Abstract
Small cell lung carcinoma (SCLC) is a highly metastatic tumor type with neuroendocrine features and a dismal prognosis. PTEN mutations and PIK3CA activating mutations have been reported in SCLC but the functional relevance of this pathway is unknown. The PTEN/PIK3CA pathway was interrogated using an AdenoCre-driven mouse model of SCLC harboring inactivated Rb and p53. Inactivation of one allele of PTEN in Rb/p53-deleted mice led to accelerated SCLC with frequent metastasis to the liver. In contrast to the high mutation burden reported in human SCLC, exome analyses revealed a low number of protein-altering mutations in mouse SCLC. Inactivation of both alleles of PTEN in the Rb/p53-deleted system led to non-metastatic adenocarcinoma with neuroendocrine differentiation. This study reveals a critical role for the PTEN/PI3-kinase pathway in both SCLC and lung adenocarcinoma and provides an ideal system to test PI3-kinase pathway inhibitors as targeted therapy for subsets of SCLC patients.
Keywords: SCLC, small cell lung cancer, PTEN, mouse model, PIK3CA
Introduction
Small cell lung carcinoma (SCLC) is a highly metastatic neuroendocrine tumor that results in the deaths of >20,000 people per year in the USA alone. It has been known that the p53 and RB tumor suppressor genes are mutated in the majority of SCLCs, and that MYC family members are frequently amplified (1, 2). Alterations in the PTEN pathway have also been reported in SCLC, through direct PTEN mutation/deletion (3, 4) or through PIK3CA activation (5). PIK3CA and/or PTEN mutations were more recently found in two recent next generation sequencing studies of SCLC (6, 7). The huge number of somatic mutations in human SCLC (6–8) necessitates the functional evaluation of key SCLC-mutated genes. As inhibition of PI3-kinase or of the downstream effectors AKT and mTOR can be achieved using targeted therapies, the importance of the PTEN pathway in SCLC is particularly critical to elucidate. Murine models for SCLC have been generated that accurately recapitulate the cardinal features of human SCLC, including recapitulating key secondary alterations (9–12). In this study, we use a mouse model to interrogate PTEN as a potential SCLC driver.
Materials and Methods
Mice
Rblox mice were obtained from Tyler Jacks (MIT). p53lox mice were generated by Anton Berns (Netherlands Cancer Institute) and obtained from the Mouse Models of Human Cancer Consortium (MMHCC). Ptenlox mice were generated by Hong Wu (UCLA) and obtained from Jackson Laboratories. All mice were maintained on a mixed genetic background. Mouse experiments were approved by the Animal Use and Care Committees at the Carnegie Institution and Fred Hutchinson Cancer Research Center.
AdenoCre SCLC model
After breeding the Ptenlox allele into the Rblox/loxp53lox/lox background, Rblox/lox; p53lox/lox; Ptenlox/+ mice were intercrossed to obtain littermate controls that differed in Pten status. Mice were infected with 1*108 pfu AdenoCre driven by the cytomegalovirus (CMV) promoter (University of Iowa Gene Transfer Core) in 75uL using intratracheal intubation as described (13). Mice were aged until moribund and the lungs were fixed in 4% paraformaldehyde (PFA) or Bouin’s solution for histological analyses. The following antibodies were used for immunohistochemistry: calcitonin gene-related peptide (CGRP) (1/2000, Sigma), synaptophysin (1/33, Invitrogen), CK19 (1/250, Abcam ab52625). CGRP immunostaining was performed on Bouin’s fixed tissue. Antigen retrieval was performed using boiling sodium citrate (pH 6.0) and samples were incubated overnight with primary antibody. We used the Vectastain ABC kit (Vector labs) for biotin-mediated signal amplification and horseradish peroxidase based detection was with 3, 3′-diaminobenzidine (DAB) (Vector labs).
Real time PCR
Total RNA from lung tumors was extracted using TRIZOL reagent (Life Technologies). cDNAs were generated using random hexamer priming and Superscript III reverse transcriptase (Life Technologies). Real time PCR was performed with Sybr Select master mix (Life Technologies) in 384-well format using an ABI 7900HT Real Time PCR System. Pten copy number was examined by designing primers to exon 5 of the murine Pten gene and comparing CT values relative to a control gene, Actb across phenol-chloroform isolated genomic DNA from tumors. All primer sequences used for real time PCR are shown in Supplemental Table 1.
Next generation sequencing to identify SCLC mutations
Mouse tumor or tail DNA was isolated following Proteinase K digest and phenol chloroform extraction. The SureSelectXT Mouse All Exon platform (Agilent Technologies) was used for exon capture and library preparation. Samples were sequenced using an Illumina HiSeq2000 with generation of 75bp paired-end reads. The Burrows Wheeler Aligner (BWA) (14) was used to align reads to the mouse mm9 reference genome. SAMtools was used to remove duplicates arising from PCR and was also used to detect variants (mismatches, and small insertions and deletions) (15). Variant positions were identified from matched pairs of tumor samples and normal tissue or cell line controls using SAMtools with computation of genotype likelihood in each sample (options -u, -D, and -S). Variant genotypes were called using bcftools (15) with Bayesian inference, per-sample genotype calling, and contrast calling to determine the likelihood of different allele frequencies between the tumor and control samples (options -c, -g, and -1). Tumor-specific mutations were identified by filtering variants with probability of allele frequency difference between matched normal and tumor samples of p < 1e-10 (i.e., PC2 > 100). Annovar was used to annotate variants (16). Protein altering mutations identified in exome analyses included non-synonymous exonic mutations as well as intronic mutations in essential splice sites that were present in the tumor but not matched normal. Exome data coverage is shown in Supplemental Table 2.
Western blotting
Cell lysates were prepared in ice-cold RIPA buffer (50mM Tris·HCl, pH 8.0, 150mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate) supplemented with protease inhibitor tablets (Roche). The following primary antibodies were used: anti-PTEN (#9559, Cell Signaling), anti-phospho Akt (Ser-473) (#4060, Cell Signaling), anti-Akt (#4691, Cell Signaling) and anti-actin (sc-1615, Santa Cruz).
Results
Despite previous reports of PTEN deletions (3, 4, 17) and PIK3CA activating mutations (5) in SCLC, the overall importance of the PTEN/PIK3CA pathway for this cancer remains unclear. Tumor dependence on mutations in the PTEN/PI3-kinase pathway may provide an avenue for SCLC treatment through therapies that target this pathway. Thus, we explored the functional importance of this pathway for SCLC. To assess the importance of the PTEN/PI3-kinase pathway for SCLC we used the Berns SCLC mouse model (9). This model uses Adenoviral Cre (AdCre) to drive Rb and p53 deletion. Resulting lung tumors arise with long latency and mimic critical features of human SCLC including neuroendocrine characteristics and metastatic spread (9). We infected Rblox/lox;p53lox/lox;Pten+/+, Rblox/lox;p53lox/lox;Ptenlox/+ and Rblox/lox;p53lox/lox;Ptenlox/lox cohorts with AdCre delivered using intratracheal intubation. Cohorts were aged and followed until the mice were moribund.
Hemizygous inactivation of Pten accelerates murine SCLC
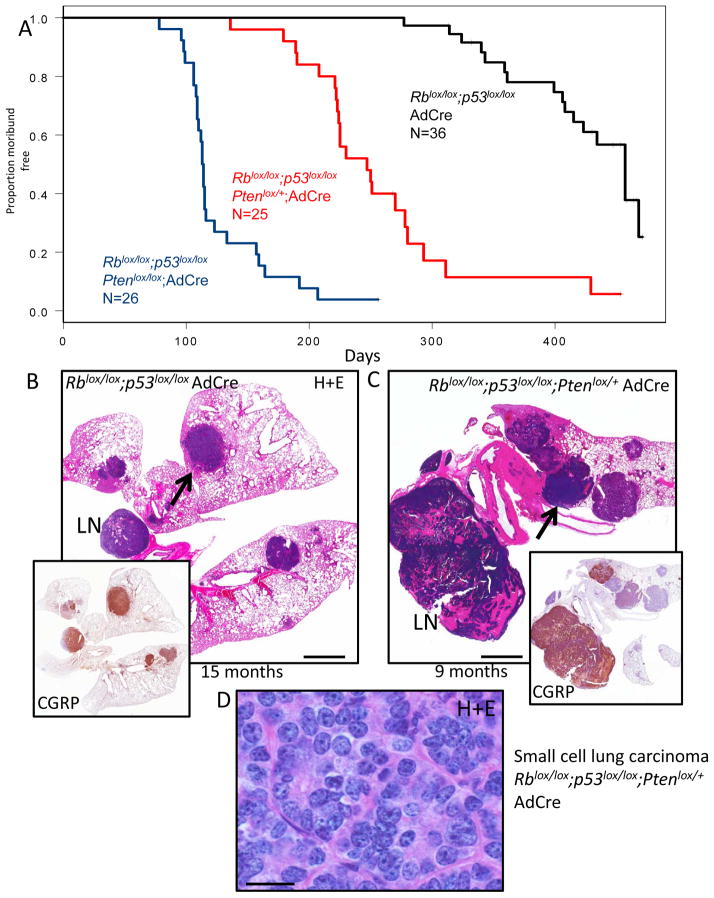
Inactivation of Rb/p53 led to morbidity from lung tumors arising with long latency. Mice in the Rb/p53 cohort became moribund with lung tumor burden at an average+/−S.D. of 387+/−57 days (Figure 1A). Tumors in the model were overall histologically similar to the SCLC tumors previously described (9). Most tumors exhibited neuroendocrine features, staining positively for neuroendocrine markers calcitonin gene related peptide (CGRP) (Figure 1B, inset) and synaptophysin (Supplemental Figure 1), although variability in staining was observed. The SCLCs were aggressive with invasion into vessels and local lymph nodes (Figure 1B, Supplemental Figure 1). We noted a minor component of acinar adenocarcinoma with neuroendocrine differentiation in some Rb/p53 deficient tumors (Supplemental Figure 1). Inactivation of one allele of Pten in the Rblox/lox;p53lox/lox background significantly accelerated tumorigenesis. Here, mice became moribund at an average +/− S.D. of 242+/−59 days (Figure 1A) and lung tumors exhibited histological features similar to the Rblox/lox;p53lox/lox model (Figure 1C, Supplemental Figure 2). Heterogeneity in CGRP and synaptophysin staining was seen within and between tumors in the Rblox/lox;p53lox/lox; Ptenlox/+ group. As in the Rblox/lox;p53lox/lox model, the major tumor component of the Rblox/lox;p53lox/lox;Ptenlox/+ mice was SCLC, with a minor component of adenocarcinoma. PCR analysis showed that both floxed alleles of Rb and p53 were recombined in 6/6 Rblox/lox;p53lox/lox;Ptenlox/+ SCLCs examined (Supplemental Figure 3A,B), and real time PCR analysis of Pten copy number was consistent with loss of Pten heterozygosity in each case (Supplemental Figure 3C). Necropsy analysis revealed gross liver metastasis in 16 of 25 of mice examined (64%) and histological analyses of liver metastases showed exclusively SCLC. The strong acceleration of SCLC in a Pten heterozygous background reveals that Pten is a critical cooperating tumor suppressor gene in SCLC.
Figure 1. Inactivation of Pten accelerates Rb/p53 mutant lung tumors.
A) Kaplan-Meier curves showing time to morbidity following AdCre delivery to Rblox/lox/p53lox/lox, Rblox/lox;p53lox/lox;Ptenlox/+ and Rblox/lox;p53lox/lox;Ptenlox/lox mice B) Hematoxylin and eosin (H+E) stain of murine SCLC (arrow) from AdCre Rblox/lox;p53lox/lox mouse at 15 months post AdCre with inset showing immunostaining for the neuroendocrine marker CGRP. Lymph node metastasis is indicated (LN). C) H+E of advanced SCLC in lymph node (LN) and lung (arrow) with CGRP immunostaining (inset) of Rblox/lox;p53lox/lox;Ptenlox/+ lung at 9 months post AdCre. Heterogeneity in CGRP staining across tumor nodules is apparent. D) High magnification (100X) image of H+E stained SCLC from Rblox/lox;p53lox/lox;Ptenlox/+ model. Scale bar for A: 2mm, D: 13 microns.
Homozygous inactivation of Pten
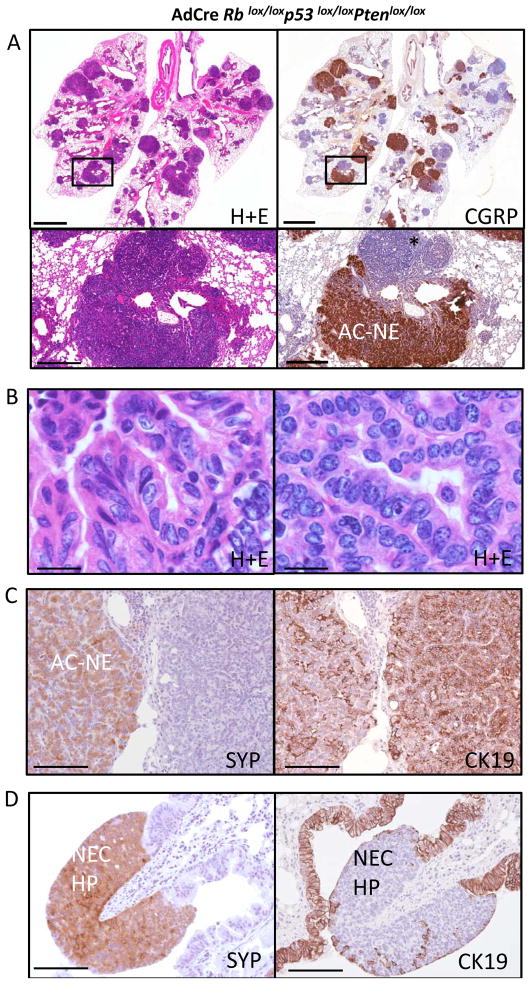
Adenoviral Cre delivered to Rblox/lox;p53lox/lox;Ptenlox/lox animals resulted in a distinct phenotype. Here, lung tumors arose extremely rapidly (average+/−S.D. of 123+/−30 days) (Figure 1A) with each lobe of the lung filled with tumors at the time of morbidity (Figure 2A). The major component of the Rblox/lox;p53lox/lox;Ptenlox/lox tumors was acinar and mixed adenocarcinoma with neuroendocrine differentiation revealed by CGRP and synaptophysin immunohistochemistry (Figure 2A,B,C). The tumors had acinar and papillary patterns of growth (Figure 2B). We also observed dysplastic and hyperplastic neuroendocrine lesions in the airways (Figure 2D), likely precursor lesions to SCLC. The adenocarcinomas, including those with neuroendocrine features, stained positively for cytokeratin 19 (CK19) (Figure 2C), while the hyperplastic neuroendocrine lesions along the airways were negative for this marker (Figure 2D). We note that SCLC tumors that arose in the Rblox/lox;p53lox/lox;Ptenlox/+ model did not stain positively for CK19 (Supplemental Figure 2). Also, while Krt7 and Krt18 mRNA expression was not significantly different between lung tumors in the Rblox/lox;p53lox/lox;Ptenlox/lox vs. Rblox/lox;p53lox/lox;Ptenlox/+ models, Krt19 levels were significantly increased in the Rblox/lox;p53lox/lox;Ptenlox/lox adenocarcinoma model (Supplemental Figure 4). Thus, despite common expression of neuroendocrine markers, the Rblox/lox;p53lox/lox;Ptenlox/+ SCLC and Rblox/lox;p53lox/lox;Ptenlox/lox adenocarcinoma models could be distinguished by histological features and by CK19/Krt19 positivity. In contrast to the Rb/p53 and Rb/p53/Pten heterozygous models, liver metastasis was not found in the Pten homozygote AdCre model. We did not perform long-term aging studies on Ptenlox/lox mice in the context of wild-type Rb and p53. However, sacrifice of 5 non-moribund Ptenlox/lox animals (wild-type for Rb and p53) at 5-months post AdCre did not reveal evidence of lung neoplasia (data not shown). Overall, these data indicate that homozygous Pten inactivation synergizes with Rb and p53 loss to promote lung adenocarcinomas with neuroendocrine differentiation. The rapid lethality from many independent adenocarcinomas likely impaired development of advanced SCLC with liver metastasis in the Pten homozygous model.
Figure 2. HomozygousPten inactivation in Rb/p53 mutant lung.
A) Hematoxylin and eosin (H+E) stain showing tumor filled lung from Rblox/lox;p53lox/lox;Ptenlox/lox mouse 3 months 20 days post AdCre (left panels). CGRP immunostaining of adjacent section showing neuroendocrine character of many tumor nodules (right panels). Boxed region shows magnified view of adenocarcinoma with neuroendocrine differentiation (Ac-NE) along with adenocarcinoma negative for CGRP (asterix). B) High-magnification (100x) view of adenocarcinoma histology C) Synaptophysin (SYP) and CK19 immunostaining of adenocarcinoma. Adjacent lesions both exhibit CK19 positivity but only tumor area to left is synaptophysin positive D) Synaptophysin (SYP) positivity and absence of CK19 immunostaining in hyperplastic neuroendocrine cells along the airway (NEC-HP). Scale bars for A (upper): 2mm; A (lower): 400 microns; B: 13 microns; C, D: 80 microns.
Molecular analyses of lung tumors
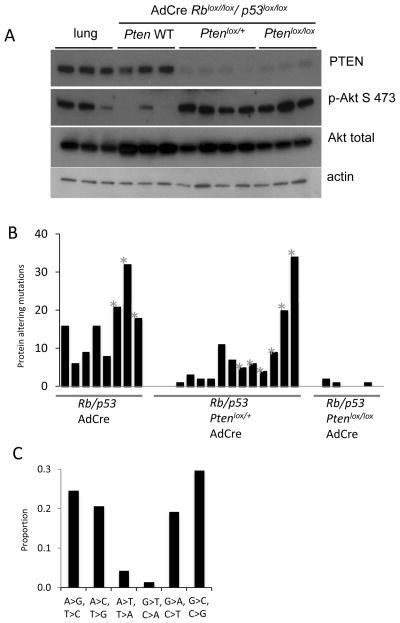
Western blot analysis of SCLCs from the Pten heterozygous model revealed complete loss of PTEN protein in 4 of 4 tumors examined (Figure 3A). This is consistent with inactivation of the remaining wild-type Pten allele (Supplemental Figure 3C). We were unable to control for the normal level of phospho-AKT in pulmonary neuroendocrine cells in these western blots, as such cells are extremely rare in the lung. However, compared to Pten wild-type mouse SCLC, Pten hemizygous and homozygous lung tumors showed increased phosphorylation of AKT at Ser 473, indicative of pathway activation (Figure 3A).
Figure 3. Analyses of murine lung neuroendocrine tumors.
A) Western blot analyses of normal lung and SCLCs from the indicated genotypes showing PTEN, Phospho Akt S473, pan-AKT and actin loading control. B) Number of protein-altering mutations in murine lung neuroendocrine tumors of the indicated genotypes. Metastatic samples are indicated (*). Rblox/lox;p53lox/lox : 8 tumors from 3 animals, Rblox/lox;p53lox/lox;Ptenlox/+ : 13 tumors from 6 animals, Rblox/lox;p53lox/lox;Ptenlox/lox : 6 tumors from 3 animals. C) Patterns of transitions and transversions in primary murine SCLC.
Secondary alterations in lung tumors
Human SCLC is a smoking associated cancer with high mutational load (6–8). In one study, an average of 175 protein-alterating mutations per SCLC tumor was reported (7). To compare the somatic mutational load in murine SCLC to human SCLC we performed whole-exome studies. In contrast to human SCLCs, the murine SCLC exome showed few protein-altering somatic mutations. We found an average of 15.8 protein altering mutations per murine SCLC in the Rblox/lox;p53lox/lox model (Figure 3B). Pten heterozygote tumors exhibited a variable and intermediate number of mutations (average 8 protein-altering mutations). There were no recurrent mutations nor mutations in known cancer genes in this small sample set. We also characterized exonic mutations in the Rblox/lox;p53lox/lox;Ptenlox/lox lung adenocarcinomas – here we found a near absence of selection for protein-altering mutations (average 0.7 mutations/tumor exome). In our murine exome analyses, 92% of the mouse tumor exome was sequenced to 10X coverage while 82% was sequenced to 20X coverage (Supplemental Table 2). In contrast to human smoking-associated SCLC (6–8), C:G>A:T transversions in murine SCLC were infrequent (Figure 3C). Thus, murine SCLC does not exhibit high numbers of point mutations typical of human smoking associated SCLC.
Discussion
PTEN/PIK3CA mutations have been described in SCLC, however, the overall importance of this pathway for SCLC is not clear. Thus, we tested the importance of this pathway using mouse genetics. We inactivated Pten in an Rb/p53 deleted mouse model of SCLC that recapitulates human SCLC in metastatic pattern and in neuroendocrine features (9). When even a single allele of Pten was inactivated, SCLC occurred with much faster kinetics. Moreover, the tumors in the Pten heterozygous model metastasized to the liver. These data definitively show that Pten is a critical tumor suppressor in a genetic mouse model of SCLC. As there are no targeted therapies for SCLC, these data may provide incentive to treat human SCLC patients with PI3-kinase or Akt inhibitors. Murine SCLC models will be ideal for assessing the therapeutic potential of this approach.
Inactivation of both Pten alleles in the Rb/p53 floxed background led to a shift in tumor spectrum. Multifocal adenocarcinomas, with the major component exhibiting neuroendocrine differentiation, led to rapid lethality. Interestingly, a subset of human adenocarcinomas that acquired resistance to targeted therapy acquired neuroendocrine characteristics and transformed into SCLC (18). In one patient, transformation to SCLC was associated with a newly acquired activating PI3-kinase mutation present in the SCLC but not in the original adenocarcinoma (18). It will be interesting to investigate whether RB, p53 and/or PTEN pathway alteration can be linked to the acquisition of neuroendocrine properties in a non-neuroendocrine cell of origin. Application of neuroendocrine promoter-driven adenoviral vectors specifically to the lung (19) will enable late-stage SCLC modeling with homozygous Pten inactivation.
We found that murine SCLCs exhibited a lower number of protein altering mutations than human SCLCs. In the AdCre Rblox/lox;p53lox/lox model, we found 15.8 protein altering mutations per tumor, a number much reduced in comparison to human SCLC. This difference is at least in part due to the fact that human SCLC typically arises in heavy smokers, leading to a high smoking-induced mutational burden. Hemizygosity for Pten in this model led to reduced somatic point mutations. The Pten hemizygous model will be particularly useful for studies of metastasis, as metastatic SCLC arises frequently and rapidly. Matched comparisons between primary and metastatic murine SCLCs may shed light on the genetic determinants of metastasis. The lower number of mutations in mouse SCLC models may facilitate study of individual SCLC-mutated genes. The high mutational burden in human SCLC is likely to lead to increased noise in similar analyses of human SCLC. Assessment of vulnerabilities to therapies associated with a specific mutation will be particularly informative using murine SCLC models given the reduced mutational complexity.
We demonstrated the critical role for the PTEN/PI3-kinase pathway in SCLC. This finding has important implications for using targeted therapies directed towards this pathway to treat the most aggressive form of lung cancer.
Supplementary Material
Implications.
The ability of PTEN inactivation to accelerate SCLC in a genetic mouse model suggests that targeting the PTEN pathway is a therapeutic option for a subset of human SCLC patients.
Acknowledgments
We are grateful to Alison Pinder for performing next generation sequencing and to Sue Knoblaugh for expert histopathology advice. We thank Tyler Jacks, Hong Wu and Anton Berns for providing valuable mouse strains used in these studies.
Grant support
This work was supported by R01CA148867 to DM and CA096823 to AYN.
Footnotes
The authors declare that no conflicts of interest exist.
References
- 1.Johnson BE, Ihde DC, Makuch RW, Gazdar AF, Carney DN, Oie H, et al. myc family oncogene amplification in tumor cell lines established from small cell lung cancer patients and its relationship to clinical status and course. J Clin Invest. 1987;79:1629–34. doi: 10.1172/JCI112999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366:1385–96. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- 3.Forgacs E, Biesterveld EJ, Sekido Y, Fong K, Muneer S, Wistuba, et al. Mutation analysis of the PTEN/MMAC1 gene in lung cancer. Oncogene. 1998;17:1557–65. doi: 10.1038/sj.onc.1202070. [DOI] [PubMed] [Google Scholar]
- 4.Yokomizo A, Tindall DJ, Drabkin H, Gemmill R, Franklin W, Yang P, et al. PTEN/MMAC1 mutations identified in small cell, but not in non-small cell lung cancers. Oncogene. 1998;17:475–9. doi: 10.1038/sj.onc.1201956. [DOI] [PubMed] [Google Scholar]
- 5.Shibata T, Kokubu A, Tsuta K, Hirohashi S. Oncogenic mutation of PIK3CA in small cell lung carcinoma: a potential therapeutic target pathway for chemotherapy-resistant lung cancer. Cancer letters. 2009;283:203–11. doi: 10.1016/j.canlet.2009.03.038. [DOI] [PubMed] [Google Scholar]
- 6.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nature genetics. 2012 doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nature genetics. 2012 doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–90. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–9. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 10.Dooley AL, Winslow MM, Chiang DY, Banerji S, Stransky N, Dayton TL, et al. Nuclear factor I/B is an oncogene in small cell lung cancer. Genes & development. 2011;25:1470–5. doi: 10.1101/gad.2046711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park KS, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nature medicine. 2011;17:1504–8. doi: 10.1038/nm.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer research. 2010;70:3877–83. doi: 10.1158/0008-5472.CAN-09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–72. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SK, Su LK, Oh Y, Kemp BL, Hong WK, Mao L. Alterations of PTEN/MMAC1, a candidate tumor suppressor gene, and its homologue, PTH2, in small cell lung cancer cell lines. Oncogene. 1998;16:89–93. doi: 10.1038/sj.onc.1201512. [DOI] [PubMed] [Google Scholar]
- 18.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–64. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.