Abstract
Targeting amino acid metabolism has therapeutic implications for aggressive brain tumors. Asparagine is an amino acid that is synthesized by normal cells. However, some cancer cells lack asparagine synthetase (ASNS), the key enzyme for asparagine synthesis. Asparaginase (ASNase) contributes to eradication of acute leukemia by decreasing asparagine levels in serum and cerebrospinal fluid. However, leukemic cells may become ASNase-resistant by up-regulating ASNS. High expression of ASNS has also been associated with biological aggressiveness of other cancers, including gliomas. Here, the impact of enzymatic depletion of asparagine on proliferation of brain tumor cells was determined. ASNase was used as monotherapy or in combination with conventional chemotherapeutic agents. Viability assays for ASNase-treated cells demonstrated significant growth reduction in multiple cell lines. This effect was reversed by glutamine in a dose-dependent manner -- as expected, because glutamine is the main amino group donor for asparagine synthesis. ASNase treatment also reduced sphere formation by medulloblastoma and primary glioblastoma cells. ASNase-resistant glioblastoma cells exhibited elevated levels of ASNS mRNA. ASNase co-treatment significantly enhanced gemcitabine or etoposide cytotoxicity against glioblastoma cells. Xenograft tumors in vivo showed no significant response to ASNase monotherapy and little response to temozolomide (TMZ) alone. However, combinatorial therapy with ASNase and TMZ resulted in significant growth suppression for an extended duration of time. Taken together, these findings indicate that amino acid depletion warrants further investigation as adjunctive therapy for brain tumors.
Keywords: Asparaginase, deamination, glutamine, glioblastoma, medulloblastoma, temozolomide
Introduction
Targeting cancer metabolism has been safely and effectively used therapeutically since the 1940’s, when folate antagonists were used in patients with leukemia (1, 2). For treatment of acute lymphoblastic leukemia (ALL), enzymatic hydrolysis of asparagine (Asn) by asparaginase (ASNase) has received the most attention (3, 4). Asn depletion contributes to clinical eradication of leukemic cells, which are believed to be insufficient in de novo asparagine biosynthesis. In addition, ASNase treatment results in depletion of glutamine (5) and other amino acids (6) in pediatric ALL patients.
In contrast, there has been less investigation on targeting amino acid metabolism in brain tumors -- mostly pre-clinical studies targeting glutamine use (7), methionine restriction (8), and methionine depletion (9). Brain cells depend on glutamine for normal neurotransmitter function, via GABA ligand – receptor interactions (10). Anti-glutamine agents as monotherapy have been tested against brain tumors in clinical settings, with limited responses (11, 12). As another approach, antimetabolite regimens exploiting glutamine utilization might improve the sensitivity of some human gliomas and medulloblastomas to chemotherapeutic agents in vitro (13, 14). Results suggest that combining anti-metabolic approaches with conventional chemotherapy might increase clinical efficacy.
The blood-brain barrier (BBB) is one possible explanation for the relative difficulty in finding efficacious drugs for brain tumors. Thus, ability to bypass the BBB may provide a therapeutic advantage. Large enzymes cannot cross the BBB. However, when ASNase deaminates Asn in the serum, Asn levels decline in the cerebrospinal fluid (CSF) (15–17). Thus, enzymatic depletion of an essential substrate in serum -- followed by decline in CSF levels -- may represent a feasible concept for brain tumor treatment.
Leukemia cells may become ASNase resistant by up-regulation of asparagine synthetase (ASNS) (18), which helps restore depleted intracellular Asn levels. There is evidence that certain aggressive solid neoplasms may be associated with ASNS overexpression, such as ovarian (19), prostate (20), and pancreatic cancers (21). In this study, we demonstrate that enzymatic amino acid depletion may synergistically enhance brain tumor cell death caused by traditional DNA damaging agents. In addition, we look at molecular mechanisms by which synergism may be occurring between ASNase and temozolomide. These studies serve as a proof of principle for altering amino acid metabolism as a potential adjunct to brain tumor chemotherapy.
Materials and Methods
Cell Lines
We primarily used 4 brain tumor cell lines (3 human and one mouse): DAOY, a pediatric medulloblastoma line with moderate temozolomide sensitivity (22), inducible alkylator resistance (23), and a well established nude mouse xeno-transplant model (24); GBM-ES and U87, both p53 and PTEN null human glioblastoma lines with reported temozolomide and radiation resistance (25); and GL-261, a mouse glioma cell line for which an in vivo model was sensitive to a ketogenic diet (26) -- suggesting metabolic vulnerability. DAOY, GL-261, and U87 cell lines were all purchased from American Type Culture Collection (ATCC; Manassas, VA, USA) and verified according to their procedures. The GBM-ES, and 235- a pediatric medulloblastoma (27) cell line was generated from patient samples collected at UCLA (via an IRB approved protocol) and authenticated using morphology analysis and growth curve analysis. All cell lines tested negative for mycoplasma.
MTS cell proliferation assays for serum cultures
ASNase ± other chemotherapeutic agents were tested against medulloblastoma and glioblastoma cell lines, in vitro, by MTS assays (28). Analysis of cell viability was performed using the CellTiter 96® AQueous One Solution Cell Proliferation Assay system (Promega, Madison, WI). Cells were seeded at 2.5×103 cells per well in 96-well plates and incubated at 37 °C in a humidified chamber with 5% CO2 for 24 h before treatment. Twenty μL of CellTiter 96® AQueous One Solution reagent was added to each well 72 h after treatment with various drug combinations. Absorbance was recorded at 490 nm after 2 h. Background wells containing neither cells nor drug were used for subtraction and final absorbance calculations. Three wells per condition were used for calculating means and standard deviations for combinations of agents ± ASNase and for glutamine rescue experiments.
Analysis of ASNS mRNA expression by real time RT-PCR
ASNS expression was measured by RT-PCR in cell lines before and after treatment with ASNase ± temozolomide (21, 29). mRNA from a housekeeping gene (18S) was used as an internal standard for the total amount of cDNA. Total RNA was extracted from cultured cells (RNeasy Mini Kit, Qiagen, Hilden, Germany). One μg of total RNA was used for first strand cDNA synthesis, using a poly-T20 primer (Transcriptor First Strand cDNA Synthesis Kit, Roche, Basel, Switzerland). The synthesized cDNA was used as a template for RT-PCR (Fast SYBR Green I Master Mix, Applied Biosystems, Foster City, CA). Primers for cDNA amplification were: ASNS-forward, CTGCACGCCCTCTATGACA, ASNS-reverse, TAAAAGGCAGCCAATCCTTCT, 18S-forward, GGACAGGATTGACAGATTGATAGC, and 18S-reverse, TGGTTATCGGAATTAACCAGACAA.
Thermal cycling consisted of 45 cycles of: denaturation for 30 s at 90 °C; annealing for 30 s at 58 °C; and extension for 40 s at 72 °C. Relative amounts of target mRNA were calculated using the comparative cycle threshold (Ct) method by normalizing levels of target mRNA Ct to levels of 18S mRNA. Relative enhancements of ASNS mRNA (expressed as a fold increase) were compared after incubating cells under different conditions (e.g., ASNase and/or temozolomide for 24, 48, or 72 h).
Neurosphere assays for ASNase sensitivity
DAOY and GBM-ES cells were plated in neurosphere (NS) culture media at low density (400 cells per well in 96-well plates), immediately after enzymatic dissociation into single cell suspensions (27, 30, 31). A separate 96-well plate for each cell line was used for NS formation. Cells were treated with different concentrations of ASNase [0.0625 to 2 International Units/mL (IU/mL); 12 wells/concentration; 100 μL/well]. Each plate also included wells with untreated cells (controls). Neurosphere cultures were supplemented weekly with an EGF/FGF + heparin mixture in 10 μL of fresh media (30). Plates were monitored closely for formation of neurospheres, which were counted after a density of 10–20 NS/well was achieved for untreated controls. This assay evaluated concentration-dependent reduction of presumably clonal neurosphere numbers.
Amino acid analysis
Serum samples from 3 mice and cell supernatants from 3 cell lines, along with control media, were sent to the Molecular Structure Facility at the University of California, Davis, for amino acid analysis (overnight on dry ice). Sample processing and amino acid levels were as described [http://msf.ucdavis.edu/aaa.html; (32)]. Briefly, an L-8900 Hitachi amino acid analyzer was used, and amino acids were quantified utilizing a lithium citrate buffer system. The analyzer uses ion-exchange chromatography to separate amino acids and a "post-column" ninhydrin reaction for detection -- sensitive to the pmol level (~100 pmol). Physiological samples were acidified with sulfosalicyclic acid to remove any intact proteins prior to analysis.
Mouse experiments
All mouse experiments complied with guidelines of the Los Angeles Biomedical Research Institute IACUC. We used SCID mice with heterotopic subcutaneous xenografts with DAOY cells [(24); Jackson Laboratory, Bar Harbor, ME; all males, 6 weeks old at the time of tumor transplantation]. Tumor cells (1x107) were injected under the skin in the right flank. Three weeks after transplantation, animals were treated according to groupings below (5 animals per group):
Untreated (DMSO only -- 10%, 50 μL, 5 days/week for 2 weeks),
ASNase only (2 IU/g Mondays, Wednesdays, Fridays for 2 weeks),
Temozolomide only (0.05 mg/g, 5 days/week for 2 weeks) and
ASNase and temozolomide at the same doses and for the same duration as above.
Tumors were measured with digital calipers 3 times a week. Animals were euthanized when tumors reached 1.5 cm in length. An ellipsoid volume formula was used to calculate tumor volumes (1/2 × length × width2) (33). Mice were weighed 2–3 times a week throughout the experiment.
Asparaginases and cell culture media
All experiments used E. coli asparaginase. For in vitro asparaginase sensitivity and combination experiments, we used DMEM medium with 862 mg/L of GlutaMAX (Gibco, Life Technologies, Grand Island, NY) and 10% fetal bovine serum. Treatment was with Elspar® asparaginase (Lundbeck Inc., Deerfield, IL). For neurosphere cultures and assays, DMEM/F12 media (with 365 mg/L of L-glutamine and 7.5 mg/L of L-asparagine; Gemini Bio-Products, Sacramento, CA) was used with a one-time addition of L-glutamine. For glutamine rescue (at 1 and 10 mM) and ASNS PCR related experiments, DMEM without L-asparagine or L-glutamine (Cellgro, Manassas, VA) was used, supplemented with 10% fetal bovine serum. ASNase was from Sigma-Aldrich (St. Louis, MO; for animal treatment) or ProSpec (Ness-Ziona, Israel; for ASNS PCR experiments). Temozolomide (CGeneTech, Indianapolis, IN) was dissolved in DMSO. The same amounts of DMSO (1%) were also used for control and ASNase treatment groups (without temozolomide).
Statistical methods
Graph pad PRISM ® 6.0 statistical software and Excel programs were used to plot and analyze data. For most in vitro effects, absorbance readings of untreated (i.e., vehicle treated) cells were used for control values. Reductions in cell growth are expressed as a percent of control. For glutamine rescue experiments, absorbance readings are shown as absolute numbers. For neurosphere assays, absolute numbers of neurospheres in untreated wells were used as 100% levels. Numbers representing treated neurospheres are reported as a percent of control.
The paired 2-tailed Student t-test was used to compare in vitro growth with DNA-damaging agents alone versus ASNase combined with these agents.
For statistical analyses of our in vivo mouse experiments, we used 2-way repeated measures Analysis of Variance (ANOVA) to compare tumor volume changes for the 2 drugs (ASNase and temozolomide). The 2 drugs were considered between-subject factors, whereas time (i.e., date) was considered a within-subject factor. A 2-way interaction between the 2 drugs, and a 3-way interaction between the 2 drugs and time, were included in the model, to test for possible synergy between the 2 drugs (34). Differences in least square means (LS-means) were computed and tested among all 4 pairs at a given time. The Tukey method was used for multiple comparison adjustments for the P-values and confidence intervals for the differences between LS-means. A residual analysis was conducted to check model assumptions. Log transformations were applied to tumor volumes for better model fitting, and to satisfy the normality assumption for ANOVA. The in vivo data analyses were done using SAS 9.3 (Cary, NC, USA). P-values <0.05 were used as the cut-off for statistical significance. The Chou-Talalay method was considered to be unfeasible for the analysis, because its main prerequisites are dose-effect curves for each drug alone and for the combination. Large sample sizes would be required to determine ED50 values in vivo. However, the nearly complete lack of response to ASNase monotherapy in mice makes generation of such dose-response curves impractical.
Relative expression levels of ASNS mRNA measured by RT-PCR were also compared to values for untreated controls. All data were corrected for differences in total mRNA by use of the 18S mRNA internal control.
Results
Asparaginase activity against brain tumor cells in vitro
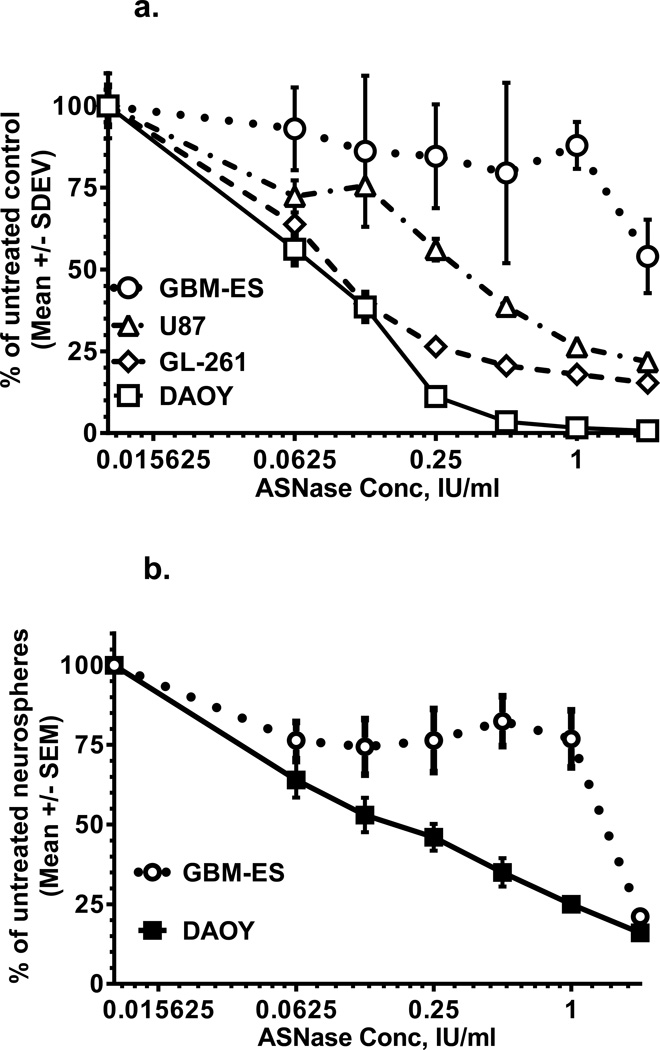
ASNase treatment in vitro resulted in dose-dependent and variable growth inhibition of 4 brain tumor cell lines, as measured by MTS assays. DAOY medulloblastoma cells were the most sensitive to ASNase, compared to the human GBM and mouse glioma lines (Fig. 1a).
Figure 1.
a. Asparaginase effects on 4 brain tumor cell lines measured by MTS assays. Medulloblastoma cells are the most sensitive, and GBM-ES cells are the most resistant.
b. Asparaginase effects on neurosphere formation are more pronounced in medulloblastoma cells compared to GBM-ES cells.
Neurosphere formation was used to evaluate in vitro inhibitory effects of ASNase monotherapy on putative stem-like brain tumor cells. We found reductions in neurosphere numbers for cells treated with different doses of ASNase (Fig. 1b). Again, DAOY medulloblastoma cells had better ASNase-sensitivity, compared to GBM-ES glioblastoma cells.
Reversal of ASNase effects by glutamine in vitro
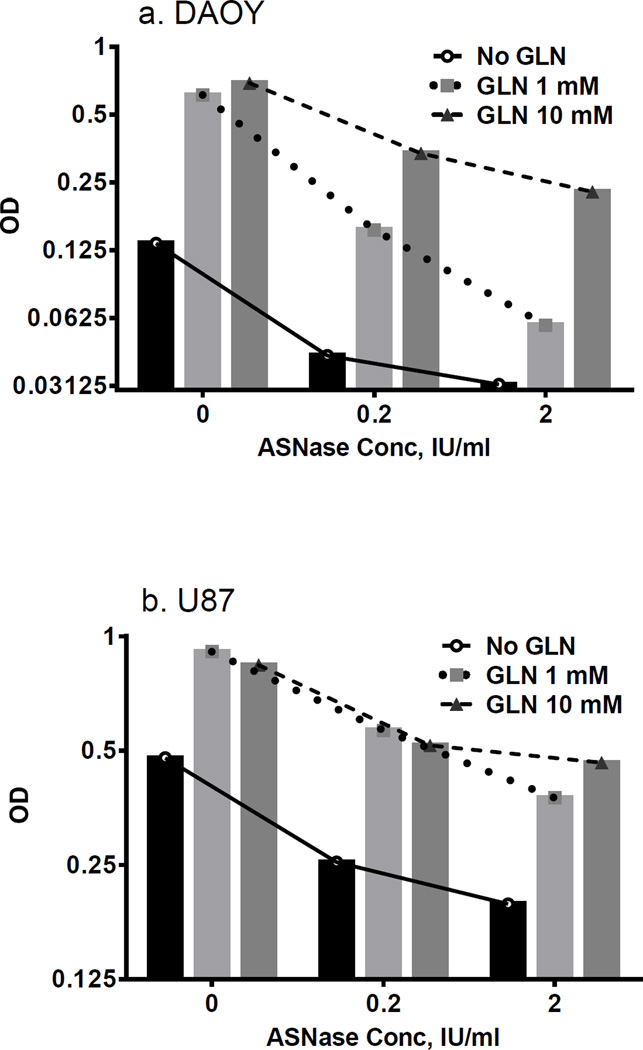
Glutamine, the main amino donor for ASNS, can enhance intrinsic asparagine synthesis and counteract ASNase effects. To test this hypothesis, we conducted “glutamine rescue” experiments, in which cells were cultured without or with glutamine (at 2 concentrations). Cells were grown with or without ASNase. Glutamine stimulated cell growth in untreated cells. Moreover, dose-dependent reversal of ASNase-induced cell inhibition was observed in medulloblastoma and GBM cell lines (Figs. 2a and 2b). Dose-dependency of glutamine rescue was more evident in DAOY cells, compared to U87.
Figure 2.
Glutamine addition rescues cells treated with ASNase. Dose-dependency of the glutamine rescue effect is pronounced in more ASNase-sensitive DAOY medulloblastoma cells (a), compared to relatively resistant U87 glioblastoma cells (b).
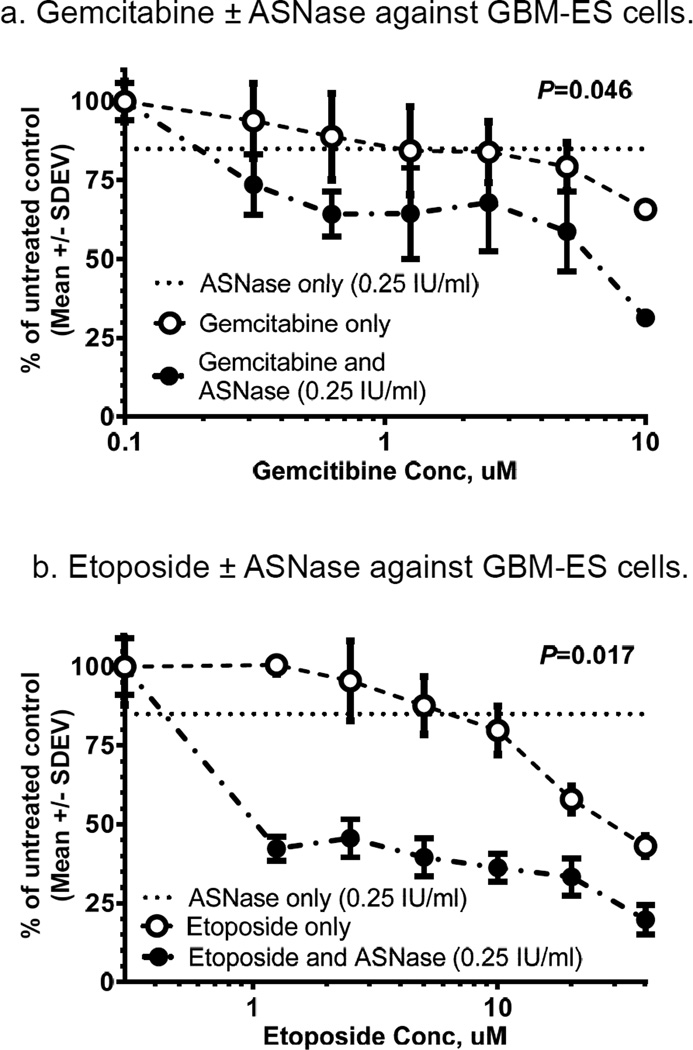
Asparaginase co-treatment enhances cytotoxicity of DNA damaging agents in vitro
We tested whether co-treatment with asparaginase potentiates cytotoxicity of agents that damage DNA or affect DNA synthesis or repair. GBM-ES cells were used, because they were the most resistant to ASNase, of the 4 lines (Fig. 1a) -- and thus most likely to reveal additive or synergistic effects. Co-treatment of GBM-ES cells with gemcitabine or etoposide (VP-16) with ASNase (0.25 IU/mL) resulted in significantly more cell reduction (20–50% as measured by MTS), compared to treatment with gemcitabine or VP-16 alone (Figs. 3a and 3b). ASNase alone (0.25 IU/mL) caused GBM-ES growth reductions of only ~15%. Dose responses for GBM-ES cells were augmented by approximately 20% when ASNase was added to gemcitabine (Fig. 3a). Considering that 0.25 IU/mL of ASNase alone induced 15% cell reduction, the result for the combination is more consistent with an additive effect, with P=0.046 (t-test). In contrast, addition of ASNase to etoposide enhanced cell reduction up to 50% or more, with P=0.017 (Fig. 3b), more suggestive of synergistic interaction.
Figure 3.
In vitro treatment of GBM-ES cells with combinations of gemcitabine ± ASNase (a) and etoposide ± ASNase (b). Both show significant augmentation of cytotoxic effects against GBM-ES by addition of ASNase.
Amino acids pre- and post-ASNase treatment
Serum amino acid levels were measured in 3 SCID mice. Two were treated with intraperitoneal ASNase (2 IU/g of body weight), and the other was a control. Retro-orbital blood samples were obtained 24 and 72 h after ASNase doses. Results showed sustained deamination of Asn 24 and 72 h after ASNase doses. Specifically, pre-treatment serum Asn levels were 37.4 nmol/mL, compared to 0 nmol/mL after 24 and 72 h. In vivo Gln levels were in the 1,000 nmol/mL range and did not decrease 24 or 72 h post-ASNase in mouse serum samples. However, glutamic acid levels doubled to 125 nmol/ml 24 h post-ASNase, returning to 68 nmol/ml at 72 h (comparable to baseline). On the other hand, aspartic acid levels were 30.9 nmol/mL before treatment, compared to 53 nmol/mL after 24 and 72 h -- compatible with more deamination of Asn to aspartic acid than for glutamine to glutamic acid.
Glutamine measurements in cell supernatants from 3 lines consistently demonstrated ≥80% glutamine deamination after 24 h of treatment with 0.5 IU/mL of ASNase, whereas aspartic and glutamic acids increased 2 and 25-fold, respectively (Supplemental Fig. 1).
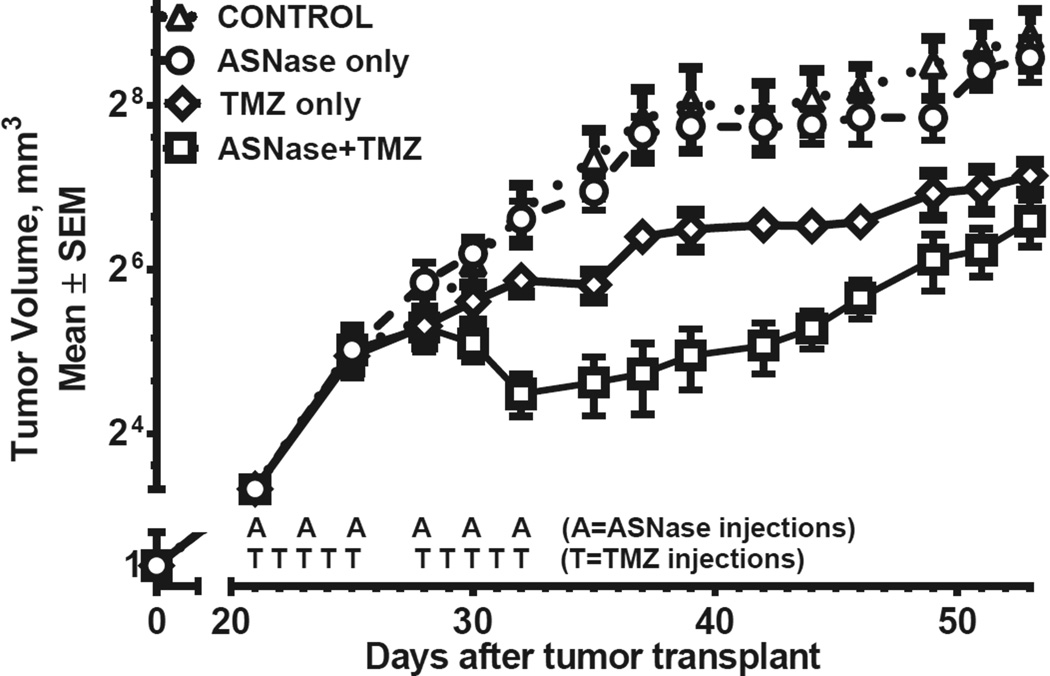
Possible synergistic effects of ASNase and temozolomide in combination in vivo (Fig. 4)
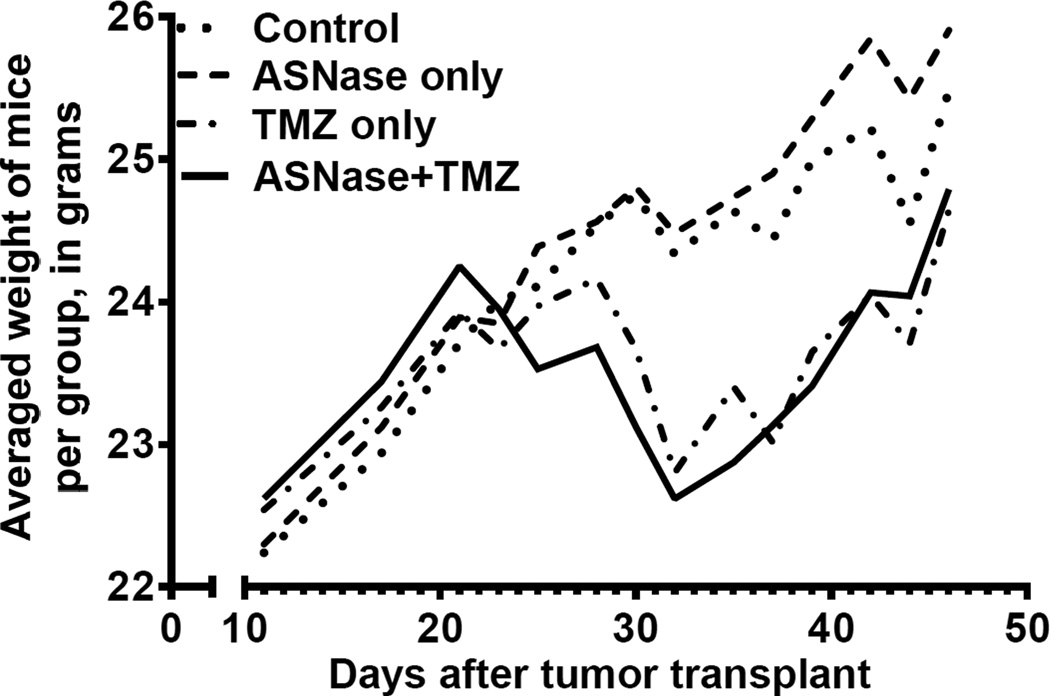
Figure 4.
In vivo tumor growth in 4 groups of animals treated with vehicle only, ASNase only, temozolomide only, or combination therapy. Each data point represents measurements of 5 tumors in SCID mice (20 mice total). Each line represents tumor growth kinetics in 5 mice. Addition of ASNase to temozolomide significantly suppressed tumor growth (P = 0.035; repeated measure ANOVA, Table 1). Note the tumor growth accelerations after cessation of therapy.
Tumor growth for controls and animals treated with ASNase alone were identical (P = 1, Table 1). Temozolomide alone provided modest suppression of tumor growth, although significantly better than for controls or ASNase alone (p<0.05). However, more pronounced growth suppression occurred with the ASNase and temozolomide combination, compared to temozolomide alone (Fig. 4, Table 1; P = 0.035). Results suggest possible synergy between the 2 agents (P = 0.055 for the interaction). On average, tumors in the combination group were half the size of tumors in the temozolomide-only group. The effect started 10 days after initiation of therapy and lasted roughly 3 weeks, when curves gradually merged. Transient decreases in tumor volume with ASNase +TMZ treatment strongly suggest cell death with these agents, in addition to slowing of growth. (Cell death is also supported by near complete eradication of DAOY cells by ASNase observed in vitro, Fig. 1).
Table 1.
Repeated measures Analysis of Variance (ANOVA) for comparison of tumor volume changes among 4 groups.
| Compared animal groups (n=5 in each group) |
Difference in LS Means* |
Confidence interval |
Adjusted P - value |
|---|---|---|---|
| Control vs ASNase only | 0.027 | −0.63 – 0.69 | 0.999 |
| Control vs TMZ only | 0.7 | 0.04 – 1.36 | 0.036 |
| Control vs ASNase and TMZ | 1.4 | 0.74 – 2.1 | 0.0001 |
| ASNase only vs TMZ only | 0.67 | 0.01 – 1.34 | 0.046 |
| ASNase only vs ASNase and TMZ | 1.38 | 0.7 – 2.04 | 0.0001 |
| TMZ Only vs ASNase and TMZ | 0.7 | 0.04 – 1.36 | 0.035 |
Least Square Means on a logarithmic scale
Weights of mice in relation to ASNase treatment
Animals were weighed at the time of tumor measurements and drug administration (Fig. 5). There were no significant weight differences with ASNase alone, compared to controls. There was 5–10% weight loss with temozolomide. However, addition of ASNase did not change weights significantly.
Figure 5.
Body weight changes in 4 treatment groups. Weight loss occurred mostly in temozolomide treated animals. Weight gain resumed after cessation of therapy.
ASNS mRNA expression in relation to ASNase sensitivity in vitro
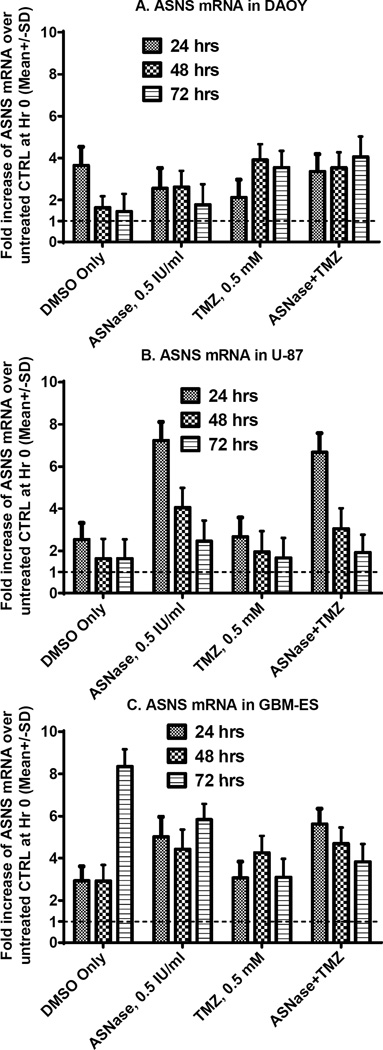
ASNS expression was measured by RT-PCR in 3 human brain tumor cell lines, at baseline and after 4 different treatments (24, 48, and 72 h following treatment; Fig. 6). ASNS mRNA expression increased in all 3 cell lines through 24–72 h (Fig. 6). However, less up-regulation of ASNS occurred in DAOY-medulloblastoma cells (which are the most sensitive to ASNase), compared to U87 and GBM-ES glioblastoma lines (Fig. 6).
Figure 6.
ASNS mRNA measured by RT-PCR in 3 human cell lines at different time points after exposure to different treatments. At 24 h of ASNase exposure, sensitive medulloblastoma cells have lower over-expression of ASNS mRNA (although ASNS mRNA production is increased over baseline), compared to cells exposed only to the vehicle (DMSO). In contrast, noticeable increases in ASNS (compared to levels in untreated cells) were observed in more resistant glioblastoma cell lines at 24 h after ASNase ±TMZ exposure. Temozolomide treatment alone induces comparable or less ASNS mRNA expression in glioblastoma lines, compared to treatment with ASNase alone.
Temozolomide appeared to induce similar (or less) ASNS mRNA over-expression in glioblastoma lines, compared to ASNase (Fig. 6). Temozolomide combined with ASNase resulted in ASNS up-regulation patterns similar to those seen for temozolomide monotherapy in medulloblastoma, and for ASNase monotherapy in glioblastoma cells.
Discussion
Asparagine in various cancers
Targeting cancer metabolism for therapeutic purposes has been one of the cornerstones of successful ALL treatment (1, 3), but the clinical role of the anti-metabolite approach for treatment of solid tumors is less established (2). Traditionally, asparagine metabolism has been considered a target for certain hematological malignancies (3). However, recent research shows that up-regulated asparagine synthetase is also associated with biological aggressiveness of ovarian (19), prostate (20) and pancreatic (21) cancers. Therefore, enzymatic amino acid depletion could conceivably become useful for a variety of malignancies (35).
Amino acid metabolism in brain tumors
In an earlier study of free amino acids in the CSF, there were lower levels of asparagine, valine, and leucine among patients with glioblastomas and low grade gliomas, compared to controls (36). Recently it was shown that certain GBMs express high levels of the enzyme that initiates catabolism of branched-chain amino acids (valine, leucine, and isoleucine), making their metabolism a possible target for GBM treatment (37). Similarly, deprivation of certain other amino acids may sensitize brain tumor cells to chemotherapy, as supported by our in vitro and in vivo data for Asn.
Interestingly, glutamine reversed growth inhibition by ASNase in our in vitro experiments. Glutamine plays a crucial role in the metabolism of neoplastic cells, including glioma cells (14). Cell rescue from ASNase by glutamine is expected, because abundant glutamine may augment de novo asparagine biosynthesis by serving as a main amino group donor for ASNS (4). Transfection of a human glioblastoma cell line with liver-type glutaminase down-regulates the DNA-repair gene, MGMT, and sensitizes cells to alkylating agents (38). It is possible that effects in our experiments might be partly explained by glutamine deamination, because ASNase also catalyzes the conversion of glutamine into glutamate (4, 5), especially in vitro (supplemental Fig. 1). Unchanged glutamine levels in mice may be partly responsible for lack of in vivo response to ASNase monotherapy (Fig. 4).
Combining ASNase with cytotoxic agents
ASNase co-treatment increased antitumor activity of various agents that interfere with DNA integrity and repair, in our in vitro GBM and in vivo medulloblastoma models. A hypothesized mechanism is temozolomide-induced DNA damage, which may limit the cell’s ability to produce sufficient ASNS to compensate for Asn depletion. However, our results do not directly support decreased ASNS mRNA expression induced by temozolomide (because temozolomide and ASNase were associated with similar ASNS mRNA expression in glioblastoma lines, compared to ASNase alone).
However, glioblastoma cells were capable of more sustained ASNS mRNA up-regulation (after 24 h of ASNase exposure) and were more resistant to both ASNase and cytotoxic monotherapy. In contrast, medulloblastoma cells had less ASNS mRNA over-expression (compared to untreated control cells at 24 h) and were more sensitive to ASNase. Efficacy of combination treatment against ASNase-resistant GBM-ES cells suggests that cytotoxics may sensitize cells to asparagine depletion. The drugs in these combinations were administered simultaneously. However, the sequence of drug administration may be relevant and should be investigated in future research.
Asparaginase and stem-like brain tumor cells
Targeting glycolysis in gliomas is another anti-metabolic approach that has been considered, but it may spare glioma stem cells (GSCs) (39). Thus, alternative anti-metabolic approaches against GSCs are needed, such as targeting amino acid metabolism. Our data confirm ASNase activity against cells transferred to neurosphere conditions, which mimic GSCs. Further in vitro and in vivo testing are needed to verify that ASNase (alone or combined with chemotherapy) can affect proliferation or survival of the putative stem-like population in brain tumors.
ASNase and temozolomide tolerability in mice
Asparaginase is not particularly myelosuppressive -- another desirable characteristic of amino acid depletion in combination therapy. Mouse weight data (as a crude measure of toxicity) tentatively support the tolerability of this combination in our model. However, use of ASNase in the treatment of leukemia has been reported to cause bleeding in the CNS, which might be a contraindication to the clinical use of this combination therapy. Further pre-clinical testing of intracranial models will allow careful monitoring for intracranial hemorrhage. In addition, fibrinogen levels may provide clinical information on this theoretical risk.
Amino acid depletion in CSF
ASNase has been clinically administered intramuscularly and intravenously. As mentioned above, ASNase does not cross the BBB. However, both routes are effective for leukemia therapy (16, 17). Therefore, Asn depletion in the CSF depends on depletion of Asn in the blood (16, 17). A disrupted BBB in brain tumors (40) may allow more penetration of enzymatic proteins into the CSF and tumor bed, with better therapeutic effects. The intrathecal route may also be an option for testing in pre-clinical models of GBM.
Enzymatic depletion of a substrate in the serum, with subsequent decreases in the CSF seems to be a feasible approach for brain tumor therapy development. One or more amino acids may be therapeutic targets for brain tumors, due to high amino acid uptake (41), which is independent of blood–brain barrier disruption (40). The uptake likely reflects high expression of amino acid transporters in gliomas (41) and serves as a basis for radiolabeled amino acid imaging for diagnosis or assessing treatment responses (42). Whether any particular amino acid has greater importance in brain tumors -- like Asn for ALL or perhaps arginine for hepatocellular carcinoma (43) -- remains to be seen. Lastly, small molecule inhibitors for ASNS that are being investigated for hematological malignancies (44) and similar anti-metabolic approaches (38) may have value in future research.
Conclusions
Pharmacological depletion of selected amino acids may serve as an adjunct to enhance cytotoxic therapy against brain tumors. An asparaginase and temozolomide combination was effective and well tolerated in our heterotopic medulloblastoma model. Design of further pre-clinical studies should include intracranial models and pharmacodynamic analysis of amino acids.
Supplementary Material
Implications.
Findings have potential impact for providing adjuvant means to enhance brain tumor chemotherapy.
Acknowledgments
Authors would like to thank the Molecular Structure Facility/Proteomics Core at the University of California, Davis, for amino acid analysis.
Financial Support: This project was supported by a seed grant from the Los Angeles Biomedical Research Institute (20673-01), a Scholars Award from the NIH/NCATS UCLA CTSI UL1TR000124, and NINDS grant NS052563.
Footnotes
Disclosure: No conflict of interest to disclose.
References
- 1.Farber S, Diamond LK, Mercer RD, Sylvester RF, Wolff JA. Temporary Remissions in Acute Leukemia in Children Produced by Folic Acid Antagonist, 4-Aminopteroyl-Glutamic Acid (Aminopterin) New England Journal of Medicine. 1948;238(23):787–793. doi: 10.1056/NEJM194806032382301. PubMed PMID: 18860765. [DOI] [PubMed] [Google Scholar]
- 2.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10(9):671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 3.Ertel IJ, Nesbit ME, Hammond D, Weiner J, Sather H. Effective Dose of l-Asparaginase for Induction of Remission in Previously Treated Children with Acute Lymphocytic Leukemia: A Report from Childrens Cancer Study Group. Cancer Research. 1979;39(10):3893–3896. [PubMed] [Google Scholar]
- 4.Avramis VI, Panosyan EH. Pharmacokinetic/Pharmacodynamic Relationships of Asparaginase Formulations: The Past, the Present and Recommendations for the Future. Clinical Pharmacokinetics. 2005;44(4):367–393. doi: 10.2165/00003088-200544040-00003. PubMed PMID: 16745802. [DOI] [PubMed] [Google Scholar]
- 5.Panosyan EHGR, Avramis IA, Seibel NL, Gaynon PS, Siegel SE, Fingert HJ, Avramis VI. Deamination of glutamine is a prerequisite for optimal asparagine deamination by asparaginases in vivo (CCG-1961) Anticancer Research. 2004;24(2C):5. [PubMed] [Google Scholar]
- 6.GRIGORYAN RS, PANOSYAN EH, SEIBEL NL, GAYNON PS, AVRAMIS IA, AVRAMIS VI. Changes of Amino Acid Serum Levels in Pediatric Patients with Higher-risk Acute Lymphoblastic Leukemia (CCG-1961) In Vivo. 2004;18(2):107–112. [PubMed] [Google Scholar]
- 7.Shelton LM, Huysentruyt LC, Seyfried TN. Glutamine targeting inhibits systemic metastasis in the VM-M3 murine tumor model. International Journal of Cancer. 2010;127(10):2478–2485. doi: 10.1002/ijc.25431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.NAJIM N, PODMORE ID, MCGOWN A, ESTLIN EJ. Methionine Restriction Reduces the Chemosensitivity of Central Nervous System Tumour Cell Lines. Anticancer Research. 2009;29(8):3103–3108. [PubMed] [Google Scholar]
- 9.Kokkinakis DM, Hoffman RM, Frenkel EP, Wick JB, Han Q, Xu M, et al. Synergy between Methionine Stress and Chemotherapy in the Treatment of Brain Tumor Xenografts in Athymic Mice. Cancer Research. 2001;61(10):4017–4023. [PubMed] [Google Scholar]
- 10.Albrecht J, Sidoryk-Węgrzynowicz M, Zielińska M, Aschner M. Roles of glutamine in neurotransmission. Neuron Glia Biology. 2010;6(04):263–276. doi: 10.1017/S1740925X11000093. [DOI] [PubMed] [Google Scholar]
- 11.Taylor SA, Crowley J, Pollock TW, Eyre HJ, Jaeckle C, Hynes HE, et al. Objective antitumor activity of acivicin in patients with recurrent CNS malignancies: a Southwest Oncology Group trial. Journal of Clinical Oncology. 1991;9(8):1476–1479. doi: 10.1200/JCO.1991.9.8.1476. [DOI] [PubMed] [Google Scholar]
- 12.Chang SM, Kuhn JG, Robins HI, Schold SC, Spence AM, Berger MS, et al. Phase II Study of Phenylacetate in Patients With Recurrent Malignant Glioma: A North American Brain Tumor Consortium Report. Journal of Clinical Oncology. 1999;17(3):984. doi: 10.1200/JCO.1999.17.3.984. [DOI] [PubMed] [Google Scholar]
- 13.Dranoff G, Elion GB, Friedman HS, Bigner DD. Combination Chemotherapy in Vitro Exploiting Glutamine Metabolism of Human Glioma and Medulloblastoma. Cancer Research. 1985;45(9):4082–4086. [PubMed] [Google Scholar]
- 14.Dranoff G, Elion GB, Friedman HS, Campbell GL, Bigner DD. Influence of Glutamine on the Growth of Human Glioma and Medulloblastoma in Culture. Cancer Research. 1985;45(9):4077–4081. [PubMed] [Google Scholar]
- 15.Hawkins DS, Park JR, Thomson BG, Felgenhauer JL, Holcenberg JS, Panosyan EH, et al. Asparaginase Pharmacokinetics After Intensive Polyethylene Glycol-Conjugated L-Asparaginase Therapy for Children with Relapsed Acute Lymphoblastic Leukemia. Clinical Cancer Research. 2004;10(16):5335–5341. doi: 10.1158/1078-0432.CCR-04-0222. [DOI] [PubMed] [Google Scholar]
- 16.Vieira Pinheiro JP, Wenner K, Escherich G, Lanvers-Kaminsky C, Würthwein G, Janka-Schaub G, et al. Serum asparaginase activities and asparagine concentrations in the cerebrospinal fluid after a single infusion of 2,500 IU/m2 PEG asparaginase in children with ALL treated according to protocol COALL-06-97. Pediatric Blood & Cancer. 2006;46(1):18–25. doi: 10.1002/pbc.20406. [DOI] [PubMed] [Google Scholar]
- 17.Panetta JCGA, Hijiya N, Hak LJ, Cheng C, Liu W, Pui CH, Relling MV. Comparison of native E. coli and PEG asparaginase pharmacokinetics and pharmacodynamics in pediatric acute lymphoblastic leukemia. Clin Pharmacol Ther. 2009;86(6):8. doi: 10.1038/clpt.2009.162. Epub 2009 Sep 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aslanian AMFB, Kilberg MS. Asparagine synthetase expression alone is sufficient to induce l-asparaginase resistance in MOLT-4 human leukaemia cells. Biochem J. 2001;357(Pt 1):8. doi: 10.1042/0264-6021:3570321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorenzi PL, Llamas J, Gunsior M, Ozbun L, Reinhold WC, Varma S, et al. Asparagine synthetase is a predictive biomarker of l-asparaginase activity in ovarian cancer cell lines. Molecular Cancer Therapeutics. 2008;7(10):3123–3128. doi: 10.1158/1535-7163.MCT-08-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sircar K, Huang H, Hu L, Cogdell D, Dhillon J, Tzelepi V, et al. Integrative Molecular Profiling Reveals Asparagine Synthetase Is a Target in Castration-Resistant Prostate Cancer. The American Journal of Pathology. 2012;180(3):895–903. doi: 10.1016/j.ajpath.2011.11.030. doi: http://dx.doi.org/10.1016/j.ajpath.2011.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui H, Darmanin S, Natsuisaka M, Kondo T, Asaka M, Shindoh M, et al. Enhanced Expression of Asparagine Synthetase under Glucose-Deprived Conditions Protects Pancreatic Cancer Cells from Apoptosis Induced by Glucose Deprivation and Cisplatin. Cancer Research. 2007;67(7):3345–3355. doi: 10.1158/0008-5472.CAN-06-2519. [DOI] [PubMed] [Google Scholar]
- 22.von Bueren AOBM, Hagel C, Heinimann K, Fedier A, Kordes U, Pietsch T, Koster J, Grotzer MA, Friedman HS, Marra G, Kool M, Rutkowski S. Mismatch repair deficiency: a temozolomide resistance factor in medulloblastoma cell lines that is uncommon in primary medulloblastoma tumours. Br J Cancer. 2012 Oct 9;107(8):1399–1408. doi: 10.1038/bjc.2012.403. Epub 2012 Sep 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman HS, Colvin OM, Kaufmann SH, Ludeman SM, Bullock N, Bigner DD, et al. Cyclophosphamide Resistance in Medulloblastoma. Cancer Research. 1992;52(19):5373–5378. [PubMed] [Google Scholar]
- 24.Jacobsen PF, Jenkyn DJ, JM P. Establishment of a human medulloblastoma cell line and its heterotransplantation into nude mice. J Neuropathol Exp Neurol. 1985;44(5):472–485. doi: 10.1097/00005072-198509000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Vlashi E, Mattes M, Lagadec C, Donna LD, Phillips TM, Nikolay P, et al. Differential Effects of the Proteasome Inhibitor NPI-0052 against Glioma Cells. Transl Oncol. 2010;3(1):50–55. doi: 10.1593/tlo.09244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stafford P, Abdelwahab MG, Kim do Y, Preul MC, Rho JM, AC S. The ketogenic diet reverses gene expression patterns and reduces reactive oxygen species levels when used as an adjuvant therapy for glioma. Nutr Metab (Lond) 2010;7:74. doi: 10.1186/1743-7075-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panosyan EH, Laks DR, Masterman-Smith M, Mottahedeh J, Yong WH, Cloughesy TF, et al. Clinical outcome in pediatric glial and embryonal brain tumors correlates with in vitro multi-passageable neurosphere formation. Pediatric Blood & Cancer. 2010;55(4):644–651. doi: 10.1002/pbc.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy JM, Thorburn A. Modulation of pediatric brain tumor autophagy and chemosensitivity. J Neurooncol. 2012;106(2):281–290. doi: 10.1007/s11060-011-0684-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irino T, Kitoh T, Koami K, Kashima T, Mukai K, Takeuchi E, et al. Establishment of Real-Time Polymerase Chain Reaction Method for Quantitative Analysis of Asparagine Synthetase Expression. The Journal of Molecular Diagnostics. 2004;6(3):217–224. doi: 10.1016/S1525-1578(10)60513-2. doi: http://dx.doi.org/10.1016/S1525-1578(10)60513-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visnyei K, Onodera H, Damoiseaux R, Saigusa K, Petrosyan S, De Vries D, et al. A Molecular Screening Approach to Identify and Characterize Inhibitors of Glioblastoma Stem Cells. Molecular Cancer Therapeutics. 2011;10(10):1818–1828. doi: 10.1158/1535-7163.MCT-11-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laks DR, Masterman-Smith M, Visnyei K, Angenieux B, Orozco NM, Foran I, et al. Neurosphere Formation Is an Independent Predictor of Clinical Outcome in Malignant Glioma. STEM CELLS. 2009;27(4):980–987. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper C, Packer N, Williams K. Methods in Molecular Biology. Humana Press; Totawa, NJ: 2000. Amino Acid Analysis Protocols; p. 159. [Google Scholar]
- 33.Reynolds CPSB, DeClerck YA, Moats RA. Assessing growth and response to therapy in murine tumor models. Methods Mol Med. 2005;111:335–350. doi: 10.1385/1-59259-889-7:335. 2005. [DOI] [PubMed] [Google Scholar]
- 34.Slinker BK. The Statistics of Synergism. Journal of Molecular and Cellular Cardiology. 1998;30(4):723–731. doi: 10.1006/jmcc.1998.0655. doi: http://dx.doi.org/10.1006/jmcc.1998.0655. [DOI] [PubMed] [Google Scholar]
- 35.Covini DTS, Bussolati O, Chiarelli LR, Pasquetto MV, Digilio R, Valentini G, Scotti C. Expanding targets for a metabolic therapy of cancer: L-asparaginase. Recent Pat Anticancer Drug Discov. 2012 Jan;7(1):4–13. doi: 10.2174/157489212798358001. [DOI] [PubMed] [Google Scholar]
- 36.Piek J, Adelt T, Huse K, WJ B. Cerebrospinal fluid and plasma aminograms in patients with primary and secondary tumors of the CNS. Infusionsther Klin Ernahr. 1987;14(2):73–77. [PubMed] [Google Scholar]
- 37.Tonjes M, Barbus S, Park YJ, Wang W, Schlotter M, Lindroth AM, et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat Med. 2013;19(7):901–908. doi: 10.1038/nm.3217. http://www.nature.com/nm/journal/v19/n7/abs/nm.3217.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szeliga M, Zgrzywa A, Obara-Michlewska M, Albrecht J. Transfection of a human glioblastoma cell line with liver-type glutaminase (LGA) down-regulates the expression of DNA-repair gene MGMT and sensitizes the cells to alkylating agents. Journal of Neurochemistry. 2012;123(3):428–436. doi: 10.1111/j.1471-4159.2012.07917.x. [DOI] [PubMed] [Google Scholar]
- 39.Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou M, et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proceedings of the National Academy of Sciences. 2011;108(38):16062–16067. doi: 10.1073/pnas.1106704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nduom EK, Yang C, Merrill MJ, Zhuang Z, Lonser RR. Characterization of the blood-brain barrier of metastatic and primary malignant neoplasms. Journal of Neurosurgery. 2013;119(2):427–433. doi: 10.3171/2013.3.JNS122226. PubMed PMID: 23621605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyagawa T, Oku T, Uehara H, Desai R, Beattie B, Tjuvajev J, et al. "Facilitated" Amino Acid Transport Is Upregulated in Brain Tumors. J Cereb Blood Flow Metab. 1998;18(5):500–509. doi: 10.1097/00004647-199805000-00005. [DOI] [PubMed] [Google Scholar]
- 42.Langen K-J, Tatsch K, Grosu A-L, Jacobs AH, Weckesser M, Sabri O. Diagnostics of Cerebral Gliomas With Radiolabeled Amino Acids. Dtsch Arztebl International. 2008;105(4):55–61. doi: 10.3238/arztebl.2008.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glazer ES, Piccirillo M, Albino V, Di Giacomo R, Palaia R, Mastro AA, et al. Phase II Study of Pegylated Arginine Deiminase for Nonresectable and Metastatic Hepatocellular Carcinoma. Journal of Clinical Oncology. 2010;28(13):2220–2226. doi: 10.1200/JCO.2009.26.7765. [DOI] [PubMed] [Google Scholar]
- 44.Richards NGJ, Kilberg MS. Annual Review of Biochemistry. Palo Alto: Annual Reviews; 2006. Asparagine synthetase chemotherapy; pp. 629–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.