Abstract
Seminal discoveries have established that epigenetic modifications are important for driving tumor progression. Polycomb group (PcG) proteins are highly conserved epigenetic effectors that maintain, by post-translational modification of histones, the silenced state of genes involved in critical biological processes including cellular development, stem cell plasticity and tumor progression. PcG proteins are found in two multimeric protein complexes called Polycomb Repressive Complexes: PRC1 and PRC2. Enhancer of zeste homolog 2 (EZH2), catalytic core subunit of PRC2, epigenetically silences several tumor suppressor genes by catalyzing the trimethylation of histone H3 at lysine 27, which serves as a docking site for DNA methyltransferases and histone deacetylases. Evidence suggests that over-expression of EZH2 is strongly associated with cancer progression and poor outcome in disparate cancers including hematological and epithelial malignancies. The regulatory circuit and molecular cues causing EZH2 deregulation vary in different cancer types. Therefore, this review provides a comprehensive overview on the oncogenic role of EZH2 during tumorigenesis and highlights the multi-faceted role of EZH2, as either a transcriptional activator or repressor depending on the cellular context. Additional insight is provided on the recent understanding of the causes and consequences of EZH2 over-expression in specific cancer types. Finally, evidence is discussed on how EZH2 has emerged as a promising target in anticancer therapy and the prospects for targeting EZH2 without affecting global methylation status. Thus, a better understanding of the complex epigenetic regulatory network controlling EZH2 expression and target genes facilitates the design of novel therapeutic interventions.
Keywords: EZH2, polycomb group, PRC2 complex, cancer, H3K27 trimethylation
INTRODUCTION
More than 5 decades ago, C. H. Waddington defined ‘Epigenetics’ as a discipline, which aim to describe changes in the development of organisms that could not be explained by the changes in DNA sequence. Local chromatin configuration at the gene promoter determines the accessibility of transcription machinery and associated proteins to bind to the specific DNA sequences. Either they directly interact with the nucleosome components or modulate the degree of compaction of nucleosome complexes into higher structures. DNA methylation at CpG islands in the promoter region and covalent chemical modification of less structured, protruding N- terminal tails of core histones by methylation, acetylation, ubiquitylation and phosphorylation at certain amino acid residues, have emerged as two integral mechanisms of epigenetic regulation that principally modulate local chromatin structure [for a review see ref(1)]. These epigenetic alterations in DNA which do not alter the nucleotide sequence are inheritable and potentially reversible unlike genetic changes. The transcription state of any gene may be predicted by deciphering the histone modification pattern at the promoter which is often referred to as ‘Histone Code’ [for a review see ref(2)].
The Polycomb Group (PcG) of proteins and their counterparts the Trithorax Group (TrxG) of proteins have long been found to be associated with the establishment of heritable gene transcription patterns. The antagonistic activities of the PcG proteins which act as epigenetic repressors and TrxG families of proteins function as epigenetic activators resulting in the maintenance of the spatial patterns of homeotic gene expression in Drosophila, throughout development and adulthood (3, 4). PcG proteins represent evolutionarily conserved multi-protein complexes which include the Polycomb repressive complexes: PRC1 and PRC2. Trimethylation at H3K27 (H3K27me3) is a distinct histone modification catalyzed by histone methyltransferase enhancer of zeste homolog 2 (EZH2), catalytic component of PRC2, involved in the regulation of homeotic (Hox) gene expression and in the early steps of X-chromosome inactivation in women (5, 6). Given the essential role of PcG proteins in establishing the repressed state of several genes during development and maintenance of embryonic stem cell (ESC) identity and pluripotency, recent studies imply that PcG proteins are often deregulated in various cancer types and their over-expression is closely associated with carcinogenesis (reviewed in (7–9).
In this review, we comprehensively discuss the role of one of the most important component of PRC2 complex viz. EZH2 in tumorigenesis of different cancer types. After evaluating the role of EZH2 in mammalian PRC2 complex, we systematically discuss its function and interaction with other epigenetic modifiers and elaborate in detail the causes and consequences of EZH2 over-expression, focusing on each cancer type. Recent literature focused on studying the molecular circuit guiding EZH2 expression and function in normal cells and its deregulation in tumor cells is also highlighted. Finally, owing to the pivotal ‘oncogenic’ role of EZH2 in the development and progression of several cancer types, recent literature highlighting the potential of EZH2 as a promising cancer target is discussed.
EZH2 and its role in PRC2 complex
The composition of PRC2 complex is dynamic, consisting of some core subunits responsible for catalyzing H3K27me3 and several accessory regulatory subunits controlling the enzymatic activity and function of the holoezyme (10, 11). The core components of mammalian PRC2 complex include: EZH1/2, suppressor of zeste 12 (SUZ12), embryonic ectoderm development (EED) and retinoblastoma associated protein 46/48 (RbAp46/48). Other accessory units that regulate PRC2 enzymatic activity and function include AEBP2, PCLs and JARID2 (Figure 1). EZH2 is the catalytic core of the PRC2 complex which catalyzes H3K27me3 repressive chromatin mark. Both genetic and biochemical evidence suggests that EED physically associates with EZH2 and histone H3 and thus functions as a scaffold protein. Molecular analysis of EZH2-EED interaction by X-ray crystallographic studies revealed that EED binding domain of EZH2 (EBD) binds to the bottom of Trp-Asp (WD) repeat domain of EED (12). SUZ12 is required for nucleosome recognition, activity and stability of PRC2 complex in both in vivo and in vitro conditions (13, 14). In addition, other PRC2 members such as RbAp46/48 is involved in histone binding and AEBP2, a zinc-finger protein, enhance the enzymatic activity of PRC2 complex (11, 15).
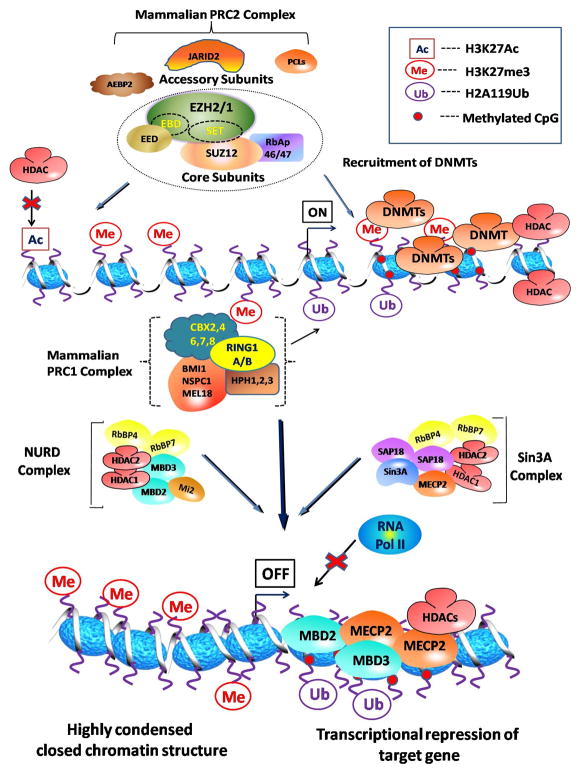
Figure 1. Epigenetic crosstalk showing functional link between PcG proteins (PRC1 and PRC2), histone deacetylases (HDACs) and DNA methyltransferases (DNMTs) during transcriptional silencing of a PRC2 target gene.
Initial deacetylation of histone H3 acetylated at Lysine 27 (H3K27Ac) by HDACs allows the catalysis of trimethylation of Lysine 27 (H3K27me3) by EZH2 containing PRC2 complex. H3K27me3 enrichment facilitate the binding of PRC1 complex that catalyze the mono-ubiquitylation of histone 2A at lysine 119 (H2AK119ub) and may also serve as a docking site for DNMTs which methylate CpG islands in the promoter region. Methylated CpG islands are then bound by a group of proteins possessing methyl DNA binding characteristics such as methyl CpG binding protein 2 (MeCP2), methyl CpG binding domain protein 1 (MBD1), which form a part of other histone modifying complexes like NURD and Sin3A. These events assist in the recruitment of HDACs and histone methyltransferase-containing protein complexes, contributing further to target gene repression. H3K27me3 and occupancy of target promoter by other histone modifying proteins lead to the formation of highly compressed chromatin structure which is inaccessible to RNA Pol II causing transcriptional silencing of target gene.
Human EZH2 is a 746 amino acid protein belonging to the histone-lysine methyltransferase family, all having a conserved SET (Su (var) 3–9, enhancer of zeste, trithorax) domain. In addition to the SET domain at the C-terminus, EZH2 contains several other functional domains such as CXC (cysteine-rich domain), ncRBD (non-coding RNA-binding domain and a DNA binding domain) required for its interaction with other PRC2 members and regulatory proteins (11, 15–17). Structural and biochemical analysis of SET domains in various histone methyltransferases (HMTase) has unraveled the molecular mechanism of histone methylation. Such studies highlighted the presence of a conserved catalytic triad—the asparagine-histidine-serine (NHS) motif responsible for recognition of the amino-acid sequence of histone peptide tail and for the binding of S-adenosyl-methionine (SAM) (18, 19). Mutation of any of these residues in the active site of EZH2 abolishes its HMTase activity. Focusing on highly evolutionarily conserved Tyr641 of EZH2, Yap et al. demonstrated that EZH2 Y641 mutant protein-containing PRC2 complexes display enhanced H3K27me3 activity on di-methylated peptides (and not on unmethylated histone peptides) as compared to wild-type containing PRC2 complexes which ultimately shifts the steady state of H3K27 in favor of trimethylation in vivo (20). The presence of wild-type EZH2-PRC2 complex was found to be mandatory for Y641 EZH2 mutant to act so that previously methylated histone substrates remain available for trimethylation. Several post-translational modifications (PTM) of EZH2 protein have also been reported to alter its H3K27me3 activity. For example, phosphorylation of EZH2 by Akt1 at serine 21 reduces H3K27me3 activity whereas phosphorylation at threonine 345 by CDK1 and CDK2 is required for maintenance of H3K27me3 repressive marks at target gene promoters (21, 22).
Notably, beside few exceptions, most invertebrates have only single copy of polycomb group protein (PcG) genes however in vertebrates multiple copies of PcG genes have been reported. Among PRC2 components, mammalian homologs EZH1 and EZH2 have been reported to be paralogs which remain an integral part of the PRC2 complex but are generally associated with contrasting H3K27me2/3 repressive roles (23). Although EZH1 was the first Ez homolog to be cloned, it is not as well characterized as its mammalian homolog EZH2 or its counterpart Drosophila Ez. Shen et al. demonstrated that EZH2 conditional knockout allele containing embryonic stem cells (EZH2−/− ESCs) sustain H3K27me3 at some developmental genes and display H3K27me1, despite global loss of di- and tri- methylation on H3K27, indicating the presence of another HMTase catalyzing methylation on H3K27 (24). They reported EZH1 to be a HMTase which is physically present in a non-canonical PRC2 complex, both in vivo and in vitro and prevents de-repression of PRC2 target genes. Margueron et al. investigated the cellular role of EZH1 in comparison to EZH2 and showed that EZH1 is ubiquitously expressed whereas the expression of EZH2 is found in proliferating tissues (23). Also, as part of similar PRC2 complexes they control an overlapping set of PRC2 target genes. Surprisingly, the functional roles of EZH1 and EZH2 were distinctly different; PRC2-EZH1 exhibit low histone lysine methyltransferase activity as compared to PRC2-EZH2, which may be attributed to their differential expression patterns and subfunctionalization of Ez during evolution (23). This indicates that PRC2-EZH2 function in establishing the cellular H3K27me3 repressive marks on PRC2 target genes while PRC2-EZH1 may help restore the repressive methylation pattern (H3K27me2/me3) of histones after demethylase activity or histone exchange that might cause the loss of their methylation mark.
EZH2 and PRC2 – dependent H3K27 trimethylation
The unstructured histone tails protruding from the nucleosome assembly are subjected to various post-translational modifications including lysine methylation which alters the physical state of chromatin and thus modulates gene expression. Lysine residue in the histone tail may be present in the unmethylated form or covalently modified by HMTases into mono, -di and –tri methylated lysine (H3K27me1, H3K27me2 and H3K27me3), with each form functionally different from the other. Methylation of histone H3 Lysine-27 by the SET domain of EZH2 (as part of PRC2 complex) has been reported to be a processive event where H3K27me3 results from mono-methylation of H3K27me2 (25). Di- and tri-methylated forms of Histone H3 Lysine 27 are associated with facultative heterochromatin region whereas mono-methylated form is associated with stably silent constitutive heterochromatin (26, 27). In embryonic stem (ES) cells, 50% of the H3 histone was reported to be dimethylated, 15% trimethylated and 15% monomethylated which add up to a total of 80% methylated histone H3 (28). H3K27me3 is a stable repressive chromatin mark catalyzed by PRC2 complexes containing either EZH1 or EZH2. H3K27me2, which is also catalyzed by PRC2, represents an intermediary product that not only serves as a substrate for subsequent H3K27me3 but might also inhibit acetylation of H3K27, which is reported to be antagonistic to PRC2-mediated gene repression (29, 30). In plants, two mono-methyltransferases namely Arabidopsis trithorax related protein 5 (ATXR5) and ATXR6 that are not orthologous to E(z) are involved in generating H3K27me1 mark but in mammals the appearance of H3K27me1 is still controversial (31). Some reports speculate that in mammals H3K27me1 might be catalyzed by an enzymatic activity distinct from PRC2 and its presence in actively transcribed genes may result from H3K27me2/3 demethylation by the demethylases UTX or JMJD3 (32). In contrast, few reports suggest that in Drosophila, generation of H3K27me1 mark is dependent on E(z) while in mammals PRC2 complex containing either EZH1 or EZH2 function redundantly control the global H3K27me1 levels. EED was also shown to be required for the H3 lysine mono-methylation (24, 33). Further experimental studies in mammals are needed to characterize PRC2 type complexes capable of HMTase activity so that the origin and function of H3K27me1 mark could be comprehensively understood.
Supported by various experimental studies in past several years, it is a well established fact that H3K27me3 enrichment is associated with gene silencing (34). Probing into the molecular mechanism reveals that this chromatin mark may serve as a docking site and facilitate the binding of PRC1 complex that catalyze the mono-ubiquitylation of histone 2A at lysine 119 (H2AK119ub) and thus maintain repressed state of the target gene (Figure 1). Also, H3K27me3 mark might indirectly regulate transcription by posing a steric hindrance to the binding of transcription machinery to the promoter region of the target gene. Presence of RNA Polymerase II (RNA Pol II) phosphorylated at serine 5 of its C-terminal tail in a wide fraction of H3K27me3-enriched promoters and low transcript levels suggests that RNA Pol II might be paused at PcG-targeted genes causing transcriptional repression (35). Studies reveal that RNA Pol II transcription complex that is recruited to the PcG target genes gets involved in early transcription but fail to proceed to the elongation stage. Kanhere et al. demonstrated that short RNAs that are transcribed from the 5′ end of polycomb target genes marked by transcription initiation marker H3K4me3, form stem-loop structure resembling PRC2 binding site could recruit and interact with PRC2 complex (36).
EZH2 and PRC2-independent function
Apart from the canonical role of EZH2 as a transcriptional repressor, recent literature reports highlight its less-known function as a transcriptional activator. In our previous review, we have comprehensively discussed the mechanism of action of EZH2 as a transcriptional inducer, where it functions independently (not as part of PRC2 complex), in endocrine-related cancers of the breast and prostate [reviewed in (37)]. Depending on the estrogen receptor (ER) status of the breast cancer cells, the EZH2 mediated target gene activation mechanism varies. In ER positive, luminal like MCF-7 cells it was demonstrated that EZH2 physically links β-catenin and TCF (ERα and Wnt signaling components) on the target gene promoters of cyclinB1 and c-Myc. The domain II of EZH2 interacts with the mediator complex and induces transcription (38). However in ER-negative MDA-MB-231 cells, EZH2 activates the transcription of NF-κB target genes TNF and IL-6 by forming a ternary complex with RelA and RelB (39). In castration resistant prostate cancer (CRPC), methylation of androgen receptor (AR) or AR-associated proteins has emerged as a potential mechanism for EZH2-mediated transcriptional induction (40). EZH2 is phosphorylated by Akt1 at serine 21 that allows it associate with AR. Furthermore, it was demonstrated that depletion of EZH2 causes a decrease in AR-associated lysine methylation at lysine 630 and 632 but the AR levels remain unchanged.
The non-canonical role of EZH2 independent of its histone methyltransferase activity was also observed in a highly aggressive lymphoid malignancy called natural killer/T-cell lymphoma (NKTL). Interestingly EZH2 over-expression in NKTL was found not to be associated with H3K27 trimethylation (41). Also, in NKTL cell lines, ectopic expression of EZH2 mutant lacking histone methyltransferase activity was found to confer growth advantage and prevent growth inhibition upon endogenous EZH2 depletion (41). Avni and colleagues showed that in differentiated T-helper cells, binding of polycomb proteins YY1, Mel-18, Ring1A, Ezh2 and Eed to the Il4 and Ifng gene loci exhibited differential pattern and induced transcription (42). In summary, recent findings support the hypothesis that EZH2 functions as a dual-faced molecule, which may function both as a transcriptional repressor or activator depending on its association with other members of the PRC2 complex and cellular context.
EZH2 and its interaction with other epigenetic modifiers
Early evidences in Drosophila and C. elegans showed that polycomb mediated H3K27me3 was deployed in transcriptional silencing but DNA methylation was absent in their chromatin, emphasized the notion that polycomb based silencing was independent from DNA methylation (43). But recent studies have completely changed this outlook and it has now become apparent that there exists a crosstalk between various epigenetic modifiers such as DNA methylation and histone modification pathways, which are mediated at the molecular level by biochemical interactions between DNA methyltransferases and SET domain containing histone methyltransferases, respectively (Figure 1). A key study by Vire et al. first described that PRC2 dependent EZH2-mediated silencing and DNA methylation systems are mechanistically linked (44). Using GST pull-down assays, they demonstrated that all three DNMTs (in vitro translated 35S-labelled DNMT1, DNMT3A and DNMT3B) can interact and bind to full length GST-EZH2. Furthermore, they showed that PRC2 members EZH2 and EED co-immunoprecipitated with all three DNMTs in HeLa nuclear extracts. The physical interaction between EZH2 and DNMTs facilitated the binding of the DNMTs to EZH2 target genes such as MYT1, WNT1 and was essential for CpG methylation of the target promoters. Interestingly, EZH2-DNMT interaction was reported to dispensable for EZH2 binding to the target gene promoter suggesting that EZH2 acts upstream of in the silencing pathway and serves as a recruiting platform for DNMTs. On the contrary, McGarvey et al. proposed a dominant role of CpG DNA methylation in gene silencing and maintenance of heritable repressive states of target gene promoters by demonstrating that EZH2 depletion neither affected the methylation status nor induced the hyper-methylated genes such as p16INK4a in U2OS cells (45). This study also suggests that EZH2 may not be essential for the maintenance of repressed state and silencing of a target gene promoter, once it becomes CpG hyper-methylated. Subsequent findings provided more inputs to the EZH2-DNA methylation link by comparing gene profiles of EZH2 target genes in normal versus cancer cells and revealed that EZH2 targets which display H3K27Me3 in normal cells are correlated to the aberrantly hyper-methylated corresponding genes in cancer cells (46, 47). This suggests that genes which acquire EZH2 catalyzed H3K27me3 marks are susceptible to subsequent DNA methylation and silencing during cellular transformation. Based on methylated CpG island microarray (MCAM) analysis of H3K27me3 modified CpG islands in PC-3 and MCF-7 cell lines, Kondo et al. proposed that DNA methylation and H3K27me3 based silencing may be independent mechanisms and overlapping genes targeted by both pathways are relatively rare (48). In conclusion, the current opinion supported by various experimental studies emphasize that DNA methylation and polycomb mediated silencing may act together in the epigenetic repression of some genes depending on the cellular context. However, there are discrepancies among studies suggesting a direct mechanistic link between these silencing pathways.
In human cells, PRC2 complex containing EZH2 has been shown to physically interact with histone deacetylases such as HDAC1 and HDAC2 (49, 50). Based on available biochemical data, HDACs are not the core subunits of PRC2 complex but they function in synergy during transient interactions in gene silencing. H3K27 acetylation has been shown to be functionally antagonistic to H3K27me3 mediated silencing and both are mutually exclusive (29, 30). Possibly, HDACs may function in deacetylation of the K27 side chains and allow PRC2-mediated methylation of ε-amino groups. Also, HDACs may alter the chromatin configuration by modulating local histone code favorable to polycomb mediated silencing by deacetylation of other histone lysines such as H3-K9, H3-K14 or H4-K8. Additionally, H3K4me3 and H3K36me2, which mark actively transcribing genes, have been shown to antagonize EZH2-mediated H3K27me3 silencing (51, 52). Although current literature reports advocate the fact that EZH2 mediated gene silencing has evolved to sense the surrounding epigenetic landscape and is linked to other epigenetic silencing mechanisms, further experimental studies are needed to address the direct mechanistic and functional link among them in driving oncogenesis.
ROLE OF EZH2 IN CANCER PROGRESSION
Accumulated evidences reveal that EZH2 deregulation and over-expression is frequently observed in a variety of cancer types including solid tumors and haematological malignancies. EZH2 levels have been reported to increase steadily as the tumor progresses and elevated EZH2 levels often directly correlated with advanced metastatic stages of cancer progression and poor prognosis. Studies suggest that EZH2 over-expression may be caused by a variety of signals or pathways, some of them are universal for all cancer types while others are more specific and limited to particular malignancies. The causes and consequences of EZH2 deregulation in different cancer types are discussed.
EZH2 and hematologic malignancies
Over-expression of polycomb group proteins particularly EZH2 has been linked to the pathogenesis of complex and heterogeneous hematologic malignancies such as myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) (53). It is well established that deregulated epigenetic factors coupled to genetic mutations contribute to their pathogenesis by causing abnormal silencing of critical tumor suppressor genes (TSGs). A study by Xu et al. demonstrated that in comparison to the patients with non-clonal cytopenia diseases, patients with MDS/AML have over-expression of EZH2, RING1 and BMI1 genes, leading to poor prognostic scoring (53). Elevated levels of DNA methyltransferase (DNMTs) and hypermethylation of tumor suppressor genes as p15, DAPK, SOCS-1 are frequently observed in MDS patients. In patients with p15INK4b methylation, EZH2 levels were found to be high as compared to patients without methylation. In a study using high-resolution chromatin immunoprecipitation (ChIP-on-chip), Paul et al. demonstrated that as compared to unmethylated p15INK4b samples, there was lower enrichment of p15INK4b locus with active chromatin mark H3K4me3 in AML cells with p15INK4b DNA methylation (54). However repressive chromatin mark H3K27me3 catalyzed by EZH2 was found at p15INK4b as well as in neighboring tumor suppressor genes p14ARF and p16INK4a, in both samples with and without p15INK4b DNA methylation. So it was concluded that the p15INK4b locus was enriched by bivalent histone marks H3K27me3/H3K4me3 in AML cells without DNA methylation. They further reported that epigenetic therapies could restore the loss of H3K4me3 and reactivate p15INK4b expression but the H3K27me3 mark is retained which ultimately causes the return of p15INK4b locus to a bivalent state.
The genomic organization and revised mapping of EZH2 gene by Cardoso et al. revealed that it is located on 7q35, a critical region associated with malignant myeloid disorders (55). Recent findings suggest that there are multiple mechanisms causing EZH2 deregulation in myeloid malignancies. Mutations in EZH2 gene have been described in myelodysplasia-MPNs (10–13%), myelofibrosis (13%), and various subtypes of MDS (9, 56–58). In myeloid neoplasms, EZH2 mutations were found to be inactivating /hypomorphic and distributed throughout the gene which includes missense, nonsense and premature stop codons (9). Histone methyltransferase activity was predicted to be lost in all EZH2 missense and nonsense mutants as the SET domain responsible for catalyzing H3K27 trimethylation was located in the C-terminal domain of the EZH2 protein (56, 57). Assessment of mutational status in 469 cases of myeloid malignancy revealed that EZH2 mutations were present in 8% cases while EED/SUZ12 mutations were reported in 3.3% cases. In addition to EZH2 mutation, decreased EZH2 expression was found be associated with hemizygous deletion (−7/del7q) involving EZH2 locus (78% of cases), diploid chromosome 7 (41% of cases) and spliceosomal U2AF1/SRSF2 mutations (63% of cases). EZH2 mutation was associated reduced H3K27me3ethylation and de-repression of EZH2 target genes which may contribute to leukemogenesis. HOXA9, one of the major downstream genes involved in stem cell renewal, was found to be elevated in cases with reduced EZH2 expression. Unlike myeloid neoplasm, in follicular lymphomas and diffused B-cell lymphomas, a heterozygous missense somatic mutation at tyrosine 641 (Y641) in the EZH2-SET domain causes a gain-of-function leading to increased H3K27me3 (59). The complex role of EZH2 in tumorigenesis is reflected by the fact that both activating and inactivating mutations in the EZH2 gene contribute to oncogenesis and malignancy.
EZH2 and prostate cancer
Prostate cancer is one of most common non-cutaneous malignancies responsible for majority of cancer-related deaths among men in the United States. Like other cancer types, interplay between genetic and epigenetic factors have been demonstrated to play a major role in the pathogenesis and progression of prostate cancer. Epigenetic alterations such as aberrant DNA methylation (hypo or hypermethylation), changes in chromatin remodeling patterns due to dysfunction in histone modifying enzymes and miRNA deregulation has been reported to be the major players in prostate carcinogenesis [reviewed in (60)]. Focusing on aberrant histone post-translational modifications, evidences indicate that critical histone modifying enzymes particularly histone deacetylases (HDACs) and histone methyltransferases (HMTs) are deregulated in prostate tumors.
In recent years, lack of success in defining useful molecular markers to evaluate disease progression and clinical outcome, had led to the application of high throughput genomic approaches such as microarray expression analysis to predict differences in the gene expression profiles of normal versus tumor samples at the molecular level. Using cDNA microarray analysis, Varambally et al. reported that increased expression of polycomb group protein EZH2 in metastatic prostate tumor samples distinguished them from organ-confined localized tumors (61). EZH2 has been found to promote prostate carcinogenesis by PRC2-mediated silencing of critical tumor suppressor genes such as DAB2IP, SLIT2, TIMP-3, and MSMB [reviewed in (37)]. However a recent study by Xu et al. highlighted that the oncogenic role of EZH2 in CRPC cells is independent of its transcriptional repressor function (40). Using androgen- dependent LNCaP cell line and its androgen-independent derivative LNCaP-abl as models to study prostate cancer progression, they have shown that EZH2 levels were higher in abl cells as compared to LNCaP cells. Also, unlike androgen dependent growth of LNCaP cells, EZH2 silencing has been demonstrated to have a more profound effect in androgen independent growth both in vitro (abl cells) and in vivo (mouse xenograft CRPC model using CWR22Rv1 cells). The study further investigated the factors causing EZH2 to switch to a gene activation function in CRPC. EZH2 phosphorylation at serine 21 by PI3K-Akt pathway was demonstrated to be crucial for EZH2 mediated androgen-independent growth, gene activation and association with AR-containing complexes.
Recent reports indicate that androgens and androgen receptor (AR) signaling pathway play critical role in regulating EZH2 levels, suggesting a potential reason for increased EZH2 levels after androgen deprivation therapy (ADT) in metastatic and hormone-refractory prostate cancer. Androgens have been shown to repress EZH2 expression (62). RB and p130-dependent pathways mediate EZH2 repression in the presence of functional AR. Using co-immunoprecipitation, Xu et al. reported that EZH2 and AR physically interact in androgen-independent abl cells and the interaction was lost in EZH2 deletion mutants either in Domain I (N-terminal protein-protein interaction domain) or in the C-terminal SET domain emphasizing the importance of these two domains for the interaction (40). They have also shown that the AR mRNA or protein levels did not change upon EZH2 depletion but there was decrease in AR- associated lysine methylation. It was further demonstrated that through their cooperative recruitment or binding, EZH2 and AR activate a set of target genes. Another study by Zhao and colleagues has shown that EZH2 and AR collaborate with each other in repressing developmental regulators involved in non-prostatic pathways (63). The androgen-receptor dependent transcriptional silencing is mediated by EZH2 in an androgen deprived environment.
In prostate cancer, various molecular mechanisms have been proposed to be responsible for EZH2 over-expression which includes EZH2 gene amplification, miR-101 deletion, and transcriptional regulation by MYC and ETS gene family members [reviewed in (37)]. Using fluorescence in situ hybridization technique, Saramaki and colleagues determined the copy number of EZH2 gene in prostate cancer cell lines LNCaP, DU145, PC-3, 22Rv1, xenografts and clinical tumors (64). The data showed increased EZH2 copy number in all the cell lines. Unlike early prostate cancer, in late-stage tumor samples EZH2 gene amplification was associated with its over-expression. Also quantitative real time RT-PCR analysis indicated that EZH2 expression was elevated in hormone-refractory prostate cancer as compared to benign prostate hyperplasia or untreated prostate cancer. In another study, Varambally et al. proposed that miR-101 negatively regulates EZH2 expression and somatic loss of one or both miR-101 genomic loci elevates EZH2 levels causing deregulation of various epigenetic pathways in prostate cancer (65). Their study revealed that there was a negative correlation between miR-101 and EZH2 levels during prostate cancer progression. An increase in EZH2 levels paralleled a concomitant decrease in miR-101 levels in human prostate cancer samples. Additionally, some studies reported that ETS transcriptional network consisting of ERG and Epithelial-specific ETS factor, ESE3 are important molecular players with opposing effects regulating EZH2 expression in prostate cancer cells (66). ESE3 have been demonstrated to suppress EZH2 expression whereas ERG competes with ESE3 for promoter occupancy and oppose its effects. Although prior studies have proposed various molecular mechanisms for EZH2 upregulation in prostate cancer cells, they were insufficient in explaining the reason for elevated EZH2 levels in primary prostate cancer and high grade prostatic intraepithelial neoplasia (PIN). Interestingly, a study by Koh et al. proposed two additional yet complementary Myc-regulated molecular mechanisms responsible for EZH2 upregulation in prostate carcinogenesis (67). Since Myc and EZH2 both are frequently over-expressed in human PIN and primary carcinoma lesions, the study demonstrates that there exists a molecular link between them. In prostate cancer, elevated Myc induces EZH2 expression by directly binding to the E-box containing promoter region of EZH2 and activating transcription. Similar findings were also reported by Salvatori et al. in acute myeloid leukemia (68). Interestingly Myc is also known to regulate miRNAs. In LNCaP and PC-3 cell lines, promoter regions of CTDSPL, CTDSP2 and CTDSP1 genes (miR-26a and miR-26b primary transcripts are embedded in the intron region) was found to be enriched by Myc, which results in transcriptional repression (67). When Myc levels are low, miR-26a/miR-26b are actively transcribed and incorporated into RISC complex. It is then targeted to the 3′UTR of EZH2 which destabilize EZH2 mRNA ultimately repressing its translation. Repression of miR-26a/miR-26b by elevated Myc levels contribute to EZH2 over-expression by increasing the stability of EZH2 mRNA due to the less availability of miR-26a/miR-26b-RISC complex binding to EZH2-3′UTR. In conclusion, elevated Myc levels enforce EZH2 over-expression by altering both transcriptional and post-transcriptional regulatory mechanisms controlling EZH2 expression in prostate cancer cells.
EZH2 and breast cancer
Recent experimental advances in breast cancer research have established EZH2 as a promising novel biomarker indicating cancer progression and disease aggressiveness. EZH2 levels have been found to increase steadily as the disease progresses from a benign tumor to clinically evident metastasis (69, 70). Highest EZH2 levels are associated with the ER-negative basal like phenotype of breast cancer cells that represents the most aggressive form of breast carcinoma characterized by nuclear polymorphism, lack of ER and BRCA1 protein (71). Using high density tissue microarray, Kleer et al. showed that as compared to normal or patients with atypical hyperplasia, EZH2 levels were significantly higher in patients with invasive breast carcinoma (69). Interestingly, Ding et al. reported that in breast tissue samples that appeared to be histologically normal but at a high-risk of developing cancer showed increased EZH2 expression and hence identified EZH2 as a potential in vivo bio-marker that may detect a precancerous state in otherwise morphologically normal breast epithelial cells (72). Both in vitro and in vivo genetic knock-down studies targeting EZH2 demonstrated that a decrease in EZH2 levels results in decreased breast cancer cell proliferation, delayed G2/M cell cycle transition, reduced breast xenograft growth and improved survival. Upregulation of EZH2 levels caused H3K27me3 mediated aberrant silencing of various essential tumor suppressor genes such as FOXC1, CDKN1C (p57KIP2), RUNX3, RKIP, CIITA, IL-6 and IL-8 [For a review see ref (37)].
Over the years, several molecular mechanisms have been proposed to address the causes and consequences of EZH2 over-expression in breast cancer cells. Bracken et al. reported that the expression of two PRC2 complex components viz. EZH2 and EED is regulated by E2F transcription factors (73). It is well established that defects in pRB-E2F pathway is a mandatory event in the pathogenesis of almost all human malignancies. E2F transcription factors are known for regulating genes such as CyclinE1, CyclinA2, CDC6, DHFR and TK1 that control entry into S-phase and DNA replication which are well studied targets of pRB (retinoblastoma) pathway. Upon pRB phosphorylation, E2F dissociates from pRB-E2F complex and the activated E2F directly binds to the promoters of EZH2 or EED containing putative E2F binding sites and transactivate their transcription. The study further confirmed that although the abrogation of EZH2 or EED expression could significantly decrease positive regulators of cell proliferation such as cyclinD1 (CCND1), cyclinE1 (CCNE1), cyclinA2 (CCNA2) and cyclinB1 (CCNB1), there was no increase in the expression of negative regulators of cell cycle viz. p14ARF. This study provides a direct link between pRB-E2F pathway mediated growth control and PcG regulated essential chromatin modifications.
In another study, Fujii et al. reported that the MEK-ERK-Elk-1 pathway, which is commonly upregulated in various cancer types, is linked to EZH2 over-expression in triple-negative and ERBB2-overexpressing breast cancer cells (74). Computational analysis revealed that EZH2 promoter harbor three Elk-1 binding motifs and other sequence elements for NF-κB, c-Myb, STAT1 and SRF (serum response factor) recruitment. Treatment with MEK inhibitor, U0126 decreased EZH2 levels in triple-negative breast cancer type MDA-MB-231 cells and ERBB2-overexpressing SKBr3 cells. Also siRNA mediated knockdown of Elk-1, caused a significant decrease in EZH2 mRNA expression that was similar to the decrease caused by U0126 treatment. Further they demonstrated that MEK inhibitor treatment significantly decreased association of phospho-Elk-1 with the EZH2 promoter in breast cancer cells. In conclusion the study suggests that MEK/ERK pathway activated via KRAS mutation, EGFR amplification and ERBB2 amplification in triple-negative and ERBB2-overexpressing cells causes EZH2 over-expression.
Under cancer-predisposed hypoxic conditions in breast tumor initiating cells (BTICs), EZH2 expression was reported to be regulated by HIF transcription factor (HIF1α). Chang et al. identified a consensus sequence for HIF response element (HRE) by promoter analysis of EZH2 promoter (75). They further demonstrated that a hypoxic microenvironment induces HIF1α binding to the HRE and transactivate EZH2 expression. Increased EZH2 expression down-regulates double-strand break repair protein RAD51 expression, which ultimately results in impaired DNA repair and accumulation of genomic abnormalities. Further they demonstrated that EZH2 mediated down-regulation of RAD51 causes expansion of self-renewing BTIC population and RAF1 amplification which further activate downstream pERK-β-catenin signaling. EZH2 mediated RAF1-ERK signaling promotes BTIC expansion and aggravates breast cancer condition.
Recent evidences indicate that post-translational modifications also play a crucial role in regulating EZH2 levels in breast cancer cells. Studies by Cha et al. have shown that Akt-mediated phosphorylation of EZH2 at a highly conserved serine 21 residue, inhibited its H3K27 methyltransferase activity (21). Although EZH2 phosphorylation had no impact on the integrity of PRC2 complex, there was marked decline in the affinity of EZH2 for Histone H3, which ultimately results in decreased H3K27me3. Interestingly in an another study Gonzalez et al. reported that elevated EZH2 in breast cancer cells induces phosphoinositide-3-kinase/Akt (PI3K/Akt) pathway by specifically activating Akt isoform1(76). Increased EZH2 levels positively correlated with elevated phospho-Akt1 (Ser473) and decreased nuclear localization of phospho-BRCA1 (Ser1423). The role of EZH2 in nuclear shutting of BRCA1 may be regarded as one of the non-PRC2 dependent EZH2 functions as discussed in previous sections. Accumulation of BRCA1 protein in the nucleus promotes tumorigenesis by causing aberrant mitosis, aneuploidy and genomic instability. The multifaceted role of EZH2 in promoting breast tumorigenesis where it may act as a transcriptional activator or repressor depending on the cellular context has been highlighted in the previous section on non-PRC2 dependent EZH2 function.
EZH2 and lung cancer
Lung cancer is a leading cause of cancer-related mortality among both men and women worldwide accounting for almost 1.4 million deaths each year (77). Small cell lung cancer (SCLC) is a highly aggressive neuroendocrine carcinoma and represents a histologic subtype which is distinct from other lung cancer types called non-small cell lung cancers (NSCLCs). SCLC has been found to be strongly correlated to cigarette smoking and accounts for approximately 15% of all lung cancer cases. Like other cancer types, accumulation of several genetic and epigenetic aberrations has been implicated in the initiation and progression of lung cancer. Focusing on the role of histone methyltransferase EZH2 in the pathogenesis of lung cancer, recent evidences suggests that EZH2 over-expression and polycomb mediated H3K27me3 promote malignancy in various lung cancer types including SCLC and NSCLCs, which is commonly associated with poor prognosis (78–80). In a recent study, Coe et al. found a strong correlation between deregulation of E2F-Rb pathway in 96% SCLC samples and EZH2 over-expression (81). Their study revealed that genomic loss of E2F-Rb pathway either by loss of RB1 or E2F amplification leads to increased EZH2 expression. Generation of ‘stem-cell like’ hypermethylator profile in SCLC tumors due to aberrant methylation of PRC2 target genes was found to be associated with elevated EZH2 levels. Additionally, the finding that lentivirus-mediated knockdown of EZH2 in two SCLC cell lines caused a significant decrease in growth as compared to empty vector controls, emphasized the pro-proliferative and oncogenic role of EZH2 in SCLC. Using microarray analysis on a genome wide scale, Aburatani and colleagues revealed that the expression levels of EZH2 and other PRC2 members such as SUZ12 and EED were significantly higher in SCLC samples in comparison to normal tissues including the lung (78). They also demonstrated that H3K27me3-mediated repression of PRC2 target genes such as JUB in both SCLC cell lines as well as clinical samples correlated with poor prognosis. Findings by Hubaux et al. highlighted the implication of EZH2 in cell death and cell cycle regulation by demonstrating that shRNA-mediated knockdown of EZH2 in SCLC cells induced apoptosis by elevating pro-apoptotic factors Puma and Bad, increased p21 protein levels and decreased fraction of cells in S or G2/M cell cycle phases.
NSCLC including squamous cell carcinoma, adenocarcinoma, large cell carcinoma and several other types which occur less frequently, account for more than 85% of all lung cancer cases and are associated with poor prognosis mainly due to late diagnosis, with an overall 5-year survival rate of approximately 11%. Immuno-histochemical analysis of 157 surgically resected NSCL samples by Kikuchi et al. revealed a strong correlation between elevated EZH2 protein levels with advanced pathologic tumor stage, moderate or poor differentiation, non-adenocarcinoma histology, high Ki-67 and cyclin E levels (79). Corroborating with the previous findings, Huqun et al. reported that there was a significant association of increased EZH2 expression with larger tumor size and shorter overall survival in NSCLC (82). These studies highlighted EZH2 protein levels as a negative prognostic indicator in NSCLC.
In recent years, studies focused on the identification of molecular mechanisms causing lung carcinogenesis have revealed that deregulation of several miRNAs such as miR-126, miR-21, miR-200c, miR-145, and miR-107, miR-185 and miR-101 contribute to tumor cell growth, invasion and apoptosis. Interestingly, Zhang et al. demonstrated EZH2 to be a direct functional target of miR-138 in NSCLC and highlighted its therapeutic importance in NSCLC treatment strategy (83). They showed that miR-138 expression was down-regulated in NSCLC tissues and cell lines A549, SPC-A1, SK-MES-1 and H460 as compared to the matched normal tissue samples from the same patients and normal bronchial epithelial cell line 16HBE, respectively. Lentivirus-mediated over-expression of miR-138 caused decreased viability, G0/G1 arrest and apoptosis in A549 and H460 cell lines. Furthermore, using bioinformatics analysis, EZH2 3′-UTR was found to harbor a highly conserved miR-138 binding site. Transfection of miR-138 over-expressing A549 and H460 cells with wild type 3′-UTR EZH2 luciferase reporter construct resulted in decreased luciferase activity as compared to cells transfected with 3′UTR EZH2 mutant plasmid construct. Also, miR-138 over-expression caused significant decrease in EZH2 mRNA and protein levels in both cells lines. Additionally, ectopic expression of EZH2 was shown to rescue miR-138 induced growth inhibition, cell cycle arrest and apoptosis in the lung cancer cells. In another study Cho et al. demonstrated an inverse correlation between miR-101 and EZH2 expression levels in lung cancer tissue samples (84). MiR-101 was shown to inhibit lung cancer cell invasion by regulating EZH2 and H3K27me3 levels. In conclusion, miRNA deregulation has emerged as a major cause of EZH2 over-expression in lung carcinomas and novel therapeutic strategies should consider targeting specific miRNAs such as miR-138, miR-101, and miR-26a in lung cancer treatment.
EZH2 and bladder cancer
Bladder cancer is one of most common malignancies in the United States and based on National Cancer Institute’s statistics it accounted for approximately 72,570 new cases and 15,210 deaths in 2013. Based on the site of origin, bladder cancer may be categorized into various types which include transitional cell carcinomas (cancer develops in the inner cells lining the bladder), squamous cell carcinoma (cancer originates in thin, flat cells) and adenocarcinoma (cancer begins in cells that produce and secrete mucus and other fluids). Transitional cell carcinomas (TCC) which account for >90% of bladder cancer are associated with superficial, non-muscle-invasive tumors, which may be treated by local transurethral resection. However the risk of recurrence is high and the non-invasive lesions may progress to more lethal and metastatic carcinoma. Although epigenetic molecular alterations involved in bladder cancer initiation and progression are not as comprehensively studied as genetic abnormalities of tumor suppressor genes or oncogenes, recent studies correlate elevated EZH2 levels to high-grade lesions and invasive bladder carcinomas (85–87). Raman et al. demonstrated increased EZH2 mRNA and protein expression in TCC compared to adjacent non-tumorous tissue (88). Takawa et al. analyzed EZH2 mRNA and protein levels in clinical bladder cancer samples by quantitative real time PCR and immuno-histochemistry (89). Their findings were in line with previous studies showing significant upregulation of EZH2 levels in tumor cells as compared to normal cells. Zhang et al. showed that inhibition of EZH2 expression by RNA interference resulted in decreased cell proliferation and G1 arrest in bladder cancer EJ cells (90). Interestingly, some studies have highlighted over-expression of E2F transcription factors including E2F3 and deregulation of pRB-E2F pathway, which directly regulate EZH2 expression, as one of the fundamental factors driving human bladder tumorigenesis (91, 92). Collectively, these findings suggest that aberration in pRB-E2F pathway may cause elevated EZH2 levels at successive stages of bladder carcinogenesis. In another study, Tang et al. revealed a novel link between p53 tumor suppressor gene and EZH2 by demonstrating that activated p53 suppress EZH2 gene expression by binding to its promoter (93). This raises an interesting question of whether alterations in p53 may lead to increased EZH2 expression in bladder cancer. In fact, Yamada et al. showed that in squamous cell carcinoma of the esophagus (SCCE), elevated EZH2 expression was associated with p53 alteration and activated p53 reduced EZH2 mRNA levels in SCCE cell lines (94). Furthermore, EZH2 was found to be a miR-144 target gene and miR-144 down-regulation in bladder cancer cells was demonstrated to restore EZH2 expression, which further resulted in the activation of Wnt/ β-Catenin pathway and subsequent cell proliferation (95).
EZH2 and ovarian cancer
Ovarian cancer develops in tissues of the ovary and can be categorized into two major types based on the tissue of origin-ovarian epithelial carcinomas (cancer forms in the cells on the surface of the ovary) or malignant germ cell tumors (cancer develops in egg cells). Epithelial ovarian cancer (EOC) accounts for majority of gynecologic cancer-related deaths in the United States. Accumulated evidences indicate that EZH2 over-expression plays a major role in the etiology and pathogenesis of EOC. Li et al. demonstrated that although elevated EZH2 levels were positively correlated with cell proliferation markers such as Ki67 and tumor grade but there was no association of EZH2 expression with tumor stage and overall or disease free survival in high-grade serous histotype EOC patients (96). Interestingly, knockdown of EZH2 expression in both in vitro and in vivo xenograft models resulted in decreased H3K27me3 levels, induced apoptosis, suppressed growth and invasion of human EOC cells. Rao et al. reported that EZH2 over-expression was absent in normal ovaries while high EZH2 expression in the ovarian carcinomas (50% cases) was positively associated with increasing histological tumor grade and advanced stage of the disease (97). Furthermore, this study underscores the potential role of EZH2 in regulating cell proliferation, migration or invasion via modulation of TGF-β1 expression and demonstrated a positive correlation between high EZH2 and TGF-β1 levels in ovarian carcinoma tissues. EZH2 knockdown was shown to reduce expression of TGF-β1 and increase E-cadherin expression either at the transcript or protein levels. Together, recent findings suggest that elevated EZH2 levels in EOC promote carcinogenesis by H3K27me3 mediated silencing of target genes which ultimately induce cell proliferation, invasiveness and suppress apoptosis.
Notably Hu et al. found that EZH2 was over-expressed in cisplatin-resistant ovarian cancer cell line A2780/DDP as compared to cisplatin-sensitive A2780 cell line (98). SiRNA mediated knockdown of EZH2 resulted in re-sensitization of A2780 cells to cisplatin and also decreased H3K27me3 levels. Similarly, Rizzo et al. reported EZH2 over-expression in ovarian tumor derived side population (SP) cells, which are stem cell like cells enriched by chemotherapy and demonstrated that siRNA knockdown of EZH2 caused loss of SP and reduced anchorage-independent growth in ovarian tumor models (99). These evidences suggest that EZH2 regulates stem cell like attributes of ovarian cancer cells, which may contribute to chemoresistance in EOCs.
In a recent study Garipov et al. examined the molecular mechanism underlying EZH2 over-expression in EOCs (100). Their study highlighted the role of NF-YA, the regulatory subunit of CCAAT-binding transcription factor NF-Y, in regulating EZH2 transcription by associating with two CCAAT boxes in the proximal EZH2 promoter in EOC cells. They also showed that there is a positive correlation between EZH2 and NF-YA expression levels in EOC and elevated NF-YA levels predicts shorter overall survival in EOC patients. Furthermore, NY-YA knockdown down-regulated EZH2 expression as well as H3K27me3 levels, induced apoptosis and reduced both in vitro and in vivo growth of human EOC cells. Consequently ectopic EZH2 expression rescued NF-YA induced apoptosis in EOC cells. In another study Guo et al. emphasized the oncogenic role of EZH2 in ovarian cancer by possible down-regulation of anti-oncogene p57 and showed that there is an inverse correlation between EZH2 and p57 mRNA expression levels in ovarian tissue samples (101). Lu et al. showed that EZH2 is an important regulator of tumor angiogenesis (102). EZH2 over-expression in EOC-associated endothelial cells was demonstrated to be a direct result of paracrine circuit involving vascular endothelial growth factor (VEGF) stimulation. EZH2 was shown to promote angiogenesis by silencing vasohibin1. Furthermore, siRNA mediated EZH2 silencing caused significant growth inhibition in tumor associated endothelial cells and reduced angiogenesis. Together, accumulated evidences propose EZH2 to be a promising novel target in anti-angiogenesis therapy.
EZH2 and skin cancer
As per the latest National Cancer Institute’s statistics, skin cancer remains as one of the most common cancers in the United States, with nearly more than 2 million people treated for non-melanoma (basal cell or squamous cell carcinoma) and 76,690 new melanoma cases each year. Melanoma is a type of cancer that develops in the melanocytes in the skin represent genetically and epigenetically complex group of malignancy characterized by deregulation of multiple tumor suppressor and oncogenic pathways, including BRAF, NRAS, PTEN, and CDKN2A (103–105). Like several other human cancers, EZH2 over-expression has been implicated in the progression of benign nevi to invasive or metastatic melanoma (106, 107). Aroni and colleagues investigated EZH2 expression in squamous cell carcinoma (SCC), and actinic keratosis (AK) (108). In this study, weak EZH2 immuno-staining was reported in AK cases (62.8%) whereas moderate expression was found in SCCs (42.1%) and 77.8% EZH2 expression levels in ‘mixed’ SCC/AK cases.
A key oncogenic event reported in majority of melanoma cases is the BRAFT1799A mutation that results in the constitutively activated BRAFV600E kinase. The epigenetic mechanism of action of BRAFV600E, which is well known to play an important role in melanoma progression, was recently studied as one of the potential mechanisms for BRAFV600E-mediated gene hypermethylation via upregulation of DNMT1 and EZH2 in melanoma cells (109). During tumor initiation, oncogenic stimuli in melanocytes exacerbate an oncogene-induced senescence, known as melanocytic nevus, which is a benign precursor of melanoma. Cells over-expressing EZH2 escape this senescence through the inhibition of p21. In contrast, EZH2 depletion, results in p21 activation and senescence induction in human melanoma cells (110). PRC2 complex inhibits the expression of several tumor suppressor miRNA in various cancer types. Luo et al. demonstrated that over-expression of miR-101 inhibited melanoma cell invasion and proliferation by down-regulation of microphthalmia-associated transcription factor (MITF) and EZH2 protein levels (111). Similarly, miR-137 was also demonstrated to have a tumor suppressive role and c-Met, YB1, MITF and EZH2 were identified as its direct targets in malignant melanoma (112). Down-regulation of miR-31 is a common event in melanoma which is associated with genomic loss in a subset of samples. Decreased miR-31 gene expression was found to be a result of epigenetic silencing by DNA methylation, and via EZH2-mediated histone methylation (113). Importantly, miR-31 has emerged as a complex player in a number of cancers and evidences suggest that miR-31 can act either as an oncomiR or a tumor-suppressive miR in a tumor type-specific manner (114). Another critical tumor suppressor gene Rap1GAP, which is down-regulated in multiple aggressive tumors including melanoma, pancreatic and thyroid cancer was demonstrated to be epigenetically silenced by promoter hypermethylation in melanoma cells (115); whereas EZH2 mediated Rap1GAP transcriptional repression by H3K27 trimethylation was reported in squamous cell carcinoma of head and neck (116).
Role of EZH2 in cancer stem cells
Cancer stem cells (CSCs) are responsible for initiation and proliferation of a tumor mass and their recent in vivo validation in certain solid tumors has attracted considerable scientific attention mainly because of their potential clinical significance (117). CSCs have been first identified in AML and later on found to be present in various solid tumors (118). Modern CSC model postulates that there exists a subset of tumor cells with stem cell-like properties; including self-renewal as well as multi-lineage differentiation capability. CSCs are characterized by their ability to seed tumor in vivo, self-renewal property and to spawn differentiated non-CSC progeny lacking tumor initiating capabilities (119).
With the emergence of CSC model, it was obvious to analyze the shared features of normal stem cells with CSCs. PcG proteins play their important role in stem cell maintenance by reversibly repressing the genes encoding transcription factors required for differentiation (120). This feature of normal stem cell was generalized for CSCs also and hypothesizes that acquisition of promoter DNA methylation at these repressed genes could lock in stem cell phenotype to initiate abnormal clonal expansion to develop cancer (121). In various cancers including breast, prostate, ovary, pancreas, lung and liver the isolated CSC populations has been reported to over-express EZH2, which is proposed to be essential for maintenance of an intact CSC population (122). In limited number of studies, epigenetic mechanism of gene silencing by EZH2 has been linked to CSC-associated features such as invasion and chemoresistance. The role of EZH2 in cancer could be related to interesting mechanism of silencing through the repression of differentiating and anti-metastatic genes. CSCs maintain their pool by suppressing cell differentiating genes such as p16, p19 and show other characteristics such as decrease of E-cadherin expression to make them metastatic (122). EZH2 mediates p16, p19 and E-cadherin silencing through histone H3K27 trimethylation. Furthermore, EZH2 has been shown to repress the Forkhead box transcription factor C1, thereby enhancing invasive potential of breast cancer cells (123). EZH2 is also a key mediator of DNA damage response in tumor cells and contribute to CSC chemo-resistance, suggesting its additional role in therapeutic resistance apart from tumorigenesis (124). Altogether, EZH2 has emerged as an important molecular component required for onset of CSC phenotype in tumor cells as well maintaining its associated features.
EZH2 AS A THERAPEUTIC TARGET
Emerging experimental evidences clearly demonstrate that EZH2 play critical roles in driving carcinogenesis, from tumor initiation to clinically evident metastasis in various cancer types. Like other epigenetic enzymes such as DNMTs and HDACs, EZH2 has emerged as a potential anticancer target in present epigenetic strategies. In recent years, search for novel small molecule EZH2 inhibitors have yielded some promising results. One of the best known and widely used EZH2 inhibitors, 3-dezaneplanocin-A (DZNep), was identified by Tan and colleagues through library based drug screening. DZNep has demonstrated a significant anti-tumor activity in various cancer types which include breast, prostate, lung, liver and brain cancer cells (125). Notably, treatment with DZNep depletes EZH2 levels and reactivates PRC2 target genes. DZNep targets S-adenosyl-L-homocysteine hydrolase (SAH) and causes SAH levels to rise, which further inhibits SAH cofactor dependent methyltransferases such as EZH2 which require SAH for catalysis. Although DZNep has exhibited some promising results in in vitro and in vivo studies, there are concerns regarding its specificity as a potential therapeutic compound as it may also affect other SAM-dependent processes. Also, questions on its short plasma half-life, effect on global methylation status and toxicity profile in animal models remains open. More recently, the quest for more specific EZH2 inhibitors has identified some molecules that are highly selective over EZH1 and other histone methyl transferases and directly inhibit EZH2 by binding to the active site through competitive inhibition with methyl group donor SAM. EPZ005687 displayed considerable specificity and selectivity when tested on tyrosine 641 or alanine 677 mutation harboring lymphoma cells (126).
Another EZH2 inhibitor El1, which also acts in a SAM-competitive manner, has been developed by Qi et al. (127). El1 has demonstrated remarkable selectivity across an HMT panel and decreased H3K27me3 levels without altering other histone H3 methylation marks. They also showed El1 mediated selective inhibition of EZH2 caused reduced proliferation, cell cycle arrest and apoptosis in DLBCL cells Y641 mutation and other EZH2 over-expressing cancer cell lines. Interestingly, Kim et al. reported an inhibition strategy of targeting EZH2 that is distinct as compared to other small-molecule inhibitors which target EZH2 catalytic site (128). They reported the dose-dependent disruption of EZH2-EED complex by stabilized α-helix of EZH2 (SAH-EZH2) peptides which inhibited H3K27me3 and reduced EZH2 protein levels. Furthermore, upon treatment with SAH-EZH2 PRC2 dependent MLL-AF9 leukemic cells were demonstrated to undergo growth arrest, monocyte-macrophage differentiation and changes in PRC2-regulated lineage-specific marker genes.
Recent studies highlighted some natural chemopreventive phytochemicals such as green tea polyphenols and curcumin as potential EZH2 inhibitors which can reactivate PRC2 silenced target genes in various cancer types. Choudhury et al. showed that epigallocatechin-3-gallate (EGCG), a major constituent of green tea and DZNep, alone or in combination, reduced PcG protein levels including EZH2 by increased ubiquitylation and proteasomal-associated degradation (129). Consistently, there was decreased H3K27me3 levels and upregulation of tumor suppressor gene expression. Hua et al. demonstrated that curcumin, a natural phytochemical present in turmeric, down-regulate EZH2 levels by modulating mitogen-activated protein kinase (MAPK) pathway in MDA-MB-435 breast cancer cells (130). Curcumin treatment showed anti-proliferative effect and induced G1 arrest in MDA-MB-435 cells. Dietary omega-3(v-3) polyunsaturated fatty acids (PUFAs) were also shown to down-regulate EZH2 expression by enhancing its proteasomal degradation, which resulted in the re-activation of EZH2 regulated tumor suppressor genes such as E-cadherin and insulin-like growth factor binding protein (131).
Since EZH2 play key roles in normal functioning of the cells by establishing the repressive state of genes that function as developmental regulators and maintain stem cell pluripotency, alternative inhibition strategies are needed which focus on the regulatory network causing EZH2 deregulation in a specific cancer type. Direct EZH2 inhibition by small molecules targeting the enzyme active site may have detrimental effects on global methylation patterns in normal cells. For example, a recent study proposed that inhibition of CDK1/2 mediated threonine 350 phosphorylation of EZH2 may emerge as an alternative therapeutic option to diminish EZH2 activity in cancer cells without affecting the global EZH2 mediated gene silencing (22). Further experimental studies may also focus on the development of ‘combined treatment strategies’ targeting the epigenetic regulatory network involving EZH2 in tumor cells.
CONCLUSION AND FUTURE PERSPECTIVES
Based on numerous in vivo and in vitro experimental evidences, it is now well established that EZH2 function as a key mediator of tumorigenesis and its over-expression is associated with invasive growth, tumor aggressiveness and poor clinical outcomes in hematological as well as epithelial cancers. Traditionally, the oncogenic role of EZH2 depends on its ability to act as a ‘transcriptional repressor’ which causes H3K27me3 based silencing of critical tumor suppressor genes and contribute to tumorigenesis. However, recent studies in hormone-refractory breast cancer and CRPC demonstrated that EZH2 is a multifaceted molecule which could switch to ‘transcriptional activator’ function and act independent of PRC2 complex on non-histone substrates. As discussed in previous sections EZH2 can be regulated through multiple mechanisms depending on cellular context and cancer type. Together, these evidences warrant the need for identification of EZH2 target genes, which are either activated or repressed, in specific cancer type so that effective therapeutic strategies could be developed. There is a scope for development of more specific EZH2 inhibitors which are highly selective and exhibit less toxicity in normal cells. As suggested by Chang et al. hypoxia inducible factor (HIF) inhibitors, which showed marked decrease in tumor growth and has been used in clinical trials, may prove effective in EZH2 inhibition because HIF1α regulates EZH2 expression in breast tumor initiating cells (75). Also, development of CDK1/2 which control EZH2 phosphorylation may emerge as an alternative treatment strategy. Notably, administration of EZH2 inhibitors by a cancer cell or tumor specific delivery mechanism may also reduce the unwanted side-effects and toxicity to the normal or stem cells. In conclusion, understanding the complex epigenetic regulatory network controlling EZH2 expression and target genes in tumor cells may help design better therapeutic intervention strategies.
Acknowledgments
This work was partially supported by funds from the United States Public Health Service Grants RO1CA115491, RO1CA108512 and R21109424 to SG. We acknowledge Shyama Prasad Mukherjee (SPM) fellowship provided to GD by the Council of Scientific and Industrial Research (CSIR), India and Fulbright-Nehru Doctoral and Professional Research fellowship provided by United States – India Educational Foundation (USIEF) for her work in the United States.
Abbreviations
- PcG
Polycomb group protein
- EZH1/2
Enhancer of zeste homolog 1/2
- H3K27me3
Histone H3 lysine 27 trimethylation
- H2AK119ub
Mono-ubiquitination of Histone 2A at Lys 119
- PRC2
Polycomb Repressive Complex 2
- DNMTs
DNA methyltransferases
- HDACs
Histone deacetylases
- CRPC
Castration resistant prostate cancer
- PcG
Polycomb group protein
- HMTase
Histone methyltransferases
- CXC
Cysteine rich domain
- SET
Su(var)3–9 enhancer of zeste trithorax domain
- EED
Embryonic ectoderm development
- RbAp46/48
Retinoblastoma associated protein 46/48
- ncRBD
non-coding RNA binding domain and a DNA binding domain
- PREs
Polycomb response elements
- ATXR5
Arabidopsis trithorax related protein 5
- ncRNA
Long non-coding RNAs
- MeCP2
Methyl CpG binding protein 2
- methyl MBD1
CpG binding protein 1
- MCAM
Methylated CpG island microarrays
- AR
Androgen receptor
- ER
Estrogen receptor
- PIN
Prostatic intraepithelial neoplasia
- ADT
Androgen deprivation therapy
- SCLC
Small cell lung cancer
- NSCLC
Non small cell lung cancer
- BTICs
Breast tumor initiating cells
- HRE
HIF response element
- PI3K/Akt
Phosphoinositide-3-kinase/Akt
- NKTL
Natural killer/T-cell lymphoma
- MDS
Myelodysplastic syndromes
- AML
Acute myeloid leukemia
- TCC
Transitional cell carcinomas
- EOC
Epithelial ovarian cancer
- SCC
squamous cell carcinoma
- AK
Actinic keratosis
- CSC
Cancer stem cell
- FOXC1
Forkhead box transcription factor C1
- DZNep
3-Dezaneplanocin-A
- SAH
S-adenosyl-L-homocysteine
- SAM
S-Adenosyl methionine
- ESC
embryonic stem cell
- MAPK
mitogen activated protein kinase
- EGCG
epigallaocatechin-3-gallate
Footnotes
Disclosure: Authors disclose no financial or commercial conflict of interest.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews Genetics. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Sims RJ, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–20. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- 3.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development (Cambridge, England) 2009;136:3531–42. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 4.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 5.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 7.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell stem cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature reviews Cancer. 2006;6:846–56. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 9.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:2613–8. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 10.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 11.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Z, Xing X, Hu M, Zhang Y, Liu P, Chai J. Structural basis of EZH2 recognition by EED. Structure (London, England : 1993) 2007;15:1306–15. doi: 10.1016/j.str.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Molecular cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 14.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. The EMBO journal. 2004;23:4061–71. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic acids research. 2009;37:2940–50. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. The EMBO journal. 2007;26:4078–88. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, Torchia J, et al. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell stem cell. 2010;6:153–66. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian C, Zhou MM. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cellular and molecular life sciences : CMLS. 2006;63:2755–63. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs SA, Harp JM, Devarakonda S, Kim Y, Rastinejad F, Khorasanizadeh S. The active site of the SET domain is constructed on a knot. Nature structural biology. 2002;9:833–8. doi: 10.1038/nsb861. [DOI] [PubMed] [Google Scholar]
- 20.Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–9. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science (New York, NY) 2005;310:306–10. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 22.Chen S, Bohrer LR, Rai AN, Pan Y, Gan L, Zhou X, et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nature cell biology. 2010;12:1108–14. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Molecular cell. 2008;32:503–18. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. The Journal of biological chemistry. 2010;285:3341–50. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Molecular cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell stem cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Molecular cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 29.Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic acids research. 2010;38:4958–69. doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development (Cambridge, England) 2009;136:3131–41. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, et al. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nature structural & molecular biology. 2009;16:763–8. doi: 10.1038/nsmb.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 33.Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, et al. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes & development. 2004;18:2973–83. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nature cell biology. 2007;9:1428–35. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 36.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Molecular cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deb G, Thakur VS, Gupta S. Multifaceted role of EZH2 in breast and prostate tumorigenesis: Epigenetics and beyond. Epigenetics : official journal of the DNA Methylation Society. 2013;8:464–76. doi: 10.4161/epi.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi B, Liang J, Yang X, Wang Y, Zhao Y, Wu H, et al. Integration of estrogen and Wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Molecular and cellular biology. 2007;27:5105–19. doi: 10.1128/MCB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Molecular cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science (New York, NY) 2012;338:1465–9. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan J, Ng SB, Tay JL, Lin B, Koh TL, Tan J, et al. EZH2 overexpression in natural killer/T-cell lymphoma confers growth advantage independently of histone methyltransferase activity. Blood. 2013;121:4512–20. doi: 10.1182/blood-2012-08-450494. [DOI] [PubMed] [Google Scholar]
- 42.Jacob E, Hod-Dvorai R, Schif-Zuck S, Avni O. Unconventional association of the polycomb group proteins with cytokine genes in differentiated T helper cells. The Journal of biological chemistry. 2008;283:13471–81. doi: 10.1074/jbc.M709886200. [DOI] [PubMed] [Google Scholar]
- 43.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutation research. 2008;647:21–9. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 44.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 45.McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer research. 2007;67:5097–102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- 46.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nature genetics. 2007;39:237–42. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nature genetics. 2007;39:232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 48.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nature genetics. 2008;40:741–50. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 49.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & development. 2002;16:2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nature genetics. 1999;23:474–8. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 51.Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Molecular cell. 2011;42:330–41. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 52.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. The Journal of biological chemistry. 2011;286:7983–9. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu F, Li X, Wu L, Zhang Q, Yang R, Yang Y, et al. Overexpression of the EZH2, RING1 and BMI1 genes is common in myelodysplastic syndromes: relation to adverse epigenetic alteration and poor prognostic scoring. Annals of hematology. 2011;90:643–53. doi: 10.1007/s00277-010-1128-5. [DOI] [PubMed] [Google Scholar]
- 54.Paul TA, Bies J, Small D, Wolff L. Signatures of polycomb repression and reduced H3K4 trimethylation are associated with p15INK4b DNA methylation in AML. Blood. 2010;115:3098–108. doi: 10.1182/blood-2009-07-233858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cardoso C, Mignon C, Hetet G, Grandchamps B, Fontes M, Colleaux L. The human EZH2 gene: genomic organisation and revised mapping in 7q35 within the critical region for malignant myeloid disorders. European journal of human genetics : EJHG. 2000;8:174–80. doi: 10.1038/sj.ejhg.5200439. [DOI] [PubMed] [Google Scholar]
- 56.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nature genetics. 2010;42:665–7. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 57.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature genetics. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 58.Jankowska AM, Makishima H, Tiu RV, Szpurka H, Huang Y, Traina F, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118:3932–41. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature genetics. 2010;42:181–5. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schulz WA, Hoffmann MJ. Epigenetic mechanisms in the biology of prostate cancer. Seminars in cancer biology. 2009;19:172–80. doi: 10.1016/j.semcancer.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 61.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 62.Bohrer LR, Chen S, Hallstrom TC, Huang H. Androgens suppress EZH2 expression via retinoblastoma (RB) and p130-dependent pathways: a potential mechanism of androgen-refractory progression of prostate cancer. Endocrinology. 2010;151:5136–45. doi: 10.1210/en.2010-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao JC, Yu J, Runkle C, Wu L, Hu M, Wu D, et al. Cooperation between Polycomb and androgen receptor during oncogenic transformation. Genome research. 2012;22:322–31. doi: 10.1101/gr.131508.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saramaki OR, Tammela TL, Martikainen PM, Vessella RL, Visakorpi T. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes, chromosomes & cancer. 2006;45:639–45. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- 65.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science (New York, NY) 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kunderfranco P, Mello-Grand M, Cangemi R, Pellini S, Mensah A, Albertini V, et al. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PloS one. 2010;5:e10547. doi: 10.1371/journal.pone.0010547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669–83. doi: 10.18632/oncotarget.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salvatori B, Iosue I, Djodji Damas N, Mangiavacchi A, Chiaretti S, Messina M, et al. Critical Role of c-Myc in Acute Myeloid Leukemia Involving Direct Regulation of miR-26a and Histone Methyltransferase EZH2. Genes & cancer. 2011;2:585–92. doi: 10.1177/1947601911416357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:268–73. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 71.Gonzalez ME, Li X, Toy K, DuPrie M, Ventura AC, Banerjee M, et al. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28:843–53. doi: 10.1038/onc.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ding L, Kleer CG. Enhancer of Zeste 2 as a marker of preneoplastic progression in the breast. Cancer research. 2006;66:9352–5. doi: 10.1158/0008-5472.CAN-06-2384. [DOI] [PubMed] [Google Scholar]
- 73.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. The EMBO journal. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujii S, Tokita K, Wada N, Ito K, Yamauchi C, Ito Y, et al. MEK-ERK pathway regulates EZH2 overexpression in association with aggressive breast cancer subtypes. Oncogene. 2011;30:4118–28. doi: 10.1038/onc.2011.118. [DOI] [PubMed] [Google Scholar]
- 75.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gonzalez ME, DuPrie ML, Krueger H, Merajver SD, Ventura AC, Toy KA, et al. Histone methyltransferase EZH2 induces Akt-dependent genomic instability and BRCA1 inhibition in breast cancer. Cancer research. 2011;71:2360–70. doi: 10.1158/0008-5472.CAN-10-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer Journal international du cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 78.Sato T, Kaneda A, Tsuji S, Isagawa T, Yamamoto S, Fujita T, et al. PRC2 overexpression and PRC2-target gene repression relating to poorer prognosis in small cell lung cancer. Scientific reports. 2013;3:1911. doi: 10.1038/srep01911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kikuchi J, Kinoshita I, Shimizu Y, Kikuchi E, Konishi J, Oizumi S, et al. Distinctive expression of the polycomb group proteins Bmi1 polycomb ring finger oncogene and enhancer of zeste homolog 2 in nonsmall cell lung cancers and their clinical and clinicopathologic significance. Cancer. 2010;116:3015–24. doi: 10.1002/cncr.25128. [DOI] [PubMed] [Google Scholar]
- 80.Lv Y, Yuan C, Xiao X, Wang X, Ji X, Yu H, et al. The expression and significance of the enhancer of zeste homolog 2 in lung adenocarcinoma. Oncology reports. 2012;28:147–54. doi: 10.3892/or.2012.1787. [DOI] [PubMed] [Google Scholar]
- 81.Coe BP, Thu KL, Aviel-Ronen S, Vucic EA, Gazdar AF, Lam S, et al. Genomic Deregulation of the E2F/Rb Pathway Leads to Activation of the Oncogene EZH2 in Small Cell Lung Cancer. PloS one. 2013;8:e71670. doi: 10.1371/journal.pone.0071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huqun, Ishikawa R, Zhang J, Miyazawa H, Goto Y, Shimizu Y, et al. Enhancer of zeste homolog 2 is a novel prognostic biomarker in nonsmall cell lung cancer. Cancer. 2012;118:1599–606. doi: 10.1002/cncr.26441. [DOI] [PubMed] [Google Scholar]
- 83.Zhang H, Zhang H, Zhao M, Lv Z, Zhang X, Qin X, et al. MiR-138 Inhibits Tumor Growth Through Repression of EZH2 in Non-Small Cell Lung Cancer. Cellular Physiology and Biochemistry. 2013;31:56–65. doi: 10.1159/000343349. [DOI] [PubMed] [Google Scholar]
- 84.Cho HM, Jeon HS, Lee SY, Jeong KJ, Park SY, Lee HY, et al. microRNA-101 inhibits lung cancer invasion through the regulation of enhancer of zeste homolog 2. Experimental and therapeutic medicine. 2011;2:963–7. doi: 10.3892/etm.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hinz S, Kempkensteffen C, Christoph F, Hoffmann M, Krause H, Schrader M, et al. Expression of the polycomb group protein EZH2 and its relation to outcome in patients with urothelial carcinoma of the bladder. Journal of cancer research and clinical oncology. 2008;134:331–6. doi: 10.1007/s00432-007-0288-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, Albadine R, Magheli A, Guzzo TJ, Ball MW, Hinz S, et al. Increased EZH2 protein expression is associated with invasive urothelial carcinoma of the bladder. Urologic oncology. 2012;30:428–33. doi: 10.1016/j.urolonc.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 87.Weikert S, Christoph F, Kollermann J, Muller M, Schrader M, Miller K, et al. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. International journal of molecular medicine. 2005;16:349–53. [PubMed] [Google Scholar]
- 88.Raman JD, Mongan NP, Tickoo SK, Boorjian SA, Scherr DS, Gudas LJ. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:8570–6. doi: 10.1158/1078-0432.CCR-05-1047. [DOI] [PubMed] [Google Scholar]
- 89.Takawa M, Masuda K, Kunizaki M, Daigo Y, Takagi K, Iwai Y, et al. Validation of the histone methyltransferase EZH2 as a therapeutic target for various types of human cancer and as a prognostic marker. Cancer science. 2011;102:1298–305. doi: 10.1111/j.1349-7006.2011.01958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang YB, Niu HT, Chang JW, Dong GL, Ma XB. EZH2 silencing by RNA interference inhibits proliferation in bladder cancer cell lines. European journal of cancer care. 2011;20:106–12. doi: 10.1111/j.1365-2354.2009.01148.x. [DOI] [PubMed] [Google Scholar]
- 91.Feber A, Clark J, Goodwin G, Dodson AR, Smith PH, Fletcher A, et al. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene. 2004;23:1627–30. doi: 10.1038/sj.onc.1207274. [DOI] [PubMed] [Google Scholar]
- 92.Oeggerli M, Tomovska S, Schraml P, Calvano-Forte D, Schafroth S, Simon R, et al. E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene. 2004;23:5616–23. doi: 10.1038/sj.onc.1207749. [DOI] [PubMed] [Google Scholar]
- 93.Tang X, Milyavsky M, Shats I, Erez N, Goldfinger N, Rotter V. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene. 2004;23:5759–69. doi: 10.1038/sj.onc.1207706. [DOI] [PubMed] [Google Scholar]
- 94.Yamada A, Fujii S, Daiko H, Nishimura M, Chiba T, Ochiai A. Aberrant expression of EZH2 is associated with a poor outcome and P53 alteration in squamous cell carcinoma of the esophagus. International journal of oncology. 2011;38:345–53. doi: 10.3892/ijo.2010.868. [DOI] [PubMed] [Google Scholar]
- 95.Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang Z, et al. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. The FEBS journal. 2013;280:4531–8. doi: 10.1111/febs.12417. [DOI] [PubMed] [Google Scholar]
- 96.Li H, Cai Q, Godwin AK, Zhang R. Enhancer of zeste homolog 2 promotes the proliferation and invasion of epithelial ovarian cancer cells. Molecular cancer research : MCR. 2010;8:1610–8. doi: 10.1158/1541-7786.MCR-10-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rao ZY, Cai MY, Yang GF, He LR, Mai SJ, Hua WF, et al. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-beta1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis. 2010;31:1576–83. doi: 10.1093/carcin/bgq150. [DOI] [PubMed] [Google Scholar]
- 98.Hu S, Yu L, Li Z, Shen Y, Wang J, Cai J, et al. Overexpression of EZH2 contributes to acquired cisplatin resistance in ovarian cancer cells in vitro and in vivo. Cancer biology & therapy. 2010;10:788–95. doi: 10.4161/cbt.10.8.12913. [DOI] [PubMed] [Google Scholar]
- 99.Rizzo S, Hersey JM, Mellor P, Dai W, Santos-Silva A, Liber D, et al. Ovarian cancer stem cell-like side populations are enriched following chemotherapy and overexpress EZH2. Molecular cancer therapeutics. 2011;10:325–35. doi: 10.1158/1535-7163.MCT-10-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Garipov A, Li H, Bitler BG, Thapa RJ, Balachandran S, Zhang R. NF-YA underlies EZH2 upregulation and is essential for proliferation of human epithelial ovarian cancer cells. Molecular cancer research : MCR. 2013;11:360–9. doi: 10.1158/1541-7786.MCR-12-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Guo J, Cai J, Yu L, Tang H, Chen C, Wang Z. EZH2 regulates expression of p57 and contributes to progression of ovarian cancer in vitro and in vivo. Cancer science. 2011;102:530–9. doi: 10.1111/j.1349-7006.2010.01836.x. [DOI] [PubMed] [Google Scholar]
- 102.Lu C, Han HD, Mangala LS, Ali-Fehmi R, Newton CS, Ozbun L, et al. Regulation of tumor angiogenesis by EZH2. Cancer cell. 2010;18:185–97. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sharpless E, Chin L. The INK4a/ARF locus and melanoma. Oncogene. 2003;22:3092–8. doi: 10.1038/sj.onc.1206461. [DOI] [PubMed] [Google Scholar]
- 104.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 105.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bachmann IM, Halvorsen OJ, Collett K, Stefansson IM, Straume O, Haukaas SA, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24:268–73. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 107.McHugh JB, Fullen DR, Ma L, Kleer CG, Su LD. Expression of polycomb group protein EZH2 in nevi and melanoma. Journal of Cutaneous Ppathology. 2007;34:597–600. doi: 10.1111/j.1600-0560.2006.00678.x. [DOI] [PubMed] [Google Scholar]
- 108.Athanassiadou AM, Lazaris AC, Patsouris E, Tsipis A, Chelidonis G, Aroni K. Significance of cyclooxygenase 2, EZH-2 polycomb group and p53 expression in actinic keratosis and squamous cell carcinomas of the skin. The American Journal of Dermatopathology. 2013;35:425–31. doi: 10.1097/DAD.0b013e318271292a. [DOI] [PubMed] [Google Scholar]
- 109.Hou P, Liu D, Dong J, Xing M. The BRAF(V600E) causes widespread alterations in gene methylation in the genome of melanoma cells. Cell Cycle (Georgetown, Tex) 2012;11:286–95. doi: 10.4161/cc.11.2.18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fan T, Jiang S, Chung N, Alikhan A, Ni C, Lee CC, et al. EZH2-dependent suppression of a cellular senescence phenotype in melanoma cells by inhibition of p21/CDKN1A expression. Molecular cancer research: MCR. 2011;9:418–29. doi: 10.1158/1541-7786.MCR-10-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Luo C, Merz PR, Chen Y, Dickes E, Pscherer A, Schadendorf D, et al. MiR-101 inhibits melanoma cell invasion and proliferation by targeting MITF and EZH2. Cancer Letters. 2013;341:240–7. doi: 10.1016/j.canlet.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 112.Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, et al. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. Journal of Investigative Dermatology. 2013;133:768–75. doi: 10.1038/jid.2012.357. [DOI] [PubMed] [Google Scholar]
- 113.Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012;3:1011–25. doi: 10.18632/oncotarget.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Perera RJ, Ray A. Epigenetic regulation of miRNA genes and their role in human melanomas. Epigenomics. 2012;4:81–90. doi: 10.2217/epi.11.114. [DOI] [PubMed] [Google Scholar]
- 115.Zheng H, Gao L, Feng Y, Yuan L, Zhao H, Cornelius LA. Down-regulation of Rap1GAP via promoter hypermethylation promotes melanoma cell proliferation, survival, and migration. Cancer Research. 2009;69:449–57. doi: 10.1158/0008-5472.CAN-08-2399. [DOI] [PubMed] [Google Scholar]
- 116.Banerjee R, Russo N, Liu M, Van Tubergen E, D’Silva NJ. Rap1 and its regulatory proteins: the tumor suppressor, oncogene, tumor suppressor gene axis in head and neck cancer. Small GTPases. 2012;3:192–7. doi: 10.4161/sgtp.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gilbertson RJ, Graham TA. Cancer: Resolving the stem-cell debate. Nature. 2012;488:462–3. doi: 10.1038/nature11480. [DOI] [PubMed] [Google Scholar]
- 118.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 119.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer research. 2006;66:9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 120.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annual review of genetics. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 121.Schuebel K, Chen W, Baylin SB. CIMPle origin for promoter hypermethylation in colorectal cancer? Nature genetics. 2006;38:738–40. doi: 10.1038/ng0706-738. [DOI] [PubMed] [Google Scholar]
- 122.Crea F, Paolicchi E, Marquez VE, Danesi R. Polycomb genes and cancer: time for clinical application? Critical reviews in oncology/hematology. 2012;83:184–93. doi: 10.1016/j.critrevonc.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 123.Du J, Li L, Ou Z, Kong C, Zhang Y, Dong Z, et al. FOXC1, a target of polycomb, inhibits metastasis of breast cancer cells. Breast cancer research and treatment. 2012;131:65–73. doi: 10.1007/s10549-011-1396-3. [DOI] [PubMed] [Google Scholar]
- 124.Wu Z, Lee ST, Qiao Y, Li Z, Lee PL, Lee YJ, et al. Polycomb protein EZH2 regulates cancer cell fate decision in response to DNA damage. Cell death and differentiation. 2011;18:1771–9. doi: 10.1038/cdd.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes & development. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature chemical biology. 2012;8:890–6. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 127.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21360–5. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nature chemical biology. 2013;9:643–50. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Choudhury SR, Balasubramanian S, Chew YC, Han B, Marquez VE, Eckert RL. (−)-Epigallocatechin-3-gallate and DZNep reduce polycomb protein level via a proteasome-dependent mechanism in skin cancer cells. Carcinogenesis. 2011;32:1525–32. doi: 10.1093/carcin/bgr171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hua WF, Fu YS, Liao YJ, Xia WJ, Chen YC, Zeng YX, et al. Curcumin induces down-regulation of EZH2 expression through the MAPK pathway in MDA-MB-435 human breast cancer cells. European journal of pharmacology. 2010;637:16–21. doi: 10.1016/j.ejphar.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 131.Dimri M, Bommi PV, Sahasrabuddhe AA, Khandekar JD, Dimri GP. Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells. Carcinogenesis. 2010;31:489–95. doi: 10.1093/carcin/bgp305. [DOI] [PMC free article] [PubMed] [Google Scholar]