Abstract
The novel antiretroviral 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) is a potent nucleoside HIV-1 reverse transcriptase (RT) inhibitor (NRTI). Unlike other FDA-approved NRTIs, EFdA contains a 3′-hydroxyl. Pre-steady-state kinetics showed RT preferred incorporating EFdA-TP over native dATP. Moreover, RT slowly inserted nucleotides past an EFdA-terminated primer, resulting in delayed chain termination with unaffected fidelity. This is distinct from KP1212, another 3′-hydroxyl-containing RT inhibitor considered to promote viral lethal mutagenesis. New mechanistic features of RT inhibition by EFdA are revealed.
Keywords: HIV, reverse transcriptase, enzyme kinetics, EFdA, polymerase
Nucleoside reverse transcriptase inhibitors (NRTIs) represent an important class of HIV-1 reverse transcriptase (RT) targeted therapy for treating HIV infection. All FDA-approved NRTIs lack a 3′-hydroxyl moiety that terminates DNA chain extension upon incorporation into the growing proviral DNA. Many NRTIs cause significant toxicity, often due to their interactions with the human mitochondrial DNA polymerase γ (pol γ) (Apostolova et al., 2011; Johnson et al., 2001; Nakata et al., 2007). A novel antiretroviral compound, 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) (Fig. 1), has shown potency several orders of magnitude superior to the NRTIs currently prescribed against HIV-1 (Nakata et al., 2007) and low toxicity (Ohrui et al., 2007; Zhang et al., 2013). We have shown that EFdA is highly favored by RT (Michailidis et al., 2009), and incorporation by pol γ is nearly negligible (Sohl et al., 2012b), indicating a favorable compromise of RT potency and low pol γ-mediated toxicity. EFdA is currently under clinical development by Merck & Co. with Yamasa Corporation, Chiba, Japan.
Fig. 1.
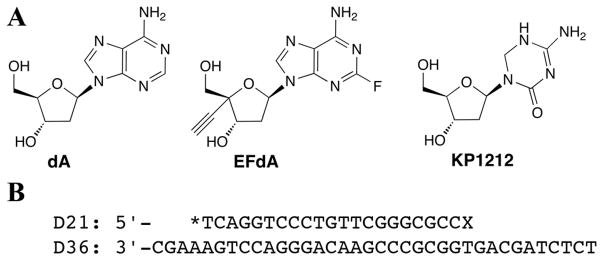
Reagents used in the kinetic studies. (A) Chemical structures of dA and and the dA analog EFdA. An additional RT inhibitor containing a 3′-hydroxyl group, the dC analog KP1212, is also shown. (B) The D21/D36 primer/template substrate used in the incorporation and extension experiments. The asterisk indicates the radiolabel at the 5′ primer end, and the “X” denotes the site of incorporation of the incoming EFdA-TP or dATP. For the extension past EFdA and phosphorolytic studies, a pre-incorprated EFdAMP present at the primer “X” site (primer-EFdAMP/template substrate) is used.
Despite the promise of EFdA, characterization of the molecular mechanism of inhibition of RT has been limited to steady-state kinetic studies (Michailidis et al., 2009; Michailidis et al., 2013; Nakata et al., 2007), which report only on the rate-limiting step (in this case, product release). Here we use pre-steady-state kinetic analysis to determine critical kinetic parameters such as the maximum rate of analog incorporation, kpol, the binding affinity of the incoming nucleotide or analog for the enzyme, Kd, and the efficiency of incorporation, kpol/Kd, combined with extension, fidelity, and ATP and pyrophosphate (PPi)-mediated resistance studies to understand the mechanism of EFdA inhibition of RT.
Single-turnover conditions with enzyme in excess of primer/template substrate were used to determine the kpol and Kd for single nucleotide or analog incorporation. A pre-incubated mixture of RT (purified as described previously) (Kerr and Anderson, 1997) and 5′-radiolabeled DNA primer/template substrate (Ray et al., 2002) were rapidly combined with excess magnesium chloride and varying concentrations of dATP or EFdA-TP (Fig. 1) using a RQF-3 rapid chemical quench (Kintek Instruments) for incubations ranging from 0.01 s to 3 s. Following reaction quenching with EDTA (0.34 M final), separation on a 20% polyacrylamide denaturing gel, and analysis via phosphorimaging (Bio-RAD Molecular Imager FX), plots of product formation versus time were generated and fit to single-exponential equations to determine the observed rates of polymerization (kobs) for varying concentrations of dATP or EFdA-TP (KaleidaGraph). Hyperbolic fits of kobs versus concentration plots yielded kpol and Kd values (Table 1).
Table 1.
Pre-steady-state rate constants for incorporation of EFdA-TP or dATP by RT and pol γ.a
| Enzyme | Nucleotide or analog | kpol (s−1) | Kd (μM) | Efficiencyb (μM−1 s−1) | Discriminationc |
|---|---|---|---|---|---|
| RT | dATP | 8.0 ± 0.7 | 3.8 ± 0.8 | 2.1 | 0.47 |
| EFdA-TP | 5.8 ± 0.3 | 1.3 ± 0.2 | 4.5 | ||
| pol γd | dATP | 220 ± 16 | 3.2 ± 0.7 | 69 | 4,300 |
| EFdA-TP | 0.29 ± 0.02 | 18 ± 4 | 0.016 |
Kinetic parameters determined from at least eight (RT), or at least seven (pol γ) single turnover experiments with varying dATP or EFdA-TP concentrations.
Efficiency = kpol/Kd
RT incorporated EFdA-TP over two-fold more efficiently than dATP (Table 1). To date, only two other RT inhibitors are preferentially inserted over the native nucleotide by RT: 2′,3′-didehydro-3′-deoxythymidine (d4T) (Vaccaro et al., 2000) and 2′,3′-didehydro-3′-deoxy-4′-ethynylthymidine (Ed4T) (Sohl et al., 2012a). Based on findings of high potency (Nakata et al., 2007; Ohrui et al., 2007) and our results indicating efficient RT incorporation, we propose low doses of EFdA can be effective in HIV patients, minimizing toxicity (Nakata et al., 2007). Additionally, while RT is extremely poor at discriminating EFdA-TP from dATP (Table 1), discrimination by pol γ is over 9,000-fold better than RT, indicating nearly negligible analog incorporation in the presence of native nucleotides. NMR studies suggest this difference in discrimination by pol γ relative to RT may come from the 4′-ethynyl and 3′-hydroxyl moieties of EFdA (Fig. 1A) which force the sugar into a Northern puckering conformation. This Northern conformation is preferred by RT for nucleotide insertion, while Southern puckering is favored by pol γ (Kirby et al., 2013; Kirby et al., 2011). In summary, these discrimination findings support the high clinical potential of EFdA in that negligible incorporation by pol γ should lessen mitochondria-based toxicity.
EFdA is not the first proposed RT inhibitor to contain a 3′-hydroxyl group; a second RT inhibitor under development, KP1212 (Harris et al., 2005), also contains this moiety (Fig. 1A). In contrast to EFdA, RT incorporated KP1212 14-fold less efficiently than dCTP (Murakami et al., 2005). Human polymerase selectivity for KP1212 was poorer as well when compared to EFdA; pol γ incorporated KP1212 26-fold less efficiently than dCTP, which is less than a two-fold difference compared to RT discrimination (Murakami et al., 2005). Thus we propose the 4′-ethynyl group imparts more influence than the 3′hydroxyl in contributing to the very high level discrimination by pol γ and highly efficient incorporation by RT.
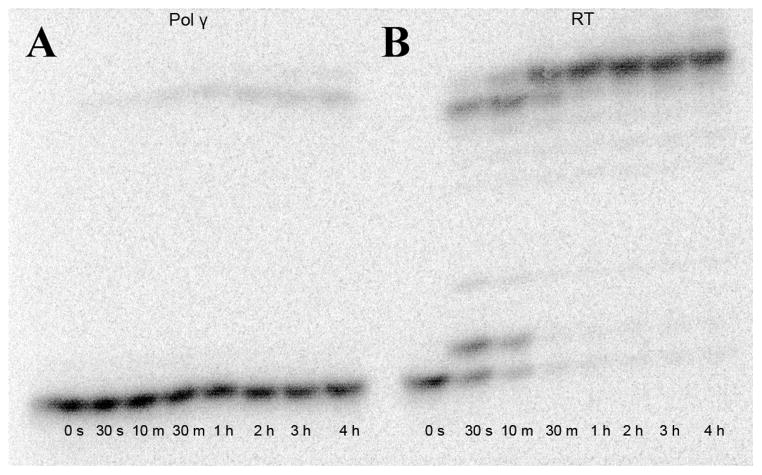
In addition to changes in incorporation efficiencies, the antiviral inhibitors containing 3′-hydroxyl groups have other unique mechanisms of action. KP-1212 propagates error-prone G to A and A to G substitutions leading to lethal viral mutagenesis (Murakami et al., 2005; Mullins et al., 2011). Entecavir, a 2′-deoxyguanosine analog used for treating hepatitis B, also supports additional primer elongation by HIV RT and has been described as a delayed chain-terminating inhibitor (Domaoal et al., 2008; Tchesnokov et al., 2008). Thus we hypothesized EFdA may also support primer elongation. To probe this, RT or exonuclease-deficient pol γ (purified as described previously) (Sohl et al., 2012a) and the DNA primer-EFdAMP/template substrate (a primer containing a pre-incorporated EFdA monophosphate (Fig. 1B), prepared as described previously (Sohl et al., 2012a)) were incubated with a mixture of dATP, dCTP, TTP, and dGTP (30 μM each) and excess magnesium chloride. Both pol γ (Fig. 2A) and RT (Fig. 2B) completed full extension (past EFdA) of the primer-EFdAMP/template substrate. However, pol γ could only extend a small amount of the primer-EFdAMP/template substrate (12% of substrate converted to product) over the course of the assay, while RT extended 88%. Remarkably, after only 30 s, RT extended 51% of the primer-EFdAMP/template, and extension was essentially complete within 1 h (Fig. 2B). By quantifying the amount of fully extended product over time, an observed rate of complete template extension, kmax, of 0.0014 ± 0.0002 s−1 was measured for RT, while the small amount of product formed by pol γ generated a kmax = 0.00024 ± 0.00004 s−1.
Fig 2.
Native nucleotide extension past pre-incorporated EFdA. (A) Extension by pol γ. The rate of formation of the final extended product, kmax, was 0.00024 ± 0.00004 s−1, with 12% of the substrate turned over to product. (B) Extension by RT. The value for kmax was 0.0014 ± 0.0002 s−1, with 88% of the substrate turned over to product. For both enzymes, the lanes represent 0 s, 30 s, 10 m, 30 m, 1 h, 2 h, 3 h, and 4 h. Experiments required 100 nM enzyme, 30 uM of each of the 4 dNTPs, and 25 nM of the primer-EFdAMP/template substrate.
The rate of incorporation of a single correct nucleotide (dCTP) past EFdA in the primer-EFdAMP/template was measured for RT and pol γ (Table 2). The kpol for RT was at least 30-fold higher than for pol γ; pol γ incorporated dCTP past EFdA so inefficiently that only an upper limit for kpol could be reliably determined (Table 2). However, RT fully extended the primer in the presence of physiologically relevant concentrations of dNTPs in a timeframe of seconds to minutes, with a more distributive versus processive mechanism (as shown by laddering, Fig. 2B). This indicates that delayed chain termination is one mechanism of inhibition of RT by EFdA.
Table 2.
Pre-steady-state rate constants for post-EFdA incorporation of a single dCTP (the next correct nucleotide) by RT and pol γa.
| Enzyme | kpol (s−1) | Kd (μM) | Efficiencyb (μM−1 s−1) |
|---|---|---|---|
| RT | 0.0064 ±0.0004 | 1.9 ± 0.4 | 0.0034c |
| pol γ | ≤0.0002d | N. D. | N. D. |
Kinetic parameters determined from nine (RT) or seven (pol γ) single turnover experiments with varying dCTP concentrations.
N.D.: not determined.
Incorporation of dCTP after AMP in a similar primer/template has a measured efficiency of 1.5 μM−1 s−1 (Kim et al., 2012).
Incorporation of dCTP after AMP in a similar primer/template has a measured kpol of 72 ± 3 s−1 (Sohl et al., 2013).
The efficiency of incorporation of nucleotides past EFdA by RT is lower than that seen with the delayed chain terminator entecavir. A 7-fold drop in efficiency is seen for the incorporation of the next correct nucleotide past entecavir (Tchesnokov et al., 2008) versus the over 1,300-fold decrease for extension efficiency past EFdA by RT (Tables 1, 2). As the koff for DNA dissociation from RT is estimated to be 0.2 s−1 (Kellinger and Johnson, 2011), we expect that traditional chain termination, in addition to delayed chain termination, likely contributes to the overall mechanism of RT inhibition by EFdA. Our findings echo our previous work proposing chain termination via translocation inhibition that identified minor extension past EFdA at comparable time points (Michailidis et al., 2009). Given that nucleotide incorporation efficiency depends on the primer/template used (Iyidogan and Anderson, 2012), we used a biologically-relevant primer/template designed to mimic the polymerization binding site (PBS) for HIV RT. Our results describing delayed chain termination represent one important example of the effects EFdA can have on RT; additional mechanisms may be factors for effective RT inhibition as well.
Slowing extension past EFdA can affect RT using several mechanisms, including (1) extremely inefficient extension, which can promote stalling and dissociation resulting in a similar outcome as chain termination or (2) error-prone extension reminiscent of KP1212 (Murakami et al., 2005) promoting viral error catastrophe. Entecavir, while serving primarily as a delayed chain terminator, has also been shown to promote some error-prone extension (Domaoal et al., 2008). Efficiencies of incorporation (Table 2, vide supra) of dCTP, the next correct nucleotide past EFdA, decreased approximately 600-fold for RT, with a ≥ 106-fold decrease in kpol for pol γ when compared to the incorporation of dATP (Table 1, 2). In comparison, the incorporation efficiency for the next correct nucleotide after KP1212 decreased approximately 3.3-fold and 4.4-fold for RT and pol γ, respectively (Murakami et al., 2005). The nearly 200-fold difference in nucleotide incorporation efficiency by RT past KP1212 versus past EFdA indicates two unique inhibition mechanisms at work: while KP1212 functions by inducing mutagenesis, EFdA substantially slows DNA chain extension.
To determine if incorporation of EFdA alters the fidelity of chain extension, we assessed misincorporation of dATP or TTP (2 mM) opposite deoxyguanine in the primer-EFdAMP/template (25 nM) by RT (100 nM) in excess magnesium chloride. These misincorporation studies gave apparent kpol (kpol_app) values of 1.3 × 10−3 s−1 for T:G and A:G by RT, which is 4- to 1,000-fold slower than typical RT misincorporation rates (Feng and Anderson, 1999), and no product formation was observed for pol (same conditions, kpol_app < 4.6 × 10−5 s−1 for T:G and A:G). Overall efficiency for misincorporation by RT was extremely low due to weak binding of these nucleotide triphosphates. Thus we conclude that significantly slowing extension past EFdA is a primary mechanism of RT inhibition, rather than a loss of fidelity as seen with KP1212.
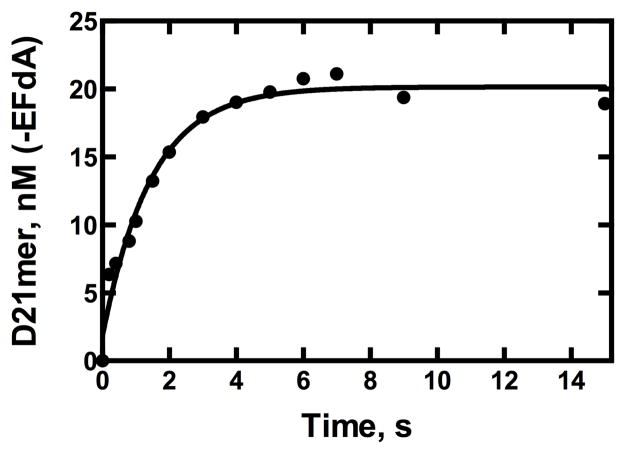
A mode of RT-mediated resistance to NRTIs is via phosphorolytic excision, in which RT uses either ATP or PPi to remove the incorporated inhibitor from the growing DNA strand. We probed the ability of RT (250 nM) to remove EFdA from the DNA primer-EFdAMP/template substrate (50 nM, Fig. 1B) in the presence either ATP (3 mM) or PPi (2 mM sodium pyrophosphate 10 hydrate) and excess magnesium chloride using a rapid chemical quench in conditions described previously (Ray et al., 2002). The removal by ATP-mediated pyrophosphorolysis was negligible, while removal with PPi was significant. We measured a rate of removal of EFdA, kremoval, of 0.69 ± 0.09 s−1 (Fig. 3). This is similar to the rate of PPi-based carbovir removal by RT (0.61 s−1) (Ray et al., 2002). Interestingly, Tchesnokov et al found that incorporating three nucleotides past entecavir by RT provided protection from ATP-mediated removal (Tchesnokov et al., 2008). While a kremoval was not reported, entecavir removal was not complete even after 1 h (Tchesnokov et al., 2008), indicating a significantly slower rate than that seen for EFdA removal. Further, the rate of extension past entecavir by RT is similar to the rate of extension past EFdA (this rate is an estimate only, as a preincorporated entecavir primer/template was not used in this study, (Domaoal et al., 2008).) This indicates that extension past entecavir is likely to be highly favored over its removal, thereby inhibiting pyrophosphorolysis-based resistance. In contrast, the rate of incorporation of the next correct nucleotide past EFdA is over two orders of magnitude slower (Table 2) than the rate of its removal (Fig. 3). This indicates that EFdA removal is significantly favored over extension. We conclude that PPi-based removal of EFdA is a possible mode of resistance.
Fig 3.
PPi-based removal of an incorporated EFdA by RT. RT (250 nM) removes EFdA from the primer-EFdAMP/template (50 nM) in the presence of PPi (2 mM) using a rapid chemical quench. A rate of removal, kremoval, was calculated to be 0.69 ± 0.09 s−1.
In summary, EFdA, a potent inhibitor of RT, functions at least in part by acting as a delayed chain terminator by slowing nucleotide extension after its highly efficient incorporation. These findings are similar to that seen with extension past entecavir by RT, although EFdA incorporation results in slower post-inhibitor incorporation efficiency and minimal changes in fidelity (Domaoal et al., 2008; Tchesnokov et al., 2008) relative to entecavir. Pyrophosphorolytic excision of EFdA by RT is a possible mode of resistance. The striking discrimination by pol γ in contrast with the preference of EFdA over dATP by RT indicates EFdA is a very promising RT inhibitor and sets an important benchmark for future NRTIs. Because of this discrimination profile, EFdA can likely be used in the clinical setting to treat HIV patients with lower doses and minimal mitochondrial-based toxicity. Understanding the mechanism of action employed by EFdA can help in the development and refinement of combination therapies for treating HIV, in addition to paving the way for the discovery of other HIV inhibitors with a similar mechanism.
Highlights.
EFdA is a novel nucleoside reverse transcriptase inhibitor (NRTI) in clinical trials.
Pre-steady-state kinetic experiments show HIV-1 reverse transcriptase (RT) prefers EFdA over dATP.
Due to a 3′-hydroxyl group on EFdA, RT can slowly incorporate additional nucleotides past it.
Phosphorolytic excision of EFdA by RT is a possible mode of resistance.
EFdA potently inhibits RT by delayed chain termination.
Acknowledgments
This work was supported by NIH grants R01 GM049551 (to K.S.A.), F32 GM099289 (to C.D.S.), and AI076119, AI099284, AI100890, and GM103368 (to S.G.S.). We would like to thank Ligong Wang for the purification of the pol γ accessory subunit.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apostolova N, Blas-Garcia A, Esplugues JV. Mitochondrial interference by anti-HIV drugs: mechanisms beyond Pol-gamma inhibition. Trends Pharmacol Sci. 2011;32:715–725. doi: 10.1016/j.tips.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Domaoal RA, McMahon M, Thio CL, Bailey CM, Tirado-Rives J, Obikhod A, Detorio M, Rapp KL, Siliciano RF, Schinazi RF, Anderson KS. Pre-steady-state kinetic studies establish entecavir 5′-triphosphate as a substrate for HIV-1 reverse transcriptase. J Biol Chem. 2008;283:5452–5459. doi: 10.1074/jbc.M707834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JY, Anderson KS. Mechanistic studies comparing the incorporation of (+) and (−) isomers of 3TCTP by HIV-1 reverse transcriptase. Biochemistry. 1999;38:55–63. doi: 10.1021/bi982340r. [DOI] [PubMed] [Google Scholar]
- Harris KS, Brabant W, Styrchak S, Gall A, Daifuku R. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antiviral Res. 2005;67:1–9. doi: 10.1016/j.antiviral.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Iyidogan P, Anderson KS. Understanding the molecular mechanism of sequence dependent tenofovir removal by HIV-1 reverse transcriptase: differences in primer binding site versus polypurine tract. Antiviral Res. 2012;95:93–103. doi: 10.1016/j.antiviral.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AA, Ray AS, Hanes J, Suo Z, Colacino JM, Anderson KS, Johnson KA. Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J Biol Chem. 2001;276:40847–40857. doi: 10.1074/jbc.M106743200. [DOI] [PubMed] [Google Scholar]
- Kellinger MW, Johnson KA. Role of induced fit in limiting discrimination against AZT by HIV reverse transcriptase. Biochemistry. 2011;50:5008–5015. doi: 10.1021/bi200204m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr SG, Anderson KS. RNA dependent DNA replication fidelity of HIV-1 reverse transcriptase: evidence of discrimination between DNA and RNA substrates. Biochemistry. 1997;36:14056–14063. doi: 10.1021/bi971385+. [DOI] [PubMed] [Google Scholar]
- Kim J, Roberts A, Yuan H, Xiong Y, Anderson KS. Nucleocapsid protein annealing of a primer-template enhances (+)-strand DNA synthesis and fidelity by HIV-1 reverse transcriptase. J Mol Biol. 2012;415:866–880. doi: 10.1016/j.jmb.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby KA, Michailidis E, Fetterly TL, Steinbach MA, Singh K, Marchand B, Leslie MD, Hagedorn AN, Kodama EN, Marquez VE, Hughes SH, Mitsuya H, Parniak MA, Sarafianos SG. Effects of substitutions at the 4′ and 2 positions on the bioactivity of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob Agents Chemother. 2013;57:6254–6264. doi: 10.1128/AAC.01703-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby KA, Singh K, Michailidis E, Marchand B, Kodama EN, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. The sugar ring conformation of 4′-ethynyl-2-fluoro-2′-deoxyadenosine and its recognition by the polymerase active site of HIV reverse transcriptase. Cell Mol Biol (Noisy-le-grand) 2011;57:40–46. [PMC free article] [PubMed] [Google Scholar]
- Michailidis E, Marchand B, Kodama EN, Singh K, Matsuoka M, Kirby KA, Ryan EM, Sawani AM, Nagy E, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. Mechanism of inhibition of HIV-1 reverse transcriptase by 4′-ethynyl-2-fluoro-2′-deoxyadenosine triphosphate, a translocation-defective reverse transcriptase inhibitor. J Biol Chem. 2009;284:35681–35691. doi: 10.1074/jbc.M109.036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidis E, Ryan EM, Hachiya A, Kirby KA, Marchand B, Leslie MD, Huber AD, Ong YT, Jackson JC, Singh K, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. Hypersusceptibility mechanism of Tenofovir-resistant HIV to EFdA. Retrovirology. 2013;10:65. doi: 10.1186/1742-4690-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JI, Heath L, Hughes JP, Kicha J, Styrchak S, Wong KG, Rao U, Hansen A, Harris KS, Laurent JP, Li D, Simpson JH, Essigmann JM, Loeb LA, Parkins J. Mutation of HIV-1 genomes in a clinical population treated with the mutagenic nucleoside KP1461. PLoS One. 2011;14:e15135. doi: 10.1371/journal.pone.0015135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami E, Basavapathruni A, Bradley WD, Anderson KS. Mechanism of action of a novel viral mutagenic covert nucleotide: molecular interactions with HIV-1 reverse transcriptase and host cell DNA polymerases. Antiviral Res. 2005;67:10–17. doi: 10.1016/j.antiviral.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Nakata H, Amano M, Koh Y, Kodama E, Yang G, Bailey CM, Kohgo S, Hayakawa H, Matsuoka M, Anderson KS, Cheng YC, Mitsuya H. Activity against human immunodeficiency virus type 1, intracellular metabolism, and effects on human DNA polymerases of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob Agents Chemother. 2007;51:2701–2708. doi: 10.1128/AAC.00277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrui H, Kohgo S, Hayakawa H, Kodama E, Matsuoka M, Nakata T, Mitsuya H. 2′-deoxy-4′-C-ethynyl-2-fluoroadenosine: a nucleoside reverse transcriptase inhibitor with highly potent activity against wide spectrum of HIV-1 strains, favorable toxic profiles, and stability in plasma. Nucleos Nucleot Nucl. 2007;26:1543–1546. doi: 10.1080/15257770701545218. [DOI] [PubMed] [Google Scholar]
- Ray AS, Basavapathruni A, Anderson KS. Mechanistic studies to understand the progressive development of resistance in human immunodeficiency virus type 1 reverse transcriptase to abacavir. J Biol Chem. 2002;277:40479–40490. doi: 10.1074/jbc.M205303200. [DOI] [PubMed] [Google Scholar]
- Sohl CD, Kasiviswanathan R, Kim J, Pradere U, Schinazi RF, Copeland WC, Mitsuya H, Baba M, Anderson KS. Balancing antiviral potency and host toxicity: identifying a nucleotide inhibitor with an optimal kinetic phenotype for HIV-1 reverse transcriptase. Mol Pharmacol. 2012a;82:125–133. doi: 10.1124/mol.112.078758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohl CD, Singh K, Kasiviswanathan R, Copeland WC, Mitsuya H, Sarafianos SG, Anderson KS. Mechanism of interaction of human mitochondrial DNA polymerase gamma with the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine indicates a low potential for host toxicity. Antimicrob Agents Chemother. 2012b;56:1630–1634. doi: 10.1128/AAC.05729-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohl CD, Kasiviswanathan R, Copeland WC, Anderson KS. Mutations in human DNA polymerase gamma confer unique mechanisms of catalytic deficiency that mirror the disease severity in mitochondrial disorder patients. Hum Mol Genet. 2013;22:1074–1085. doi: 10.1093/hmg/dds509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov EP, Obikhod A, Schinazi RF, Gotte M. Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J Biol Chem. 2008;283:34218–34228. doi: 10.1074/jbc.M806797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro JA, Parnell KM, Terezakis SA, Anderson KS. Mechanism of inhibition of the human immunodeficiency virus type 1 reverse transcriptase by d4TTP: an equivalent incorporation efficiency relative to the natural substrate dTTP. Antimicrob Agents Chemother. 2000;44:217–221. doi: 10.1128/aac.44.1.217-221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Parniak MA, Mitsuya H, Sarafianos SG, Graebing PW, Rohan LC. Preformulation studies of EFdA, a novel nucleoside reverse transcriptase inhibitor for HIV prevention. Drug Dev Ind Pharm. 2013 doi: 10.3109/03639045.2013.809535. [DOI] [PMC free article] [PubMed] [Google Scholar]