Abstract
Recycling old drugs, rescuing shelved drugs and extending patents' lives make drug repositioning an attractive form of drug discovery. Drug repositioning accounts for approximately 30% of the newly US Food and Drug Administration (FDA)-approved drugs and vaccines in recent years. The prevalence of drug-repositioning studies has resulted in a variety of innovative computational methods for the identification of new opportunities for the use of old drugs. Questions often arise from customizing or optimizing these methods into efficient drug-repositioning pipelines for alternative applications. It requires a comprehensive understanding of the available methods gained by evaluating both biological and pharmaceutical knowledge and the elucidated mechanism-of-action of drugs. Here, we provide guidance for prioritizing and integrating drug-repositioning methods for specific drug-repositioning pipelines.
Keywords: drug repositioning, drug repurposing, computational methods, lotus leaves flowchart (LLF), fishbone flowchart, efficient repositioning pipelines
Teaser
Drug repositioning reuses old drugs for new indications. This article provides a guide for understanding the existing drug-repositioning methods and customizing them into new efficient drug-repositioning pipelines. A repositioned drug does not need the initial 6–9 years typically required for the development of new drugs, but instead goes directly to preclinical testing and clinical trials, thus reducing risk and costs [1]. Repositioning or repurposing drugs has been implemented in several ways. One of the well-known examples is sildenafil citrate (brand name: Viagra), which was repositioned from a common hypertension drug to a therapy for erectile dysfunction [2]. Similarly, off-label use of FDA-approved drugs for cancer medical practice is also popular. The National Comprehensive Cancer Network (NCCN) estimates off-label use accounts for 50–75% of drugs or biologic therapies for cancer in the USA [3]. It has been reported that 78% and 75% of patients with breast or lung cancer, respectively received FDA-approved drugs, although 68% and 95% of these drugs, respectively, were used for off-label indications not approved by the FDA [4]. Obviously, these examples were serendipitously identified and these repositioning strategies lack guidance and information to support clinical decision.
Pharmaceutical companies rely on traditional drug discovery methods to seek repositioning opportunities. Among the 75 agents (50 small molecules and 25 biologics) approved between 1999 and 2008, 28 first-in-class small molecules were discovered by phenotypic drug screening and 17 were identified by target-based methods [5,6], accounting for more than 50% of the FDA-approved small molecules and biologics. Phenotypic drug-screening approaches discover drug candidates from libraries serendipitously. Alternatively, target-based methods improve the repositioning process by including known target information into drug-repositioning studies.
Nevertheless, the low knowledge content of elucidated mechanisms for traditional drug-repositioning methods makes it hard to satisfy unmet medical needs by successfully repositioning a large number of existing or shelved drugs. Computational methods are able to alleviate this problem by high-level integration of available knowledge and elucidation of unknown mechanisms. These computational methods significantly improve the discovery process in which new indications for a drug or new drugs for a disease can be identified. They take advantage of the methods and tools available in chemoinformatics [7–9], bioinformatics [10–14], network biology [15–17] and systems biology [18–20] to make full use of known targets, drugs and disease biomarkers or pathways, thus leading to the development of proof-of-concept methods and the design of clinical studies with accelerated timelines. Accordingly, computational drug-repositioning methods can be classified into target-based, knowledge-based, signature-based, pathway- or network-based, and targeted-mechanism-based methods, as shown in Figure 1. These methods focus on different orientations defined by available information and elucidated mechanisms, such as drug oriented, disease oriented and treatment oriented. These computational drug-repositioning methods enable researchers to examine nearly all drug candidates and test on a relatively large number of diseases within significantly shortened time lines.
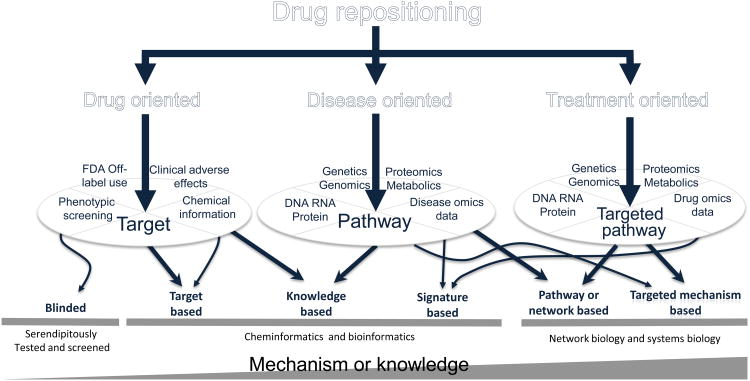
Figure 1.
Lotus leaves flowchart (LLF) for categorization of existing drug-repositioning methods. Drug repositioning takes advantage of different potential avenues to repurpose drugs for new indications, including drug, disease and treatment oriented. These avenues were developed according to the availability of biological and pharmaceutical knowledge and requirement of understanding the mechanisms of action of drugs. Traditional phenotype-based screening methods do not need prior knowledge, and the repositioned drugs are just serendipitously tested. Targeted-based methods need specific knowledge about the targets, such as 3D protein structures, whereas knowledge-based methods require the knowledge about the drugs or diseases, such as adverse effects, FDA approval labels, records of clinical trials and published disease biomarkers (potential targets) or disease pathways. Signature-based methods mainly make use of gene signatures defined by ‘-omics’ data (for diseases, drug treatments, or both). Pathway- or network-based methods generally use pathway analysis or network biology methods to discover essential pathways from genetic, genomic, proteomic and metabolic data of diseases to find new targets for repositioned drugs. More advanced drug-repositioning methods, such as targeted mechanism-based methods, aim to discover mechanisms of action of drugs by identification of off-targets or targeted pathways of treated drugs using drug omics data (before and after drug treatments). Details of these methods are in Table 1 (main text). Integrated knowledge and elucidated mechanisms of drug actions increases with the complexity of modeling methods.
In recent years, the number of drug-repositioning methods has dramatically increased. It is essential to better understand these existing methods and prioritize them based on specific studies. Application of an efficient drug-srepositioning pipeline to a specific study needs identification of feasible methods based on available information of the drugs or diseases of interest. In this review, we link existing drug-repositioning methods with their integrated biological and pharmaceutical knowledge and discuss how to customize a new drug-repositioning pipeline for specific studies.
Prioritize available drug-repositioning methods
Figure 1 is a top-down flowchart that we developed to better understand orientations, integrated information types, categories and complexities of existing drug-repositioning methods. We call this flowchart a lotus leaves flowchart (LLF). It enables better understanding of repositioning methods from the top down while customizing new repositioning pipelines from the bottom up. As an example, if one wants to reposition drugs for an orphan disease, one needs to identify how much pharmaceutical or biological knowledge is available for this disease and whether understanding the mechanisms of action of repositioned drugs is necessary. There are several options to do such drug repositioning. Option 1: when little information is available for the disease, phenotypic screening or FDA off-label use would be the best option. Option 2: if there exists one protein biomarker for the disease, target-based or knowledge-based methods should be prioritized for the study. Option 3: if there is more disease information available, either knowledge-based or signature-based methods can be deployed to integrate available disease pathways or disease omics data (i.e., omics data generated from diseases) into the drug-repositioning process. Lastly, option 4: if treatment omics data (i.e., omics data generated from drug treatment) are available, it is possible to use signature-based or targeted-mechanism-based methods to elucidate unknown targeted mechanisms, such as off-targets and targeted signaling pathways.
It is easy to see that the development of an efficient drug-repositioning pipeline is a process of tradeoff among purposes, methods and available information. Here, we introduce the repositioning methods shown in the LLF to facilitate understanding the purpose, the integrated information and the complexities of these methods. The LLF flowchart will be helpful for scientists and researchers to better understand existing computational methods and customize these methods into their own pipelines for drug-repositioning studies.
Blinded search or screening methods
Blinded drug-repositioning methods do not include pharmaceutical or biological information and are less likely to help elucidate any mechanisms of action of drugs. Most of them depend on serendipitous identification from tests aimed at specific diseases and drugs [3,4,21]. The advantage of these methods, which include FDA off-label use and phenotypic screening, is that they have high flexibility for application to a large number of drugs or diseases. This explains why the phenotypic screening method was used in the discovery of 28 of 75 small molecules and biologics approved by the FDA between 1999 and 2008.
Target-based methods
Target-based drug-repositioning methods comprise in vitro and in vivo high-throughput and/or high-content screening (HTS/HCS) of drugs for a protein or a biomarker of interest [7–9] and in silico screening of drugs or compounds from drug libraries [7,22], such as ligand-based screening or docking [23,24]. Compared with blinded methods, targeted-based methods significantly improve the likelihood of drug discovery because most targets link directly with the disease mechanisms. Integration of target information into the drug repositioning process ensures a higher possibility of finding useful drugs compared with traditional blinded methods. The advantage of targeted-based methods, such as docking, is that these methods enable researchers to screen nearly all drugs or compounds with known chemical structure information (e.g., SMILES[LM1]) within a few days. This is why so many pharmaceutical companies, including Genentech and Melior, have been using these methods to find new indications.
Knowledge-based methods
Knowledge-based drug-repositioning methods are those applying bioinformatics or cheminformatics approaches to include the available information of drugs, drug–target networks [10–14], chemical structures of targets and drugs [14], clinical trial information (adverse effects) [25,26], FDA approval labels [27], signaling or metabolic pathways [28], and so on, into drug-repositioning studies. The information content of blinded and target-based methods are poor and they cannot be used to identify new mechanisms beyond the known targets. By contrast, knowledge-based methods incorporate known information into predicting unknown mechanisms, such as unknown targets for drugs, unknown drug–drug similarities and new biomarkers for diseases. The advantage of knowledge-based methods is that they include a large amount of known information into the drug-repositioning process to improve its prediction accuracy. For example, THOMSON REUTERS™ has used this strategy to do drug repositioning based on its rich volumes of accumulated prior knowledge. Moreover, these methods have been applied to repurpose known drugs to pediatric hematology oncology. Blatt and Corey describe how the knowledge in the Harriet Lane Handbook (HLH) of the Johns Hopkins School of Medicine (compiled based on perceived interest to the general pediatric practitioners) and information acquired by searching PubMed and Google.com might also be helpful to repurpose drugs for children [29].
Signature-based methods
Signature-based drug-repositioning methods make use of gene signatures derived from disease omics data with or without treatments [30–37] to discover unknown off-targets or unknown disease mechanisms. As the advancement of microarray and next generation sequencing techniques speed up the generation of vast volumes of genomics data pertinent for drug-repositioning studies, gene signatures can be used to discover unknown mechanisms. One can easily access such genomics data in publicly available databases, such as NCBI-GEO (http://www.ncbi.nlm.nih.gov/geo/), SRA [LM2](http://www.ncbi.nlm.nih.gov/Traces/sra/), CMAP [38], and CCLE [39]. More details on these databases are shown in Table 2. The advantage of signature-based methods is that they are useful to uncover unknown mechanisms of action of molecules and drugs. Compared with knowledge-based methods, signature-based methods involve more molecular-level mechanisms, such as the significantly changed genes, by using computational approaches.
Table 2. Databases used for drug-repositioning studies.
| Fields | Databases[LM6] | Website[LM7] | Refs |
|---|---|---|---|
| Chemical structure | PubChem | http://pubchem.ncbi.nlm.nih.gov | |
| Collaborative Drug Discovery Vault | https://www.collaborativedrug.com | ||
| Drugbank | [51] | ||
| TTD | [52] | ||
| PharmGKB | [53] | ||
| DrugMap Central | [43] | ||
| ChemSpider | http://www.chemspider.com | ||
| ChemFrog | http://www.chemfrog.com | ||
| ChemDB | http://www.chemdb.com | ||
| iScienceSearch | http://cwmglobalsearch.com/gs/Default.aspx | ||
| Chemicalize (ChemAxon) | http://www.chemicalize.org | ||
| DistilBio | http://distilbio.com | ||
| Target 3D structure | PDB | http://www.rcsb.org | |
| OCA | http://oca.weizmann.ac.il/oca-bin/ocamain | ||
| OPM (membrane proteins) | http://opm.phar.umich.edu | ||
| Proteopedia | http://proteopedia.org | ||
| TOPSAN | http://www.topsan.org | ||
| Drug-target information | Drugbank | [51] | |
| TTD | [52] | ||
| PharmGKB | [53] | ||
| DrugMap Central | [43] | ||
| MATADOR (manually annotated) | http://matador.embl.de | ||
| SuperTarget | [54] | ||
| STITCH | [55] | ||
| GLIDA | [56] | ||
| PDSP Ki | [57] | ||
| BindingDB | [58] | ||
| Adverse effects and clinical trial information | SIDER | [59] | |
| FAERS (US FDA) | http://www.fda.gov/Drugs/ | ||
| Adverse Reaction Database (Canada) | http://www.hc-sc.gc.ca | ||
| IDIS | http://itsnt14.its.uiowa.edu | ||
| Clinicaltrial.gov | http://clinicaltrials.gov | ||
| DrugMap Central | [43] | ||
| FDA label information | FDALABEL(US FDA) | ||
| DailyMed (US FDA) | |||
| SPL (US FDA) | |||
| DrugMap Central | [43] | ||
| Pathway information | NCI PID | [60] | |
| KEGG | [61] | ||
| BioCarta | http://www.biocarta.com | ||
| Reactome | http://www.reactome.org | ||
| PathwayCommons | [62] | ||
| DrugMap Central | [43] | ||
| Protein interaction information | HPRD | [63] | |
| BioGRID | [64] | ||
| STRING | [65] | ||
| PathwayCommons | [62] | ||
| MIPS | [66] | ||
| IntAct | [67] | ||
| DIP | [68] | ||
| Molecular omics data | NCBI-GEO | http://www.ncbi.nlm.nih.gov/geo/ | |
| SRA | http://www.ncbi.nlm.nih.gov/Traces/sra/) | ||
| Stanford Microarray Database | http://smd.princeton.edu | ||
| ArrayExpress | http://www.ebi.ac.uk/arrayexpress/) | ||
| PUMAdb | http://puma.princeton.edu | ||
| CellMiner (for NCI-60) | http://discover.nci.nih.gov/cellminer/ | ||
| Oncomine | https://www.oncomine.org | ||
| CCLE | [39] | ||
| Genetic data or information | dbSNP | http://www.ncbi.nlm.nih.gov/projects/SNP/) | |
| SRA | http://www.ncbi.nlm.nih.gov/Traces/sra/), | ||
| OMIM | [69] | ||
| Drug omics data | CMAP | [38] | |
| CCLE | [39] | ||
| NCBI-GEO | http://www.ncbi.nlm.nih.gov/geo/) | ||
| SRA | http://www.ncbi.nlm.nih.gov/Traces/sra/) |
Pathway- or network-based methods
Pathway- or network-based drug-repositioning methods utilize disease omics data, available signaling or metabolic pathways and protein interaction networks to reconstruct disease-specific pathways that provide the key targets for repositioned drugs [15–17]. The advantage of these methods is that they are helpful in narrowing general signaling networks from a large number of proteins down to a specific network with a few proteins (or targets). A recent study of drug repositioning addressed distinct signaling mechanisms of metastatic subtypes of breast cancer [17]. Neither knowledge-based nor signature-based methods can address these repositioning results because the subtype signaling mechanisms are hard to elucidate from existing breast cancer pathways or the gene signatures.
Targeted mechanism-based methods
Targeted mechanism-based drug-repositioning methods integrate treatment omics data, available signaling pathway information and protein interaction networks to delineate the unknown mechanisms of action of drugs [18–20]. The era of precision medicine motivates such drug-repositioning studies. For instance, drug resistance remains an unresolved issue in cancer therapy. Although patients respond well to a drug initially, they often acquire resistance to that drug after a few months of treatment. This indicates that deriving a successful drug treatment needs additional information about the mechanisms of action of drugs to find better drug targets. Systems biology approaches are promising in addressing this challenge. The advantage of these methods is that their goals are not only to discover the mechanisms related to diseases or drugs, but also to identify those directly related to treatments of drugs to specific diseases. Owing to the difficulties in deriving effective computational models, there are only a few studies on these targeted mechanism-based methods [18–20] that developed elegant computational models to predict the drug effects and related targeted pathways. Comprehensive overviews of these drug-repositioning methods are given in Tables 1 and 2.
Table 1. Details of available drug-repositioning methods.
| Fields | Methods | Categories | Mechanism or knowledge complexity | Approach summaries with references | Examples |
|---|---|---|---|---|---|
| Drug-oriented | |||||
| FDA off-label use | Blinded | Serendipitously tested | Extremely low | Clinical decisions [3,4,21] | Sildenafil citrate (erectile dysfunction); rituximab (breast cancer); fluorouracil (lung cancer) and etoposide (bladder cancer) |
| Phenotypic screening | Blinded | Screening | Extremely low | In vivo and in vitro HTS/HCS screening [46–49] | HDECC inhibitors for stem-like lung cancer cells; widely used in GSK and Novartis |
| Phenotypic screening | Target based | Screening | Low | In vivo and in vitro HTS/HCS screening [7–9] | Widely used in Genentech |
| Target 3D structure, chemical structure information of drugs and ligands | Target based | Cheminformatics | Low | In silico screening [7,22], ligand based and docking [23,24] | MLR-1023 for diabetes (Melior) |
| Drug-target information, chemical structure information of targets and drugs | Knowledge based | Bioinformatics, Cheminformatics | Moderate | Drug–target prediction [10–14] | Simvastatin and Ketoconazole (breast cancer) |
| Adverse effects (clinical trial information) | Knowledge based | Bioinformatics | Moderate | Using correlation to define disease adverse effect associations [25], and adverse effects to define drug similarity [26] | |
| FDA approval labels and adverse effects | Knowledge based | Bioinformatics | Moderate | Using principal component analysis to define drug similarity measurement [27] | |
| Disease-oriented | |||||
| Available Pathway information | Knowledge based | Bioinformatics | Moderate | Discovery of disease mechanism and address of key targets [28] | Vismodegib for skin cancer |
| Disease omics data | Signature based | Bioinformatics | Moderate | From gene signature (most changed genes) in disease of interest to identify key targets [30] | |
| Genetics data | Signature based | Bioinformatics | Moderate | Genome-wide association study analysis to identify key targets [31] | |
| Disease omics data, available pathway information, and protein interaction network | Pathway or network based | Network biology | High | Reconstruction of disease-specific pathways and networks to identify key targets [15–17] | Sunitinib and dasatinib for breast cancer brain metastases |
| Treatment-oriented | |||||
| Drug omics data | Signature based | Bioinformatics | Moderate | Connectivity Map, linking diseases with treated drugs by using gene signatures [32–34] | Sirolimus for patients with acute lymphoblastic leukemia with dexamethasone resistance |
| Signature and network based | Bioinformatics, network biology | Moderate | Using gene signatures to define the distances among drugs and then use community structure to classify drug [50] | Fasudil (a Rho-kinase inhibitor) for neurodegenerative disorders | |
| Disease omics and drug omics data | Signature based | Bioinformatics | High | Using both drug and disease gene signatures to define similarities between drugs and diseases [35–37] | Cimetidine for lung cancer and topiramate for Inflammatory bowel disease |
| Drug omics data, disease pathway and protein interaction network | Targeted-mechanism based | Network biology and systems biology | Extremely high | Elucidating targeted pathways for each treated drugs to define potential repositioning score [18–20] | Daunorubicin and clomifene for breast cancer |
Customize and generate new drug-repositioning pipelines
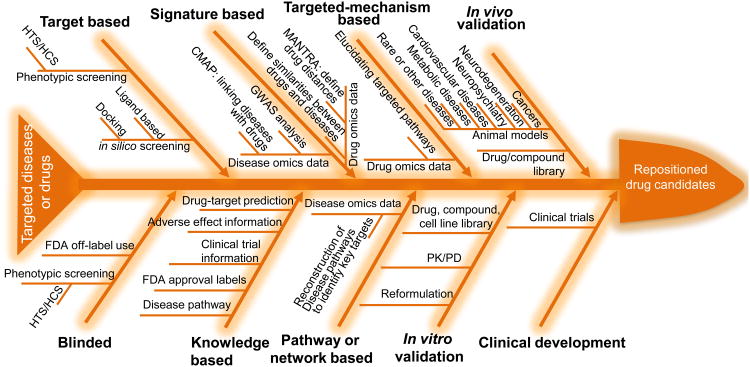
The low-risk and low-cost drug-repositioning strategies have been widely used to identify new clinical opportunities for old drugs. Accordingly, numerous strategies have been developed from drug-repositioning studies. Based on the LLF, we use a fishbone flowchart to present the existing methods with preclinical and clinical validations (Figure 2). The fishbone flowchart helps readers to understand the existing drug-repositioning pipelines and shows how to generate new drug-repositioning pipelines.
Figure 2.
Fishbone flowchart of drug-repositioning pipelines. Developed drug-repositioning pipelines comprise at least one of these methods (i.e., blinded, target based, knowledge based, signature based, pathway or network based, and targeted-mechanism based), preclinical studies (in vitro and/or in vivo validations) and clinical development (testing the new indications identified from the preclinical development). Development of a new drug-repositioning pipeline for a drug or a disease of interest should evaluate the priorities of these repositioning methods based on the available information of the drug or the disease. For example, an infectious disease that only has limited available signaling information related to cell wall and cytoplasmic membrane proteins would lead to high priority of target-based drug-repositioning studies focusing on these cell wall and cytoplasmic membrane proteins. Many drug-repositioning pipelines can reverse the order of the listed methods in the fishbone flowchart and make use of them flexibly. As an example, several existing drug-repositioning pipelines first consider pathway- or network-based drug-repositioning methods to reconstruct disease pathways and then use knowledge-based or targeted-based methods to identify candidate drugs. The fishbone provides all relevant components of general drug-repositioning pipelines, and one can customize specific drug-repositioning pipelines according to the available knowledge and information of targeted drugs or diseases. Abbreviations: FDA, Food and Drug Administration; GWAS, genome-wide association study; HTS/HCS, high-throughput and/or high-content screening; PK/PD, pharmacokinetics/pharmacodynamics.
A general fishbone flowchart of existing methods
The fishbone flowchart includes general methods used for drug-repositioning studies. Starting with a disease or a drug, one can choose the feasible methods determined by the available information or prior knowledge about the disease or drug. Then, during the validation stages, one can decide which in vitro and in vivo validations are needed for further testing of the newly identified indication(s). Eventually, the repositioned drugs will enter into clinical trials to evaluate their efficacy and performance in patients. Here, we take advantage of already published pipelines to discuss how to select the best drug-repositioning methods for specific studies.
Example pipeline 1: neglected tropical diseases (malaria) + phenotypic screening methods + drug libraries + in vivo validations ⇒ Astemizole
Orphan or rare diseases are those diseases that affect small numbers of people compared with the general population (<200 000 patient population per year in the USA) [40,41]. The orphan and/or rare diseases in developing regions of Africa, Asia and the Americas are also known as neglected tropical diseases. Most of these diseases receive less treatment and research funding, which results in little information about the disease mechanisms. According to the fishbone flowchart in Figure 2, the methods involving little information would be useful to reposition drugs for these diseases. In one such study using phenotypic screening, Chong et al. screened 1937 FDA-approved drugs and 750 drugs that were either approved for use abroad or undergoing phase II clinical trials for inhibition of Plasmodium falciparum growth. They identified a drug, astemizole, as an antimalarial agent by testing the drug using two mouse models of malaria [42].
Example pipeline 2: distinct breast cancer metastases + knowledge-based methods + pathway- or network-based methods + drug libraries + in vitro and in vivo validations + clinical trials ⇒ sunitinib for brain metastasis
We remain unclear about the pathways or mechanisms responsible for breast cancer metastasis to brain, bone and lung, leading to challenges in repositioning drugs for these cancer subtypes. Knowledge-based methods alone cannot solve this repositioning issue because these methods provide only general or canonical breast cancer signaling pathways instead of those specific to various types of metastasis. In a recent study, knowledge-based and network-based methods were combined to reconstruct the signaling networks for these metastatic breast cancer subtypes so that drug repositioning for each type was feasible to implement [17]. The knowledge-based method in this study deployed newly discovered signaling network elements, called cancer signaling bridges [18], to identify general known signaling information for breast cancer, whereas the network-based method used a mathematical model to address the specific signaling networks for subtypes of metastatic breast cancer. By checking the known targets in a recently developed drug database, DrugMap Central [43], 15, nine and two drug candidates were repositioned for brain, lung and bone metastases, respectively. For breast cancer brain metastasis, in vitro and in vivo validations were used to test the efficacies of these 15 drug candidates and two drugs were identified (sunitinib and dasatinib); the efficacy of sunitinib for breast cancer brain metastasis is now being tested in a phase II clinical trial (ClinicalTrials.gov ID: NCT00570908).
Example pipeline 3: treatment omics data (164 drug compounds) + disease omics data (100 diseases) + signature-based methods + in vivo validations ⇒ Cimetidine for lung adenocarcinoma
To test a large number of diseases for a specific drug or a large number of drugs for a specific disease, it is difficult to unify the needed computational approaches because the available information for different diseases or drugs varies. For example, to use target-based methods to reposition drugs for 100 diseases, one would have to know the biomarkers or available pathways for each of these diseases. The knowledge needed for this type of drug repositioning might be unavailable or difficult to derive from the literature or available databases. However, one can derive the gene signatures for these diseases from the publicly available genomics data. Together with the drug signatures identified from treatment omics data, Sirota et al. considered signature-based methods to evaluate the drug–disease scores [44]. Based on the drug–disease score map, they validated cimetidine for lung adenocarcinoma using tumor xenograft experiments.
Example pipeline 4: treatment omics data (>1000 drug–dose pairs) + targeted mechanism-based methods + in vitro validations ⇒ repositioned drugs with targeted mechanisms
In example pipeline 3, there is an issue in the gene signatures for 164 drugs: the treatment gene signatures for the 100 diseases are only based on treatments on three to five cancer cell lines in the CMAP database. Thus, when applying these drug gene signatures to other diseases, it is hard to ensure that the disease–drug scores are not biased by the limited treatment information. Another issue encountered with signature-based methods is in evaluating the accuracy of the large number of predictions on disease–drug associations. By integrating the targeted mechanisms into the drug-repositioning process, new computational approaches for predicting the efficacies of repositioned drugs were tested [18]. It was confirmed that the analysis could accurately predict clinical responses to more than 90% of drugs approved by the FDA and more than 75% of experimental clinical drugs that were tested. The high accuracy of prediction ensures more favorable repositioning results for disease–drug associations. To keep the accuracies of treatment information, only drugs for three cancer cell lines were repositioned that were used for treatments in CMAP. Moreover, the importance of the identified targeted pathways was addressed by explaining the differences in treatment responses.
Concluding remarks
Drug-repositioning studies are dependent on the prior knowledge and available information from specific studies to select and determine appropriate repositioning methods. Establishing accurate and efficient drug-repositioning pipelines for specific studies requires the prioritization of existing computational methods based on the available knowledge or the development of new computational methods. In this review, we described the available drug-repositioning methods according to the categorization of their integrated knowledge and information. We introduced the LLF to characterize existing repositioning methods and presented them in a generalized drug-repositioning pipeline (Fishbone flowchart). These flowcharts are powerful tools to understand the existing drug-repositioning methods and customize them into new drug-repositioning pipelines for specific studies.
Still, many challenges remain for cost-effective drug-repositioning studies. Not every existing drug-repositioning study can be generalized to a new study, especially those including computational methods. One has to evaluate carefully the available drug-repositioning methods according to the prior knowledge and available information of the study of interest and determine which is the best for that study. Another issue for the available drug-repositioning studies for diseases with low knowledge and low complexity-of-mechanism is that they have relatively low success rates. The complexities and richness of information available to drug-repositioning studies largely determine their success rates; obviously, knowledge-based and signature-based methods are more likely to identify more successful repurposed drugs than are blinded search or screening methods. As an example, we mentioned drug repositioning for pediatric population in the ‘Knowledge-based drug-repositioning methods’ section. The existing studies did not consider the blinded search or screening methods for drug repositioning for pediatric diseases. Instead, the drug knowledge from HLH, PubMed and Google was used in the drug repositioning targeting this special population [29], because knowledge-based methods consider the factor of patient variability in the drug-repositioning process.
In the era of precision medicine, it is important to delineate disease mechanisms, such as signaling pathways, or treatment mechanisms, such as off-targets and targeted pathways, to explain the mechanisms of action of drugs. This also leads to the application of drug repositioning to new indications for individual patients. Mechanism-based repositioning approaches are able to consider fully the heterogeneity and complexity of patients while reducing the inefficacy and toxicity caused by patient variability [45]. We would like to emphasize that drug-repositioning studies have to be solidly grounded on science to be successful. Toward better drug repositioning, the field needs better development of more in-depth mechanistic computational methods or models that can readily be customized into drug-repositioning pipelines that integrate computational and experimental methods seamlessly to ensure high success rates of repositioned drugs.
Understanding the existing drug repositioning methods with a top-down flowchart
Prioritizing repositioning methods using their integrated knowledge and information
A general drug repositioning pipeline with a fishbone flowchart
Guidance for Customizing and generating new drug repositioning pipelines
Acknowledgments
This research is funded by the National Institutes of Health (NIH) U54 CA149196, National Cancer Institute (NCI) CA121225, a John S. Dunn Research Foundation grant and a T.T. & W.F. Chao Foundation grant to S.T.C.W. The authors would like to thank Rebecca Danforth for proofreading the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 2.Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol Sci. 2013;34:267–272. doi: 10.1016/j.tips.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Pfister DG. Off-label use of oncology drugs: the need for more data and then some. J Clin Oncol. 2012;30:584–586. doi: 10.1200/JCO.2011.38.5567. [DOI] [PubMed] [Google Scholar]
- 4.Burstein HJ. Off-label use of oncology drugs: too much, too little, or just right? J Natl Compr Canc Netw. 2013;11:505–506. doi: 10.6004/jnccn.2013.0066. [DOI] [PubMed] [Google Scholar]
- 5.Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- 6.Hurle MR, et al. Computational drug repositioning: from data to therapeutics. Clin Pharmacol Ther. 2013;93:335–341. doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- 7.Swamidass SJ. Mining small-molecule screens to repurpose drugs. Brief Bioinform. 2011;12:327–335. doi: 10.1093/bib/bbr028. [DOI] [PubMed] [Google Scholar]
- 8.Crisman TJ, et al. Understanding false positives in reporter gene assays: in silico chemogenomics approaches to prioritize cell-based HTS data. J Chem Inf Model. 2007;47:1319–1327. doi: 10.1021/ci6005504. [DOI] [PubMed] [Google Scholar]
- 9.Feng BY, et al. A high-throughput screen for aggregation-based inhibition in a large compound library. J Med Chem. 2007;50:2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- 10.Yildirim MA, et al. Drug-target network. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 11.Zhao S, Li S. Network-based relating pharmacological and genomic spaces for drug target identification. PLoS ONE. 2010;5:e11764. doi: 10.1371/journal.pone.0011764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng F, et al. Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput Biol. 2012;8:e1002503. doi: 10.1371/journal.pcbi.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alaimo S, et al. Drug-target interaction prediction through domain-tuned network-based inference. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt307. LM3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinnings SL, et al. Drug discovery using chemical systems biology: repositioning the safe medicine comtan to treat multi-drug and extensively drug resistant tuberculosis. PLoS Comput Biol. 2009;5:e1000423. doi: 10.1371/journal.pcbi.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ideker T, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 16.Segal E, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 17.Hong Z, et al. Novel modeling of cancer cell signaling pathways enables systematic drug repositioning for distinct breast cancer metastases. Cancer Res. doi: 10.1158/0008-5472.CAN-12-4617. (in press)[LM4] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin GX, et al. A novel method of transcriptional response analysis to facilitate drug repositioning for cancer therapy. Cancer Res. 2012;72:33–44. doi: 10.1158/0008-5472.CAN-11-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin GX, et al. An enhanced Petri-net model to predict synergistic effects of pairwise drug combinations from gene microarray data. Bioinformatics. 2011;27:I310–I316. doi: 10.1093/bioinformatics/btr202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iskar M, et al. Characterization of drug-induced transcriptional modules: towards drug repositioning and functional understanding. Mol Syst Biol. 2013;9:662. doi: 10.1038/msb.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephen R, et al. Off-label use of oncology drugs in a community oncology EMR database. Pharmacoepidemiol Drug Safety. 2009;18:S62. [Google Scholar]
- 22.Ekins S, et al. In silico pharmacology for drug discovery: methods for virtual ligand screening and profiling. Br J Pharmacol. 2007;152:9–20. doi: 10.1038/sj.bjp.0707305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doman TN, et al. Molecular docking and high-throughput screening for novel inhibitors of protein tyrosine phosphatase-1B. J Med Chem. 2002;45:2213–2221. doi: 10.1021/jm010548w. [DOI] [PubMed] [Google Scholar]
- 24.Kolb P, et al. Docking and chemoinformatic screens for new ligands and targets. Curr Opin Biotechnol. 2009;20:429–436. doi: 10.1016/j.copbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L, Agarwal P. Systematic drug repositioning based on clinical side-effects. PLoS ONE. 2011;6:e28025. doi: 10.1371/journal.pone.0028025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campillos M, et al. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 27.Bisgin H, et al. Investigating drug repositioning opportunities in FDA drug labels through topic modeling. BMC Bioinformatics. 2012;13(Suppl. 15):S6. doi: 10.1186/1471-2105-13-S15-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An SM, et al. Stem cell signaling as a target for novel drug discovery: recent in the WNT and Hedgehog pathways. Acta Pharmacol Sin. 2013;34:777–783. doi: 10.1038/aps.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blatt J, Corey SJ. Drug repurposing in pediatrics and pediatric hematology oncology. Drug Discov Today. 2013;18:4–10. doi: 10.1016/j.drudis.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Haeberle H, et al. Identification of cell surface targets through meta-analysis of microarray data. Neoplasia. 2012;14:666–669. doi: 10.1593/neo.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol. 2012;30:317–320. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 32.Lamb J, et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 33.Lamb J. The connectivity map: using gene-expression profiling to identify new therapeutics and potential adverse drug effects. Chem Res Toxicol. 2007;20:2018–2018. [Google Scholar]
- 34.Qu XYA, Rajpal DK. Applications of Connectivity Map in drug discovery and development. Drug Discov Today. 2012;17:1289–1298. doi: 10.1016/j.drudis.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 35.Dudley JT, et al. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lussier YA, Chen JL. The emergence of genome-based drug repositioning. Sci Transl Med. 2011;3:96ps35. doi: 10.1126/scitranslmed.3001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirota M, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 39.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu ZC, et al. In silico drug repositioning: what we need to know. Drug Discov Today. 2013;18:110–115. doi: 10.1016/j.drudis.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 41.Ekins S, et al. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov Today. 2011;16:298–310. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 42.Chong CR, et al. A clinical drug library screen identifies astemizole as an antimalarial agent. Nat Chem Biol. 2006;2:415–416. doi: 10.1038/nchembio806. [DOI] [PubMed] [Google Scholar]
- 43.Fu C, et al. DrugMap Central: an on-line query and visualization tool to facilitate drug repositioning studies. Bioinformatics. 2013;29:1834–1836. doi: 10.1093/bioinformatics/btt279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sirota M, et al. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li YY, Jones SJ. Drug repositioning for personalized medicine. Genome Med. 2012;4:27. doi: 10.1186/gm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia X, et al. Image-based chemical screening identifies drug efflux inhibitors in lung cancer cells. Cancer Res. 2010;70:7723–7733. doi: 10.1158/0008-5472.CAN-09-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kondoh Y, Osada H. High-throughput screening identifies small molecule inhibitors of molecular chaperones. Curr Pharm Des. 2013;19:473–492. [PubMed] [Google Scholar]
- 48.Carnero A. High throughput screening in drug discovery. Clin Transl Oncol. 2006;8:482–490. doi: 10.1007/s12094-006-0048-2. [DOI] [PubMed] [Google Scholar]
- 49.Toledo-Sherman LM, Chen D. High-throughput virtual screening for drug discovery in parallel. Curr Opin Drug Discov Dev. 2002;5:414–421. [PubMed] [Google Scholar]
- 50.Iorio F, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci U S A. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wishart DS, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36(Database issue):D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu F, et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40(Database issue):D1128–D1136. doi: 10.1093/nar/gkr797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hewett M, et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30:163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gunther S, et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36(Database issue):D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuhn M, et al. STITCH 3: zooming in on protein–chemical interactions. Nucleic Acids Res. 2012;40(Database issue):D876–D880. doi: 10.1093/nar/gkr1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okuno Y, et al. GLIDA: GPCR-ligand database for chemical genomic drug discovery. Nucleic Acids Res. 2006;34(Database issue):D673–D677. doi: 10.1093/nar/gkj028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roth BL, et al. The multiplicity of serotonin receptors: uselessly diverse molecules or an embarrassment of riches? Neuroscientist. 2000;6:252–262. [Google Scholar]
- 58.Nicola G, et al. BindingDB: a protein-ligand database for drug discovery. Protein Sci. 2012;21:208–208. [Google Scholar]
- 59.Kuhn M, et al. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6 doi: 10.1038/msb.2009.98. LM5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaefer CF, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keshava Prasad TS, et al. Human Protein Reference Database: 2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41(Database issue):D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pagel P, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 67.Kerrien S, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40(Database issue):D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xenarios I, et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]