Abstract
The gas-phase oxidation of methionine residues is demonstrated here using ion/ion reactions with periodate anions. Periodate anions are observed to attach to varying degrees to all polypeptide ions irrespective of amino acid composition. Direct proton transfer yielding a charge reduced peptide ion is also observed. In the case of methionine and, to a much lesser degree, tryptophan containing peptide ions, collisional activation of the complex ion generated by periodate attachment yields an oxidized peptide product (i.e., [M+H+O]+), in addition to periodic acid detachment. Detachment of periodic acid takes place exclusively for peptides that do not contain either a methionine or tryptophan side-chain. In the case of methionine containing peptides, the [M+H+O]+ product is observed at a much greater abundance than the proton transfer product (viz., [M+H]+). Collisional activation of oxidized Met-containing peptides yields a signature loss of 64 Da from the precursor and/or product ions. This unique loss corresponds to the ejection of methanesulfenic acid from the oxidized methionine side chain and is commonly used in solution-phase proteomics studies to determine the presence of oxidized methionine residues. The present work shows that periodate anions can be used to ‘label’ methionine residues in polypeptides in the gas-phase. The selectivity of the periodate anion for the methionine side chain suggests several applications including identification and location of methionine residues in sequencing applications.
Keywords: Ion/ion reaction, methionine oxidation, tandem mass spectrometry
INTRODUCTION
The oxidation of methionine residues in polypeptides/proteins has important roles in aging and age-related degenerative diseases.[1] Methionine oxidation is a common chemical degradation pathway and results in the modification of the methionine side chain, where the thioether group (i.e., –CH2SCH3) is converted to a sulfoxide derivative (i.e., CH2SOCH3).[2,3] The sulfoxide derivative can be further oxidized to a sulfone (i.e., CH2SO2CH3); however, the sulfone is rarely observed in biological systems as it requires a significantly stronger oxidizing agent.[3] The single oxidation reaction has far-reaching implications in biological applications. Oxidation of free methionine to the sulfoxide derivative inhibits its methyl donating capabilities.[4,5] Methionine oxidation in calmodulin, a calcium-binding messenger protein, results in the decreased function of calcium signaling, ultimately leading to loss of calcium homeostasis in aged brains.[6,7] Additionally, oxidation of Met-35 in β-amyloid peptide contributes to the insolubility and overall stability of the peptide, specifically the C-terminal region, which has implications in Alzheimer's disease.[8,9,10] There is also evidence to suggest that methionine oxidation is important for cellular regulation. The methionine sulfoxide [Met(O)] functional group can be reduced under physiological conditions by methionine sulfoxide reductase, which has a role in the activation-deactivation cycle of various signaling proteins as well as the maintenance of homeostatic balance in physiological systems.[11,12]
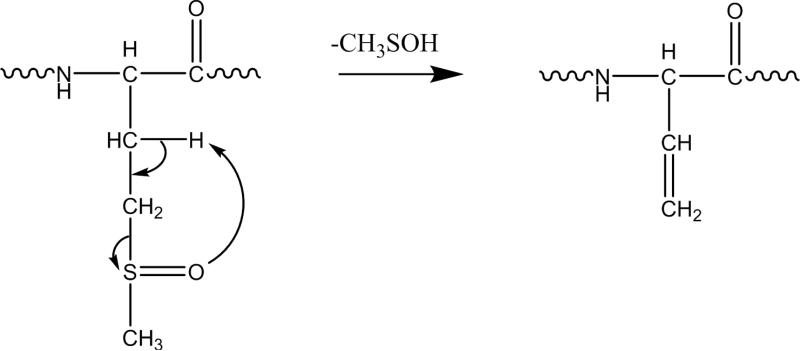
Mass spectrometry (MS) has been used extensively to characterize Met(O) in proteins and peptides. Tandem MS (MS/MS) is useful in identifying the presence of this modification and locating the site of modification via gas-phase fragmentation.[13] Ion trap collision-induced dissociation (CID) of peptide or protein ions containing Met(O) produces a neutral loss of 64 Da, corresponding to loss of methanesulfenic acid (CH3SOH), from the precursor and/or product ions containing the oxidized methionine residue.[13,14,15,16,17] The rearrangement mechanism for the neutral loss of 64 Da is shown in Scheme 1 below.[18,19,20,21] This neutral loss is unique to Met(O) and is useful in differentiation between Met(O) and phenylalanine, which have the same nominal mass, during MS-peptide sequencing. The loss of methanesulfenic acid has been shown to be a low-energy CID pathway; therefore, dominant loss of 64 Da is readily observed allowing straightforward identification of the presence of oxidized methionine in the precursor ion. Electron capture dissociation (ECD) has also been used to characterize peptides containing oxidized methionine and provides complementary information to CID.[22] ECD of Met(O)-containing peptides retains the oxidation modification and yields sequence-informative c and z• ions.
Scheme 1.
Mechanism of rearrangement of the oxidized methionine side chain to produce loss of methanesulfenic acid (64 Da).[18-21]
Recently, it has been shown that chemical reactions commonly performed in solution to derivatize peptides and proteins can also be conducted in the gas-phase via reactions of oppositely charged ions.[23] The gas-phase reactions can be driven at rates ranging from 1-100 s-1, with typical reaction time-scales on the order of 100 ms for reaction efficiencies of tens of percent. In addition to short reaction times relative to analogous condensed-phase derivatization approaches, the gas-phase approach is amenable to mass-selection of reactants and obviates addition of reagents to the sample solution and, as a result, avoids concomitant complication of the mass spectrum of the analyte.[23] The extent of modification (i.e., the number of modifications per analyte ion) can also be varied via control of reagent ion number and reaction time, as well as via use of the ion parking technique.[24] Gas-phase ion/ion reactions can occur either through long-range transfer of small charged particles (e.g., protons or electrons) or through the formation of a relatively long-lived complex.[41] Covalent modifications of biomolecules via ion/ion reactions proceed through the formation of a long-lived complex. Recent demonstrations of covalent modification in the gas-phase via ion/ion reactions include the use of NHS ester derivatives in cross-linking [25,26] and covalent labeling [27,28,29] studies, the use of 4-formyl-1,3-benzenedisulfonic acid (FBDSA) to tag peptide ions [30,31] and increase sequence coverage obtained from CID experiments, [32,33] and the use of N-cyclohexyl-N’-(2-morpholinoethyl)carbodiimide (CMC) in covalent labeling experiments. [34] Gas-phase ion/ion reactions have been useful in the characterization of proteins [35] and oligonucleotides,[36,37] for manipulating charge states, or for creating different precursor ion types to study via gas-phase fragmentation.
Here, we describe the selective oxidation of methionine residues present in peptide cations using periodate monoanions. Upon ion/ion reaction, a long-lived complex is generated corresponding to the attachment of periodate to a multiply protonated peptide. Activation of the long-lived complex yields a covalently modified peptide ion, indicated by the addition of an oxygen atom to the methionine residue in the protonated peptide, represented as [M+H+O]+. Formation of the oxidized species is the dominant pathway for methionine-containing peptides following activation of the long-lived complex. This pathway is also observed for tryptophan-containing peptides, but at a significantly lesser abundance than Met-containing peptides. Peptides lacking tryptophan and methionine residues do not form the oxidized species. Further activation of oxidized Met-containing peptides yields a signature loss of 64 Da from the precursor and product ions, while this 64 Da loss is not observed in peptides lacking methionine residues. This is the first reported demonstration of selective gas-phase oxidation of peptides via ion/ion reaction.
EXPERIMENTAL SECTION
Materials
Methanol and glacial acetic acid were purchased from Mallinckrodt (Phillipsburg, NJ). KGAILMGAILR was synthesized by CPC Scientific (San Jose, CA). Substance P was synthesized by Bachem (Bubendorf, Switzerland). Anti-inflammatory peptide I was synthesized by AnaSpec (Fremont, CA). GLSDGEWQQVLNVWGK was synthesized by SynPep (Dublin, CA). ARACAKA, ARAWAKA, and GRGMGRGMGRL were synthesized by Pepnome Limited (Shenzhen, China). ARAMAKA was synthesized by NeoBioLab (Cambridge, MA). Angiotensin II and sodium periodate were purchased from Sigma Aldrich (St. Louis, MO). All peptide stock solutions for positive nanoelectrospray were prepared in a 49.5/49.5/1 (v/v/v) solution of methanol/water/acetic acid at an initial concentration of ~1 mg/mL and diluted 100-fold prior to use. The periodate solution was prepared in a 50/50 (v/v) solution of methanol/water at a concentration of ~1 mg/mL and diluted 10-fold prior to use. For all solution-phase oxidations 5 μL of the prepared periodate solution was added to an equivalent volume of peptide solution.
Mass Spectrometry
All experiments were performed on a QTRAP 4000 hybrid triple quadrupole/linear ion trap mass spectrometer (AB Sciex, Concord, ON, Canada), previously modified for ion/ion reactions.[38] Multiply protonated peptides and singly charged anion reagent populations were sequentially injected into the instrument via alternately pulsed nanoelectrospray (nESI).[39] The peptide cations and periodate anions were independently isolated in the Q1-mass filter prior to injection into the q2 reaction cell. The opposite polarity ions were allowed to react for a defined mutual storage reaction time of 1000 ms. The ion/ion reaction products were then transferred to Q3, where the complex was subjected to further characterization via MSn and mass analysis using mass-selective axial ejection (MSAE).[40]
RESULTS AND DISCUSSION
Selective oxidation of methionine residues with periodate
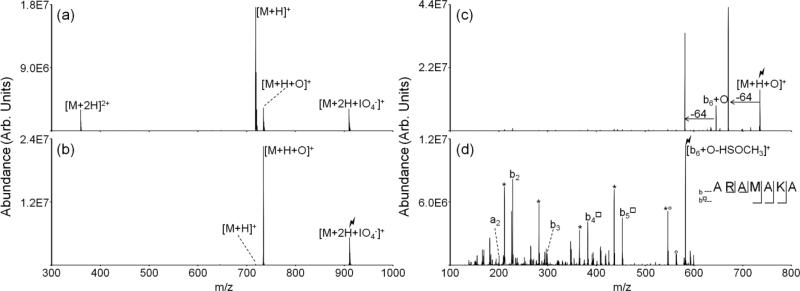
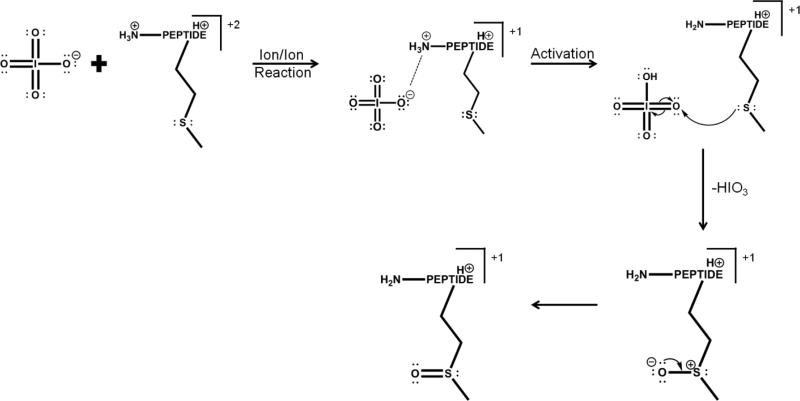
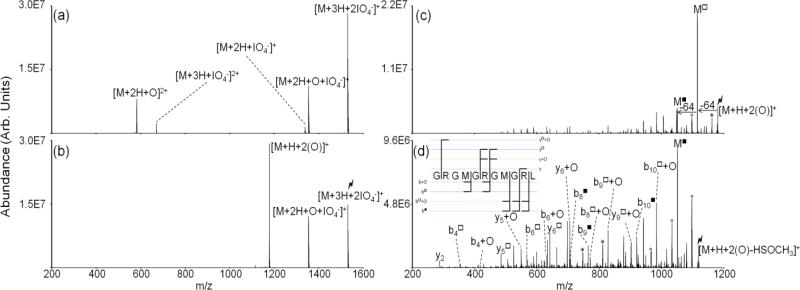
Peptide dications containing methionine residues (i.e., doubly protonated ARAMAKA, KGAILMGAILR, MHRQETVDC, RPKPQQFFGLM, GSNKGAIIGLM) were subjected to ion/ion reactions with periodate monoanions. Figure 1 illustrates the oxidation of doubly protonated ARAMAKA via ion/ion reaction. Upon mutual storage of the peptide cations and periodate anions, direct proton transfer from the peptide cation to the reagent anion or formation of a long-lived complex, [M+2H+IO 4-]+, is observed (Figure 1(a)).[41] The complex decomposes via one of two pathways upon activation. One pathway results in proton transfer from the peptide cation to the periodate anion, which yields loss of neutral periodic acid (i.e., HIO4) and the charge-reduced species, [M+H]+. A second pathway is outlined in Scheme 2 and results in covalent modification of the methionine residue to produce the oxidized species, [M+H+O]+.[42,43] The latter species is also observed in Figure 1(a) and arises from collisional activation of the complex upon transfer from the reaction cell to Q3. The generation of [M+H+O]+ ions from collisional activation of the complex has been observed to be the favored pathway for methionine-containing peptides (see Figure 1(b)). The reaction is presumed to proceed via nucleophilic attack by the sulfur atom on one of the neutral oxygen atoms on the periodate reagent resulting in oxidation of the methionine side-chain and loss of neutral iodic acid (i.e., HIO3). The net result is oxidation of the methionine side chain to yield the sulfoxide form. The extent to which the oxidation takes place in the complex prior to collisional activation versus being driven by collisional heating of the complex is unclear.
Figure 1.
Spectra illustrating gas-phase covalent modification of ARAMAKA, including (a) ion/ion reaction between doubly protonated peptide cation and periodate anion, (b) CID of the isolated ion/ion complex producing the [M+H+O]+ species, (c) MS3 of the oxidized peptide, and (d) MS4 of the [b6+O-64]+ identifying the site of modification as the methionine residue. Degree symbols (°) denote water losses whereas asterisks (*) denote ammonia losses. Squares (□) denote fragments that have lost the modified methionine side chain, e.g., y □8 corresponds to [y8+O-HSOCH3]+. The lightning bolt ( ) is used to denote the ion that has been subjected to CID.
) is used to denote the ion that has been subjected to CID.
Scheme 2.
Proposed mechanism for ion/ion reaction between periodate anion and a doubly cationic methionine-containing peptide to form the oxidized species. Adapted from references 42 and 43.
Collisional activation of the oxidized [M+H+O]+ species produces dominant neutral losses of 64 Da from precursor or product ions (Figure 1(c)). This corresponds to the loss of methanesulfenic acid (HSOCH3) via the rearrangement shown in Scheme 1. For the oxidized [M+H+O]+ species produced via ion/ion reaction between doubly protonated ARAMAKA and periodate anion, the 64 Da losses from the precursor and b6 ions are the most abundant species in the CID spectrum. The b6+O ion corresponds to a lysine cleavage that is the dominant cleavage site upon activation of the [M+H]+ species (i.e., the b6 ion dominates the CID spectrum of the singly protonated peptide). The unique 64 Da loss can be used to localize the site of oxidation. Figure 1(d) demonstrates the localization of the oxidation to the methionine residue in the peptide ARAMAKA via activation of the 64 Da loss from the b6+O ion, i.e., [b6+O-HSOCH3]+. A series of b-ions, b2-b5, is observed. The presence of the non-modified b2 and b3-ions and modified b4□ and b5□ ions further confirms oxidation of the methionine residue. The open square (□) indicates loss of methanesulfenic acid from an oxidized methionine side chain, e.g., b4□ corresponds to [b4+O-HSOCH3]+.
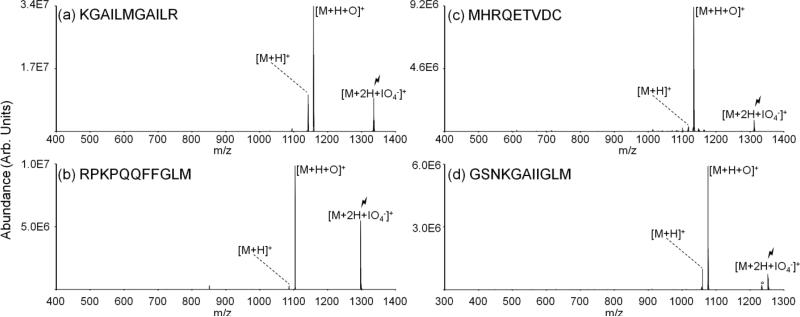
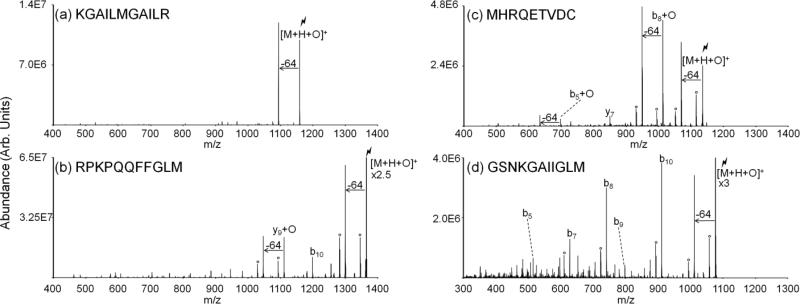
Collisional activation of complexes produced via gas-phase reactions between periodate anions and methionine-containing peptide cations examined to date ([M+2H+IO4-]+) predominantly yields the oxidized species ([M+H+O]+). Figure 2 provides results of the collisional activation of the [M+2H+IO4-]+ species for four different methionine-containing peptides. In each case, the abundance of the oxidized species produced via covalent modification ([M+H+O]+) is greater than the abundance of the charge-reduced species produced via proton transfer from the peptide to the periodate anion (Figure 2(a)-(d)). The peptide KGAILMGAILR has the lowest abundance ratio of [M+H+O]+ to [M+H]+ of the four peptides shown, with the [M+H+O]+ ion at approximately three times the abundance of [M+H]+ ion. Both substance P (RPKPQQFFGLM) and MHRQETVDC show almost exclusive production of the [M+H+O]+ ion. The 11-residue β-amyloid peptide fragment results in a [M+H+O]+ to [M+H]+ ratio intermediate to the other systems discussed. This efficient production of the oxidized [M+H+O]+ species is beneficial as it leaves enough signal for further analysis and modification localization via MSn.
Figure 2.
Spectra illustrating activation of ion/ion complexes produced via reactions between periodate anions and doubly protonated (a) KGAILMGAILR, (b) substance P, (c) MHRQETVDC, and (d) an eleven-residue segment of β-amyloid peptide.
Figure 3 summarizes the fragmentation of the respective oxidized [M+H+O]+ species generated from the peptides KGAILMGAILR, substance P (RPKPQQFFGLM), MHRQETVDC, and an eleven-residue peptide derived from β-amyloid peptide GSNKGAIIGLM. The neutral loss of 64 Da is a prominent pathway in the CID product ion spectra of peptides with oxidized methionine residues. The neutral loss of methanesulfenic acid is unique to methionine residues oxidized to the sulfoxide form and thus can be utilized to determine the presence of oxidized methionine residues in peptides and proteins. Upon ion trap CID of oxidized KGAILMGAILR produced via gas-phase ion/ion reaction (i.e., [KGAILMGAILR+H+O]+), the loss of 64 Da is the sole ion observed (Figure 3(a)). For substance P, the 64 Da loss is also predominant, though other fragment ions are observed (Figure 3(b)). This loss of 64 Da is observed from the substance P oxidized y9 species as well. Cleavage of the peptide bond on the C-terminal side of aspartic acid upon activation is well-known to be facile [44] and competes with the neutral 64 Da loss in the CID product ion spectrum for the peptide MHRQETVDC to form the b8+O ion. Neutral 64 Da losses from the parent ion and oxidized b5 species for MHRQETVDC are also observed (Figure 3(c)). The oxidation of the methionine residue in β-amyloid peptide under physiological conditions has been shown to have implications in aggregation associated with Alzheimer's disease. The eleven-residue peptide GSNKGAIIGLM contains the methionine residue of the β-amyloid peptide and has been shown to have some of the same biological properties as the full-length β-amyloid peptide including the formation of fibrils, free radicals, and neurotoxicity.[8,45,46] The neutral loss of 64 Da gives rise to the second most abundant peak observed in the CID spectrum, which allows facile identification of an oxidized methionine residue somewhere in the peptide (Figure 3(d)). All of the major product ions that originate from backbone cleavages are N-terminal fragments and, as a result, no sequence-related product ions show an additional loss of 64 Da, which is indirect evidence for modification at the C-terminus. For example, non-modified b10 ion indicates cleavage between the leucine and methionine residue thus eliminating all residues except for methionine as the modification site. Similarly, further MSn of oxidized species for KGAILMGAILR and substance P localize the oxidation to methionine as well (Supplemental Figure 1).
Figure 3.
Ion trap CID of the [M+H+O]+ species for reactions between periodate anion and doubly protonated (a) KGAILMGAILR, (b) substance P, (c) MHRQETVDC, and (d) β-amyloid peptide residues 25-35. Degree symbols (°) denote water losses whereas asterisks (*) denote ammonia losses. The lightning bolt ( ) is used to denote the ion that has been subjected to CID.
) is used to denote the ion that has been subjected to CID.
Non-methionine containing peptides
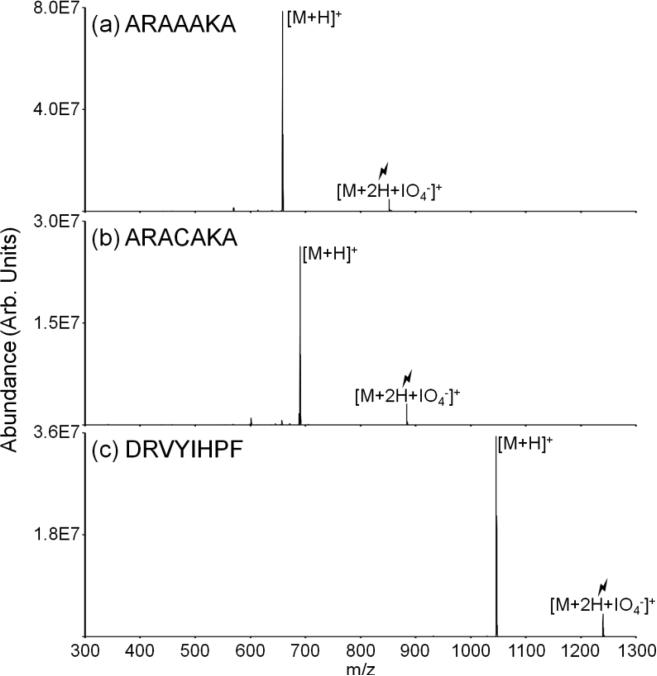
Spectra derived from collisional activation of various ion/ion complexes ([M+2H+IO4-]+) for peptides lacking methionine residues, e.g., ARAAAKA, ARACAKA, and angiotensin II (DRVYIHPF) are shown in Figure 4. For all nonmethionine containing peptides studied, dominant production of the charge-reduced species (i.e., [M+H]+) is observed upon collisional activation of the complex with periodate. The peptides ARAAAKA and ARACAKA differ from the previous peptide shown, ARAMAKA, by the replacement of the methionine residue with either an alanine or cysteine residue. Activation of the [M+2H+IO4-]+ species for both ARAAAKA (Figure 4(a)) and ARACAKA (Figure 4(b)) peptides produces the [M+H]+ ion and yields no evidence for the oxidized product, [M+H+O]+, in contrast with activation of the [M+2H+IO4-]+ species for ARAMAKA (Figure 1(b)). The peptide ARACAKA was examined because cysteine residues are oxidized by periodate in solution. However, the lack of oxidation observed upon activation of the [M+2H+IO4-]+ ion demonstrates the selectivity of the gas-phase ion/ion chemistry for methionine residues. Doubly protonated angiotensin II (DRVYIHPF) peptide cations were also subjected to ion/ion reactions with periodate anions. Angiotensin II contains tyrosine and histidine residues,[50] which are both oxidized in solution by periodate. Similarly to ARAAAKA and ARACAKA, no evidence for formation of [M+H+O]+ is observed upon activation of the [M+2H+IO4-]+ ion and the charge-reduced [M+H]+ is formed essentially exclusively. Cysteine residues are often protected to prevent disulfide formation. These modifications are susceptible to oxidation in solution and, similar to oxidized methionine, yield neutral losses of the oxidized side chain upon gas-phase fragmentation.[47,48,49] Peptides ions containing S-methyl cysteine residues react similarly to methionine-containing peptides. Upon ion/ion reaction the [M+H+O]+ species is formed upon activation of the complex and CH3SOH is lost upon further activation of the oxidized species. Conversely, no oxidation was observed upon activation of the [M+2H+IO4-]+ complex formed via ion/ion reactions between cysteine-containing peptides protected with S-acetylamido (Acm) protecting groups and periodate anions. Other protecting groups with electron-donating properties similar to those of methyl groups may also undergo oxidation upon ion/ion reaction with periodate anions.
Figure 4.
Spectra illustrating activation of ion/ion complexes produced via reactions between periodate anions and doubly protonated (a) ARAAAKA, (b) ARACAKA, and (c) angiotensin II.
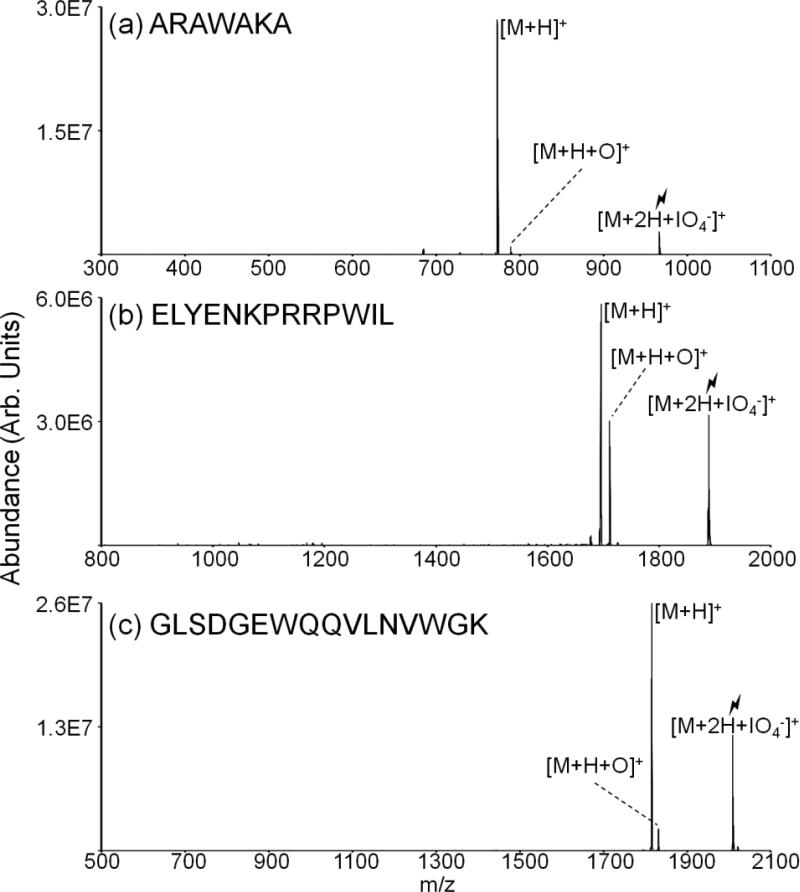
Tryptophan is also oxidized to a small extent via gas-phase ion/ion reactions of tryptophan-containing peptides with periodate anions (Figure 5). However, for all peptides studied, the [M+H]+ ion is observed in greater abundance than the [M+H+O]+ ion upon activation of the [M+2H+IO4-]+ ion. For the tryptophan analog of ARAMAKA, viz., ARAWAKA, the ratio of [M+H]+ to [M+H+O]+ is the reverse of that observed for ARAMAKA (compare Figure 1(b) with Figure 5(a)). While the relative abundance of the [M+H+O]+ ion for Trp-11 neurotensin (Figure 5(b)) is greater than that observed for reactions with ARAWAKA, the extent of oxidation remains much less for tryptophan-containing peptides than the methionine-containing peptides. Supplemental Figure 1(d) demonstrates the localization of the oxidation to the tryptophan residue for ARAWAKA via activation of the [M+H+O]+ species. The peptide GLSDGEWQQVLNVWGK is a tryptic peptide from myoglobin and, although it contains two tryptophan residues, the [M+H+O]+ species is produced in extremely low abundance. When a peptide cation containing both methionine and tryptophan residues is subjected to ion/ion reaction with periodate anions, oxidation of methionine is the dominant process observed, with no peaks indicating oxidation of the tryptophan residue (Supplemental Figure 2). Activation of the [M+H+O]+ species for tryptophan-containing peptides does not contain the 64 Da losses predominant for methionine-containing peptides making them easily distinguishable (Supplemental Figure 1(d)).
Figure 5.
Spectra illustrating activation of ion/ion complexes produced via reactions between periodate anions and doubly protonated (a) ARAWAKA, (b) Trp-11 Neurotensin, and (c) GLSDGEWQQVLNVWGK.
Solution-phase versus gas-phase oxidation
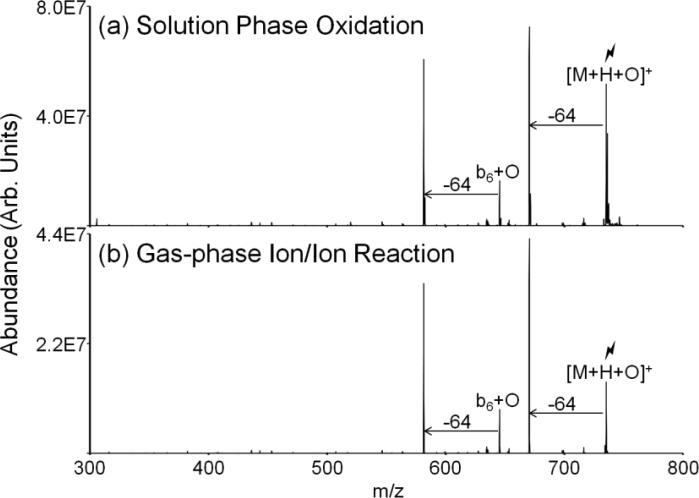
The gas-phase dissociation behavior of solution and gas-phase oxidized peptides is compared in Figure 6. Peptides were oxidized in solution via addition of an aqueous solution of sodium periodate. The product ion spectra for solution and gas-phase oxidations were identical, as shown for the peptide ARAMAKA in Figure 6. The product ion spectra show dominant 64 Da losses from both the oxidized peptide and the b6 ion. This experiment indicates the gas-phase chemistry mimics that of the solution-phase. While periodate oxidizes methionine residues faster than other amino acids, it does also modify other amino acids in the peptide, specifically cysteine, histidine, tyrosine, and tryptophan.[50] In the gas-phase, the oxidation appears to be more selective for methionine under the ion/ion reaction conditions used here, based on the lack of reactivity noted for other side-chains except for the minimal reactivity observed for tryptophan.
Figure 6.
Comparison of solution-phase and gas-phase oxidation of the peptide ARAMAKA. (a) CID of the [M+H+O]+ species produced via solution-phase reaction with sodium periodate, and (b) MS3 of the [M+H+O]+ from dissociation of the [M+2H+IO4-]+ species produced by the gas-phase ion/ion reaction of doubly protonated ARAMAKA with periodate anion.
Modification of multiple methionine residues via addition of multiple periodate anions
Multiple modifications to peptide cations containing more than one methionine residue have been observed via sequential ion/ion reactions between the peptide cation and periodate anions. This approach can be useful in determining the number of methionine residues in a peptide. For example, triply protonated GRGMGRGMGRL was subjected to ion/ion reactions under conditions in which sequential reactions are likely (see Figure 7). Peaks indicating the addition of one periodate anion to form a complex of the form [M+3H+IO4-]2+ and two periodate anions to form a complex of the form [M+3H+2IO4-]+ are observed (Figure 7(a)). Proton transfer also occurs as evidenced by the presence of the [M+2H+IO4-]+ species. Some fragmentation of the complexes due to energetic transfer conditions from the collision quadrupole to Q3 is also observed, as illustrated by the peaks corresponding to [M+2H+O]2+ and [M+2H+O+IO4-]+. The most abundant species observed is the addition of two periodate anions to form the [M+3H+2IO4-]+ species illustrating the high efficiency with which the reaction proceeds.
Figure 7.
a) Ion/ion reaction between doubly protonated GRGMGRGMGRL and periodate anions. Product ion spectra derived from b) ion trap CID of [M+3H+2IO4-]+, c) further activation of [M+H+2(O)]+, and d) activation of the 64 Da loss from the doubly oxidized species,[M+H+2(O)-HSOCH3]+. Degree symbols (°) denote water losses whereas asterisks (*) denote ammonia losses. Open squares (□) denote fragments that have lost one modified methionine side chain while closed squares (■) denote fragments that have lost two modified methionine side chains, e.g., y8□ corresponds to [y8 +O-HSOCH ++O-HSOCH3]+ y8■ corresponds to [y8+O-2HSOCH3]+. The lightning bolt ( ) is used to denote the ion that has been subjected to CID.
) is used to denote the ion that has been subjected to CID.
The breakup of the [M+3H+2IO4-]+ complex in principle can proceed via either of the pathways previously described for the doubly protonated methionine-containing peptides with one periodate anion: viz., proton transfer to yield the charge-reduced species with concomitant neutral loss of periodic acid or covalent modification to oxidize the methionine side chain with concomitant loss of neutral iodic acid, for each of the periodate anions added onto the peptide. Activation of the [M+3H+2IO4-]+ complex exclusively yields sequential losses of iodic acid to produce the doubly oxidized species [M+2(O)+H]+ (parentheses around the O atom are used for clarity) (see Figure 7(b)). Activation of the doubly oxidized [M+2(O)+H]+ species of the peptide GRGMGRGMGRL produces nominal losses of 64 and 128 Da (Figure 7(c)). The first 64 Da loss can presumably arise from either of the two methionine residues in GRGMGRGMGRL. This 64 Da loss is the most abundant peak in the spectrum which would allow easy identification of the presence of at least one methionine residue during analysis of an unknown peptide. The nominal loss of 128 Da can arise from sequential losses of 64 Da, which was confirmed by activation of the 64 Da loss product (Figure 7(d)). The presence of the peak due to loss of two 64 Da neutrals in the product ion spectrum obtained from activation of the doubly oxidized [M+2(O)+H]+ indicates the presence of a second methionine in the peptide. The presence of the other fragments in Figure (d) can also be used to further confirm the oxidation of both methionine residues in GRGMGRGMGRL. The non-modified y2 ion is the only observed fragment not containing a methionine residue and indicates that neither of the last two residues is modified. Fragment ions containing only one methionine residue are present as either containing one oxygen atom, e.g., b4+O, b6+O, y5+O, and y6+O, or as having lost methanesulfenic acid from an oxidized methionine residue, e.g., b4□, b6□, y5□, and y6□. Only fragment ions containing both methionine residues are present as either loss of methanesulfenic acid with an added oxygen atom, e.g., b8□8+O, b9□+O, b10□+O, and y10□+Om, or loss of two methanesulfenic acid groups, e.g.,b8■, b9■, and b10■ where the closed square (■) indicates loss of two modified methionine side chains (e.g., b10■ denotes [b +2(O)-2HSOCH3]+).
CONCLUSIONS
The selective gas-phase oxidation of methionine residues has been demonstrated using ion/ion reactions with periodate anions. Ion trap CID of complexes comprised of methionine-containing peptide cations and the periodate anion results in formation of an oxidized species. The reaction presumably proceeds via nucleophilic attack by the sulfur atom on one of the neutral oxygen atoms on the periodate reagent. This reaction results in oxidation of the methionine side-chain and loss of neutral iodic acid, HIO3. Peptide cations lacking methionine residues predominantly transfer a proton to the periodate anion to form the charge-reduced species and an absence of oxidation is generally observed, although minor oxidation is observed for peptides containing tryptophan residues. The ion trap CID spectra of the oxidized species produced via gas-phase ion/ion reaction and solution-phase addition of periodate are identical. Solution-phase and gas-phase dissociation behavior both produce losses of 64 Da from precursor and product ions consistent with loss of methanesulfenic acid from the sulfoxide derivative of the methionine side chain. The unique 64 Da loss can be utilized to localize the oxidation to the methionine residue via MSn experiments. These results demonstrate a novel ion/ion reaction for the selective “labeling” of methionine and, to a lesser degree, tryptophan.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health under Grant GM 45372.
References
- 1.Berlett BS, Stadtman ER. Protein Oxidation in Aging, Disease, and Oxidative Stress. J. Biol. Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 2.Mulinacci F, Poirier E, Capelle MAH, Gurny R, Arvinte T. Influence of methionine oxidation on the aggregation of recombinant human growth hormone. Eur. J. of Pharm. And Biopharm. 2013;85:42–52. doi: 10.1016/j.ejpb.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Vogt W. Oxidation of methionyl residues in proteins – tools, targets, and reversal. Free Radical Biology and Medicine. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 4.Paik WK, Kim S. Protein Methylation - Chemical, Enzymological, and Biological Significance. Adv. Enzymol. 1975;42:277–286. doi: 10.1002/9780470122877.ch5. [DOI] [PubMed] [Google Scholar]
- 5.Bonvini E, Bougnoux P, Stevenson HC, Miller P, Hoffman T. Activation of the Oxidative Burst in Human-Monocytes is Associated With Inhibition of Methionine- Dependent Methylation of Neutral Lipids and Phospholipids. J. Clin. Invest. 1984;73:1629–1637. doi: 10.1172/JCI111369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao J, Yin D, Yao Y, Williams TD, Squier TC. Progressive Decline in the Ability of Calmodulin Isolated from Aged Brain to Activate the Plasma Membrane Ca- ATPase. Biochemistry. 1998;37:9536–9548. doi: 10.1021/bi9803877. [DOI] [PubMed] [Google Scholar]
- 7.Gao J, Yin DH, Yao Y, Sun H, Qin Z, Schoneich C, Williams TD, Squier TC. Loss of Conformational Stability in Calmodulin Upon Methionine Oxidation. Biophys. J. 1998;74:1115–1134. doi: 10.1016/S0006-3495(98)77830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watson AA, Fairlie DP, Craik DJ. Solution structure of methionine-oxidized amyloid beta-peptide (1-40). Does oxidation affect conformational switching? Biochemistry. 1998;37:12700–12706. doi: 10.1021/bi9810757. [DOI] [PubMed] [Google Scholar]
- 9.Kuo Y-M, Webster S, Emmerling MR, De Lima N, Roher AE. Irreversible Dimerization/Tetramerization and Post-translational Modifications Inhibit Proteolytic Degradation of A β-Peptides of Alzheimer's Disease. Biochim. Biophys. 1998;1406:291–296. doi: 10.1016/s0925-4439(98)00014-3. [DOI] [PubMed] [Google Scholar]
- 10.Palmblad M, Westlind-Danielsson A, Bergquist J. Oxidation of Methionine 35 Attenuates Formation of Amyloid β-Peptide 1-40 Oligomers. J. Biol. Chem. 2002;277:19506–19510. doi: 10.1074/jbc.M112218200. [DOI] [PubMed] [Google Scholar]
- 11.Stadtman ER, Moskovitz J, Berlett BS, Levine RL. Cyclic Oxidation and Reduction of Protein Methionine Residues is an Important Antioxidant Mechanism. Mol. Cell. Biochem. 2002:234–235. 3–9. [PubMed] [Google Scholar]
- 12.Hoshi T, Heinemann SH. Regulation of Cell Function by Methionine Oxidation and Reduction. J. Physiol. 2001;531:1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schey KL, Finley EL. Identification of Peptide Oxidation by Tandem Mass Spectrometry. Acc. Chem. Res. 2000;33:299–306. doi: 10.1021/ar9800744. [DOI] [PubMed] [Google Scholar]
- 14.Lagerwerf FM, van de Weert M, Heerma W, Haverkamp J. Oxidation of Oxidized Methionine Peptides. Rapid Commun. Mass Spectrom. 1996;10:1905–1910. doi: 10.1002/(SICI)1097-0231(199612)10:15<1905::AID-RCM755>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Reid GE, Roberts KD, Kapp EA, Simpson RJ. Statistical and Mechanistic Approaches to Understanding the Gas-Phase Fragmentation Behavior of Methionine Sulfoxide Containing Peptides. J. Proteome Res. 2004;3:751–759. doi: 10.1021/pr0499646. [DOI] [PubMed] [Google Scholar]
- 16.Qin J, Chait BT. Identification and characterization of posttranslational modifications of proteins by MALDI ion trap mass spectrometry. Anyl. Chem. 1997;69:3995–4001. doi: 10.1021/ac970489n. [DOI] [PubMed] [Google Scholar]
- 17.Jiang XY, Smith JB, Abraham EC. Identification of a MS-MS fragment diagnostic for methionine sulfoxide. J. Mass Spectrom. 1996;31:1309–1310. [Google Scholar]
- 18.Turecek F, Drinkwater DE, McLafferty FW. Gas-Phase Chemistry of CH3SOH, CH2+SHOH, CH3SO•, and •CH2SOH by Neutralization-Reionization Mass Spectrometry. J. Am. Chem. Soc. 1989;111:7696. [Google Scholar]
- 19.O'Hair RAJ, Reid GE. Neighboring group versus cis-elimination mechanisms for side chain loss from protonated methionine, methionine sulfoxide and their peptides. Eur. J. Mass Spectrom. 1999;5:325–334. [Google Scholar]
- 20.Amunugama M, Roberts KD, Reid GE. Mechanisms for the Selective Gas-phase Fragmentation Reactions of Methionine Side Chain Fixed Charge Sulfonium Ion Containing Peptides. J. Am. Soc. Mass Spectrom. 2006;17:1631–1642. doi: 10.1016/j.jasms.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 21.Lioe H, Laskin J, Reid GE, O'Hair RAJ. Energetics and Dynamics of the Fragmentation Reactions of Protonated Peptides containing Methionine Sulfoxide or Aspartic Acid via Energy- and Time-Resolved Surface Induced Dissociation. J. Phys. Chem. A. 2007;111:10580–10588. doi: 10.1021/jp073040z. [DOI] [PubMed] [Google Scholar]
- 22.Guan Z, Yates NA, Bakhtiar R. Detection and Characterization of Methionine Oxidation in Peptides by Collision-Induced Dissociation and Electron Capture Dissociation. J. Am. Soc. Mass Spectrom. 2003;14:605–613. doi: 10.1016/S1044-0305(03)00201-0. [DOI] [PubMed] [Google Scholar]
- 23.Prentice BM, McLuckey SA. Gas-Phase Ion/Ion Reactions of Peptides and Proteins: Acid/Base, Redox, and Covalent Chemistries. Chem. Commun. 2013;49:947–965. doi: 10.1039/c2cc36577d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLuckey SA, Reid GE, Wells JM. Ion Parking During Ion/ion Reactions in Electrodynamic Ion Traps. Anal. Chem. 2002;74:336–346. doi: 10.1021/ac0109671. [DOI] [PubMed] [Google Scholar]
- 25.Mentinova M, McLuckey SA. Intra- and Inter-Molecular Cross-Linking of Peptide Ions in the Gas-Phase: Reagents and Conditions. J. Am. Soc. Mass Spectrom. 2011;22:912–921. doi: 10.1007/s13361-011-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb IK, Mentinova M, McGee WM, McLuckey SA. Gas-Phase Intramolecular Protein Crosslinking via Ion/Ion Reactions: Ubiquitin and a Homobifunctional sulfo- NHS Ester. J. Am. Soc. Mass Spectrom. 2013;24:733–743. doi: 10.1007/s13361-013-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mentinova M, McLuckey SA. Covalent Modification of Gaseous Peptide Ions with N Hydroxysuccinimide Ester Reagent Ions. J. Am. Chem. Soc. 2010;132:18248–18257. doi: 10.1021/ja107286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mentinova M, Barefoot NZ, McLuckey SA. Solution versus Gas-Phase Modification of Peptide Cations with NHS-Ester Reagents. J. Am. Soc. Mass Spectrom. 2012;23:282–289. doi: 10.1007/s13361-011-0291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGee WM, Mentinova M, McLuckey SA. Gas-Phase Conjugation to Arginine Residues in Polypeptide Ions via N-Hydroxysuccinimide Ester-based Reagent Ions. J. Am. Chem. Soc. 2012;134:11412–11414. doi: 10.1021/ja304778j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han H, McLuckey SA. Selective Covalent Bond Formation in Polypeptide Ions via Gas-Phase Ion/Ion Reaction Chemistry. J. Am. Chem. Soc. 2009;131:12884–12885. doi: 10.1021/ja904812d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassell KM, Stutzman JR, McLuckey SA. Gas Phase Bio-conjugation of Peptides via Ion/Ion Charge Inversion: Schiff Base Formation on the Conversion of Cations to Anions. Anal. Chem. 2010;82:1594–1597. doi: 10.1021/ac902732v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stutzman JR, McLuckey SA. Ion/Ion Reactions of MALDI-Derived Peptide Ions: Increased Sequence Coverage via Covalent and Electrostatic Modification upon Charge Inversion. Anal. Chem. 2012;84:10679–10685. doi: 10.1021/ac302374p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stutzman JR, Hassel KM, McLuckey SA. Dissociation Behavior of Tryptic and Intramolecular Disulfide-linked Peptide Ions Modified in the Gas Phase via Ion/Ion Reactions. Int. J. Mass Spectrom. 2012;312:195–200. doi: 10.1016/j.ijms.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prentice BM, Gilbert JD, Stutzman JR, Forrest WP, McLuckey SA. Gas- Phase Reactivity of Carboxylic Acid Functional Groups with Carbodiimides. J. Am. Soc. Mass Spectrom. 2013;24:30–37. doi: 10.1007/s13361-012-0506-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Huang T-Y, McLuckey SA. Top-Down Protein Identification/Characterization of a priori Unknown Proteins via Ion Trap Collision- Induced Dissociation and Ion/Ion Reactions in a Quadrupole/Time-of-Flight Tandem Mass Spectrometer. Anal. Chem. 2009;81:1433–1441. doi: 10.1021/ac802204j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang TY, Liu J, McLuckey SA. Top-down Tandem Mass Spectrometry of tRNA via Ion Trap Collision-Induced Dissociation. J. Am. Soc. Mass Spectrom. 2010;21:890–898. doi: 10.1016/j.jasms.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 37.Huang T-Y, Liu J, Liang X, Hodges BDM, McLuckey SA. Collision-Induced Dissociationof Intact Duplex and Single-Stranded SiRNA Anions. Anal. Chem. 2008;80:8501–8508. doi: 10.1021/ac801331h. [DOI] [PubMed] [Google Scholar]
- 38.Xia Y, Wu J, Londry FA, Hager JW, McLuckey SA. Mutual Storage Mode Ion/Ion Reactions in Hybrid Linear Ion Trap. J. Am. Soc. Mass Spectrom. 2005;16:71–81. doi: 10.1016/j.jasms.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 39.Liang W, Xia Y, McLuckey SA. Alternately Pulsed Nano-electrospray Ionization/Atmospheric Pressure Chemical Ionization for Ion/Ion Reactions in an Electrodynamic Ion Trap. Anal. Chem. 2006;78:3208–3212. doi: 10.1021/ac052288m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Londry FA, Hager JW. Mass selective axial ion ejection from a linear quadrupole ion trap. J. Am. Chem. Soc. 2003;14:1130–1147. doi: 10.1016/S1044-0305(03)00446-X. [DOI] [PubMed] [Google Scholar]
- 41.Wells JM, Chrisman PA, McLuckey SA. Formation and Characterization of Protein-protein Complexes in Vacuo. J. Am. Chem. Soc. 2003;125:7238–7249. doi: 10.1021/ja035051l. [DOI] [PubMed] [Google Scholar]
- 42.Ruff F, Kucsman A. Mechanism of the Oxidation of Sulfides with Sodium Periodate. J. Chem. Soc. Perkin Trans. 1985;2:683–687. [Google Scholar]
- 43.Ruff F, Fabian A, Farkas O, Kucsman A. Mechanism for the Oxidation of Sulfides and Sulfoxides with Periodates: Reactivity of the Oxidizing Species. Eur. J. Org. Chem. 2009:2102–2111. [Google Scholar]
- 44.Gu C, Tsaprailis G, Breci L, Wysocki VH. Selective gas-phase cleavage at the peptide bond terminal to aspartic acid in fixed-charge derivatives of asp-containing peptides Anal. Chem. 2000;72:5804–5813. doi: 10.1021/ac000555c. [DOI] [PubMed] [Google Scholar]
- 45.Hensley K, Carney JM, Mattson MP, Aksenova M, Harris M, Wu JF, Floyd RA, Butterfield DA. A Model for Beta-Amyloid Aggregation and Neurotoxicity Based on Free-Radical Generation by the Peptide - Relevance to Alzheimer Disease. Proc. Natl. Acad. Sci. U.S.A. 1994;91:3270–3274. doi: 10.1073/pnas.91.8.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pike CJ, Walencewicz-Wasserman AJ, Kosmoski J, Cribbs DH, Glabe CG, Cotman CW. Structure-Activity Analyses of Beta Amyloid Peptides - Contributions Of the Beta 25-35 Region to Aggregation and Neurotoxicity. J. Neurochem. 1995;64:253–265. doi: 10.1046/j.1471-4159.1995.64010253.x. [DOI] [PubMed] [Google Scholar]
- 47.Froelich JM, Reid GE. Mechanisms for the Proton Mobility Dependent Gas-Phase Fragmentation Reactions of S-alkyl Cysteine Sulfoxide-Containing Peptide Ions. J. Am. Soc. Mass Spectrom. 2007;18:1690–1705. doi: 10.1016/j.jasms.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 48.Steen H, Mann M. Similarity between condensed phase and gas phase chemistry: Fragmentation of peptides containing oxidized cysteine residues and its implications for proteomics. J. Am. Soc. Mass Spectrom. 2001;12:228–232. doi: 10.1016/S1044-0305(00)00219-1. [DOI] [PubMed] [Google Scholar]
- 49.Chowdhury SM, Munske GR, Ronald RC, Bruce JE. Evaluation of low energy CID and ECD fragmentation behavior of mono-oxidized thio-ether bonds in peptides. J. Am. Soc. Mass Spectrom. 2007;18:493–501. doi: 10.1016/j.jasms.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clamp JR, Hough L. Periodate Oxidation of Amino Acids with Reference to Studies on Glycoproteins. Biochem. J. 1965;94:17–24. doi: 10.1042/bj0940017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.