Abstract
Resistance to RAF inhibitors such as vemurafenib and dabrafenib is a major clinical problem in the treatment of melanoma. Mutant BRAF melanoma patients that progress on RAF inhibitors have limited treatment options and drug removal from resistant tumors may elicit multiple effects. A frequent mechanism of resistance to RAF inhibitors is caused by expression of mutant BRAF splice variants. RAF inhibitor-resistant cell lines, generated in vivo, were tested as to whether or not mutant BRAF splice variants confer a fitness advantage in the presence of RAF inhibitor. Critically, cells expressing distinct mutant BRAF splice variants grow more efficiently in vitro and in vivo in the presence of the vemurafenib analog, PLX4720, compared to in the absence of inhibitor. PLX4720-treated BRAF splice variant-expressing cells exhibited levels of phospho-ERK1/2 comparable to untreated parental cells. In addition, a reduction in phospho-ERK1/2 levels following treatment with the MEK inhibitor, trametinib (GSK1120212) phenocopied the fitness benefit provided by PLX4720. These data indicate that mutant BRAF splice variant-expressing melanoma cells are benefited by defined concentrations of RAF inhibitors.
Implications
The present study provides evidence that RAF inhibitor-resistant melanoma cells benefit from continued therapy.
Keywords: BRAF, Drug resistance, ERK1/2, Melanoma, Vemurafenib
INTRODUCTION
Dysregulated extracellular regulated kinase 1/2 (ERK1/2) signaling has been linked to a number of malignancies including melanoma. Approximately 50% of melanoma patients present with a mutation in the serine-threonine kinase BRAF; most mutations alter codon 600 from a valine to glutamic acid (V600E) (1). The V600E mutation leads to a constitutively active kinase form of BRAF, which aberrantly signals to its downstream target, mitogen-activated protein kinase kinase (MEK) (2). Activated MEK phosphorylates ERK1/2, which in turn phosphorylates transcription factors that control a number of proliferative and anti-apoptotic targets.
The landscape of treatment options for patients suffering from metastatic BRAFV600E melanoma drastically changed with the introduction of targeted RAF inhibitors (3). Vemurafenib/PLX4032 and dabrafenib, which are both FDA-approved drugs, act as ATP-competitive inhibitors of BRAFV600E. RAF inhibitors achieve clinical response rates of approximately 50% and the median progression-free survival is 5–7 months (4, 5). However, most of the patients who initially respond will develop resistance to the treatment and ultimately succumb to the disease (5). RAF inhibitor resistance is currently a major challenge for the melanoma field.
Multiple mechanisms of RAF inhibitor resistance have been demonstrated, most of which funnel through re-activation of ERK1/2 (reviewed in (6)). Previous work has identified multiple mechanisms of resistance including upregulation of the receptor tyrosine kinases (RTKs) PDGFRβ and IGF-1R, BRAF amplification, COT expression, mutation of RAS isoforms, and mutations in MEK1 and MEK2 (7–16). However, the most frequent mechanism identified to date is the expression of BRAFV600E splice variants lacking the RAS binding domain (7, 9). Despite RAS-binding defects, these variants retain RAF kinase activity in the presence of vemurafenib due to their enhanced homodimerization properties (7, 9).
The treatment options for patients progressing on vemurafenib or dabrafenib are limited. Disease progression is often too rapid for immune treatments such ipilimumab to elicit effects and, hence, standard chemotherapies are usually administred (17–19). In order to provide a preclinical basis for possible second-line treatment strategies for patients progressing on RAF inhibitor, the effect of inhibitor withdrawl or a “drug holiday” has been investigated. Cessation of targeted inhibitor treatment has been suggested to exploit the adaptive mechanisms that cells may utilize to preserve ERK1/2 signaling (20). In one study, a drug holiday was shown to be efficacious for a melanoma cell line in which RAF inhibitor resistance was mediated by BRAF copy number amplification (21). In contrast, an independent study showed that BRAFV600E melanoma patients progressing on vemurafenib or dabrafenib experienced rapid disease progression after cessation of the treatment regimen, suggesting that presence of RAF inhibitor was slowing growth of the resistant cells (8). These data seemingly oppose each other and suggest that many factors may be involved in determining the response of resistant cells to RAF inhibitors. To further examine this, we tested the effects of a drug holiday on the growth and signaling of in vivo-derived RAF inhibitor resistant cell lines that express BRAFV600E splice variants.
MATERIALS AND METHODS
Cell culture
1205Lu ERK1/2 reporter cells were generated by transducing cells with UAS/EGFP firefly luciferase and UbC/GAL4-ELK1 constructs. Full procedures are described in (7). PRT #3 and PRT #4 cells express the BRAF V600E splice variants, ΔEx3–10 and ΔEx 2–8, respectively (7). Cells were cultured in 37°C humidified chamber supplemented with 5% CO2 in MCDB 153 media containing 20% Leibovitz-L15 media, 2% fetal bovine serum, 0.2% sodium bicarbonate, and 5µg/mL insulin. PRT lines were maintained in 1µM PLX4720.
Inhibitors
PLX4720 and PLX4720 rodent chow was provided by Dr. Gideon Bollag (Plexxikon Inc., Berkeley, CA). Trametinib was purchased from Selleck (Houston, TX).
Proliferation assay
Cells (1.4 × 104) were seeded in 6 well dishes. Medium and indicated drugs were replenished every 2 days. After nine days, plates were washed in PBS and fixed in buffered formalin with 0.2% crystal violet. Plates were then scanned and percent area coverage was quantified using ImageJ software. Displayed images are representative and the associated graphs are normalized composites of at least 3 independent experiments. Error bars are +/− standard error of the mean (SEM). Significance was determined with the use of two-tailed student’s t-test assuming unequal variance. p values less than 0.05 were considered significant.
Xenografts growth assay
PRT cells (1.0 × 106) were injected intradermally on the backs of female athymic mice (Nu/J Jackson; sixteen mice per cell line). Eight of the 16 mice for each cell line were then randomly chosen to be treated with vehicle AIN-76A chow, and the other half was treated with 417 mg/kg PLX4720 laced AIN-76A chow. Tumor volume measurements were taken every 2–3 days starting on day 2 with digital calipers. Tumor volume was determined utilizing the volume = (length × width2) × 0.52 formula. Maximal tumor size was limited by appearance of skin necrosis requiring euthanasia. Data were analyzed using mixed effects model. Analyses were performed using SAS software and p values less than 0.05 were considered significant.
Western blotting
Blotting was performed as previously described (22). Antibodies used were: Actin #A2066 from Sigma (St. Louis, MO), ERK2 #sc-1647, MEK1 #sc-219 from Santa Cruz Biotechnology (Santa Cruz, CA), Sprouty2 #07-524 from Millipore (Billerica, MA), and pERK1/2 #9101, pMEK1/2 #9121, p21Cip1 #2946, p27Kip1 #2552 from Cell Signaling (Danvers, MA). Chemiluminence was visualized with VersaDoc MultiImager and quantified using Quantity-One software (Bio-Rad, Hercules, CA).
S-phase entry analysis
Cells (2.0 × 105) were seeded in triplicate on 6-well plates. Cells were then treated with either PLX4720 or DMSO (vehicle) for a total of 88 hours. The thymidine analog, EdU was added at a final concentration of 10µM for final 16 hours. EdU incorporation was measured using the Click-it EdU Alexa Fluor 647 Flow Cytometry assay kit was utilized as per manufacturer’s instructions (Molecular Probes, Grand Island, NY). EdU staining was quantified on BD FacsCalibur and data was analyzed with FlowJo software. Data points are normalized averages of triplicates from three experimental replicates. Error bars are +/− SEM. p < 0.05 was considered significant.
RESULTS
BRAFV600E splice variant-expressing melanoma cells grow more efficiently in the presence of RAF inhibitor
We have previously generated several RAF inhibitor-resistant variants of BRAFV600E 1205Lu melanoma cells through continued treatment with PLX4720 of xenografts in athymic mice (7). BRAF splice variants are expressed in approximately 40% the PLX4720 resistant tumor (PRT) cell lines and their expression is required for RAF inhibitor resistance (7). Since mutant BRAF splice variant harboring cells may represent a subpopulation of cells that has grown out during RAF inhibitor treatment due to a fitness advantage, we assessed whether the variant-expressing PRT cells displayed enhanced growth in the presence of RAF inhibitor.
In our previous work, we have shown that the expression of BRAF variants in PRT #3 and PRT #4 cells is necessary for RAF inhibitor resistance and also that no alterations in NRAS, MEK1 and MEK2 were detected (7). Additionally, Western blot analysis revealed no obvious up-regulation of Akt signaling that can provide an ERK1/2 pathway-independent mode of resistance (Supplemental Fig. 1). We subjected these two cell lines, PRT #3 (expresses BRAFV600E ΔEx 3–10) and PRT #4 (expresses BRAFV600E ΔEx 2–8) to a 2D crystal violet proliferation assay in the presence of increasing concentrations of the RAF inhibitor, PLX4720. In parental cells, PLX4720 concentrations of 50 nM and higher significantly decreased the proliferation of parental cells in a dose dependent manner (Fig. 1A, B). The proliferation of PRT #3 and PRT #4 cells was not significantly different to the vehicle treatment at low (10 - 100nM) PLX4720 concentrations; however, both PRT lines showed enhanced proliferation at intermediate PLX4720 concentrations. PRT #3 cells displayed a significant proliferation advantage with 1µM PLX4720 treatment while increasing the dose to 5µM or above led to a significant reduction in growth. PRT #4 cells demonstrated significant growth advantages at both 500nM and 1µM PLX4720, while the 10µM dosage significantly slowed cell proliferation. Thus, both PRT cells lines showed enhanced proliferation in an intermediate concentration of PLX4720 which can be overcome by further increasing the dose of RAF inhibitor. To further support these findings, cells were assayed for S-phase entry via EdU incorporation. In parental 1205Lu cells, 1µM PLX4720 treatment inhibited S-phase entry as expected (Fig. 1C). In contrast, PRT #3 and PRT #4 cells displayed a significant increase in S-phase entry following 1µM PLX4720 treatment.
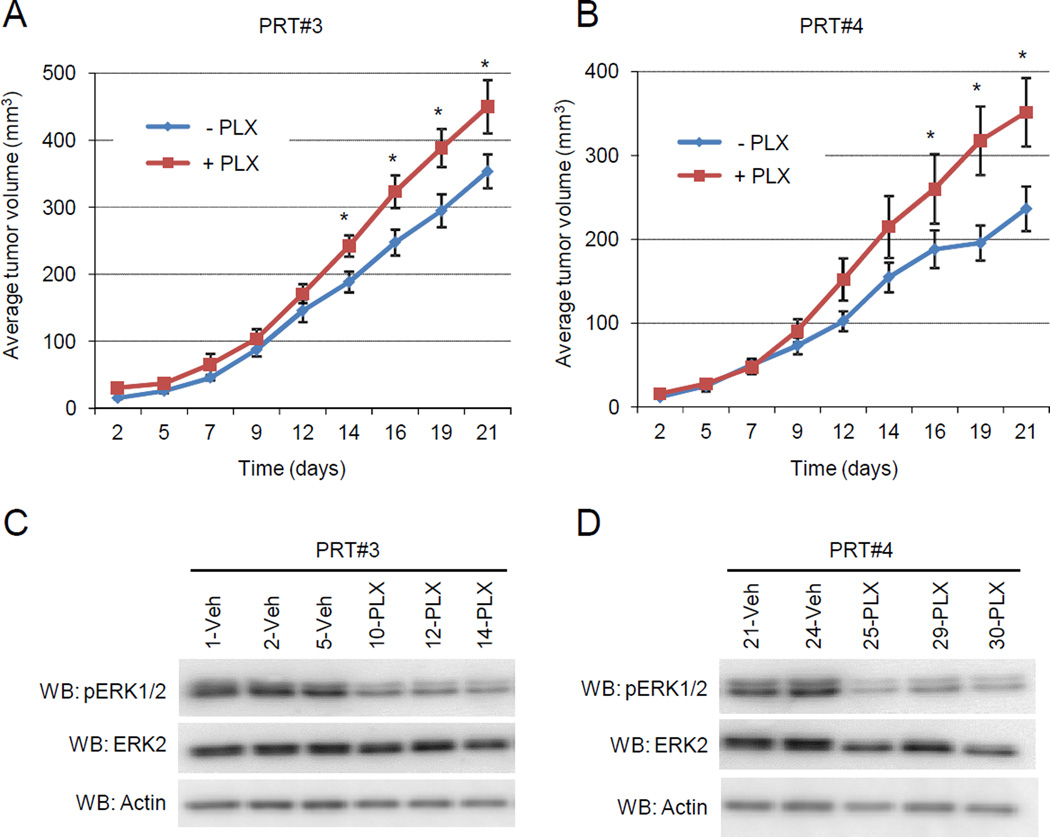
Fig. 1. Growth benefit conferred by resistant cells treated with RAF inhibitor.
(A) 1.4×104 of the indicated cell line were seeded and treated with DMSO or different concentrations of PLX4720 (10nM, 50nM, 100nM, 500nM, 1µM, 5µM or 10µM). Media was changed every 2 days, for 9 days of total treatment. Plates where then PBS washed, and fixed/stained with 0.25% crystal violet in buffered formalin. (B) Quantification of percent area coverage, each cell line normalized to DMSO condition. (C) 2.0 × 105 of all three cell lines were seeded in triplicate and drug treated with DMSO or 1µM PLX4720 for a total of 88hrs. A final concentration of 10µM EdU was allowed to incorporate for 16hrs prior to processing. S-phase entry of each cell line is normalized to DMSO condition, and data points are indicative of 3 experimental repeats. P values < 0.05 indicated with *, and error bars are +/− SEM.
We also questioned if these cells possess altered invasion properties with drug treatment. PRT cells invasion assays were performed with the use of Matrigel coated Boyden chambers with either DMSO or 1µM PLX4720. Serum containing medium (with DMSO or 1µM PLX4720) was used as a chemoattractant. After 24 hours, there was no significant difference in the invasive potential of PRT #3 and PRT #4 cells in the presence of RAF inhibitor (Supplemental Fig. 2). Taken together these data suggest the resistant cells show enhanced proliferation at intermediate RAF inhibitor doses (1µM PLX4720), while still being susceptible to an increased dosage. Furthermore, while we observed an increase rate of proliferation of the PRT cells under drug treatment, we did not detect altered invasion properties.
BRAFV600E splice variant-expressing cell xenografts grow more effectively with RAF inhibitor-treatment
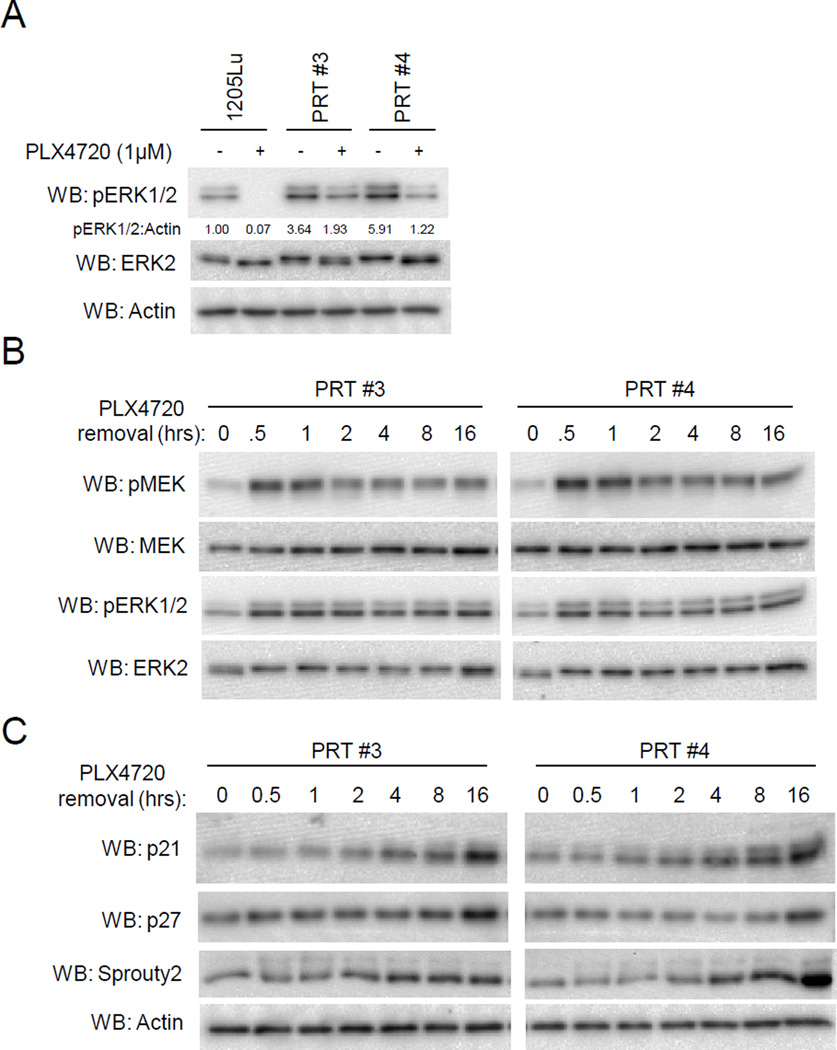
Since PRT #3 or PRT #4 cells were generated through in vivo selection to PLX4720, we wanted to examine if the benefit of PLX4270 treatment observed in vitro would be replicated in vivo. In previous studies, we have shown that the growth of parental cell xenografts in athymic mice is effectively blocked by PLX4720 treatment but continued treatment gave rise to the resistant cell lines, PRT #3 and PRT #4 which are able to growth in the presence of PLX4720 (7). Here, we determined the effect of PLX4720 withdrawal on PRT #3 and PRT #4 xenografts by dividing mice into two cohorts: one receiving vehicle chow; the other receiving PLX4720-laced chow. When tumor growth was measured over 21 days, PLX4720 treatment produced a significant increase in the mean tumor size in both PRT #3 and PRT #4 xenografts compared to the vehicle-treated (drug-removed) xenografts (Fig. 2A, 2B). To determine the possible cause of this enhanced growth, tumors were harvested from mice in each of the cohorts and cell cultures generated. Tumor cells from the indicated mouse were cultured short term (P2) in tissue culture plates maintaining the presence or absence of 1µM PLX4720 to mimic the in vivo setting. Western blotting of lysates revealed an increased state of ERK1/2 phosphorylation in the vehicle-treated PRT cells compared to PLX4720-treated PRT cells (Fig. 2C, 2D). These in vivo data support the notion that RAF inhibitor may increase the proliferation of mutant BRAF splice variant-expressing cells by maintaining a lower phospho-ERK steady state.
Fig. 2. Accelerated growth of PRT #3 and PRT #4 in vivo with RAF inhibitor treatment.
(A,B) Intradermal injections of 1.0 x106 cells on the back of 7 week old female Nu/J mice was performed with PRT #3 and PRT #4 (16 mice for each cell line). 8 mice from each of the cell lines were then randomly chosen to be fed chow laced with 417mg/kg PLX4720 or vehicle chow. Tumor measurements with digital calipers were taken over a 21 day time course. Graphs are depicting mean tumor growth in mm3 for PRT #3 (A) and PRT #4 (B). An asterisk (*) on the indicated day represents a p value < 0.05. The error bars represent +/− SEM. (C) PRT #3 xenografts were harvested and single cell suspensions were generated for in vitro testing. Tumor cells arising from the indicated mouse number were treated with either DMSO or 1µM PLX4720 in an attempt to maintain in vivo conditions. Once sufficient cell numbers were obtained (approximately 3 days), they were seeded with or without 1µM PLX4720 and lysed for western blot analysis. (D) As for (C), except PRT #4 xenografts were analyzed.
Phospho-ERK1/2 is elevated in PRT cell lines after RAF inhibitor removal
We further explored the phospho-ERK1/2 status by utilizing PRT #3 and PRT #4 cells in in vitro studies. When cultured in 1µM PLX4720, PRT #3 and PRT #4 displayed levels of phospho-ERK1/2 that were comparable to untreated levels in parental cells (Fig. 3A). However, overnight removal of PLX4720 strongly increased the ERK1/2 activation in both PRT #3 and PRT #4 cells (Fig. 3A). Drug removal was also associated with a morphological change to an elongated and spindle-like appearance, consistent with morphology changes observed during elevated ERK1/2 signaling (23) (Supplemental Fig. 3). In time course experiments, PLX4720 removal from PRT #3 and PRT #4 cell cultures demonstrated enhanced phosphorylation of MEK and ERK1/2 by 30 minutes, and the elevated phosphorylation state was maintained throughout a 16 hour time course (Fig. 3B). Consistent with aberrantly high ERK1/2 activity inhibiting cell cycle progression (24–26), enhanced levels of cyclin dependent kinase (CDK) inhibitors p21Cip1 and p27Kip1 were observed following PLX4720 withdrawal (Fig. 3C). In addition, the ERK1/2 target Sprouty2 (27) was also shown to accumulate correlating with elevated ERK1/2 function. These data suggest that the fitness deficit caused by removal of RAF inhibitor in mutant BRAF splice variant cells may be due to hyperactivation of ERK1/2 and upregulation CDK inhibitor proteins.
Fig. 3. Phospho-ERK1/2 levels are hyperactivated in PRT #3 and PRT #4 cell lines after RAF inhibitor withdrawal.
(A) Parental 1205Lu cells were seeded overnight in drug-free medium, and PRT #3 and PRT #4 cells were seeded in 1µM PLX4720. The next day, cells were washed and then administered either 1µM PLX4720 or DMSO for a 16 hour treatment. Western blots were performed to assay ERK1/2 activation status. (B) PRT #3 and PRT #4 cells were cultured in PLX4720 (1µM) and then medium changed to drug-free medium. Cell lysates were harvested at the indicated time and analyzed by western blotting for levels of phosphorylated MEK as well as phosphorylated ERK1/2. (C) Similar to (B), cells were lysed after culturing in drug-free medium for the indicated time, and the CDK inhibitors p21Cip1 and p27Kip1 were analyzed by western blot, as well as the putative ERK target sprouty2.
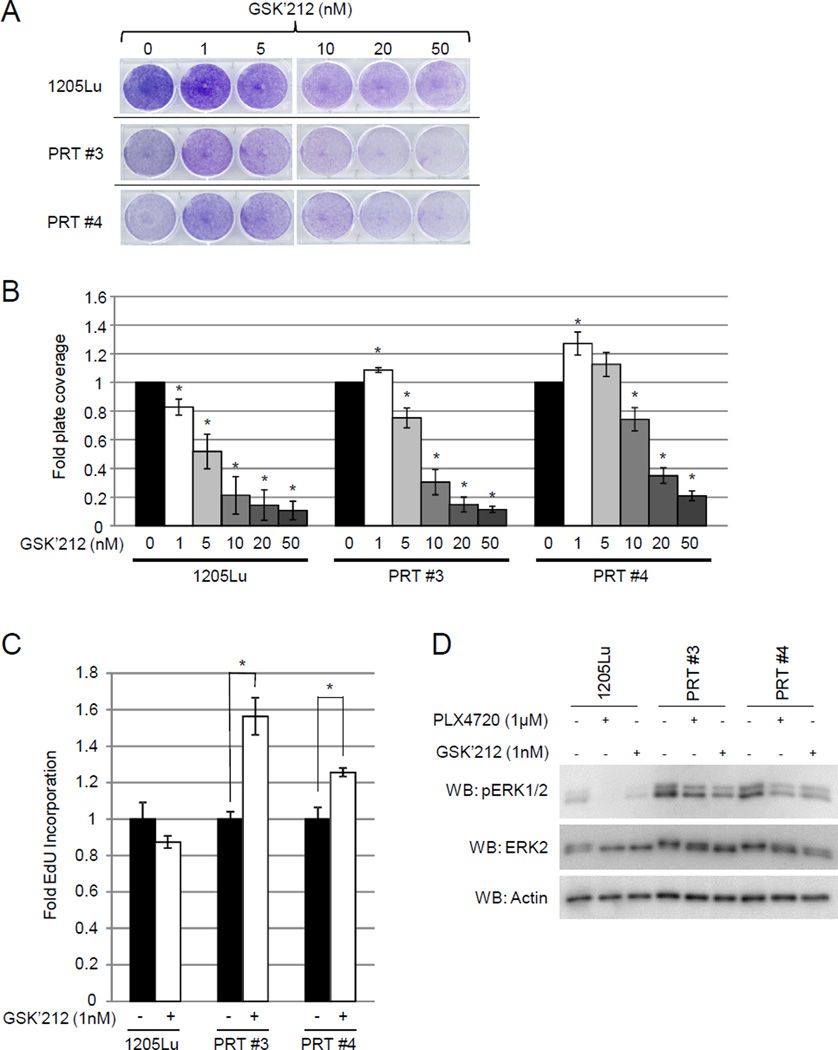
Trametinib (GSK’212) phenocopies the effects of PLX4720 treatment in PRT cell lines
Our data suggest that cells harboring BRAF splice variants possess a higher basal phospho-ERK1/2 level when compared to non-expressing cells, and that intermediate doses of PLX4720 are able to reduce phospho-ERK1/2 to levels comparable to those observed in parental cells. Since the MEK inhibitor, trametinib (GSK1120212, “GSK’212”), is FDA-approved in BRAFV600E/K melanoma and the combination of RAF inhibitor and MEK inhibitor is being tested in the clinic (28), we tested if GSK’212 would elicit a similar effect to PLX4720 treatment. PRT #3 and PRT #4 cells as well as parental 1205Lu cells were subjected to proliferation assays in the presence of an escalating dose of GSK’212 (Fig. 4A, 4B). In both PRT cell lines, a low dose (1nM) of GSK’212 was able to increase cell growth (PRT #3 p <0.0132, PRT #4 p < 0.029), similar to the effects we observed with a 1µM dosage of PLX4720 (Fig. 1). This was further highlighted by EdU incorporation of PRT #3 and PRT #4 where S-phase entry was significantly higher in cells treated with 1nM GSK’212 (Fig. 4C). Additionally, western blot analysis demonstrated that 1nM GSK’212 reduced the ERK1/2 phosphorylation to similar levels as 1µM PLX4720 treatment, suggesting that both of these drugs reduces the unfavorable ERK hyperactivation in non-treated BRAF variant-expressing resistant cells (Fig. 4D).
Fig. 4. Low dose MEK inhibitor is able to provide growth advantage to PRT #3 and PRT #4.
(A) 1.4×104 of the indicated cell line were seeded and treated with DMSO or different concentrations of trametinib (GSK’212) (1nM, 5nM, 10nM, 20nM, or 50nM). Medium was changed every 2 days for 9 days of total treatment. Cells were then fixed and stained with 0.25% crystal violet in buffered formalin. (B) Quantification of percent area coverage is displayed normalized to each cell line’s DMSO condition. Error bars are +/− SEM three replicates and * indicates a p value of < 0.05. (C) 2.0 × 105 of parental 1205Lu, PRT #3, and PRT #4 were seeded normal culture media, and next day cells were treated with either DMSO, or 1nM GSK’212 for a total of 88hrs. S-phase entry as measured via EdU incorporation of parental 1205Lu, PRT #3, and PRT #4 cells was then performed. Error bars are +/− SEM of three experimental repeats, and * indicates a p value < 0.05. (D) Cells were seeded overnight in normal growth medium and then treated with either DMSO, 1µM PLX4720, or 1nM GSK’212 overnight. Cell lysates were obtained and analyzed for ERK1/2 phosphorylation via western blotting.
We further explored the effect of GSK’212 by combining it with PLX4720. We compared parental 1205Lu cells, PRT #3, and PRT #4 in 2D growth assays when these cells were challenged with GSK’212 monotherapy, or the combination of GSK’212 with 1µM PLX4720. As expected the combination of GSK’212 and PLX4720 was more effective than GSK’212 monotherapy at inhibiting the growth of parental 1205Lu cells (Supplemental Fig. 4A). Moreover, the combination of drugs slowed the growth of PRT #3 and #4 cells. At 1, 5, and 50nM GSK’212/PLX4720 combination a significant reduction in growth of PRT #3 was observed compared to GSK’212 monotherapy (p = 0.021, p = 0.041, and p = 0.018, respectively). Similarly, a significant reduction of growth under combination treatment was observed in PRT #4 at 10 and 20nM GSK’212 concentrations (p = 0.006 and p = 0.035, respectively). In addition, western blots were performed after overnight treatments at similar drug concentrations to assess ERK1/2 activation status (Supplemental Fig 4B). As anticipated, the phosphorylation of ERK1/2 was ablated at lower concentrations of GSK’212 when combined with treatment of PLX4720.
Taken together, these data suggest that GSK’212 is able to provide the same benefit afforded to proliferation and S-phase entry of the PRT cell lines as PLX4720 treatment. Similar to PLX4720, GSK’212 also was able to substantially reduce cell growth and ERK1/2 phosphorylation at higher concentrations, (~5nM and higher) suggesting that both drugs are able to suppress phospho ERK1/2 levels below the homeostatic threshold. Additionally, the combination of PLX4720 and GSK’212, elicited a more potent reduction in proliferation and ERK1/2 phosphorylation when compared to GSK’212 alone.
DISCUSSION
While the FDA approval of vemurafenib and dabrafenib have dramatically impacted on the treatment of patients suffering from metastatic melanoma, a lasting response for these patients has been elusive. When patients begin to progress on RAF inhibitors, difficult decisions must be made for further treatments. Here, we demonstrate that removal of RAF inhibitor slows the growth of BRAFV600E splice variant-expressing, RAF inhibitor-resistant cells. This effect is associated with decreased S-phase entry, hyperelevated levels of phosphorylated ERK1/2, and stabilization of the CDK inhibitors, p21Cip1 and p27Kip1. These data are in line with a recent publication that demonstrates a “drug holiday” induces the regression of established resistant tumors (21). This group was able to demonstrate that a BRAFV600E amplified RAF inhibitor resistant cell line possessed a higher level of cell viability under RAF inhibitor treatment compared to the absence of inhibitor.
At first glance, these data may suggest that cessation of RAF inhibitor treatment in relapsed patients would be beneficial; however, other studies have implicated that removal of RAF inhibitor in relapsing patients causes a rapid increase in disease burden (8). In one facet of a study conducted by Carlino and colleagues investigated the effects of RAF inhibitor withdrawal on a patient-derived BRAF splice variant with exons 4–10 deleted. It was reported that these cells had increased BrdU positivity and proliferation after removal of the RAF inhibitor supporting the notion that continued drug treatment is in the patient’s best interest even after disease progression had been observed. Other studies have examined the effects of re-challenging tumors with RAF inhibitors. One report examines two patients both of which were progressing on and therefore removed from RAF inhibitor treatment (29). Subsequent re-challenge with RAF inhibitor led to a significant reduction in tumor burden as well as an increase in overall well-being as measured by Kamofsky score in both patients (29). This report suggests that RAF inhibitor resistant tumors can become re-sensitized after a drug free interval. It also further highlights the apparent heterogeneity of tumor response observed after RAF inhibitor cessation.
The in vitro and xenograft model studies in this work add to this notion of heterogeneity in cellular response from drug removal in acquired RAF inhibitor resistant cells. Our experimental system utilized two in vivo-derived, RAF inhibitor-resistant cell lines, both of which harbor a BRAF splice variant lacking the RAS binding domain as the mediator of RAF inhibitor resistance (7). We found that both of these cell lines experience a growth advantage in the presence of RAF inhibitor. Our data also suggest that there is a “window” of ERK1/2 activity that seems to be beneficial to the BRAF variant expressing resistant tumor cells. When RAF inhibitor concentration is too low (or absent), ERK1/2 phosphorylation is hyperelevated and may contribute to the stabilization of CDK inhibitors p21Cip1 and p27Kip1. Conversely, when the RAF inhibitor concentration is high enough, RAF activity is indeed blocked leading to a loss of ERK1/2 signaling and ultimately cell death. It is also possible that this window may also be achieved by low doses of the BRAF and MEK inhibitor combination.
Presently, it is unknown why the patients in the Carlino study had a different response to RAF inhibitor cessation compared to the results reported here. It is possible that upon RAF inhibitor withdrawal, the magnitude of ERK1/2 reactivation in the patient-derived resistant tumors is not as high as the level we observed in our in vivo-derived BRAFV600E splice variant RAF inhibitor resistant cells. If this were the case, perhaps the patients' tumors would not possess ERK1/2 signaling strong enough to elicit stabilization of the CDK inhibitors we observed in our studies. It may be interesting to stratify the response of relapsed tumors to RAF inhibitor withdrawal based on the strength of the initial RAF inhibitor dosage. This might give insights to the apparent heterogeneity of relapse tumor response to RAF inhibitor removal. Additionally, further studies examining what specifically elicits a cellular benefit or detriment after removal of RAF inhibition should be investigated, as this may yield better understanding for possible options for relapsing patients.
Supplementary Material
ACKNOWLDEGEMENTS
These studies were supported by grants from the National Institutes of Health (R01-CA160495), Department of Defense (W81XWH-11-1-0385), and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to A.E. Aplin). E.J. Hartsough and K. Basile were funded in part by the National Cancer Center and the Joanna M Nicolay Melanoma Foundation, respectively. We thank members of the Aplin laboratory for critical feedback on this article.
Footnotes
Disclosure: There are no potential conflicts of interest
REFERENCES
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Conner SR, Scott G, Aplin AE. Adhesion-dependent activation of the ERK1/2 cascade is by-passed in melanoma cells. J Biol Chem. 2003;278:34548–34554. doi: 10.1074/jbc.M305797200. [DOI] [PubMed] [Google Scholar]
- 3.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartsough E, Shao Y, Aplin AE. Resistance to RAF Inhibitors Revisited. J Invest Dermatol. 2013 doi: 10.1038/jid.2013.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basile KJ, Abel EV, Dadpey N, Hartsough EJ, Fortina P, Aplin AE. In vivo MAPK reporting reveals the heterogeneity in tumoral selection of resistance to RAF inhibitors. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-13-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlino MS, Gowrishankar K, Saunders CA, Pupo GM, Snoyman S, Zhang XD, et al. Antiproliferative effects of continued mitogen-activated protein kinase pathway inhibition following acquired resistance to BRAF and/or MEK inhibition in melanoma. Mol Cancer Ther. 2013;12:1332–1342. doi: 10.1158/1535-7163.MCT-13-0011. [DOI] [PubMed] [Google Scholar]
- 9.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villanueva J, Infante JR, Krepler C, Reyes-Uribe P, Samanta M, Chen HY, et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep. 2013;4:1090–1099. doi: 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abel EV, Basile KJ, Kugel CH, 3rd, Witkiewicz AK, Le K, Amaravadi RK, et al. Melanoma adapts to RAF/MEK inhibitors through FOXD3-mediated upregulation of ERBB3. J Clin Invest. 2013;123:2155–2168. doi: 10.1172/JCI65780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saenger YM, Wolchok JD. The heterogeneity of the kinetics of response to ipilimumab in metastatic melanoma: patient cases. Cancer Immun. 2008;8:1. [PMC free article] [PubMed] [Google Scholar]
- 18.Pennock GK, Waterfield W, Wolchok JD. Patient responses to ipilimumab, a novel immunopotentiator for metastatic melanoma: how different are these from conventional treatment responses? Am J Clin Oncol. 2012;35:606–611. doi: 10.1097/COC.0b013e318209cda9. [DOI] [PubMed] [Google Scholar]
- 19.Wolchok JD, Hoos A, O'Day S, Weber JS, Hamid O, Lebbe C, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. 2009;15:7412–7420. doi: 10.1158/1078-0432.CCR-09-1624. [DOI] [PubMed] [Google Scholar]
- 20.Suda K, Mitsudomi T. Unintentional Weakness of Cancers: The MEK-ERK Pathway as a Double-Edged Sword. Mol Cancer Res. 2013;11:1125–1128. doi: 10.1158/1541-7786.MCR-13-0228. [DOI] [PubMed] [Google Scholar]
- 21.Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu R, Aplin AE. alphaB-crystallin is mutant B-RAF regulated and contributes to cyclin D1 turnover in melanocytic cells. Pigment Cell Melanoma Res. 2010;23:201–209. doi: 10.1111/j.1755-148X.2010.00668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woods D, Parry D, Cherwinski H, Bosch E, Lees E, McMahon M. Raf-induced proliferation or cell cycle arrest is determined by the level of Raf activity with arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5598–5611. doi: 10.1128/mcb.17.9.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen D, Heath V, O'Garra A, Johnston J, McMahon M. Sustained activation of the raf-MEK-ERK pathway elicits cytokine unresponsiveness in T cells. J Immunol. 1999;163:5796–5805. [PubMed] [Google Scholar]
- 26.Petti C, Molla A, Vegetti C, Ferrone S, Anichini A, Sensi M. Coexpression of NRASQ61R and BRAFV600E in human melanoma cells activates senescence and increases susceptibility to cell-mediated cytotoxicity. Cancer Res. 2006;66:6503–6511. doi: 10.1158/0008-5472.CAN-05-4671. [DOI] [PubMed] [Google Scholar]
- 27.Ozaki K, Kadomoto R, Asato K, Tanimura S, Itoh N, Kohno M. ERK pathway positively regulates the expression of Sprouty genes. Biochem Biophys Res Commun. 2001;285:1084–1088. doi: 10.1006/bbrc.2001.5295. [DOI] [PubMed] [Google Scholar]
- 28.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seghers AC, Wilgenhof S, Lebbe C, Neyns B. Successful rechallenge in two patients with BRAF-V600-mutant melanoma who experienced previous progression during treatment with a selective BRAF inhibitor. Melanoma Res. 2012;22:466–472. doi: 10.1097/CMR.0b013e3283541541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.