-2 is expressed specifically in the pharynx. (J) The bav-1 food-avoidance phenotype can be rescued by expressing genomic chs-2 under its own promoter. Error bars represent s.e.m.
To gain more insight into the bav-1 phenotype, I mapped the mutation and identified the bav-1 locus by SNP-mapping and Illumina sequencing. The bav-1 (mbd1) allele was associated with a G→A 3′ splice site mutation at position 3846 of the gene for chitin synthase 2, chs-2. chs-2 is one of two chitin synthase genes in C. elegans (Veronico et al., 2001; Zhang et al., 2005b) and is expressed specifically in the pharynx (Fig. 2I and references as above). Secreted chitin lines the lumen walls of the pharynx and the pharyngeal grinder (the worm's teeth). RNAi knockdown of chs-2 causes animals to arrest as L1 larvae (Zhang et al., 2005b). Expressing chs-2 under its own promoter fully rescued the bav-1 food-avoidance phenotype (Fig. 2J). Taken together with my studies of other feeding-defective mutants, these data suggest that chs-2 mutants are chronically undernourished as a result of defects in pharyngeal function, and that this evokes aversion to the bacteria that the animals are growing on.
Food avoidance is mediated via the sensory neurons AWC and AWB, and jointly by ASJ and ASK
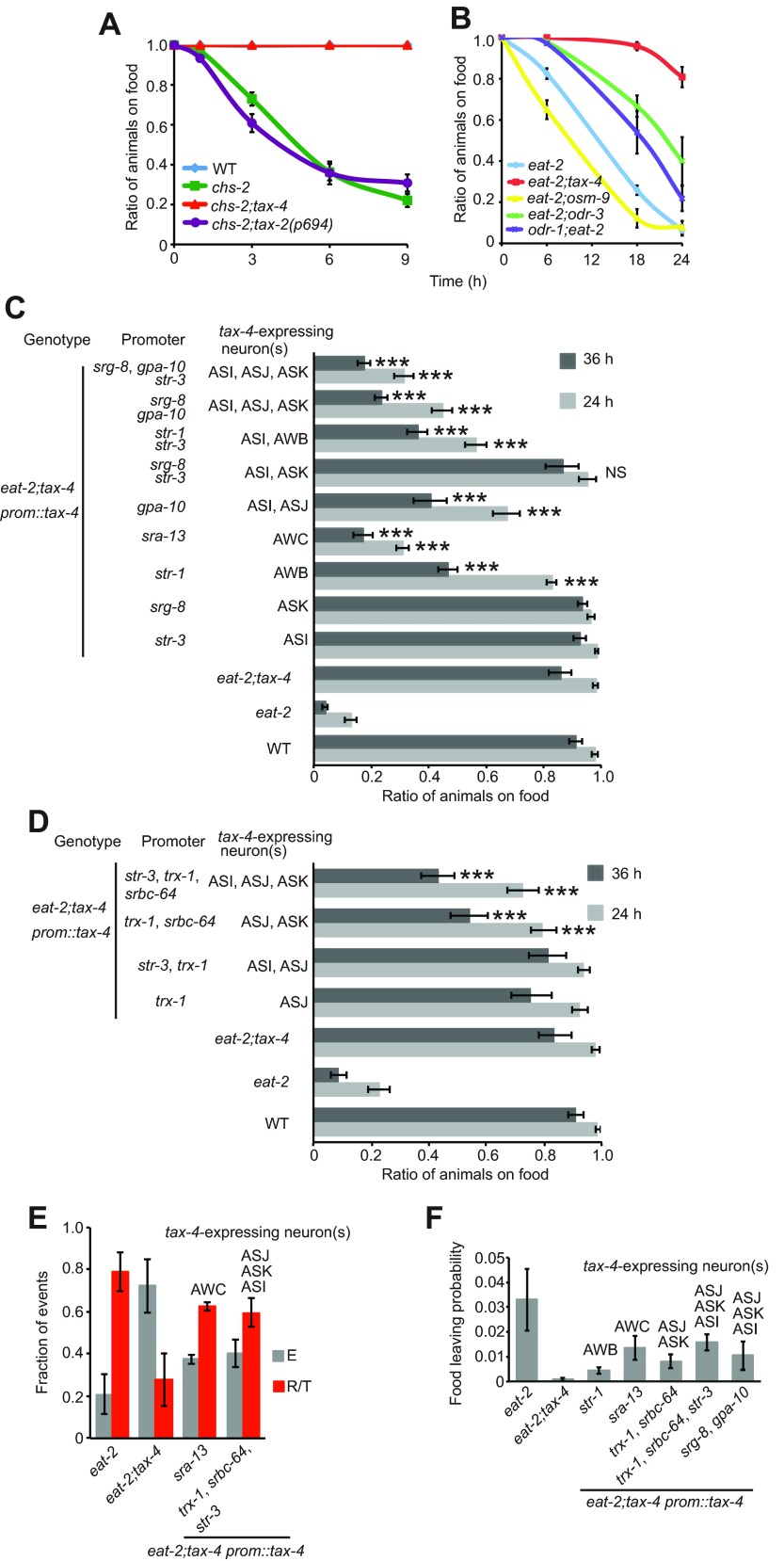
Although eat-2 and chs-2 animals accumulated off the bacterial food they nevertheless remain close to it, often forming a ring just outside the bacterial lawn (Fig. 1F). This behavior suggested that the animals retained some long-range attraction to food. By contrast, the increased reversals and turns when chronically underfed worms approached food indicated that some aversive cues from the food dominate in the balance between attraction and avoidance at short range. In C. elegans, food-associated cues like odors and tastants are sensed predominantly by sensory neurons in the amphids. To dissect food-avoidance behavior in chronically underfed animals at a sensory level, I constructed double mutants defective in eat-2 or chs-2 and in genes required for specific neural functions. Mutants in tax-4, encoding an α-subunit of a cGMP-gated ion channel, are defective in the function of 14 neuron pairs (Komatsu et al., 1996). tax-4 mutations suppressed food avoidance in both eat-2 and chs-2 mutant animals (Fig. 3A,B). The tax-2 gene encodes the β-subunit of the TAX-4-containing cGMP-channel and functions in the same 14 neurons. For tax-2, a promoter mutation, tax-2(p694), is available that selectively disrupts tax-2 expression in six of the 14 neurons (Coburn and Bargmann, 1996; Coates and de Bono, 2002; Bretscher et al., 2011). The tax-2(p694) mutation failed to suppress chs-2 food avoidance, suggesting that one or more of the remaining tax-2/tax-4-expressing neurons promoted this behavior. These include the AWB and AWC olfactory neurons, and the ASG, ASI, ASJ and ASK gustatory neurons. I used cell-specific promoters to express tax-4 cDNA in these neurons singly or in combination (except ASG, for which no cell-specific promoter has been described), and asked where tax-4 expression was required to restore food-avoidance behavior to eat-2;tax-4 mutant animals. tax-4 expression in AWC neurons almost fully rescued the avoidance phenotype (Fig. 3C). Partial rescue was observed when tax-4 expression was restored to AWB neurons and there was a strong rescue of the avoidance phenotype when tax-4 was expressed in a combination of three neurons: ASI, ASJ and ASK with the gpa-10 and srg-8 promoters (Fig. 3C). As the gpa-10 promoter drives expression in ASI and ASJ (and several more non-tax-4-expressing neurons (Jansen et al., 1999), I sought to test all neurons individually. There was no rescue of avoidance behavior when tax-4 was individually expressed in ASI, ASJ or ASK (Fig. 3C,D) or in combinations in ASI and ASJ or ASI and ASK. Food-avoidance behavior was only restored in eat-2;tax-4 animals with combined expression in ASJ and ASK (Fig. 3D). Similarly, the strong phenotype in eat-2;tax-4 animals in reducing reversals and turns was restored upon expressing the tax-4 transgene in AWC or a combination of ASJ and ASK (and ASI) (Fig. 3E). As tax-4 also regulates food leaving (Milward et al., 2011), the food-leaving behavior of eat-2;tax-4 was analyzed. In agreement with our previous findings, tax-4 suppressed eat-2 food leaving. Partial rescue was observed when tax-4 expression was restored in the sensory neurons that rescued food avoidance of eat-2;tax-4 (Fig. 3F), indicating that AWB, AWC and ASJ and ASK likely promote food-leaving behavior (Fig. 3F). Hence, tax-4 and cGMP signaling promote both food leaving and food avoidance in feeding-defective animals.
Fig. 3.
Food avoidance is regulated by the AWC, AWB, ASJ and ASK neurons. (A) Food avoidance of chs-2 mutants is suppressed by a tax-4(null) mutation but not by tax-2(p694) promoter deletion. (B) The food-avoidance phenotype of eat-2 mutants was fully suppressed by mutations in tax-4, and partially suppressed by mutations in odr-3 and odr-1. By contrast, a mutation in osm-9 enhanced food aversion of eat-2 animals. (C,D) Food avoidance can be restored to eat-2;tax-4 animals by cell-specific expression of tax-4 cDNA in AWC. Expressing tax-4 cDNA in AWB, or in ASI, ASJ and ASK partially restores food-avoidance behavior in eat-2;tax-4 animals. Each genotype was assayed at least six times. Error bars represent s.e.m. *P<0.05, **P<0.01, ***P<0.005, Student's t-test. (E) Rescue of food-induced reversals and turns in eat-4;tax-4 animals expressing tax-4 cDNA in the indicated neurons (scored manually at 24 h when animals are attempting to re-enter the lawn). Number of attempts scored per genotype: eat-2, N=68; eat-2;tax-4, N=62; eat-2;tax-4 psra-13::tax-4, N=80; eat-2;tax-4 ptrx-1, psrbc-64, pstr-3::tax-4, N=79. (F) Food leaving is partially restored in eat-2;tax-4 animals when tax-4 is expressed in AWB, AWC and ASJ, ASK and ASI neurons.
To investigate further the involvement of AWB and AWC neurons, I asked whether disrupting odr-1 or odr-3 suppressed eat-2 food avoidance (Fig. 3B). ODR-1 encodes a transmembrane guanylyl cyclase necessary for odorant responses mediated by AWC and AWB neurons (L'Etoile and Bargmann, 2000). ODR-3 encodes a Gα protein and is required in several sensory neurons including AWC (Roayaie et al., 1998; Jansen et al., 1999; Lans et al., 2004). Mutations in odr-1 or odr-3 partially suppressed eat-2 food-avoidance behavior, in agreement with the tax-4 rescue data. In summary, our results suggest that AWC and AWB olfactory neurons are important for feeding-defective worms to avoid food. A separate set of chemosensory neurons, ASJ and ASK, previously implicated in taste responses, also acts to promote food avoidance.
Regulation of avoidance behavior by HEN-1 and TGF-β/DBL-1 signaling
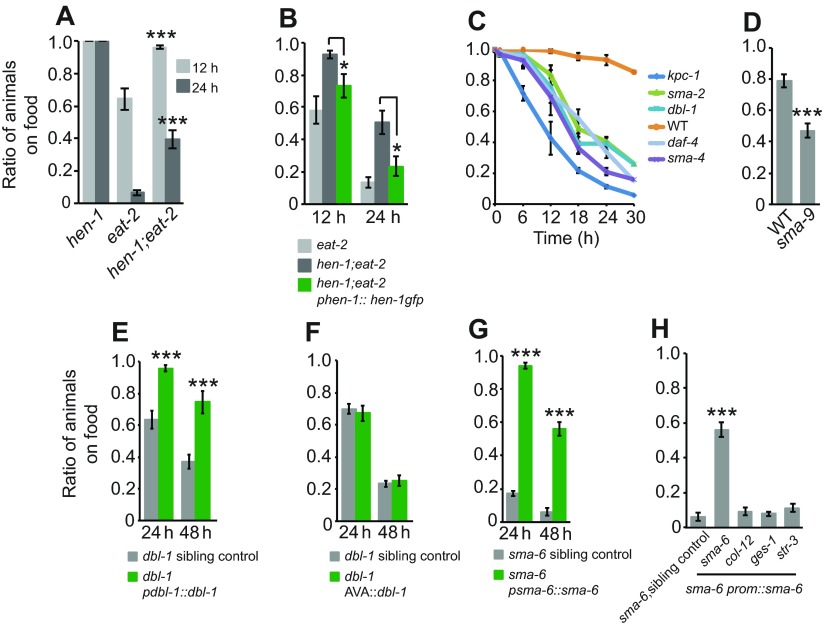
Food that is attractive to well-fed animals repels chronically underfed animals, suggesting that the animals' perception of food is altered in response to their reduced nutritional state. This altered perception likely involves integration of metabolic state and food cues. HEN-1 is a secreted protein with an LDL-a motif that, together with its putative receptor SCD-2, regulates integration of sensory stimuli in the AIA interneuron (Ishihara et al., 2002; Shinkai et al., 2011). hen-1 mutant animals fail to integrate conflicting sensory cues and are defective in associative learning (Ishihara et al., 2002). I therefore tested whether HEN-1 function is required for the food-avoidance behavior displayed by chronically underfed animals. hen-1 suppressed the food-avoidance phenotype of eat-2 mutants (Fig. 4A), although the mutant animals eventually accumulated off food. This phenotype could be rescued by expressing hen-1 from its own promoter in hen-1;eat-2 animals (Fig. 4B). These data indicate that hen-1 promotes food avoidance in undernourished animals, but that its role is not essential.
Fig. 4.
HEN-1 and the DBL-1/Sma pathway regulate food-avoidance behavior. (A,B) Disrupting hen-1, which was previously implicated in sensory integration of attractive and aversive cues, suppressed food aversion in eat-2 animals (A). A hen-1 transgene can restore avoidance to hen-1;eat-2 animals (B). (C) Mutants defective in components of the DBL-1/Sma pathway avoid food. (D) Animals defective in sma-9, the downstream transcription factor of the DBL-1/Sma pathway, show a high off-food distribution but a weaker phenotype than other mutants in the pathway. (E) dbl-1 expression under its own promoter but not in the AVA interneuron restores WT behavior (F). (G) sma-6 animals show food aversion, which can be rescued by expressing sma-6 under its own promoter (G), but not under a promoter that express selectively in the hypodermis (pcol-12), intestine (pges-1) or ASI neurons (pstr-3) (H). H shows the 48 h time point. Each genotype was assayed at least six times. Error bars represent s.e.m. *P<0.05, **P<0.01, ***P<0.005, Student's t-test.
Avoidance behavior to palatable food by chronically underfed animals could share cellular and neuronal mechanisms with avoidance behavior to harmful (Pujol et al., 2001; Zhang et al., 2005a; Schulenburg and Ewbank, 2007) or indigestible food (Andrew and Nicholas, 1976; Shtonda and Avery, 2006). Naive C. elegans are normally attracted to pathogenic bacteria such as Serratia marcescens but learn to avoid the bacteria (Pujol et al., 2001; Zhang et al., 2005a). Similarly, C. elegans learn to avoid pathogenic Pseudomonas aeruginosa (Zhang et al., 2005a) and it was recently reported this olfactory learning relies on the TGF-β/DBL-1 pathway (Zhang and Zhang, 2012). I therefore analyzed how animals defective in components of the DBL-1 pathway responded to food palatable to wild-type animals. All mutants analyzed in the pathway behave similar to chronically underfed worms (Fig. 4C). Mutants in dbl-1, encoding the TGF-β ligand of the Sma/Mad pathway (Suzuki et al., 1999), or in its cognate receptors DAF-4 and SMA-6 (Estevez et al., 1993; Krishna et al., 1999), left and avoided food (Fig. 4C,G). Mutants for the downstream signal transducers SMA-2 (R-Smad), SMA-4 (Co-Smad) (Brenner, 1974; Savage et al., 1996) and the transcription factor SMA-9 (Liang et al., 2003) also avoided food (Fig. 4C,D). Moreover, mutants lacking the Kex-2/subtilisin-like pro-protein convertase KPC-1, predicted to process TGF-β like pro-proteins (Gómez-Saladin et al., 1997), left and avoided the food strongly. Thus, the DBL-1 pathway acts to suppress food avoidance. This is in contrast to its role in aversive olfactory learning of pathogenic bacteria, where it promotes avoidance of harmful food.
DBL-1 is secreted from the AVA interneurons to promote aversive learning of pathogenic bacteria (Zhang and Zhang, 2012), whereas its receptor SMA-6 acts in the hypodermis and in the chemosensory neuron ASI (Zhang and Zhang, 2012). AVA expression of DBL-1 failed to restore wild-type behavior to dbl-1 mutants in our assays (Fig. 4E,F). Expressing the SMA-6 receptor, using previously reported strains able to rescue sma-6 aversive olfactory learning defects (Zhang and Zhang, 2012), in hypodermis (using the col-12 promoter) or in ASI neurons (from the str-3 promoter) did not rescue sma-6 food-avoidance behavior (Fig. 4G,H). sma-6 is also expressed in the pharynx (Yoshida et al., 2001), but pharyngeal expression of sma-6 also failed to restore wild-type behavior (supplementary material Fig. S3). Together, these data suggest that the TGF-β/DBL-1 pathway suppresses food avoidance in this paradigm and imply that this function is distinct from its role in aversive learning.
Avoidance behavior is enhanced by increased population size and suppressed in the absence of functional nutrient sensors
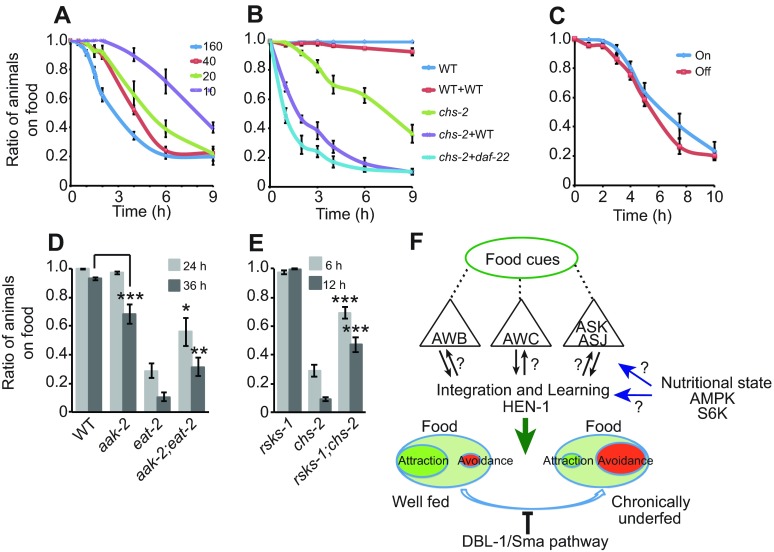
Caenorhabditis elegans feed on the E.coli strain OP-50 in the lab and one possibility is that avoidance is a food source peculiarity and not, as hypothesized, linked to the reduced feeding status of the animal. To test this, similar avoidance assays were performed using the E. coli strain HB101, described as a good quality food source (Shtonda and Avery, 2006). As predicted, the feeding-defective eat-2 and chs-2 mutant animals as well as sma-6 mutants, but not wild-type animals, avoided HB101 similarly to OP-50 (supplementary material Fig. S4). This further supports the link between feeding status and interpretation of food quality. The value of a food patch is influenced not only by its nutritional quality (perceived or real) but also by the presence of competitors on that patch. Animal responses to food cues are therefore likely to be influenced by the population density on the patch. Consistent with this, previous work has shown that population density modulates olfactory plasticity in C. elegans (Yamada et al., 2010). To test the effect of animal number in the avoidance assay, I used both fewer (10 and 20 animals per plate) and more (160 animals per plate) animals than in the standard assay. chs-2 animals accumulated off food in all conditions, but the response dynamics were accelerated by increasing population size (Fig. 5A).
Fig. 5.
Food-avoidance behavior is affected by population size and mutations in nutritional state sensors. (A) Food avoidance by chs-2 mutants is accelerated by increased population density (from 10 to 160 animals per assay plate). (B) Conditioning the food enhances avoidance behavior of chs-2 mutants. daf-22 mutants, which are defective in the production of some ascaroside pheromones, including dauer pheromone, condition food similar to WT animals. (C) There is no difference in avoidance whether the chs-2 animals come from plates with ~100% off-food distribution (pre-incubated overnight on fresh food) versus ~100% on-food distribution (pre-incubated 1 h on fresh food). See Materials and methods for details. (D) Disrupting AMPK, encoded by the aak-2 gene, suppresses food aversion of eat-2 mutants. (E) A mutation in S6 kinase, rsks-1, suppresses chs-2 food avoidance. Each genotype was assayed at least six times. Error bars represent s.e.m. *P<0.05, **P<0.01, ***P<0.005, Student's t-test. (F) Model of food aversion in response to chronically reduced feeding/nutritional status. AWC, AWB, ASJ and ASK neurons promote food avoidance in chronically food-deprived animals. AMPK and S6 kinase appear to be involved in sensing nutritional state and promoting food avoidance in chronically underfed animals. The TGF-β/DBL-1 pathway inhibits this food-avoidance behavior via an as yet unknown anatomical location.
As increasing population density enhanced avoidance behavior, I tested whether conditioning the food also enhanced avoidance. I conditioned food by incubating 50 animals overnight on a thick food patch, removing these animals and then using the plate as a test plate. Avoidance in chs-2 mutant animals was strongly enhanced when the bacterial lawn was conditioned (Fig. 5B). This response did not depend on dauer pheromone as daf-22 mutants, which are unable to produce the potent ascarosides (Golden and Riddle, 1985; Butcher et al., 2009; Pungaliya et al., 2009), conditioned the food to a similar degree as wild-type animals. Taken together, these data suggest that population density modulates the development of bacterial avoidance. Whether this reflects the action of unidentified pheromones or of bacterial metabolites is unclear.
To test whether animals that have already developed food-avoidance behavior responded differently when subsequently presented with fresh food, I compared the behavior of chs-2 animals pre-incubated on fresh food for 1 h (all animals remain on food, ‘on’) with that of animals kept on the same lawn overnight (90–100% of animals off food, ‘off’) when presented with a fresh bacterial lawn. The response to fresh food was indistinguishable across the two conditions (Fig. 5C). These data suggest that fresh food somehow resets the animal's behavioral state.
If the animal's perception of its metabolic state is important for its response to food, interfering with nutrient state sensors should alter food-leaving and food-avoidance behavior. Two major conserved cellular nutrient-sensing pathways are mediated by AMP-activated protein kinase, AMPK, and the TOR-kinase pathway (Lindsley and Rutter, 2004). In C. elegans, the aak-2 gene encodes one of the two catalytic α-subunits of AMPK (Apfeld et al., 2004) and the let-363 gene encodes TOR-kinase (Long et al., 2002). As mutations in let-363 cause developmental arrest (Long et al., 2002), I used mutants in its downstream target p70 S6-kinase, rsks-1 (Pan et al., 2007). The food avoidance of eat-2 mutant animals was significantly suppressed by a mutation in aak-2 (Fig. 5D). Similarly, double mutants with rsks-1 strongly suppressed chs-2 food avoidance (Fig. 5E). These data suggest that interfering with the animals' ability to assess its nutritional state affects its behavioral responses to food. Food avoidance in response to underfeeding thus relies on intact nutrient sensors, and is strongly affected by population density on the food source.
DISCUSSION
Caenorhabditis elegans that are chronically undernourished because of defects in their alimentary tract develop aversion to a bacterial food source that is attractive to wild-type animals. This plastic response involves animals reversing and turning away from food despite their malnourished state. The behavior involves the AWC and AWB olfactory neurons, and the ASK and ASJ chemosensory neurons, and is also regulated by the nutrient state sensors AMP kinase and S6 kinase.
Avoidance by C. elegans of normally attractive food has been reported previously (Melo and Ruvkun, 2012). This study inactivated core cellular processes (e.g. mitochondrial function) using RNAi and it was observed that in many cases animals distributed off food more than wild-type. The authors did not distinguish between food-leaving and food-aversive behavior, although only a small subset of the RNAi-treated animals accumulated off food (i.e. had an aversion score higher than 0.5). These authors suggested this response to be part of a pathogen defense mechanism. Given my results, I suggest that the observed food-leaving/food-aversive behavior induced by RNAi treatment may reflect an altered sense of nutritional state. Consistent with this, some of the RNAi treatments reduced pharyngeal pumping (Melo and Ruvkun, 2012).
While it is known that nutritional state influences foraging, the mechanisms involved are poorly understood. I show here that in C. elegans, two different cellular nutrient sensors, the 5′-AMP-activated protein kinase (AMPK) and the ribosomal S6 kinase, a target of TOR kinase (TORC1), promote food avoidance behavior in chronically underfed C. elegans. AMP kinase is activated by increases in the intracellular AMP/ATP ratio, a hallmark of metabolic stress. TOR signaling, and therefore S6 kinase activity, is stimulated by intracellular amino acids, and promotes anabolic metabolism. Decreased food avoidance in C. elegans AMP kinase mutants could reflect a compromised ability to perceive poor nutritional state. The phenotype of C. elegans S6 kinase mutants could reflect reduced energy demand: rsks-1 mutants grow more slowly and have smaller broods than wild-type animals (Pan et al., 2007). However, given the complex functions of these nutrient sensors, we cannot exclude other or additional explanations. aak-2 mutant animals exhibit slightly elevated pharyngeal pumping, suggesting increased food intake (Cunningham et al., 2012). Inactivating AMPK in Drosophila melanogaster also causes hyperphagia (Johnson et al., 2010). A role for S6 kinase in a different kind of food-choice paradigm has been described in Drosophila. Downregulating ribosomal S6 kinase triggers well-fed D. melanogaster larvae to exploit hard-to-get food that is normally rejected by well-fed animals but accepted by food-deprived ones (Wu et al., 2005). This phenotype suggests that flies with reduced S6 kinase activity have increased motivated foraging and feeding. In adult flies, S6 kinase acts in unidentified neuronal pathways to regulate nutrient preference: downregulating S6-kinase induces a preference for a protein-rich yeast diet (Ribeiro and Dickson, 2010; Vargas et al., 2010). Both AMPK and TOR (mTORC1) regulate food intake in mammals (Pimentel et al., 2013). Activation of TOR kinase in the hypothalamus decreases food intake whilst activation of AMPK increases food intake. The mechanisms by which the AMPK and TOR pathways influence feeding behavior are not fully understood and cell-specific roles remain to be dissected (Kola, 2008).
The neuronal mechanisms promoting food-avoidance behavior in chronically underfed worms are reminiscent of those involved in avoidance of pathogenic food. Both behaviors involve the olfactory neurons AWC and AWB, which respond to changes in odor concentrations via a cGMP signaling cascade (Ha et al., 2010). The AWC neurons normally direct attractive responses, but can in some circumstances mediate repulsive responses (Tsunozaki et al., 2008), as in the underfed animals described here. In well-fed animals, AWC (with ASK) drives area-restricted search behavior (Wakabayashi et al., 2004; Gray et al., 2005), which allows animals to remain in an area where they last found food. The AWB neurons mediate avoidance of noxious odors (Bargmann, 2006) and direct avoidance of certain bacterial products (Pradel et al., 2007). However, there are some differences between food aversion to normally palatable food and aversive learning. Animals with defects in the TGF-β/DBL-1 pathway fail to learn to avoid pathogenic bacteria (Zhang and Zhang, 2012) and have reduced naive preference. I find that animals defective in DBL-1 and its downstream signaling effectors behave very similar to chronically underfed animals: they leave and avoid good food, indicating that the DBL-1 pathway acts to suppress food avoidance. This behavior could be connected to defects in naive preference as it is clearly distinct from aversive learning given neither the ligand DBL-1 nor the receptor SMA-6 act in the same cells and tissues as in learning. My data rather suggest an endocrine signaling mechanism and it will be interesting to identify the cellular focus where dbl-1 and sma-6 function.
The other set of neurons we define that promotes food-avoidance behavior includes the ciliated gustatory neurons ASJ and ASK. Neural imaging shows that ASK neurons respond to food-conditioned media by tonically reducing their cytosolic Ca2+ levels (Wakabayashi et al., 2004). Ca2+ levels in ASK are transiently inhibited by ascarosides (Macosko et al., 2009), pheromones that drive dauer development (Golden and Riddle, 1982; Jeong et al., 2005) and mediate male attraction to hermaphrodites (Srinivasan et al., 2008; Jang et al., 2012). Like ASK, ASJ neurons transmit pheromone responses and promote dauer development (Schackwitz et al., 1996). ASK is a major post-synaptic target of ASJ.
Is food avoidance in chronically underfed animals a learnt response? Disrupting hen-1, which has been implicated in learning and integration of antagonistic sensory stimuli suppresses the aversion phenotype of eat-2 but it is not required as the animals eventually accumulate off food. The Drosophila homolog of the hen-1 receptor, Alk, has been proposed to spare the CNS during nutrient restriction by activating PI3-kinase (normally activated by the insulin receptor) (Cheng et al., 2011) as well as being implicated in learning (Gouzi et al., 2011). This suggests a wider role for this pathway. It is possible that C. elegans HEN-1 signaling also has dual roles.
Food avoidance is strongly affected by population density and this effect seems not to be mediated via dauer pheromones. Interestingly, it was recently reported that survival of L1 larvae during starvation is density dependent, and that this effect is not mediated by ascarosides/dauer pheromone but rather by an unknown starvation signal (Artyukhin et al., 2013). It is possible that this proposed starvation signal also promotes food leaving and avoidance in underfed adult animals. My preliminary data suggest that even a very distantly related species, Pristionchus pacificus, is able to condition food to enhance food avoidance of chs-2 (B.O., unpublished) which could further support my finding that dauer pheromone is not responsible for the conditioning effect. It will be interesting to find the molecular identity of this signal. Exudates from C. elegans have been reported to contain a large variety of compounds, some of which are known to be aversive to C. elegans (Ward, 1973; Kaplan et al., 2009).
In summary, C. elegans that are chronically underfed as a result of defects in their alimentary tract avoid high quality and abundant food. The behavior is dependent on cGMP signaling via TAX-4 in the odor-sensing neurons AWC and AWB. This illustrates that AWC can support aversive behavior in a wider context than previously recognized. Sensory input from the chemosensory ASJ and ASK neurons also promotes food-avoidance behavior. As population density affects avoidance, it will be interesting to find how C. elegans interpret population density and whether this is mediated via ASJ and/or ASK. Avoidance behavior is suppressed when metabolic state sensing is disrupted, when animals are defective in sensory integration and in well-fed animals through the DBL-1 pathway. This study illustrates how long-term feeding experiences can alter foraging and could potentially lead to insights into eating disorders such as anorexia nervosa.
MATERIALS AND METHODS
Materials
For strains and rescue constructs see supplementary material Table S1 and below.
Behavioral assays
For food-avoidance assays, regular NGM plates were seeded 2 days prior to assay with 200 μl of OP50 (CGC) grown overnight in 2×YT. The rich 2×YT medium allows growth of a thick food patch. Forty young adults were picked (as much as possible from outside food to avoid transfer of food between the growth plate and the assay plate) to each assay plate and placed in the center of the food lawn. The animals were left to forage and at each time point the ratio of worms on food versus total worms was calculated. Feeding-defective assays were performed similar to methods described previously (Milward et al., 2011) with the exception that the worms were left to settle for 1 h and then assayed immediately (i.e. in high food condition). Food responses (reversals and turns) were scored manually at 24 h on low peptone plates (to increase leaving events of N2 wild-type animals). Low peptone plates contain 5% of the bactopeptone found in NGM medium (Milward et al., 2011). Animals approaching the bacterial lawn from off food either entered food (E), or hesitated, reversed or turned away from food to remain off food (R/T). To condition the food, 50 adults of indicated genotype were left on food overnight and subsequently removed to generate a conditioned assay plate.
For the HB101 experiment in supplementary material Fig. S4, the animals were grown on OP-50 and assayed on a 2 day old lawn on HB101 in 2×YT.
Mapping and cloning of bav-1
Linkage analysis and SNP mapping located bav-1 to the left arm of chromosome II, left of SNP, F46F5. Recombination suppression in this genomic area precluded finer mapping. This left a region of 809 kb where bav-1 could be located. Genomic DNA was prepared from bav-1 mutants and sequenced on an Illumina GAIIx machine to ~16× coverage. The bioinformatics analysis of the sequence data identified two polymorphisms in non-intergenic regions within these 809 kbp. These changes were re-sequenced with standard dideoxy sequencing and one change was confirmed. The change was a G→A point mutation at position 3846 of the chs-2 gene (www.wormbase.org). This point mutation changes the 3′ splice acceptor site of intron 6 from CAG to CAA. For rescue experiments, 2.0 kb of the chs-2 promoter was cloned (by nested PCR) into a pDEST Gateway vector containing the 3′ UTR of the unc-54 gene (5′ primer including an Nhe I site: ATATGCTAGCTGAGAATTGCTTGTTTGCG; 3′ primer including a Xho I site: ATATCTCGAGGTTTAATAGAAGCTCAGAAATT) and the chs-2 gene was amplified from genomic DNA with nested PCR (5′ primer including a Sac I site: ATCGGAGCTCAAACATGATGAACACATTGGACCATC; 3′ primer including a KpnI site: ATCGGGTACCTTAAAATCTTCTCAGCTCCACGTC) and ligated into a Gateway pENTRY vector containing an intercistronic region (from the gpd-2 gpd-3 operon) followed by GFP upstream of the attL2 site [both the pDEST and the pENTRY vectors are described elsewhere (Coates and de Bono, 2002)]. A pEXP clone was generated by LR-clonase recombination (Invitrogen). The construct was injected into bav-1 mutants (outcrossed three times) and GFP expression in the pharynx as well as rescue of the behavioral phenotype was observed.
Transgenic strain generation
The tax-4 expression constructs were generated using either two-way gateway or multisite gateway cloning.
Primers for psra-13 pDEST cloning
A 918 bp fragment upstream of the sra-13 gene, two nucleotides upstream of the ATG was cloned as an Nhe 1 and Xho 1 fragment into a modified pDEST vector containing the 3′ UTR of unc-54 (primers: 5′-TAAGCTAGCGAATTCGTGAAAAAGTCCACCG-3′ and reverse 5′-TTACTCGAGTAGTGGAAATATCTGAGTTAGTTG-3′).
The tax-4 pENTRY clone
The ptrx-1::tax-4 construct was a gift from C. Chen and K. E. Busch. A 1 kb fragment of the trx-1 promoter had been amplified with the following primers: forward GGGGACAACTTTGTATAGAAAAGTTG(=attB4)-agaatggatacctgatcattc and reverse GGGGACTGCTTTTTTGTACAAACTTG(attB1r)-gatgaaatacaagtgtagaaaattc.
The psrbc-64::tax-4 construct was a kind gift from C. Chen and M. de Bono. A 1.8 kb fragment of the srbc-64 promoter had been amplified with the following primers: forward GGGGACAACTTTGTATAGAAAAGTTG(=attB4)-gttttctaaaaatgagatattactagtggtttgattgctaaacc and reverse GGGGACTGCTTTTTTGTACAAACTTG(attB1r)-cagactgtgacaagaaaactgaaatatcaaaaaaaaaagg.
All tax-4 rescue constructs were injected at 40–50 ng μl−1. Three independent lines were tested for all rescue experiments.
For hen-1 rescue, hen-1;eat-2 animals were injected with a genomic hen-1gfp construct (a gift from T. Ishihara).
Dye filling
DiO labeling was done in the presence of food (1:200 dilution of stock solution 2 mg ml−1 in DMF) for 16–18 h rotating at room temperature. After labeling, the worms were moved to freshly seeded plates and mounted and examined after 3 h.
Chemoattraction/avoidance and osmotic avoidance tests
Attraction to NaCl, benzaldehyde and diacetyl (1:1000) and avoidance of 1-octanol, 2-nonanone and 8 mol l−1 glycerol was essentially done as described elsewhere (Hart, 2006).
Fat staining
Fixed worms were stained with Nile Red [essentially as described previously (Brooks et al., 2009)] with the exception of the fixation, which was done in 1% paraformaldehyde and three cycles of freeze-thawing.
Supplementary Material
ACKNOWLEDGEMENTS
I am grateful to Howard Baylis, Mario de Bono and members of the de Bono lab for reviewing the manuscript. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). I also want to thank everyone who generously provided strains and constructs: Yun Zhang, Simon Tuck, Takeshi Ishihara, Changchun Chen and Mario de Bono. I also would like to thank Ralf Schnabel for depositing the strain GE337 with CGC and noting its bacterial avoidance behavior. Finally, I thank Stefanie Reichelt for help with microscopy and Simon Tuck for a short visit to his lab to learn fat staining.
FOOTNOTES
Competing interests
The author declares no competing financial interests.
Funding
This work was funded by the Medical Research Council, United Kingdom. Deposited in PMC for immediate release.
Supplementary material
Supplementary material available online at http://jeb.biologists.org/lookup/suppl/doi:10.1242/jeb.099929/-/DC1
References
- Andrew P. A., Nicholas W. L. (1976). Effect of bacteria on dispersal of Caenorhabditis elegans (Rhabditidae). Nematologica 22, 451-461 [Google Scholar]
- Apfeld J., O'Connor G., McDonagh T., DiStefano P. S., Curtis R. (2004). The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 18, 3004-3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artyukhin A. B., Schroeder F. C., Avery L. (2013). Density dependence in Caenorhabditis larval starvation. Sci. Rep. 3, 2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L. (1993). The genetics of feeding in Caenorhabditis elegans. Genetics 133, 897-917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I. (2006). Chemosensation in C. elegans. C. elegans Research Community, WormBook; http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. J., Kodama-Namba E., Busch K. E., Murphy R. J., Soltesz Z., Laurent P., de Bono M. (2011). Temperature, oxygen, and salt-sensing neurons in C. elegans are carbon dioxide sensors that control avoidance behavior. Neuron 69, 1099-1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks K. K., Liang B., Watts J. L. (2009). The influence of bacterial diet on fat storage in C.elegans. PLoS ONE 4, e7545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher R. A., Ragains J. R., Li W., Ruvkun G., Clardy J., Mak H. Y. (2009). Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc. Natl. Acad. Sci. USA 106, 1875-1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. Y., Bailey A. P., Leevers S. J., Ragan T. J., Driscoll P. C., Gould A. P. (2011). Anaplastic lymphoma kinase spares organ growth during nutrient restriction in Drosophila. Cell 146, 435-447 [DOI] [PubMed] [Google Scholar]
- Coates J. C., de Bono M. (2002). Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature 419, 925-929 [DOI] [PubMed] [Google Scholar]
- Coburn C. M., Bargmann C. I. (1996). A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans. Neuron 17, 695-706 [DOI] [PubMed] [Google Scholar]
- Cunningham K. A., Hua Z., Srinivasan S., Liu J., Lee B. H., Edwards R. H., Ashrafi K. (2012). AMP-activated kinase links serotonergic signaling to glutamate release for regulation of feeding behavior in C. elegans. Cell Metab. 16, 113-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. W., Somerville D., Lee R. Y., Lockery S., Avery L., Fambrough D. M. (1995). Mutations in the Caenorhabditis elegans Na,K-ATPase alpha-subunit gene, eat-6, disrupt excitable cell function. J. Neurosci. 15, 8408-8418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez M., Attisano L., Wrana J. L., Albert P. S., Massagué J., Riddle D. L. (1993). The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature 365, 644-649 [DOI] [PubMed] [Google Scholar]
- Golden J. W., Riddle D. L. (1982). A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218, 578-580 [DOI] [PubMed] [Google Scholar]
- Golden J. W., Riddle D. L. (1985). A gene affecting production of the Caenorhabditis elegans dauer-inducing pheromone. Mol. Gen. Genet. 198, 534-536 [DOI] [PubMed] [Google Scholar]
- Gómez-Saladín E., Luebke A. E., Wilson D. L., Dickerson I. M. (1997). Isolation of a cDNA encoding a Kex2-like endoprotease with homology to furin from the nematode Caenorhabditis elegans. DNA Cell Biol. 16, 663-669 [DOI] [PubMed] [Google Scholar]
- Gouzi J. Y., Moressis A., Walker J. A., Apostolopoulou A. A., Palmer R. H., Bernards A., Skoulakis E. M. (2011). The receptor tyrosine kinase Alk controls neurofibromin functions in Drosophila growth and learning. PLoS Genet. 7, e1002281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. M., Hill J. J., Bargmann C. I. (2005). A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102, 3184-3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha H. I., Hendricks M., Shen Y., Gabel C. V., Fang-Yen C., Qin Y., Colón-Ramos D., Shen K., Samuel A. D., Zhang Y. (2010). Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron 68, 1173-1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart A. C., Behavior (2006). WormBook (ed. The C. elegans Research Community, WormBook ) doi/10.1895/wormbook.1.87.1, http://www.wormbook.org
- Ishihara T., Iino Y., Mohri A., Mori I., Gengyo-Ando K., Mitani S., Katsura I. (2002). HEN-1, a secretory protein with an LDL receptor motif, regulates sensory integration and learning in Caenorhabditis elegans. Cell 109, 639-649 [DOI] [PubMed] [Google Scholar]
- Jang H., Kim K., Neal S. J., Macosko E., Kim D., Butcher R. A., Zeiger D. M., Bargmann C. I., Sengupta P. (2012). Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron 75, 585-592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G., Thijssen K. L., Werner P., van der Horst M., Hazendonk E., Plasterk R. H. (1999). The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat. Genet. 21, 414-419 [DOI] [PubMed] [Google Scholar]
- Jeong P. Y., Jung M., Yim Y. H., Kim H., Park M., Hong E., Lee W., Kim Y. H., Kim K., Paik Y. K. (2005). Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature 433, 541-545 [DOI] [PubMed] [Google Scholar]
- Johnson E. C., Kazgan N., Bretz C. A., Forsberg L. J., Hector C. E., Worthen R. J., Onyenwoke R., Brenman J. E. (2010). Altered metabolism and persistent starvation behaviors caused by reduced AMPK function in Drosophila. PLoS ONE 5, e12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F., Badri D. V., Zachariah C., Ajredini R., Sandoval F. J., Roje S., Levine L. H., Zhang F., Robinette S. L., Alborn H. T., et al. (2009). Bacterial attraction and quorum sensing inhibition in Caenorhabditis elegans exudates. J. Chem. Ecol. 35, 878-892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola B. (2008). Role of AMP-activated protein kinase in the control of appetite. J. Neuroendocrinol. 20, 942-951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H., Mori I., Rhee J. S., Akaike N., Ohshima Y. (1996). Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron 17, 707-718 [DOI] [PubMed] [Google Scholar]
- Krishna S., Maduzia L. L., Padgett R. W. (1999). Specificity of TGFbeta signaling is conferred by distinct type I receptors and their associated SMAD proteins in Caenorhabditis elegans. Development 126, 251-260 [DOI] [PubMed] [Google Scholar]
- L'Etoile N. D., Bargmann C. I. (2000). Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron 25, 575-586 [DOI] [PubMed] [Google Scholar]
- Lans H., Rademakers S., Jansen G. (2004). A network of stimulatory and inhibitory Galpha-subunits regulates olfaction in Caenorhabditis elegans. Genetics 167, 1677-1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Lints R., Foehr M. L., Tokarz R., Yu L., Emmons S. W., Liu J., Savage-Dunn C. (2003). The Caenorhabditis elegans schnurri homolog sma-9 mediates stage- and cell type-specific responses to DBL-1 BMP-related signaling. Development 130, 6453-6464 [DOI] [PubMed] [Google Scholar]
- Lindsley J. E., Rutter J. (2004). Nutrient sensing and metabolic decisions. Comp. Biochem. Physiol. 139B, 543-559. [DOI] [PubMed] [Google Scholar]
- Long X., Spycher C., Han Z. S., Rose A. M., Muller F., Avruch J. (2002). TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 12, 1448-1461 [DOI] [PubMed] [Google Scholar]
- Macosko E. Z., Pokala N., Feinberg E. H., Chalasani S. H., Butcher R. A., Clardy J., Bargmann C. I. (2009). A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458, 1171-1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay J. P., Raizen D. M., Gottschalk A., Schafer W. R., Avery L. (2004). eat-2 and eat-18 are required for nicotinic neurotransmission in the Caenorhabditis elegans pharynx. Genetics 166, 161-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J. A., Ruvkun G. (2012). Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149, 452-466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milward K., Busch K. E., Murphy R. J., de Bono M., Olofsson B. (2011). Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108, 20672-20677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörck C., Rauthan M., Wågberg F., Pilon M. (2004). pha-2 encodes the C. elegans ortholog of the homeodomain protein HEX and is required for the formation of the pharyngeal isthmus. Dev. Biol. 272, 403-418 [DOI] [PubMed] [Google Scholar]
- Pan K. Z., Palter J. E., Rogers A. N., Olsen A., Chen D., Lithgow G. J., Kapahi P. (2007). Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6, 111-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel G. D., Ropelle E. R., Rocha G. Z., Carvalheira J. B. (2013). The role of neuronal AMPK as a mediator of nutritional regulation of food intake and energy homeostasis. Metabolism 62, 171-178 [DOI] [PubMed] [Google Scholar]
- Pradel E., Zhang Y., Pujol N., Matsuyama T., Bargmann C. I., Ewbank J. J. (2007). Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104, 2295-2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provenza F. D. (1996). Acquired aversions as the basis for varied diets of ruminants foraging on rangelands. J. Anim. Sci. 74, 2010-2020 [DOI] [PubMed] [Google Scholar]
- Pujol N., Link E. M., Liu L. X., Kurz C. L., Alloing G., Tan M. W., Ray K. P., Solari R., Johnson C. D., Ewbank J. J. (2001). A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. 11, 809-821 [DOI] [PubMed] [Google Scholar]
- Pungaliya C., Srinivasan J., Fox B. W., Malik R. U., Ludewig A. H., Sternberg P. W., Schroeder F. C. (2009). A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 106, 7708-7713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C., Dickson B. J. (2010). Sex peptide receptor and neuronal TOR/S6K signaling modulate nutrient balancing in Drosophila. Curr. Biol. 20, 1000-1005 [DOI] [PubMed] [Google Scholar]
- Roayaie K., Crump J. G., Sagasti A., Bargmann C. I. (1998). The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron 20, 55-67 [DOI] [PubMed] [Google Scholar]
- Savage C., Das P., Finelli A. L., Townsend S. R., Sun C. Y., Baird S. E., Padgett R. W. (1996). Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor beta pathway components. Proc. Natl. Acad. Sci. USA 93, 790-794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackwitz W. S., Inoue T., Thomas J. H. (1996). Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17, 719-728 [DOI] [PubMed] [Google Scholar]
- Schulenburg H., Ewbank J. J. (2007). The genetics of pathogen avoidance in Caenorhabditis elegans. Mol. Microbiol. 66, 563-570 [DOI] [PubMed] [Google Scholar]
- Shinkai Y., Yamamoto Y., Fujiwara M., Tabata T., Murayama T., Hirotsu T., Ikeda D. D., Tsunozaki M., Iino Y., Bargmann C. I., et al. (2011). Behavioral choice between conflicting alternatives is regulated by a receptor guanylyl cyclase, GCY-28, and a receptor tyrosine kinase, SCD-2, in AIA interneurons of Caenorhabditis elegans. J. Neurosci. 31, 3007-3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtonda B. B., Avery L. (2006). Dietary choice behavior in Caenorhabditis elegans. J. Exp. Biol. 209, 89-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J., Kaplan F., Ajredini R., Zachariah C., Alborn H. T., Teal P. E., Malik R. U., Edison A. S., Sternberg P. W., Schroeder F. C. (2008). A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 454, 1115-1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Yandell M. D., Roy P. J., Krishna S., Savage-Dunn C., Ross R. M., Padgett R. W., Wood W. B. (1999). A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development 126, 241-250 [DOI] [PubMed] [Google Scholar]
- Tsunozaki M., Chalasani S. H., Bargmann C. I. (2008). A behavioral switch: cGMP and PKC signaling in olfactory neurons reverses odor preference in C. elegans. Neuron 59, 959-971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas M. A., Luo N., Yamaguchi A., Kapahi P. (2010). A role for S6 kinase and serotonin in postmating dietary switch and balance of nutrients in D. melanogaster. Curr. Biol. 20, 1006-1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronico P., Gray L. J., Jones J. T., Bazzicalupo P., Arbucci S., Cortese M. R., Di Vito M., De Giorgi C. (2001). Nematode chitin synthases: gene structure, expression and function in Caenorhabditis elegans and the plant parasitic nematode Meloidogyne artiellia. Mol. Gen. Genet. 266, 28-34 [DOI] [PubMed] [Google Scholar]
- Wakabayashi T., Kitagawa I., Shingai R. (2004). Neurons regulating the duration of forward locomotion in Caenorhabditis elegans. Neurosci. Res. 50, 103-111 [DOI] [PubMed] [Google Scholar]
- Ward S. (1973). Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc. Natl. Acad. Sci. USA 70, 817-821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Zhang Y., Xu J., Shen P. (2005). Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc. Natl. Acad. Sci. USA 102, 13289-13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Hirotsu T., Matsuki M., Butcher R. A., Tomioka M., Ishihara T., Clardy J., Kunitomo H., Iino Y. (2010). Olfactory plasticity is regulated by pheromonal signaling in Caenorhabditis elegans. Science 329, 1647-1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Morita K., Mochii M., Ueno N. (2001). Hypodermal expression of Caenorhabditis elegans TGF-beta type I receptor SMA-6 is essential for the growth and maintenance of body length. Dev. Biol. 240, 32-45 [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhang Y. (2012). DBL-1, a TGF-β, is essential for Caenorhabditis elegans aversive olfactory learning. Proc. Natl. Acad. Sci. USA 109, 17081-17086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lu H., Bargmann C. I. (2005a). Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179-184 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Foster J. M., Nelson L. S., Ma D., Carlow C. K. (2005b). The chitin synthase genes chs-1 and chs-2 are essential for C. elegans development and responsible for chitin deposition in the eggshell and pharynx, respectively. Dev. Biol. 285, 330-339 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.