To the Editor
ATRX is a member of the SWI/SNF family of chromatin remodelers, originally identified as mutated in patients with Alpha Thalassemia/Mental Retardation, X-linked syndrome. The protein product contains several highly conserved domains, including an ADD (ATRX-DNMT3-DNMT3L) domain that binds methylated histone H3 at lysine 9 and an ATPase domain responsible for its remodeling activities (Ratnakumar & Bernstein, 2013). Recently, whole genome sequencing studies identified ATRX mutations in multiple tumors, including those of neural crest cell origin: neuroblastoma, low-grade glioma and glioblastoma (Cheung et al., 2012; Heaphy et al., 2011a; Jiao et al., 2011; Kannan et al., 2012; Schwartzentruber et al., 2012). ATRX alterations encompass point mutations throughout the coding region as well as large N terminal deletions. While mechanistically unclear, ATRX mutations result in loss of protein as assessed by immunohistochemistry (IHC) and often correlate with alternative lengthening of telomeres (ALT) (Cheung et al., 2012; Heaphy et al., 2011a; Kannan et al., 2012; Schwartzentruber et al., 2012).
To our knowledge, an investigation of ATRX in cutaneous melanoma is currently lacking. Our previous studies have demonstrated that decreased expression of histone variant macroH2A drives melanoma cell proliferation and metastasis (Kapoor et al., 2010), and that ATRX interacts with macroH2A to negatively regulate its association with chromatin (Ratnakumar et al., 2012). Taken together with recent reports of decreased ATRX protein in neural crest cell-derived tumors, we hypothesized that ATRX function might be compromised in melanoma.
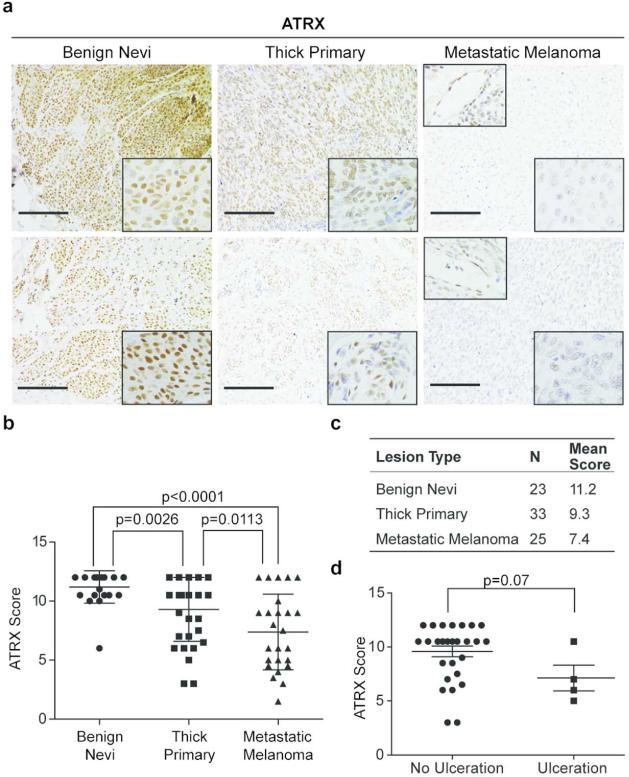
In order to test this hypothesis, we performed IHC on a panel of 23 benign nevi, 33 primary melanoma (≥1.0 mm deep) and 25 metastatic melanoma specimens that were formalin fixed paraffin embedded (FFPE) (Figure 1a, c). Slides were evaluated by two dermatopathologists in a blinded fashion using a scoring system based on number of positive nuclei and staining intensity (inter-rater correlation r=0.693, p<0.0001; see Supplemental Methods for details). As depicted in Figure 1a, ATRX protein expression is appreciably reduced with increased malignancy. Benign nevi showed a higher proportion and intensity of nuclear staining when compared to metastatic lesions (Figure 1a, b; p<0.0001). Furthermore, ATRX protein expression was reduced between benign nevi and primary melanoma, with heterogeneous staining observed in the latter (p=0.0026; Figure 1a, b), and between primary and metastatic melanoma (p=0.0113; Figure 1b). This suggests a potential step-wise loss of ATRX expression during melanoma progression.
Figure 1. Loss of ATRX protein expression is associated with melanoma progression.
(a) IHC for ATRX in representative benign nevi, thick primary, and metastatic melanoma tissue. Images were taken at 20x magnification; insets (bottom right) show nuclei at 40x magnification. Insets (top left) show ATRX positive stain of endothelial cells within metastatic specimens. Scale bar 100 μm. (b) IHC scores of benign nevi, thick primary, and metastatic melanoma from two independent pathologists. Each tissue section was quantified based on number of positively stained cells (1-4) multiplied by stain intensity (1-3) to generate a score. (c) Table summarizing number of total samples and average IHC score per lesion. (d) ATRX protein levels vs. presence of ulceration in primary melanoma specimens. All statistical significance assessed using two-side Mann-Whitney U test, p value indicated. Mean +/- SD.
We further examined whether ATRX levels in primary melanoma correlated with clinicopathologic predictors of prognosis. ATRX staining did not correlate with depth of the lesion (data not shown), however the primary melanomas examined in our cohort were of Breslow thickness greater than 1.0mm (average depth 5.6 mm), and thus quite aggressive. We did however find an inverse correlation with the presence of ulceration, a poor prognostic factor (Figure 1d). Because our study is retrospective with a small sample size, we note that any correlations, or lack thereof, are preliminary.
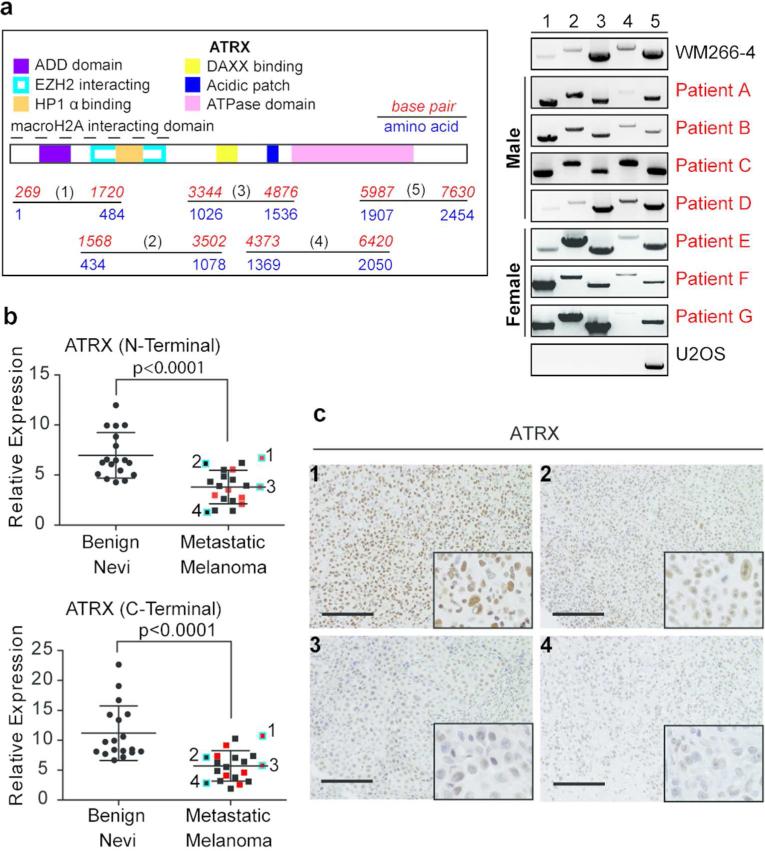
Because structural variations of ATRX exist in neuroblastoma and osteosarcoma (Cheung et al., 2012; Lovejoy et al., 2012), we determined whether such alterations are present in metastatic melanoma. Using a technique to detect structural variations of ATRX, we performed qualitative reverse transcriptase (RT)-PCR of cDNA derived from a cohort of fresh frozen metastatic melanoma samples (n=7). Due to the large ATRX coding region, we amplified the cDNA into five fragments ranging from 1.5-2 kilobase pairs. Because ATRX is located on the X chromosome, we analyzed both male and female patients for potential effects due to gene dosage. Our analysis shows that the ATRX gene product is intact in all metastatic melanomas assayed, as evidenced by appropriately sized bands within each sample (Figure 2a). The cell line WM266-4 derived from a melanoma metastasis served as a positive control for PCR amplicons, as it is devoid of ATRX mutations (Cancer Cell Line Encyclopedia at http://cbioportal.org). The osteosarcoma cell line U2OS, which has large deletions of the ATRX locus (Lovejoy et al., 2012) was used to ensure our assay worked effectively. This analysis suggested that decreased ATRX protein level in metastatic melanoma is unlikely the result of large genomic alterations.
Figure 2. ATRX mRNA levels are decreased in metastatic melanoma.
(a) Illustration of ATRX with domains and five amplicons depicted (left). ATRX cDNA was amplified as indicated for analysis of putative deletions in metastatic melanoma specimens (right). WM266-4 and U2OS were used as controls. (b) qRT-PCR analysis of ATRX from fresh frozen benign nevi and metastatic melanoma lesions. N- and C-terminal primers were used. Melanoma specimens analyzed in (a) are depicted in red and in (c) are highlighted in blue and numbered. Expression levels were normalized to GAPDH and statistical significance was derived using unpaired Student's t-test, p-values indicated. Mean +/- SD. (c) ATRX IHC in representative metastatic melanoma tissues from (b). Images were taken at 20x magnification; insets (bottom right) show nuclei at 40x magnification. Scale bar 100 μm.
We next queried whether diminished ATRX protein in metastatic disease was due to transcriptional regulation. We performed qPCR analysis on a cohort of 18 fresh frozen benign nevi and 20 metastatic melanoma tumors, including those samples analyzed for deletions (Figure 2a; indicated in red in Figure 2b). Using both N- and C- terminal primers for ATRX, we found a statistically significant loss of ATRX mRNA levels in metastatic melanoma as compared to benign tissue (p<0.0001; Figure 2b). We next performed IHC on a subset of these tumors, for which FFPE tissue was available (indicated in blue in Figure 2b). The level of ATRX protein indeed corroborated our qPCR findings (Figure 2c). Collectively, these results indicate that ATRX loss occurs, at least in part, by transcriptional repression resulting in loss of protein expression in late stage disease.
Collectively, we demonstrate that ATRX loss correlates with melanoma progression. Using two independent cohorts (FFPE and fresh frozen; total of 119 tissues), we found a significant decrease of both mRNA and protein levels of ATRX in metastatic melanoma. While it remains to be tested in a prospective study, ATRX may serve as a biomarker to predict prognosis of disease. Though we did not find evidence of large genomic alterations in a subset of melanoma patients, we do not exclude the possibility of ATRX mutations in melanoma. In fact, a 4-7.5% rate of mutation in cutaneous melanoma is reported by TCGA, Broad and Yale studies (http://www.cbioportal.org). Interestingly, these mutations are distributed throughout the ATRX coding region and do not correlate with decreased mRNA levels (Supplemental Figure S1). This suggests that multiple mechanisms underlie ATRX dysregulation in melanoma – transcriptional regulation as described here and point mutations that may result in loss of protein expression, as reported for other tumor types (Cheung et al., 2012; Kannan et al., 2012; Schwartzentruber et al., 2012).
While ATRX staining did not anti-correlate with macroH2A levels (data not shown), we previously showed that macroH2A is transcriptionally silenced by DNA methylation in malignant melanoma and thus might not be regulated at the level of chromatin deposition (Kapoor et al., 2010). The mechanism by which ATRX transcription is suppressed in melanoma may also be through epigenetic silencing (e.g. DNA methylation or histone modifications), or by microRNA mediated regulation (Pacurari et al., 2013). Finally, we posit that investigating the chromatin landscape of tumors that have lost ATRX expression should provide insight into the mechanism(s) by which ATRX loss drives melanoma progression.
Supplementary Material
Acknowledgements
The authors thank Daniel Choi and Dan Hasson for critical reading of this manuscript, and Hsan-Au Wu for assistance with figures. The authors thank the Michael Donovan and Ruijin Shi from the Mount Sinai Biorepository Cooperative, Miguel Segura, Iman Osman, and Eva Hernando (New York University), and Qin Yu (Mount Sinai) for specimens. The authors thank Madelaine Haddican, Shelbi Jim On, and Giselle Singer in the Department of Dermatology for assistance with benign nevi collection, and Mark Lebwohl for his continued support. This work is supported by ASA and MRF Medical Student Grant awards to S.H., an NCI T32-CA078207 to L.F.D, a Ph.D. fellowship from CONACyT (239663) to D.V-G. and a Hirschl/Weill-Caulier Charitable Trust Award and NCI/NIH R01CA154683 to E.B.
Abbreviations
- ATRX
Alpha Thalassemia/Mental Retardation, X-linked
- IHC
immunohistochemistry
Footnotes
Conflicts of Interest
The authors state no conflict of interest.
References
- Cheung NK, Zhang J, Lu C, et al. Association of age at diagnosis and genetic mutationsin patients with neuroblastoma. JAMA : the journal of the American Medical Association. 2012;307:1062–71. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011a;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199, 203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Inagaki A, Silber J, et al. Whole-exome sequencing identifies ATRX mutation as a key molecular determinant in lower grade glioma. Oncotarget. 2012;3:1194–203. doi: 10.18632/oncotarget.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Goldberg MS, Cumberland LK, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–9. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Li W, Reisenweber S, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS genetics. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacurari M, Addison JB, Bondalapati N, et al. The microRNA-200 family targets multiple non-small cell lung cancer prognostic markers in H1299 cells and BEAS 2B cells. International journal of oncology. 2013;43:548–60. doi: 10.3892/ijo.2013.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar K, Bernstein E. ATRX: the case of a peculiar chromatin remodeler. Epigenetics. 2013;8:3–9. doi: 10.4161/epi.23271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnakumar K, Duarte LF, LeRoy G, et al. ATRX-mediated chromatin association of histone variant macroH2A1 regulates alpha-globin expression. Genes & development. 2012;26:433–8. doi: 10.1101/gad.179416.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.