Abstract
Nonsense-Mediated mRNA Decay (NMD) is a regulatory pathway that functions to degrade transcripts containing premature termination codons (PTCs) and to maintain normal transcriptome homeostasis. Nonsense and frameshift mutations that generate PTCs cause approximately one-third of all known human genetic diseases and thus NMD has a potentially important role in human disease. In genetic disorders in which the affected genes carry PTC-generating mutations, NMD acts as a double-edge sword. While it can benefit the patient by degrading PTC-containing mRNAs that encode detrimental, dominant-negative truncated proteins, it can also make the disease worse when a PTC-containing mRNA is degraded that encodes a mutant but still functional protein. There is evidence that the magnitude of NMD varies between individuals, which, in turn, has been shown to correlate with both clinical presentations and the patients’ responses to drugs that promote read-through of PTCs. In this review, we examine the evidence supporting the existence of inter-individual variability in NMD efficiency and discuss the genetic factors that underlie this variability. We propose that inter-individual variability in NMD efficiency is a common phenomenon in human populations and that an individual’s NMD efficiency should be taken into consideration when testing, developing, and making therapeutic decisions for diseases caused by genes harboring PTCs.
Keywords: Nonsense-mediated mRNA decay, inter-individual NMD efficiency, regulation of NMD, expression quantitative trait loci, copy number variation, Staufen-mediated mRNA decay, miR-128, miR-125
1 Introduction
NMD is an RNA degradation pathway originally discovered by virtue of its role as a quality control mechanism that we now know also serves to regulate normal patterns of gene expression. With regard to its role in quality control, NMD is an RNA surveillance pathway that degrades transcripts harboring PTCs. Such aberrant transcripts can arise as a consequence of mutations (nonsense or frameshift), alternative or aberrant mRNA splicing, errors in transcription, and programmed gene rearrangements. NMD is a crucial RNA surveillance mechanism since it rapidly degrades mRNAs that would otherwise translate truncated proteins with potentially harmful dominant-negative effects (Nicholson et al., 2010).
Approximately one third of all human genetic disorders of known etiology are caused by genes with germline or de novo mutations that generate PTCs (Frischmeyer and Dietz, 1999). Thus, NMD has the potential to influence the outcome of a large fraction of human diseases. In most positions, PTCs trigger NMD, which is beneficial for disease outcome if the mRNA encodes a truncated deleterious protein. But some mutant mRNAs with PTCs encode truncated proteins that retain partial function and therefore, by degrading such mRNAs, NMD can actually worsen disease outcome (Khajavi et al., 2006). Thus, NMD is a double-edged sword that can either benefit the patient or promote disease.
Recent evidence suggests that variation in the efficiency of NMD can potentially lead to different clinical outcomes in patients, even if they carry the same PTC-generating mutation (Kerr et al., 2001; Nguyen et al., 2012; Resta et al., 2006). For example, two patients who carry the same PTC-generating mutation in the X-chromosome linked DMD gene exhibit markedly different phenotypes: one has Duchene Muscular Dystrophy (DMD) (Mendelian Inheritance in Man, MIM 310200) whereas the other has much less severe Becker Muscular Dystrophy (BMD) phenotype (MIM 300376) (Kerr et al., 2001). When examining muscle biopsy taken from the patient with BMD, the investigators noted a moderate expression of DMD protein, suggesting that NMD was weak in this patient, allowing accumulation of PTC-containing mRNA and hence translation of the truncated, but still functional DMD protein (Kerr et al., 2001). As another potential example of this phenomenon, embryonic tissues from two spontaneously aborted foetuses with Roberts Syndrome (MIM 268300) differed significantly in their ability to downregulate the same mutant PTC-containing transcript emanating from the causative gene, ESCO2, in a manner that inversely correlated with the length of the survival of the two foetuses (Resta et al., 2006). As these examples illustrate, NMD efficiency appears to vary from individual to individual, which may be an important modifier of some disease phenotypes.
While the mechanism and regulation of NMD have been studied extensively (Huang and Wilkinson, 2012; Karam et al., 2013; Nicholson et al., 2010; Schoenberg and Maquat, 2012; Schweingruber et al., 2013), relatively little is known about the factors underlying variable NMD efficiency. This review aims to focus on this less well-explored aspect of NMD. We will first briefly recapitulate the general functions of NMD and its underlying mechanism. We will then discuss its involvement in human genetic diseases, particularly with regard to the role of inter-individual variation in NMD efficiency. Known factors that influence NMD efficiency, including stress, microRNAs, and feedback regulation will be detailed. We will close by discussing how recent findings concerning NMD may lead to the generation of modalities that suppress or enhance NMD for therapeutic purposes.
2 Nonsense-Mediated RNA Decay
NMD requires the action of several proteins, many of which are in well-defined complexes. The first NMD factors discovered—up-frameshift-1 (Upf1), Upf2, and Upf3—were identified in a genetic screen in Saccharomyces cerevisiae (Leeds et al., 1991; Leeds et al., 1992). Orthologs of these three NMD genes, as well as four other NMD genes—SMG1, SMG5, SMG6, and SMG7—were later discovered in Caenorhabditis elegans through another genetic screen. Mutations in these NMD factor genes in C. elegans resulted in morphological defects in the male bursa or the hermaphrodite vulva, leading them to be named suppressor of morphological defects on genitalia (Smg) genes (Cali et al., 1999; Hodgkin et al., 1989; Pulak and Anderson, 1993). Subsequently, human orthologs of these genes were identified based on sequence conservation, as well as new NMD genes (SMG8, SMG9 and DHX34) using other methods (Applequist et al., 1997; Denning et al., 2001; Longman et al., 2007; Lykke-Andersen et al., 2000; Mendell et al., 2000; Ohnishi et al., 2003; Perlick et al., 1996; Serin et al., 2001; Yamashita et al., 2009; Yamashita et al., 2001) (Table 1). The UPF1, UPF2, and UPF3 proteins are considered to be the core NMD factors and are highly conserved (Applequist et al., 1997; Serin et al., 2001).
Table 1.
Proteins involved in the formation and function of the NMD and EJC complexes.
| Group | Factor | Aliases | MIM # | Cellular localization | Role in NMD | References |
|---|---|---|---|---|---|---|
| NMD factors | UPF1 | RENT1 | 601430 | Shuttling protein, but mainly in cytoplasm | Joins to EJCs when PTC was recognized, mediates translational repression and recruitment of downstream degradation machinery | (He and Jacobson, 1995; Hosoda et al., 2005; Isken et al., 2008; Kashima et al., 2006; Maderazo et al., 2000; Nazarenus et al., 2005) |
| UPF2 | RENT2 | 605529 | Perinuclear (cytoplasmic) | Joins EJC when the mRNP is exported to the cytoplasm, recruits and promotes phosphorylation of UPF1 | (Hosoda et al., 2005; Le Hir et al., 2001; Lykke-Andersen et al., 2000; Mendell et al., 2000; Serin et al., 2001) | |
| UPF3B | RENT3B, UPF3X | 300298 | Shuttling protein, mainly in nucleus | Joins EJC via a contiguous surfaced formed by EIF4A3/Y14/MAGOH, recruits UPF2 to EJC | (Gehring et al., 2005; Gehring et al., 2003; Kadlec et al., 2004; Kim et al., 2001a; Le Hir et al., 2001; Serin et al., 2001) | |
| UPF3A | RENT3A, UPF3 | 605530 | Shuttling protein, mainly in nucleus | Similar to UPF3B but less efficient, compete for binding to UPF2 for stabilization | (Chan et al., 2009; Kim et al., 2001a; Kunz et al., 2006; Le Hir et al., 2001; Serin et al., 2001) | |
| SMG1 | ATX | 607032 | Cytoplasm and nucleus | Promotes phosphorylation of UPF1 | (Denning et al., 2001; Kashima et al., 2006; Morita et al., 2007; Yamashita et al., 2001) | |
| SMG5 | EST1B | 610962 | Cytoplasm and nucleus | Involved in dephosphorylation of UPF1, mediates exonucleotic decay pathway | (Chiu et al., 2003; Lejeune et al., 2003; Ohnishi et al., 2003; Okada-Katsuhata et al., 2012; Unterholzner and Izaurralde, 2004) | |
| SMG6 | EST1A | 610963 | Cytoplasm | Mediates endonucleotic decay pathway | (Eberle et al., 2009; Huntzinger et al., 2008; Kashima et al., 2010) | |
| SMG7 | EST1C | 610964 | Cytoplasm | Involved in dephosphorylation of UPF1, mediates exonucleotic decay pathway | (Chiu et al., 2003; Fukuhara et al., 2005; Lejeune et al., 2003; Ohnishi et al., 2003; Okada-Katsuhata et al., 2012; Unterholzner and Izaurralde, 2004) | |
| SMG8 | 613175 | n.a. | Forms complex with SMG1 to inhibit its kinase activity before SMG1 joins EJC | (Yamashita et al., 2009) | ||
| SMG9 | 613176 | n.a. | Forms complex with SMG1 to inhibit its kinase activity before SMG1 joins EJC | (Yamashita et al., 2009) | ||
| NBAS | NAG, SMGL1 | 608025 | n.a. | Required for NMD in C. elegans and D. rerio through unknown mechanism | (Longman et al., 2007) | |
| DHX34 | SMGL2 | n.a. | n.a. | Required for NMD in C. elegans and D. rerio through unknown mechanism | (Longman et al., 2007) | |
| EJC factors | RBM8A | Y14 | 605313 | Shuttling protein, mainly in nucleus | Directly interacts with EIF4A3 and MAGOH to form the contiguous surface for UPF3B interaction | (Fribourg et al., 2003; Gehring et al., 2003; Kataoka et al., 2000; Kim et al., 2001b; Lau et al., 2003; Shi and Xu, 2003) |
| MAGOH | 602603 | Shuttling protein, mainly in nucleus | Binds strongly to RBM8A, EIF4A3 and mRNA, interacts directly with UPF3B | (Kataoka et al., 2001; Lau et al., 2003; Shi and Xu, 2003; Zhao et al., 2000) | ||
| MAGOHB | n.a. | n.a. | Paralog of MAGOH with almost identical sequence conservation. Binds directly to RBM8A, EIF4A3 and UPF3B to activate NMD. | (Singh et al., 2013) | ||
| CASC3 | BTZ, MLN51 | 606504 | Mostly cytoplasm | Binds to mRNA directly and interacts with EIF4A3 | (Baguet et al., 2007; Degot et al., 2004; Gehring et al., 2005; Gehring et al., 2009a; Palacios et al., 2004) | |
| EIF4A3 | 608546 | Nucleus | Binds to mRNA directly, provides anchor for the remaining EJC components | (Chan et al., 2004; Gehring et al., 2005; Gehring et al., 2009a; Palacios et al., 2004) | ||
| RNPS1 | 606447 | Nuclear speckles | Binds to mRNA directly, mediates an alternative NMD pathway independent of some EJC factors | (Gehring et al., 2005; Le Hir et al., 2001; Le Hir et al., 2000; Lykke-Andersen et al., 2001) | ||
| WIBG | PYM | n.a. | Cytoplasm | Forms complex with MAGOH-RBM8A, acts to disassemble EJC and negatively regulates NMD | (Bono et al., 2004; Gehring et al., 2009b) |
Abbreviations: n.a., not analyzed, not available; MIM, Medelian Inheritance in Man.
In many eukaryotes, including mammals, the UPF2 and UPF3 proteins associate with the exon junction complex (EJC), a set of four proteins—Y14, MAGOH, eIF4AIII, and MLN51 (Table 1)—that are deposited ~20 to 24 nucleotides (nt) upstream of the exon-exon junctions during pre-mRNA splicing (Kim et al., 2001a; Le Hir et al., 2001; Le Hir et al., 2000; Lejeune et al., 2002). Recent studies have shown that most, but not all, spliced exon-exon junctions have an EJC (Sauliere et al., 2010; Sauliere et al., 2012; Singh et al., 2012). The position of the stop codon relative to the last exon-exon junction (i.e., where the most 3′ EJC is deposited) dictates whether NMD will be triggered or not in many mammalian mRNAs. If the stop codon lies in the last exon, this allows the ribosome to strip off all EJCs before translation termination and thereby prevent EJC-dependent NMD. However, if a natural stop codon or a PTC generated by mutation lie in a middle exon, the EJC(s) downstream of the stop codon will typically escape removal by the EJC. As a consequence, these EJCs will have an opportunity to interact with NMD factors recruited upon translation termination (e.g., UPF1), leading to mRNA decay by mechanisms that are still under investigation (Figure 1). A notable exception to this “downstream exon-exon junction” rule are mRNAs in which the stop codon is ~55 nt or closer to the last exon-exon junction (Nagy and Maquat, 1998). Most of these mRNAs are immune to NMD, most likely because the dimensions of the EJC and the ribosome lead the former to be stripped off by the latter. For a more complete discussion of models of NMD action, see recent reviews (Huang and Wilkinson, 2012; Karam et al., 2013; Nicholson et al., 2010; Schoenberg and Maquat, 2012).
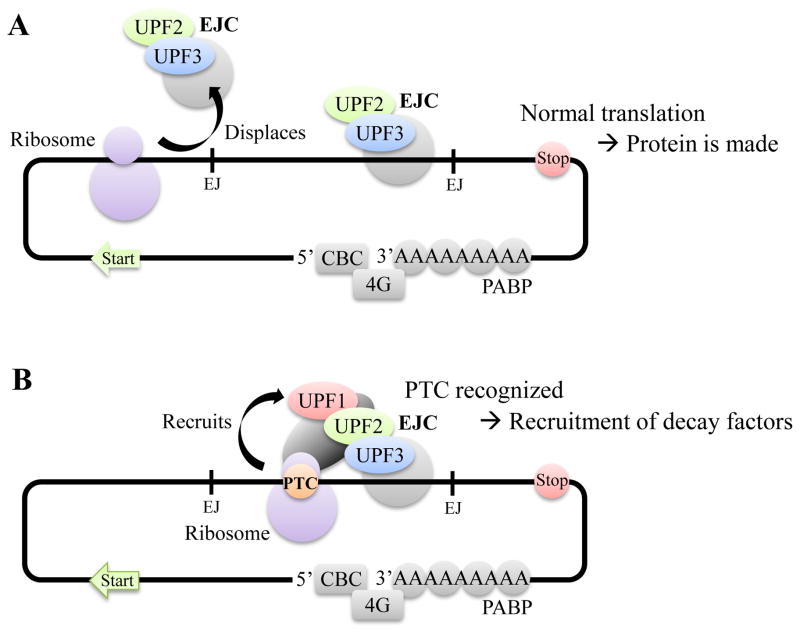
Figure 1. Current simplified model of canonical NMD.
In the nucleus and during pre-mRNA splicing process, the EJCs are deposited by the spliceosomes ~ 20–24 nt upstream of the exon-exon junction (EJ). EJC is made up of several proteins and is associated with the UPF3 proteins. The mRNP (mRNA-protein complex) is then exported to the cytoplasm, at which point the EJC-UPF3 complex acquires UPF2 via direct interaction with UPF3 proteins.
A. During the translation process, the ribosome reads along the mRNA and displaces all downstream EJCs before terminating at the normal stop codon. Full-length protein is made.
B. If a stop codon is introduced in a context that would be considered premature, the translating ribosome stalls at the PTC and recognizes downstream EJC. Here, another complex, which contains UPF1, is recruited, bringing together the interaction of the three “core” components of NMD UPF1, UPF2 and UPF3. This subsequently leads to UPF1 being phosphorylated, recruiting further downstream exo- and endonucleotic decay factors to degrade the mRNA.
Notable exceptions to the 55-nt boundary rule exist. Two examples are Immunoglobulin (Ig) and T-cell receptor (TCR) transcripts, both of which are transcribed from genes that undergo programmed rearrangement events, a process that frequently generates frameshifts. As a consequence of these frameshifts, non-productively rearranged Ig and TCR genes give rise to transcripts with PTCs that are degraded by NMD (Baumann et al., 1985). The downregulation of PTC-bearing Ig and TCR transcripts by NMD is typically extremely robust (>50-fold) when the stop codon is >55 nt upstream of the last exon-exon junction, however stop codons closer than this can also trigger NMD in some transcripts, albeit not as efficiently (Buhler et al., 2004; Wang et al., 2002a). In further support of this, an in vivo study was recently published that showed that a knock-in allele of a TCR-β gene that lacks all introns in the region that acquires PTCs as a result of programmed rearrangement gives rise to mRNAs that are more modestly downregulated by naturally derived PTCs than does a knock-in TCR-β gene allele that is identical except that it contains these introns (Mahowald et al., 2011). These data suggest that there is fail-safe EJC-independent mechanism that collaborates with an EJC-dependent NMD mechanism to degrade aberrant TCR-β transcripts.
Further support for the existence of different NMD branches with different co-factor requirements comes from in vitro studies. Gehring et al. (2005) uncovered two NMD branches that are not affected by RNAi-mediated knockdown of UPF2/RNPS1 or EJC components, respectively, suggesting that there are alternative NMD branches independent of these factors (Gehring et al., 2005). Using the same approach, Chan et al. (2009) obtained evidence for a UPF3-independent branch of NMD that is not affected by depletion of UPF3A (an autosomal gene also known as UPF3) and UPF3B (an X-chromosome linked gene also known as UPF3X), the two known UPF3 paralogs that exist in mammals (Chan et al., 2009; Chan et al., 2007). Confirmation of a UPF3B-independent branch of NMD comes from recent studies in Upf3b-null mice (Huang et al., 2011). Little is known about the specificity of these alternative NMD branches, but it is likely that they sometimes act independently and sometimes in concert to regulate the expression of different subsets of genes in different tissue types and developmental stages.
3 NMD as a crucial regulator of the transcriptome
Over the past decade, it has become clear that the role of NMD goes well beyond its originally defined function as quality control mechanism that degrades aberrant mRNAs. NMD is now considered a fundamental developmental regulator of the transcriptome. Genome-wide analysis of cell lines depleted of NMD factors, as well as NMD-deficient mice, have shown that 3 to 20% of the transcriptome is deregulated as a consequence of compromised NMD (Chan et al., 2009; He et al., 2003; Mendell et al., 2004; Rehwinkel et al., 2005; Tani et al., 2012; Weischenfeldt et al., 2008; Weischenfeldt et al., 2012; Wittmann et al., 2006). It is not clear yet what proportion of these are direct NMD targets. One example of a direct NMD target is PTBP2 mRNA, which encodes a RNA-binding protein that promotes the neural differentiation program. In neural progenitor cells, PTBP2 mRNA contains a PTC introduced by the exclusion of exon 10 and thus it is rapidly degraded by NMD. However, upon differentiation of progenitor cells into neurons, the expression of the splicing repressor PTBP1 that inhibits exon 10 inclusion is lost, which as a consequence restores the reading frame of PTBP2 transcript so that it is now immune to NMD. This allows functional PTBP2 protein to be made, which in turn activates a cascade of events required for neuronal maturation (Makeyev et al., 2007). Other notable examples of conserved NMD substrates are the mRNAs encoding most SR protein family members (Lareau et al., 2007). SR proteins are RNA-binding proteins that regulate many post-transcriptional events, including RNA splicing, mRNA export, and translation, and thus the ability of NMD to regulate their levels is likely to have profound effects on fundamental processes, including growth and development. Consistent with this, knockout (KO) mice of Upf1, Upf2, Magoh, and Smg1 NMD genes all suffer from early embryonic lethality and marked developmental abnormalities (McIlwain et al., 2010; Medghalchi et al., 2001; Silver et al., 2010; Weischenfeldt et al., 2008). Partial and complete loss of function mutations in NMD factor genes in man that do not lead to embryonic lethality are associated with various forms of neuro-developmental conditions and congenital anomalies (Albers et al., 2012; Laumonnier et al., 2010; Lynch et al., 2012; Nguyen et al., 2013; Tarpey et al., 2007; Xu et al., 2013), a topic that will be described in greater detail in Section 4.
What are the features in normal mRNAs that prompt them to be degraded by NMD? The current consensus is that the context of the stop codon defining the end of the main open reading frame (ORF) dictates whether NMD is triggered or not (Nagy and Maquat, 1998). These features include (i) a natural stop codon >55 nt upstream of an exon-exon junction (Nagy and Maquat, 1998), (ii) alternative splicing (AS) events that cause a frameshift that results in the generation of an in-frame stop codon >55 nt upstream of an exon-exon junction (Horikawa et al., 2000; Wang et al., 2002b), (iii) leaky translation due to differential usage of internal ribosomal entry sites (Welch and Jacobson, 1999), and (iv) long 3′ UTRs (Kebaara and Atkin, 2009). In addition, ORFs 5′ of the main reading frame—so-called upstream ORFs (uORFs)—can, in some cases, elicit NMD; perhaps because their stop codons recruit termination factors that can, in turn interact with downstream NMD-promoting factors, such as the EJC (Ruiz-Echevarria and Peltz, 2000; Silva et al., 2006).
4 NMD factors associated with genetic diseases
The most significant evidence supporting the involvement of NMD in human genetic disorders comes from patients with neuro-developmental disorders and intellectual disability (ID) that carry mutations in the UPF3B gene (MIM 300676). Studies of several families with multiple affected individuals have demonstrated that mutations in UPF3B cause ID in these patients (Addington et al., 2011; Laumonnier et al., 2010; Lynch et al., 2012; Tarpey et al., 2007; Xu et al., 2013). Many of the patients, some of them also singletons, present with comorbidities including schizophrenia or autism, suggesting that loss of UPF3B might contribute to these psychiatric disorders (Addington et al., 2011; Laumonnier et al., 2010; Lynch et al., 2012; Tarpey et al., 2007). A summary of the clinical features associated with UPF3B mutations and an in-depth discussion of the role of NMD in psychiatric disorders has recently been compiled by Laumonnier et al. (2013).
In addition to UPF3B mutations, heterozygous deletions of genomic regions that include UPF2 were recently shown to be associated with ID and neuro-developmental disorders (Nguyen et al., 2013). In these patients, compromised NMD led to deregulation of 5–10% of the transcriptome, as determined using their lymphoblastoid cell lines (LCL). Several of these deregulated genes were further validated using neuronal cell models (Jolly et al., 2013), and as such, they may explain at least some aspects of the patients’ clinical phenotypes (Nguyen et al., 2012; Nguyen et al., 2013). Further support for the involvement of NMD in psychiatric disorders comes from the identification of a de novo missense mutation in UPF2 in a patient with schizophrenia, although the functional consequences of the mutation have not been experimentally tested (Gulsuner et al., 2013).
NMD has also been linked with another human disease: Thrombocytopenia with Absent Radius (TAR) Syndrome (MIM 274000). This is a debilitating disease that was recently shown to be caused by mutations in the RBM8A gene, which encodes the core EJC factor RBM8A; also known as Y14 (Albers et al., 2012). TAR typically results from compound heterozygosity for a deletion (de novo or inherited) of RBM8A (on one allele) and a deleterious regulatory single nucleotide polymorphisms (SNP) in its promoter or its intron (on the other allele) (Albers et al., 2012). The main clinical hallmarks of patients with TAR syndrome are reduction in the number of platelets and skeletal anomalies with variable severity, ranging from absence of radii to upper limbs, with or without lower limb defects. Intriguingly, a small proportion of these patients (~7%) have compromised cognitive function, which may be due to the thinning of the corpus callosum and cysts, as identified by MRI (MacDonald et al., 1994; Rosenfeld et al., 2012). The small genomic region deleted in these patients, also termed the 1q21.1 proximal region, was found to be significantly associated with developmental delay and congenital anomalies without hallmarks of TAR syndrome (Rosenfeld et al., 2012). This suggests that at least some of the 16 genes in the region, including RBM8A, may play role in normal cognitive development. The evidence supporting the notion that RBM8A has such a role is that it encodes a protein that is part of the same protein complex as UPF3B, which causes intellectual disability when absent or mutated (Tarpey et al., 2007).
A recent survey of copy number variations (CNVs) encompassing 18 known NMD and EJC genes in ~57,000 patients and ~20,000 controls identified several of these genes as being significantly associated with neurological diseases. Both copy number gains and losses of regions encompassing UPF2 or RBM8A were associated with various forms of neuro-developmental disorders, with or without congenital anomalies (Nguyen et al., 2013). Also associated with these conditions were copy number losses of genomic regions encompassing UPF3A and copy number gains in genomic regions encompassing SMG6, EIF4A3 or RNPS1. While the molecular pathology of these CNVs have not yet been investigated in humans, it is worth noting that overexpression of Rbm8a in a mouse model has been shown to increase anxiety-like behavior, impair social skills and decrease immobile time (Alachkar et al., 2013). Mice overexpressing Rbm8a also display enhanced frequency of miniature excitatory postsynaptic currents (Alachkar et al., 2013). Together, these studies suggest that either too much or too little of a given NMD factor can cause neurological or psychiatric disorders. However, it remains to be determined whether overexpression of NMD factors causes disease because it affects the magnitude of NMD or because this triggers NMD-independent deleterious effects.
The clinical presentations of known patients with NMD factor mutations and CNVs vary considerably. For example, there has been no consistent diagnosis for males from the ten families identified with UPF3B mutations, apart from ID (Addington et al., 2011; Laumonnier et al., 2010; Lynch et al., 2012; Tarpey et al., 2007; Xu et al., 2013). Broad and variable clinical expressivity, even due to identical mutations in the same gene, is not unusual, but the underlying mechanism in most cases is not known. Pertaining to this, molecular analysis of a brother pair who carry exactly the same mutation in UPF3B revealed that the UPF3B protein paralog, UPF3A, is expressed at a level that inversely correlates with the extent of their transcriptome deregulation and the severity of their neurological symptoms (Nguyen et al., 2012). This is potentially important since UPF3A has been shown to be stabilized and to compensate for the loss or depletion of UPF3B (Chan et al., 2009; Nguyen et al., 2012). Thus, UPF3A could be a modifier of the clinical phenotype of UPF3B patients. What determines differential UPF3A protein levels in these patients remains to be elucidated.
Similar to patients with UPF3B mutations, patients carrying CNVs encompassing other NMD and EJC genes show a wide range of clinical presentations. Of note, some of these CNVs are also found in apparently normal individuals (at much lower frequencies) or were, in some instances, inherited from unaffected parents, suggesting that they are incompletely penetrant (Nguyen et al., 2013). Such variation in penetrance is not unusual and is observed in many genetic disorders (Girirajan and Eichler, 2010). In the case of NMD factor genes, we hypothesize that their variable copy number alters the magnitude of NMD efficiency, which can make a non-penetrant individuals fall below the “clinically relevant” threshold and thus present with a disease, as will be further discussed in Section 7.
5 Inter-individual variation in NMD efficiency may affect phenotype
Several lines of evidence suggest that differences in NMD efficiency between individuals can modify the outcome of genetic diseases. In addition to the above-mentioned examples of variable responses to PTC-eliciting mutations in DMD or ESCO2 (Section 1), NMD efficiency has also been suggested to vary among individuals with nonsense mutations in the CTFR gene (Kerem et al., 2008; Linde et al., 2007; Linde and Kerem, 2008). Two “PTC read-through” drugs, Gentamicin and PTC124, have been used with some success in clinical trials to inhibit reading of the PTCs in the CFTR mRNA in these patients and thereby restore translation of some full-length CFTR protein (Keeling and Bedwell, 2011). Of note, there has been some controversy as to whether PTC124 is a bona fide read-through inhibitor; several studies have obtained evidence that while PTC124 is active in the luciferase-based reporter screen used to identify it (Welch et al., 2007), this is due to an off-target effect (McElroy et al. (2013) and references therein). Regardless of this concern, it is interesting that patients who had higher basal level of the mutant transcript responded better to treatment with either Gentamicin or PTC124 (Kerem et al., 2008; Linde et al., 2007; Linde and Kerem, 2008). While not explicitly tested, it is possible that such patients had intrinsically inefficient NMD, which resulted in higher amounts of mRNA available to be translated, and hence higher levels of functional CFTR protein (Kerem et al., 2008; Linde et al., 2007; Linde and Kerem, 2008).
Variable NMD efficiency also appears to be an important factor contributing to the diversity of normal phenotypic traits in humans. In a genome-wide survey of loss-of-function (LOF) variants in human protein coding genes, MacArthur et al. (2012) found on average ~100 heterozygous LOF variants per personal genome, with ~20 genes being completely inactivated (MacArthur et al., 2012). A large proportion of these LOF variants consisted of naturally occurring mutations that introduce a PTC as a result of a deletion or a small duplication and/or aberrant splicing leading to frameshift. However, they observed only a small proportion (25%) of the mRNAs predicted to be degraded by NMD were consistently downregulated by NMD in most individuals, at least as determined by qPCR analysis of 119 lymphoblastoid cell lines from individuals with and without LOF variants (MacArthur et al., 2012). This experimental evidence suggested considerable inter-individual variability in NMD efficiency, especially since there was a large standard deviation in many of the mRNA levels. It is possible that the PTC-containing mRNAs escaping NMD in these individuals could produce truncated proteins with dominant-negative effects, which could in turn contribute to phenotypic variation. In further support of inter-individual variability of NMD, nonsense and frameshift mutations that generate NMD-immune PTCs in the nuclear factor 1 X-type (NFIX) gene are responsible for the lethal Marshall-Smith Syndrome (MIM 602535), whereas mutations in NFIX that generate NMD-sensitive PTCs cause Sotos-like Syndrome (MIM 614753), which is a milder disease (Malan et al., 2010). Likewise, a patient with a mutation causing the generation of an NMD-immune PTC in the latent TGFβ-binding protein-4 (LTBP4) gene exhibited severe malformation of the gastrointestinal tract, a symptom not typically associated with LTBP4 mutations, as they normally cause a mild disease called Type-1 Recessive Cutis Laxa (MIM 613177) (Callewaert et al., 2013). There are several other examples of disease severity being modified by NMD through downregulation of mRNAs leading to production of dominant-negative truncated proteins, as discussed in several reviews (Holbrook et al., 2004; Khajavi et al., 2006).
Taken together, these studies demonstrate that inter-individual variation in NMD efficiency might be relatively common and thus is relevant for health and disease (Figure 2). In addition to point mutations and CNVs that impact NMD genes (Addington et al., 2011; Albers et al., 2012; Laumonnier et al., 2010; Lynch et al., 2012; Nguyen et al., 2013; Tarpey et al., 2007; Xu et al., 2013), there are several other factors that have been discovered that can influence NMD efficiency, which will be discussed below.
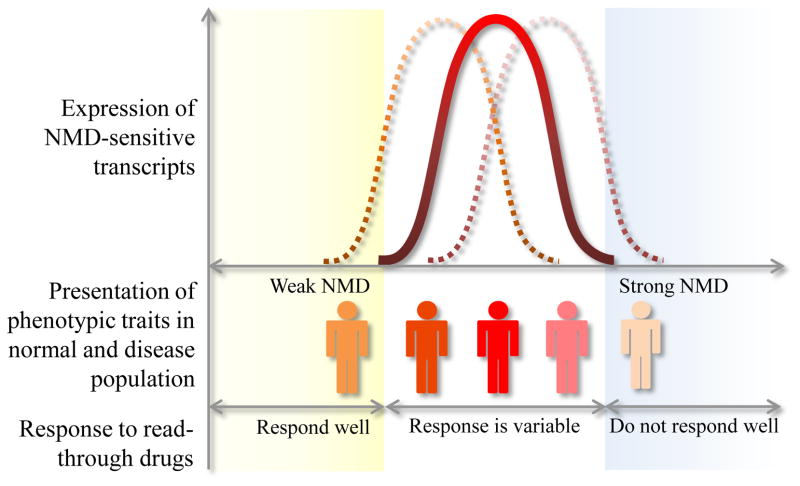
Figure 2. Variable NMD efficiency modifies phenotypic traits.
Variation in NMD efficiency could modify the presentation of clinical phenotypes and response to PTC read-through drug treatment.
6 Sources of inter-individual variation in NMD efficiency
6.1 Expression quantitative trait loci
Expression quantitative trait loci (eQTL) are important factors that contribute to the phenotypic diversity within and among different human population (Dixon et al., 2007; Stranger et al., 2012). These loci typically are SNPs or CNVs that are associated with gene expression changes. The current SNP-eQTL map of the human genome has been extensively drawn from several large-scale studies involving hundreds of samples (Dixon et al., 2007; Stranger et al., 2012). With regard to NMD, multiple SNPs have been found to be highly associated with the expression of SMG7 (P < 1.0e−8, Table 2) (Dixon et al., 2007; Stranger et al., 2012), which encodes a protein essential for NMD that promotes the dephosphorylation of UPF1 (Page et al., 1999). Likewise, a frequent SNP rs2428212 with minor allele frequency (MAF) of 0.23 (i.e., the frequency of the less common allele is 23% in the population) was found to be associated with the expression of UPF3B (P < 1.0e−8, Table 2) (Castagne et al., 2011). These SNP-eQTL map in cis with SMG7 and UPF3B, suggesting that they directly regulate their expression, but it cannot be excluded that they are in linkage disequilibrium (i.e. close by) with the bona fide regulatory SNPs. While these studies suggest that cis-regulatory SNPs cause only a few cases of inter-individual variation in NMD efficiency, the actual number of NMD factors subjected to this type of regulation is likely to be higher than reported so far, as published studies have been conducted on a limited number of cell types (e.g., LCLs and blood monocytes) and only examined common SNPs with frequencies greater than 5%. Since SNP-eQTL are often tissue- and population-specific (Fu et al., 2012; Stranger et al., 2012), it would not be surprising if there were many additional layers of NMD factor eQTL variability that are responsible for NMD factor variability among different individuals.
Table 2.
SNP-eQTL and SNP-pQTL associated with the expression and efficiency of NMD.
| NMD Factors | SNPs | MAF | Reference | Effects | Experimentally validated |
|---|---|---|---|---|---|
| SMG7 | rs1044879, rs10911353, rs2702180, rs2275675, rs2296164, rs2274064, rs6662844, rs4047801, rs4652800 | >0.05 | Dixon et al. (2007) | Either increased or decreased SMG7 expression | No |
| rs2702182, rs2702178, rs2702180, rs10911353, rs12117885, rs12032165, rs12144253 | >0.05 | Stranger et al. (2012) | Either increased or decreased SMG7 expression | No | |
| UPF3B | rs2428212 | 0.23 | Castagne et al. (2011) | Either increased or decreased UPF3B expression | No |
| RBM8A | rs139428292 | 0.03 | Albers et al. (2012) | Allele A - Reduced RBM8A expression | Yes |
| rs201779890 | 0.0042 | Albers et al. (2012) | Allele C - Reduced RBM8A expression | No | |
| MAGOH | rs6673692 | 0.46 | Wu et al. (2013) | 10% change in MAGOH protein expression | No |
Abbreviation: MAF, Minor Allele Frequency
While, as described above, most studies have focused on relatively common SNPs, two rare SNPs associated with RBM8A expression have been reported: rs139428292 (MAF = 0.03) and the intronic rs201779890 (MAF = 0.0042), both of which were found to be significantly enriched in patients with TAR syndrome (Table 2). Since inheritance of either one of these SNPs predisposes the carrier to TAR syndrome, there is tremendous interest in defining their molecular mechanism (Albers et al., 2012). Allele A of rs139428292 was experimentally shown to decrease the binding of transcription factor EVI1 to the promoter of RBM8A, thereby providing a likely explanation for reduced RBM8A expression (Albers et al., 2012). Allele C of rs201779890 is also associated with a reduction in RBM8A expression, but the functional role of this SNP, if any, is not known. More such low frequency SNPs will hopefully be captured as more personal genomes are generated. Together, this may provide a more complete picture of how mutations can alter the expression of NMD and EJC factors, and thereby influence the transcriptome under the control of the NMD pathway.
In addition to being associated with gene expression, SNPs associated with protein levels—referred to as SNP protein-QTL (SNP-pQTL)—are beginning to be defined. In a recent study, the level of ~5000 proteins in LCLs from 95 HapMap individuals were examined alongside with their genotypes (Wu et al., 2013). Among the few hundred SNP-pQTL identified, one [rs6673692 (MAF = 0.46)] was found to be significantly associated with the expression of MAGOH, a core member of the EJC (Table 1 and 2). While the variation in MAGOH level was only modest (~10%), it is possible that this elicits sizeable physiological effect, based on the observed defects in neural stem cell division and brain size in heterozygous Magoh mice (Silver et al., 2010).
When interpreting the effect of simple nucleotide and CNV variation on inter-individual variability in NMD efficiency, it is important to bear in mind that NMD efficiency does not necessarily correlate with the level of NMD factors. If a given NMD factor is not rate limiting in a given cell type at a particular developmental stage, then mutations that increase its expression or modestly reduce its levels would be predicted to have no impact on the magnitude of NMD. On the other hand, if a NMD factor is rate limiting, then even a modest increase or decrease in its level could lead to major changes in the transcriptome. For example, Huang et al. (2011) found that modest overexpression of SMG1 increased NMD efficiency in HeLa cells, while modest overexpression of several other NMD factors did not, suggesting that SMG1 is the only rate-limiting NMD factor in these cells (Huang et al., 2011). UPF1 overexpression increases the magnitude of NMD in U2OS osteosarcoma cells, indicating that UPF1 is rate limiting for NMD in these cells (Gardner, 2008). Evidence that many NMD factors are not rate limiting in vivo was observed by Zetoune et al. (2008), who observed that while the magnitude of NMD differed significantly between different mouse tissues (as indicated by the ratio of PTC+ to PTC-Men1 transcripts in a Men1 heterozygote), this did not correlate in an obvious way with NMD factor mRNA levels (Zetoune et al., 2008).
6.2 Competition with the Staufen-mediated mRNA decay pathway
Similar to NMD, Staufen-mediated mRNA decay (SMD) pathway is an RNA decay pathway used by cells to regulate gene expression. SMD shares with NMD the requirement for UPF1, however, SMD differs from NMD in other respects, including that it is independent of splicing and the EJC (Hosoda et al., 2005; Kim et al., 2005). SMD is a translation-dependent mechanism that is triggered when the RNA-binding protein, Staufen, binds to the 3′ UTR of its target mRNA. This leads to recruitment of UPF1, followed by rapid degradation of the transcript via mechanisms that may be similar to that which attacks NMD substrates (Kim et al., 2005; Kim et al., 2007; Maquat and Gong, 2009).
SMD and NMD are competitive pathways by virtue of the fact that both depend on UPF1. One line of evidence for this is that the SMD factors, STAU1 and its paralog STAU2, bind to the same region of UPF1 protein as the NMD factor UPF2 (Gong et al., 2009; Park et al., 2013). As a result of this, STAU1 and STAU2 bind to UPF1 in a mutually exclusive manner with UPF2. Another line of evidence is that down-regulating STAU1 enhances NMD efficiency, whereas down-regulating UPF2 increases SMD efficiency (Gong et al., 2009). This is believed to be an important mechanism utilized by cells to fine tune the expression of specific transcripts that drive specific developmental fates. As evidence for this, differentiation of C2C12 myoblast-like proliferative cells into multinucleated myotubes was shown to reciprocally regulate the efficiency of SMD and NMD, leading to decreased expression of the anti-muscle differentiation factor PAX3 and increased expression of the pro-muscle differentiation factor MYOGENIN, respectively (Gong et al., 2009).
Interestingly, available evidence suggests that nucleotide variations in STAU2 may be common in normal human populations. Seven partial CNVs (3 gains and 4 losses) spanning various exons of STAU2 have been recorded in the Database of Genomic Variants (DGV), which houses CNV data obtained from large number of controls from published studies (http://dgv.tcag.ca/dgv/app/home). These partial CNVs have breakpoints within the coding region of STAU2 and, therefore are likely to alter the reading frame of STAU2 transcripts, leading to haploinsufficiency. Although no SNP-eQTL was found to be associated with STAU2 mRNA expression, there are also over 50 small CNVs that reside within the introns of STAU2, which could also affect its transcription or processing by disrupting transcriptional enhancers or splicing cis elements, respectively. While disruption of STAU2 expression or function may be partially compensated by STAU1, it is known that depletion of STAU2 disrupts both SMD and NMD, even in the presence of STAU1, at least in cell lines (Park et al., 2013). Given the competitive relationship of SMD and NMD, it will be important to determine how and to what extent mutations of SMD genes impact NMD and whether this has clinically measurable and significant effects.
6.3. MicroRNA-mediated regulation of NMD
Micro-RNAs (miRNAs) are small (~22 nucleotide) non-coding RNAs that typically repress gene expression via complementary base-pairing of their “seed” sequence with the 3′ UTR of their target mRNAs. Transcripts bound by miRNAs are targeted for degradation, translationally repressed, or both (Okamura, 2012). Currently, there are over 2000 human miRNAs recorded in the miRbase database (Kozomara and Griffiths-Jones, 2011). Together, these miRNAs could target a large portion of the genes in the transcriptome; however, due to the high false positive rates of different prediction programs, experimental validations are necessary to determine the exact number of genes affected (Thomson et al., 2011). To identify miRNAs that regulate NMD, Bruno et al. (2011) performed a bioinformatics search for miRNAs predicted to target NMD factors. They identified two identical miRNAs generated from independent gene loci, miR-128-1 and miR-128-2, as potential regulators of the core NMD factor UPF1 and the EJC factor MLN51, which they confirmed experimentally (Bruno et al., 2011). The ability of miR-128 to repress UPF1 and MLN51 was functionally relevant, as miR-128 was shown to be capable of regulating the magnitude of NMD (Bruno et al., 2011). miR-128 is highly upregulated during both brain development and neuronal maturation, which raised the possibility that this downregulates NMD efficiency, which, in turn, would increase the stability of NMD substrate mRNAs in neurons. In support of this, the investigators identified several NMD substrates encoding neural-relevant proteins that are upregulated during brain development in vivo (Bruno et al., 2011). That this is physiologically relevant is supported by finding that forced expression of miR-128 in neural stem cells upregulates scores of mRNAs encoding proteins with functions in neural differentiation and function (Bruno et al., 2011).
Indirect genomic evidence suggests the existence of inter-individual variation in the expression of mir-128. This comes from the host genes of mir128-1 and mir128-2, R3HDM1 and ARPP21, respectively. There are several partial CNVs spanning regions of both R3HDM1 (2 gains, 3 losses) and ARPP21 (4 gains, 2 losses) (as recorded in the DGV). Since mir128-1 and mir128-2 are located in the introns of these genes, it is reasonable to speculate that alterations in the copy number of these loci will lead to alterations in the levels of these miRNAs. Intriguingly, it was recently reported that there are 20 CNV gains (complete duplications) of ARPP21 in a large cohort of both controls and patients with neuro-developmental conditions, though not being significantly enriched in any particular population (Cooper et al., 2011). Recently, another miRNA—miR-125—was shown to regulate NMD through its ability to repress the expression of the NMD factor SMG1 (Wang et al., 2013). Like miR-128, miR-125 is transcribed and processed from two independent loci. Genomic evidence suggests variability in the expression of these two miRNAs, as there are 11 CNVs (6 gains, 5 losses) encompassing one of them (miR-125a) and 5 CNVs (3 gains, 2 losses) encompassing the other (miR-125b), based on the DGV and published cohorts of patients and controls (Cooper et al., 2011). In coming years it will be interesting to elucidate the complexity of the networks involving NMD and the miRNAs controlling neural differentiation and function.
6.4 Negative feedback loops
Given the importance of NMD in maintaining cellular homeostasis, it is not surprising that self-regulatory feedback mechanisms have evolved to govern this process. The first hint of NMD feedback regulation in mammalian cells was noted by Mendel et al. (2004) and Chan et al. (2007), who found that SMG5 mRNA was significantly upregulated in HeLa cells upon depletion of UPF1 or UPF3B, respectively. This raised the possibility that the upregulation of NMD factors could rescue NMD function when NMD is perturbed. More recently, two other studies confirmed this hypothesis and provided further details of this regulation (Huang et al., 2011; Yepiskoposyan et al., 2011). These studies showed that not only SMG5, but also UPF1, UPF2, UPF3B, SMG1, SMG6 and SMG7 are up-regulated in HeLa cells upon inhibition of NMD. Interestingly, all of these NMD factor mRNAs contain “NMD-inducing features” (long 3′ UTRs and/or uORFs), some of which where empirically shown to trigger NMD (Huang et al., 2011; Yepiskoposyan et al., 2011). Together, these data suggest that a broad-acting buffering mechanism exists that stabilizes the mRNAs encoding most NMD factors in response to NMD perturbation.
Studies conducted by Huang et al. (2011) revealed that this NMD feedback loop acts in a cell- and tissue-specific manner and is also developmentally regulated. For example, the investigators found that loss of the NMD factor Upf3b in mice only upregulated NMD factors mRNAs in specific tissues and cell types. Among the tissues tested, the spleen was particularly active in “compensating” for the loss of Upf3b, as evidenced by it upregulating the NMD factor transcripts Upf1, Upf2, Smg1, Smg6 and Smg7. This is interesting in light of the fact that the spleen houses T cells and B cells, which express antigen receptors generated from mRNAs that frequently contain PTCs as a result of error-prone programmed gene rearrangements, as described in Section 2. The brain was also found to be subjected to NMD feedback regulation, as Smg1 mRNA levels were increased in response to loss of Upf3b (Huang et al., 2011).
The NMD feedback regulatory response is highly conserved. Evidence suggests that this mechanism extends to Drosophila melanogaster, Arabidopsis thaliana, and humans (Chan et al., 2009; Huang et al., 2011; Kerenyi et al., 2008; Nguyen et al., 2012; Rehwinkel et al., 2005; Saul et al., 2009). This may be clinically relevant since such feedback response could alleviate environment toxicants that are known to inhibit NMD (see the next Section). In addition, such feedback regulation may be the reason why patients lacking a functional UPF3B gene are able to survive and maintain some behavioural and cognitive functions (Chan et al., 2009; Nguyen et al., 2012). While the selective forces that led to the generation of this feedback regulation is not known, it is reasonable to suspect that one functional benefit that led to its formation was limiting the variability in NMD function. By providing a buffer to ensure that NMD efficiency is within an acceptable range, this feedback system maintains transcriptome homeostasis (Figure 3).
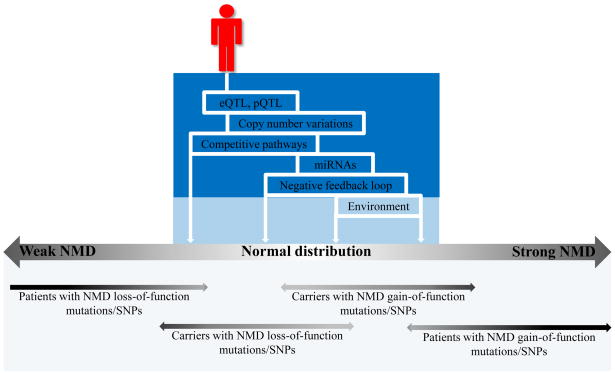
Figure 3. Multiple factors influencing NMD efficiency of each individual.
Different combination of genetic and non-genetic factors determines the NMD efficiency of each individual (top panel). Individual’s NMD efficiency may lie outside the normal distribution, in which case it will result in a detectable phenotype i.e. ID as seen in patients with NMD or EJC CNVs (Bottom panel).
6.3 Environmental factors
The efficiency of NMD can also be influenced by external factors. It has been shown that cellular stress elicited by amino-acid starvation, oncogene overexpression and hypoxia can rapidly inhibit NMD activity (Gardner, 2008; Mendell et al., 2004; Wang et al., 2011a). A common downstream consequence of these cellular stresses is increased phosphorylation of the translation initiation factor eIF2α, which has been empirically shown to inhibit NMD (Gardner, 2008). An obvious explanation for why eIF2α phosphorylation inhibits NMD is that it reduces the ability of nonsense codons to be recognized by the translation apparatus, but there is also evidence that this is not the case (Gardner, 2008, 2010; Wang et al., 2011a). Importantly, NMD inhibition upregulates many transcripts necessary for stress response, including the mRNAs encoding ATF3, ATF4, and CHOP, all of which are known to allow cells better cope with stress (Gardner, 2008). While an increased capability of dealing with stress is beneficial for cells, NMD inhibition may also have negative consequences. For example, there is evidence that NMD normally serves to repress tumor formation and thus inhibition of NMD would be predicted to promote tumor progression (Wang et al., 2011b). Indeed, inhibition of NMD leads to upregulation of many transcripts necessary for tumour progression (Wang et al., 2011b). It will be important in the future to determine whether inter-individual variability in NMD efficiency influences the likelihood of tumor progression. While the notion that NMD inhibits tumor formation is intriguing, we note that very few mutations have been reported in NMD genes in tumours in the Database of Somatic Mutations in Cancer, or Cosmic Database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/), (unpublished observation). Thus, it is unclear whether changes in NMD function or efficiency are major contributing factors that drive tumor formation in humans.
7 Perspective
As we have discussed in this Review, it is becoming apparent that NMD efficiency varies between individuals and that this efficiency can be influenced by both genetic and non-genetic actors. Both the full spectrum of these factors and the interplay between these factors is likely to dictate a given individual’s net magnitude of NMD (Figure 3). We propose that variable NMD efficiency provides a plausible explanation for the variable clinical presentations and drug responses of patients carrying the same PTC mutation (Section 5). It may also explain apparently normal clinical presentations of some carrier parents (Section 4). Thus, we suggest that inter-individual differences in NMD efficiency should be taken into consideration when treating human diseases caused by genes with PTC-generating mutations and in particular those treatments incorporating PTC read-through drugs.
More studies are needed in order to fully appreciate the impact of inter-individual variability in NMD efficiency. Firstly, it is necessary to determine the extent of inter-individual variability in NMD efficiency in disease and non-disease cohorts. As readout, the levels of established canonical NMD targets ought to be determined in well-defined sets of tissue types from different population groups. This can be facilitated by the large number of microarray and RNA-Seq datasets currently available from the HapMap depository (http://hapmap.ncbi.nlm.nih.gov/) and expression studies of different disease cohorts, e.g. autism (Voineagu, 2012). Secondly, it will be necessary to establish the physiological consequences (both functional and clinical) of copy number gains or losses of NMD factor genes in patients with neuro-developmental conditions (Section 4). Thus far, the evidence in support of a cause-and-effect relationship has been limited to studies in manipulated cell lines; e.g., HeLa cells. In the future, it will be important to perform whole transcriptome and proteome analyses on primary cells, iPS-derived neurons and other cell types from individuals with such CNVs (Nguyen et al., 2013).
Although it may or may not be possible to establish a broadly applicable and “clinically relevant” threshold of NMD, the information on individual’s NMD efficiency will form another facet of personalized therapy approaches, including those aimed at suppressing NMD and others aimed at enhancing the magnitude of NMD. At present, only NMD-inhibitory drugs are being tested. This should be expanded to include NMD-enhancing drugs since it is clear that compromised or ‘weak’ NMD efficiency can also be clinically relevant (section 4). We suggest that one target of such NMD-enhancement therapy be the NMD factor, UPF3A, since its levels positively correlate with less severe symptoms in ID patients (Nguyen et al., 2012). The recent evidence for the intricate and dynamic mechanisms that regulate NMD (Karam et al. (2013) and references therein) should be taken into account when developing and clinically applying both NMD-enhancing and –inhibiting drugs. Thus, it will be important to develop tissue- and NMD factor-specific approaches to reduce side effects and maximize therapeutic efficacy. This follows from the evidence that (i) NMD efficiency is variable, (ii) different NMD components are probably rate-limiting factors in different tissues, and (iii) mRNAs differ with regard to the branch of NMD they respond to (Chan et al., 2007; Gehring et al., 2005; Metze et al., 2013). Because “out-of-range” NMD efficiency can impair neuronal development and function (Section 4), drug dose should be carefully titrated to adjust the efficiency of NMD to be within the “normal” range (Figure 3). Thus, future clinical trials should include comprehensive mental assessments of the participants after both short and long term administration of the drugs.
Research into the existence, scale and implications of inter-individual variation in NMD efficiency is in its infancy. This subject represents both an opportunity and a considerable challenge. An increased understanding of the factors controlling NMD efficiency will be essential for the better management and treatment of human diseases caused by genes with protein truncating mutations, and thus provide another example of “personalized medicine.” Given that NMD is, to one extent or another, involved in about one-third of all currently known human genetic disorders, manipulation of the efficiency of NMD has the potential to have a large impact on human disease and human well being, in general.
Highlights.
Nonsense-mediated mRNA decay (NMD) is a crucial regulator of normal development.
We examine evidence supporting inter-individually variable NMD and its relevance to disease.
Sources of variable NMD include eQTL, miRNA, SMD and negative feedback loop.
NMD efficiency should be considered when manipulating it for therapeutic purposes.
Acknowledgments
This study was supported by the Australian NH&MRC project grant 628952 (to JG) and the National Institutes of Health grant GM058595 (to MW). JG is supported by NH&MRC Senior Principal Research Fellowship 1041920. LSN was supported by the MS McLeod Foundation PhD Scholarship. The manuscript was written by LSN and edited by MW and JG.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N, Miller R, Tossell J, Bakalar J, Germain G, Gochman P, Long R, Rapoport JL, Rouleau GA. A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry. 2011;16:238–239. doi: 10.1038/mp.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alachkar A, Jiang D, Harrison M, Zhou Y, Chen G, Mao Y. An EJC Factor RBM8a Regulates Anxiety Behaviors. Current molecular medicine. 2013;13:887–899. doi: 10.2174/15665240113139990019. [DOI] [PubMed] [Google Scholar]
- Albers CA, Paul DS, Schulze H, Freson K, Stephens JC, Smethurst PA, Jolley JD, Cvejic A, Kostadima M, Bertone P, Breuning MH, Debili N, Deloukas P, Favier R, Fiedler J, Hobbs CM, Huang N, Hurles ME, Kiddle G, Krapels I, Nurden P, Ruivenkamp CA, Sambrook JG, Smith K, Stemple DL, Strauss G, Thys C, van Geet C, Newbury-Ecob R, Ouwehand WH, Ghevaert C. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat Genet. 2012;44:435–439. doi: 10.1038/ng.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applequist SE, Selg M, Raman C, Jack HM. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguet A, Degot S, Cougot N, Bertrand E, Chenard MP, Wendling C, Kessler P, Le Hir H, Rio MC, Tomasetto C. The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci. 2007;120:2774–2784. doi: 10.1242/jcs.009225. [DOI] [PubMed] [Google Scholar]
- Baumann B, Potash MJ, Kohler G. Consequences of frameshift mutations at the immunoglobulin heavy chain locus of the mouse. EMBO J. 1985;4:351–359. doi: 10.1002/j.1460-2075.1985.tb03636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Ebert J, Unterholzner L, Guttler T, Izaurralde E, Conti E. Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep. 2004;5:304–310. doi: 10.1038/sj.embor.7400091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, Pfaff SL, Wilkinson MF. Identification of a MicroRNA that Activates Gene Expression by Repressing Nonsense-Mediated RNA Decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M, Paillusson A, Muhlemann O. Efficient downregulation of immunoglobulin mu mRNA with premature translation-termination codons requires the 5′-half of the VDJ exon. Nucleic Acids Res. 2004;32:3304–3315. doi: 10.1093/nar/gkh651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali BM, Kuchma SL, Latham J, Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaert B, Su CT, Van Damme T, Vlummens P, Malfait F, Vanakker O, Schulz B, Mac Neal M, Davis EC, Lee JG, Salhi A, Unger S, Heimdal K, De Almeida S, Kornak U, Gaspar H, Bresson JL, Prescott K, Gosendi ME, Mansour S, Pierard GE, Madan-Khetarpal S, Sciurba FC, Symoens S, Coucke PJ, Van Maldergem L, Urban Z, De Paepe A. Comprehensive clinical and molecular analysis of 12 families with type 1 recessive cutis laxa. Hum Mutat. 2013;34:111–121. doi: 10.1002/humu.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagne R, Zeller T, Rotival M, Szymczak S, Truong V, Schillert A, Tregouet DA, Munzel T, Ziegler A, Cambien F, Blankenberg S, Tiret L. Influence of sex and genetic variability on expression of X-linked genes in human monocytes. Genomics. 2011;98:320–326. doi: 10.1016/j.ygeno.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, 2nd, Wilkinson MF. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu SY, Serin G, Ohara O, Maquat LE. Characterization of human Smg5/7a: a protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, McCracken E, Niyazov D, Leppig K, Thiese H, Hummel M, Alexander N, Gorski J, Kussmann J, Shashi V, Johnson K, Rehder C, Ballif BC, Shaffer LG, Eichler EE. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degot S, Le Hir H, Alpy F, Kedinger V, Stoll I, Wendling C, Seraphin B, Rio MC, Tomasetto C. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J Biol Chem. 2004;279:33702–33715. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP. Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J Biol Chem. 2001;276:22709–22714. doi: 10.1074/jbc.C100144200. [DOI] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WO. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol. 2003;10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- Fu J, Wolfs MG, Deelen P, Westra HJ, Fehrmann RS, Te Meerman GJ, Buurman WA, Rensen SS, Groen HJ, Weersma RK, van den Berg LH, Veldink J, Ophoff RA, Snieder H, van Heel D, Jansen RC, Hofker MH, Wijmenga C, Franke L. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LB. Nonsense-mediated RNA decay regulation by cellular stress: implications for tumorigenesis. Mol Cancer Res. 2010;8:295–308. doi: 10.1158/1541-7786.MCR-09-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009a;7:e1000120. doi: 10.1371/journal.pbio.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009b;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010;19:R176–187. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Nimgaonkar VL, Go RCP, Savage RM, Swerdlow NR, Gur RE, Braff DL, King MC, McClellan JM. Spatial and Temporal Mapping of De Novo Mutations in Schizophrenia to a Fetal Prefrontal Cortical Network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE, del Bosque-Plata L, Oda Y, Yoshiuchi I, Colilla S, Polonsky KS, Wei S, Concannon P, Iwasaki N, Schulze J, Baier LJ, Bogardus C, Groop L, Boerwinkle E, Hanis CL, Bell GI. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- Huang L, Lou CH, Chan W, Shum EY, Shao A, Stone E, Karam R, Song HW, Wilkinson MF. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–961. doi: 10.1016/j.molcel.2011.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3:807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt315. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol. 2004;11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- Karam R, Wengrod J, Gardner LB, Wilkinson MF. Regulation of nonsense-mediated mRNA decay: Implications for physiology and disease. Biochim Biophys Acta. 2013;1829:624–633. doi: 10.1016/j.bbagrm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, Izaurralde E. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes Dev. 2010;24:2440–2450. doi: 10.1101/gad.604610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G. Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-dependent exon-exon junction complex. EMBO J. 2001;20:6424–6433. doi: 10.1093/emboj/20.22.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Kebaara BW, Atkin AL. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:2771–2778. doi: 10.1093/nar/gkp146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Bedwell DM. Suppression of nonsense mutations as a therapeutic approach to treat genetic diseases. Wiley Interdiscip Rev RNA. 2011;2:837–852. doi: 10.1002/wrna.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerem E, Hirawat S, Armoni S, Yaakov Y, Shoseyov D, Cohen M, Nissim-Rafinia M, Blau H, Rivlin J, Aviram M, Elfring GL, Northcutt VJ, Miller LL, Kerem B, Wilschanski M. Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet. 2008;372:719–727. doi: 10.1016/S0140-6736(08)61168-X. [DOI] [PubMed] [Google Scholar]
- Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 2008;27:1585–1595. doi: 10.1038/emboj.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr TP, Sewry CA, Robb SA, Roberts RG. Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum Genet. 2001;109:402–407. doi: 10.1007/s004390100598. [DOI] [PubMed] [Google Scholar]
- Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14:1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001a;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 2001b;20:2062–2068. doi: 10.1093/emboj/20.8.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1015–1022. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Lau CK, Diem MD, Dreyfuss G, Van Duyne GD. Structure of the Y14-Magoh core of the exon junction complex. Curr Biol. 2003;13:933–941. doi: 10.1016/s0960-9822(03)00328-2. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Nguyen LS, Jolly L, Raynaud M, Gecz J. The role of the UPF3B gene and nonsense-mediated mRNA decay in Autism Spectrum Disorders. In: Patel VB, Preedy VR, Marin CR, editors. The Comprehensive Guide to Autism. Springer; 2013. [Google Scholar]
- Laumonnier F, Shoubridge C, Antar C, Nguyen LS, Van Esch H, Kleefstra T, Briault S, Fryns JP, Hamel B, Chelly J, Ropers HH, Ronce N, Blesson S, Moraine C, Gecz J, Raynaud M. Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol Psychiatry. 2010;15:767–776. doi: 10.1038/mp.2009.14. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Ishigaki Y, Li X, Maquat LE. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J. 2002;21:3536–3545. doi: 10.1093/emboj/cdf345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Linde L, Boelz S, Nissim-Rafinia M, Oren YS, Wilschanski M, Yaacov Y, Virgilis D, Neu-Yilik G, Kulozik AE, Kerem E, Kerem B. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest. 2007;117:683–692. doi: 10.1172/JCI28523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde L, Kerem B. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet. 2008;24:552–563. doi: 10.1016/j.tig.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Longman D, Plasterk RH, Johnstone IL, Caceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21:1075–1085. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Lynch SA, Nguyen LS, Ng LY, Waldron M, McDonald D, Gecz J. Broadening the phenotype associated with mutations in UPF3B: Two further cases with renal dysplasia and variable developmental delay. Eur J Med Genet. 2012;55:476–479. doi: 10.1016/j.ejmg.2012.03.010. [DOI] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB, Albers CA, Zhang ZD, Conrad DF, Lunter G, Zheng H, Ayub Q, DePristo MA, Banks E, Hu M, Handsaker RE, Rosenfeld JA, Fromer M, Jin M, Mu XJ, Khurana E, Ye K, Kay M, Saunders GI, Suner MM, Hunt T, Barnes IH, Amid C, Carvalho-Silva DR, Bignell AH, Snow C, Yngvadottir B, Bumpstead S, Cooper DN, Xue Y, Romero IG, Wang J, Li Y, Gibbs RA, McCarroll SA, Dermitzakis ET, Pritchard JK, Barrett JC, Harrow J, Hurles ME, Gerstein MB, Tyler-Smith C. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald MR, Schaefer GB, Olney AH, Patton DF. Hypoplasia of the cerebellar vermis and corpus callosum in thrombocytopenia with absent radius syndrome on MRI studies. Am J Med Genet. 1994;50:46–50. doi: 10.1002/ajmg.1320500111. [DOI] [PubMed] [Google Scholar]
- Maderazo AB, He F, Mangus DA, Jacobson A. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol Cell Biol. 2000;20:4591–4603. doi: 10.1128/mcb.20.13.4591-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald GK, Mahowald MA, Moon C, Khor B, Sleckman BP. Out-of-frame T cell receptor beta transcripts are eliminated by multiple pathways in vivo. PLoS One. 2011;6:e21627. doi: 10.1371/journal.pone.0021627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan V, Rajan D, Thomas S, Shaw AC, Louis Dit Picard H, Layet V, Till M, van Haeringen A, Mortier G, Nampoothiri S, Puseljic S, Legeai-Mallet L, Carter NP, Vekemans M, Munnich A, Hennekam RC, Colleaux L, Cormier-Daire V. Distinct effects of allelic NFIX mutations on nonsense-mediated mRNA decay engender either a Sotos-like or a Marshall-Smith syndrome. Am J Hum Genet. 2010;87:189–198. doi: 10.1016/j.ajhg.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Gong C. Gene expression networks: competing mRNA decay pathways in mammalian cells. Biochem Soc Trans. 2009;37:1287–1292. doi: 10.1042/BST0371287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy SP, Nomura T, Torrie LS, Warbrick E, Gartner U, Wood G, McLean WH. A lack of premature termination codon read-through efficacy of PTC124 (Ataluren) in a diverse array of reporter assays. PLoS Biol. 2013;11:e1001593. doi: 10.1371/journal.pbio.1001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, Mak TW. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A. 2010;107:12186–12191. doi: 10.1073/pnas.1007336107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol. 2000;20:8944–8957. doi: 10.1128/mcb.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Metze S, Herzog VA, Ruepp MD, Muhlemann O. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA. 2013;19:1432–1448. doi: 10.1261/rna.038893.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Yamashita A, Kashima I, Ogata K, Ishiura S, Ohno S. Distant N- and C-terminal domains are required for intrinsic kinase activity of SMG-1, a critical component of nonsense-mediated mRNA decay. J Biol Chem. 2007;282:7799–7808. doi: 10.1074/jbc.M610159200. [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Nazarenus T, Cedarberg R, Bell R, Cheatle J, Forch A, Haifley A, Hou A, Wanja Kebaara B, Shields C, Stoysich K, Taylor R, Atkin AL. Upf1p, a highly conserved protein required for nonsense-mediated mRNA decay, interacts with the nuclear pore proteins Nup100p and Nup116p. Gene. 2005;345:199–212. doi: 10.1016/j.gene.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Nguyen LS, Jolly L, Shoubridge C, Chan WK, Huang L, Laumonnier F, Raynaud M, Hackett A, Field M, Rodriguez J, Srivastava AK, Lee Y, Long R, Addington AM, Rapoport JL, Suren S, Hahn CN, Gamble J, Wilkinson MF, Corbett MA, Gecz J. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol Psychiatry. 2012;17:1103–1115. doi: 10.1038/mp.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LS, Kim HG, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, Layman LC, Shaffer LG, Gecz J. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet. 2013;22:1816–1825. doi: 10.1093/hmg/ddt035. [DOI] [PubMed] [Google Scholar]
- Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- Okada-Katsuhata Y, Yamashita A, Kutsuzawa K, Izumi N, Hirahara F, Ohno S. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2012;40:1251–1266. doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K. Diversity of animal small RNA pathways and their biological utility. Wiley Interdiscip Rev RNA. 2012;3:351–368. doi: 10.1002/wrna.113. [DOI] [PubMed] [Google Scholar]
- Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- Park E, Gleghorn ML, Maquat LE. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci U S A. 2013;110:405–412. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ, Jr, Dietz HC. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci U S A. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resta N, Susca FC, Di Giacomo MC, Stella A, Bukvic N, Bagnulo R, Simone C, Guanti G. A homozygous frameshift mutation in the ESCO2 gene: evidence of intertissue and interindividual variation in Nmd efficiency. J Cell Physiol. 2006;209:67–73. doi: 10.1002/jcp.20708. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA, Traylor RN, Schaefer GB, McPherson EW, Ballif BC, Klopocki E, Mundlos S, Shaffer LG, Aylsworth AS. Proximal microdeletions and microduplications of 1q21.1 contribute to variable abnormal phenotypes. Eur J Hum Genet. 2012;20:754–761. doi: 10.1038/ejhg.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Peltz SW. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell. 2000;101:741–751. doi: 10.1016/s0092-8674(00)80886-7. [DOI] [PubMed] [Google Scholar]
- Saul H, Elharrar E, Gaash R, Eliaz D, Valenci M, Akua T, Avramov M, Frankel N, Berezin I, Gottlieb D, Elazar M, David-Assael O, Tcherkas V, Mizrachi K, Shaul O. The upstream open reading frame of the Arabidopsis AtMHX gene has a strong impact on transcript accumulation through the nonsense-mediated mRNA decay pathway. Plant J. 2009;60:1031–1042. doi: 10.1111/j.1365-313X.2009.04021.x. [DOI] [PubMed] [Google Scholar]
- Sauliere J, Haque N, Harms S, Barbosa I, Blanchette M, Le Hir H. The exon junction complex differentially marks spliced junctions. Nat Struct Mol Biol. 2010;17:1269–1271. doi: 10.1038/nsmb.1890. [DOI] [PubMed] [Google Scholar]
- Sauliere J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonqueze O, Audic Y, Paillard L, Roest Crollius H, Le Hir H. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19:1124–1131. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zund D, Yamashita A, Muhlemann O. Nonsense-mediated mRNA decay - mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829:612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol Cell Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Xu RM. Crystal structure of the Drosophila Mago nashi-Y14 complex. Genes Dev. 2003;17:971–976. doi: 10.1101/gad.260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AL, Pereira FJ, Morgado A, Kong J, Martins R, Faustino P, Liebhaber SA, Romao L. The canonical UPF1-dependent nonsense-mediated mRNA decay is inhibited in transcripts carrying a short open reading frame independent of sequence context. RNA. 2006;12:2160–2170. doi: 10.1261/rna.201406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, Liaw HJ, Myung K, Walsh CA, Gaiano N, Pavan WJ. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13:551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KK, Wachsmuth L, Kulozik AE, Gehring NH. Two mammalian MAGOH genes contribute to exon junction complex composition and nonsense-mediated decay. RNA Biol. 2013;10:1291–1298. doi: 10.4161/rna.25827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Montgomery SB, Dimas AS, Parts L, Stegle O, Ingle CE, Sekowska M, Smith GD, Evans D, Gutierrez-Arcelus M, Price A, Raj T, Nisbett J, Nica AC, Beazley C, Durbin R, Deloukas P, Dermitzakis ET. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol. 2012;9:1370–1379. doi: 10.4161/rna.22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarpey PS, Nguyen LS, Raymond FL, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, O’Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Hills K, Jones D, Mironenko T, Perry J, Varian J, West S, Widaa S, Teague J, Dicks E, Butler A, Menzies A, Richardson D, Jenkinson A, Shepherd R, Raine K, Moon J, Luo Y, Parnau J, Bhat SS, Gardner A, Corbett M, Brooks D, Thomas P, Parkinson-Lawrence E, Porteous ME, Warner JP, Sanderson T, Pearson P, Simensen RJ, Skinner C, Hoganson G, Superneau D, Wooster R, Bobrow M, Turner G, Stevenson RE, Schwartz CE, Futreal PA, Srivastava AK, Stratton MR, Gecz J. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Voineagu I. Gene expression studies in autism: moving from the genome to the transcriptome and beyond. Neurobiol Dis. 2012;45:69–75. doi: 10.1016/j.nbd.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Wang D, Wengrod J, Gardner LB. Overexpression of the c-myc oncogene inhibits nonsense-mediated RNA decay in B lymphocytes. J Biol Chem. 2011a;286:40038–40043. doi: 10.1074/jbc.M111.266361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zavadil J, Martin L, Parisi F, Friedman E, Levy D, Harding H, Ron D, Gardner LB. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol. 2011b;31:3670–3680. doi: 10.1128/MCB.05704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Jiang B, Jia C, Chai B, Liang A. MicroRNA 125 represses nonsense-mediated mRNA decay by regulating SMG1 expression. Biochem Biophys Res Commun. 2013;435:16–20. doi: 10.1016/j.bbrc.2013.03.129. [DOI] [PubMed] [Google Scholar]
- Wang J, Gudikote JP, Olivas OR, Wilkinson MF. Boundary-independent polar nonsense-mediated decay. EMBO Rep. 2002a;3:274–279. doi: 10.1093/embo-reports/kvf036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Vock VM, Li S, Olivas OR, Wilkinson MF. A quality control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts by a mechanism requiring UPF2 and translation. J Biol Chem. 2002b;277:18489–18493. doi: 10.1074/jbc.M111781200. [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Waage J, Tian G, Zhao J, Damgaard I, Jakobsen JS, Kristiansen K, Krogh A, Wang J, Porse BT. Mammalian tissues defective in nonsense-mediated mRNA decay display highly aberrant splicing patterns. Genome Biol. 2012;13:R35. doi: 10.1186/gb-2012-13-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, Wilde RG, Karp G, Takasugi J, Chen G, Jones S, Ren H, Moon YC, Corson D, Turpoff AA, Campbell JA, Conn MM, Khan A, Almstead NG, Hedrick J, Mollin A, Risher N, Weetall M, Yeh S, Branstrom AA, Colacino JM, Babiak J, Ju WD, Hirawat S, Northcutt VJ, Miller LL, Spatrick P, He F, Kawana M, Feng H, Jacobson A, Peltz SW, Sweeney HL. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- Welch EM, Jacobson A. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 1999;18:6134–6145. doi: 10.1093/emboj/18.21.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, Tang H, Snyder M. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhang L, Tong P, Xun G, Su W, Xiong Z, Zhu T, Zheng Y, Luo S, Pan Y, Xia K, Hu Z. Exome sequencing identifies UPF3B as the causative gene for a Chinese non-syndrome mental retardation pedigree. Clin Genet. 2013;83:560–564. doi: 10.1111/cge.12014. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, Kurata R, Hirano H, Anderson P, Ohno S. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Muhlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetoune AB, Fontaniere S, Magnin D, Anczukow O, Buisson M, Zhang CX, Mazoyer S. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 2008;9:83. doi: 10.1186/1471-2156-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XF, Nowak NJ, Shows TB, Aplan PD. MAGOH interacts with a novel RNA-binding protein. Genomics. 2000;63:145–148. doi: 10.1006/geno.1999.6064. [DOI] [PubMed] [Google Scholar]