Abstract
Grass carp reovirus (GCRV) is a causative agent of haemorrhagic disease in grass carp that drastically affects grass carp aquaculture. Here we report a novel GCRV isolate isolated from sick grass carp that induces obvious cytopathic effect in CIK cells and name it as GCRV096. A large number of GCRV 096 viral particles were found in the infected CIK cells by electron microscope. The shape, size and the arrangement of this virus were similar to those of grass carp reovirus. With the primers designed according to GCRV 873 genome sequences, specific bands were amplified from sick grass carp and the infected CIK cells. The homology rates among vp4, vp6 and vp7 gene in GCRV 096 and those of some GCRV isolates were over 89%. In this study, the sequences of vp4, vp6 and vp7 were used to analyse sequence variation, phylogenetic relationships and genotypes in twenty five GCRV isolates. The results indicated these twenty five GCRV isolates should be attributed to four genotypes. And there were no obvious characteristics in the geographical distribution of GCRV genotype. The study should provide the exact foundation for developing more effective prevention strategies of grass carp haemorrhagic disease.
Keywords: Grass carp reovirus (GCRV), Identification, Phylogenetic relationship, Genotype
Introduction
Grass carp reovirus (GCRV), which is known as a member of the Aquareovirus genus and the Reoviridae family, can cause serious haemorrhagic disease in grass carp (Chen and Jiang 1983) and obvious cytopathic effect (CPE) on many cell lines from fish (Zuo et al. 1986; Lu et al. 1990). To date, a number of various GCRV isolates have been isolated from diseased grass carp around the world, including GCRV 873, GCRV 875, GCRV HZ08, GCRV GD108, AGCRV and others (Fang et al. 2002; Chi et al. 2011; Ye et al. 2012; Zeng et al. 2013). These isolates are distinct not only in their levels of virulence and cell culture characteristics, but also in their antigenicity (Fang et al. 2002; Mohd Jaafar et al. 2008; Zhang et al. 2010a).
GCRV is a double-stranded RNA virus that is assigned to the Aquareovirus C species. The genome of GCRV is known to consist of 11 segments of dsRNA contained in a core surrounded with a double-layered icosahedral capsid (Rangel et al. 1999). To our knowledge, there are few published reports about the serotype and genotype of GCRV. Furthermore, there are no uniform criteria for virus genotyping. One of the virus genotyping methods is based on the analysis of the nucleotide sequence.
So far, some gene sequences of GCRV isolates have been reported (Mohd Jaafar et al. 2008; Rangel et al. 1999; Fang et al. 2000; Su et al. 2010; Attoui et al. 2002; Fan et al. 2010). vp4, vp6 and vp7 gene in GCRV encode major outer capsid proteins and are conservative. Moreover, there are many variable sites and informative sites between sequences of vp4, vp6 and vp7 gene in different GCRV isolates. Considering vp4, vp6 and vp7 gene as molecular makers, we investigated sequence variation characteristics, the phylogenetic relationships and genotypes of twenty five GCRV isolates to find the evolutive characteristic of GCRV in the study. In this study, a new GCRV isolate was found and identified from diseased grass carp. This study provides the theoretical basis for the prevention and treatment of haemorrhagic disease in grass carp.
Materials and methods
Virus and cells
GCRV 096 was isolated from the diseased grass carp in Xiaogan, Hubei Province and stored in our laboratory. A widely used GCRV sensitive cell steain, grass carp kidney cells (CIK) were purchased from shenzhen inspection and quarantine bureau in China. CIK is GCRV sensitive cell and are widely used in the related study of GCRV (Ye et al. 2012; Zhang et al. 2010b; Ma et al. 2011).
Some GCRV isolates were examined in the present study, which were identified in previous studies (Mohd Jaafar et al. 2008; Rangel et al. 1999; Fang et al. 2000; Su et al. 2010; Attoui et al. 2002; Fan et al. 2010). Table 1 presents information about the specific names of twenty five GCRV isolates, their abbreviations, locations where they were collected, the genes of GCRV and their GenBank accession numbers.
Table 1.
Names of GCRV isolates, abbreviations, localities, genes of GCRV used in this study and their GenBank accession numbers
| Names | Abbreviations | Localities | Genes | GenBank accession numbers |
|---|---|---|---|---|
| AGCRV PB01-155 | 155 | America | vp4, vp6, vp7 | EF589103, EF589105, EF589107 |
| AGCRV | ARV | America | vp4, vp6, vp7 | NC010589, NC010593, NC010594 |
| GCRV 096 | 096 | Hubei, China | vp4, vp6, vp7 | JN206664, HQ452490, JN206665 |
| GCRV 104 | 104 | Hubei, China | vp6 | HM234682 |
| GCRV 097 | 097 | Shanxi, China | vp4 | GQ469997 |
| GCRV 873 | 873 | Hunan, China | vp4, vp6, vp7 | AF403392, AF260512, AF260513, |
| GCRV 875 | 875 | Hubei, China | vp6, vp7 | AF403412, AF403409 |
| GCRV 876 | 876 | Jiangxi, China | vp6, vp7 | AF403413, AF403410 |
| GCRV 991 | 991 | Hunan, China | vp6, vp7 | AF403414, AF403411 |
| GCRV GD108 | 108 | Guangdong, China | vp4, vp6, vp7 | HQ231208, HQ231205, HQ231203 |
| GCRV HeNan988 | 988 | Henan, China | vp4, vp6 | KC847325, KC847328 |
| GCRV HN12 | H12 | China | vp6 | KC130075 |
| GCRV HS11 | H11 | China | vp6 | KC130076 |
| GCRV HuNan794 | 794 | Hunan, China | vp4, vp6 | KC238681, KC238684 |
| GCRV HZ08 | H08 | Zhejiang, China | vp4, vp6, vp7 | GQ896337, GU350746, GU350744 |
| GCRV JS12 | J12 | China | vp6 | KC130077 |
| GCRV NC11 | N11 | China | vp6 | KC130078 |
| GCRV QC11 | Q11 | China | vp6 | KC130079 |
| GCRV QY12 | Q12 | China | vp6 | KC130080 |
| GCRV YX11 | Y11 | China | vp6 | KC130081 |
| GCRV ZS11 | Z11 | China | vp6 | KC130082 |
| GCRV 106 | 106 | China | vp4, vp6 | KC201171, KC201174 |
| GCRV 918 | 918 | China | vp4, vp6 | KC201182, KC201185 |
| GCRV JX01 | J01 | Jiangxi, China | vp7 | JQ042807 |
| GCRV JX02 | J02 | Jiangxi, China | vp7 | JX263303 |
| Bovine rotavirus B223 | Bovine | vp4, vp6, vp7 | D13394, AF317128, X57852 |
Virus culture and transmission electron microscopy observation
Cell culture, viral infection and propagation determination were performed as previously described (Fang et al. 1989). GCRV 096 particles were extracted with the differential centrifugation at 250-6000 g, and the supernatant was then ultracentrifuged at 35,000 g at 4°C for 2.5 h. The purified virus pellet was resuspended in phosphate-buffered saline (PBS) with pH 7.4 and then stored at -70°C for the further use. After removing the cell culture medium from the confluent monolayer cell, the monolayer cell was rinsed two times with the PBS buffer and the virus was added with the adsorption for one hour at room temperature. Then, after aspirating off the virus, the maintain solution (M199 containing 2% FBS) was added. The infected CIK cells were incubated at 28°C and observed daily.
Electron microscopic section of the infected CIK cells with CPE was made and observed in transmission electron microscope.
RT-PCR amplification
With viral RNA kit (Takara, Dalian, China), the GCRV 096 genome RNA was extracted from purified GCRV 096 virus. The cDNA of GCRV 096 genome RNA was acquired with RT-PCR kit (Takara, Dalian, China) using the random primers and M-MLV reverse transcriptase.
According to the genome sequence of GCRV 873, primers for GCRV 096 vp4, vp6 and vp7 gene amplification were designed based on homologous sequence in GCRV 873: 5″-CACTTCGCACTCTCTCTACAATG-3′ and 5′-AGTACGACACTTCCCGCCGTT-3′, 5′-TGTGATGGCACAGCGTCAG-3′ and 5′-GTTAGA CGAACATCGCCTGC-′3, 5′-TCACCACGATGCCACTTCAC-3′ and 5′-CGGTGCTTAATCGGATGGCT-3′, respectively. Primers were also designed based on homologous sequence from GCRV GD108 and GCRV HZ08 for vp4, vp6 and vp7 gene: 5′-ACTTACGGCCACTATCATGG-3′ and 5′-TCGGTGTACACGACCTAAG-3′, 5′-CTTTGAGTCGACGCACGTAT-3′ and 5′-CCGTCGGGTGGATTAGGTC-3′, 5′-TCTACTGCCAAGATGGCCAC-3′ and 5′-GCACGCACCTTACTTACAGCA-3′. The PCR cycling conditions were an initial denaturation at 95°C for 3 min followed by 30 cycles consisting of 94°C for 30 s, 55°C for 60 s and 72°C for 70 s, and a final extension step of 30 min at 72°C. The composition of the PCR system (50 μl) includes 33 μl sterile water, 3 μl dNTP (each is 2.5 mmol/L), 10 pmol/L primer for 2 μl each, 10 × buffer for 5 μl (containing Mg++), DNA for 100 ng and Taq polymerase for 0.25 μl (5 U/μl). The aimed genes were purified using Gel Extraction Kit (Takara, Dalian, China) from gelose gel and connected with pMD18-T vector at 16°C, then transformed to DH5α E.coli. The recombined plasmid was verified by sequencing.
Gene sequence analysis of GCRV isolates
Sequences of vp4, vp6 and vp7 genes were aligned by using the Clustal V method in DNAstar software. Subsequently, the alignment was manually adjusted. Variable sites, information sites, genetic distances, and homologic rates of segments were calculated with MEGA5.1 (Tamura et al. 2007) and DnaSP5.10 (Rozas and Rozas 1999) software.
Phylogenetic relationships of GCRV isolates
Evolutionary models of vp4, vp6 and vp7 gene in GCRV were separately simulated in ModelTest3.7 (Posada and Crandall 1998). Subsequently, phylogenetic trees were restructured with simulation results. Using bovine rotavirus B223 as the outgroup, maximum parsimony (MP) trees, maximum likelyhood (ML) trees, and UPGMA trees were constructed with MEGA 4.1 (Tamura et al. 2007) software. MP trees were also built in PAUP4.0 (Swofford 1998) by running the heuristic search with TBR branch swapping, 100 random addition sequence replications, and non-parameter bootstrap resampling procedures to get the coincidence of the resultant MP trees. Bayesian analysis were performed with MrBayes3.12 (Huelsenbeck and Ronquist 2001) using the general-time-reversible + gamma + invariants (GTR + G + I) model of sequence evolution and four Markov Chain Monte Carlo (MCMC) sampling to assess phylogenetic relationships. We set the parameters in MrBayes as follows: nst = 6, rate = gamma, basefreq = estimate, generations = 10,000,000, and the posterior probability and branches of the phylogeny were summed by burnin = 500 and contype = allcompat.
Sequence variation analysis of vp4, vp6 and vp7 genes in GCRV isolated to the same genotype
Sequences of vp4, vp6 and vp7 genes in GCRV isolated to the same genotype were aligned by using the Clustal V method in DNAstar software. Alignment was manually adjusted. Variable sites were analysed.
Results
Virus infection in sensitive cellls and particle identification
Three days after the culture of the CIK cells infected by GCRV 096, CPE phenomenon was observed and the shedding and apoptosis occurred in most of the CIK cells five days after the infection. While, the controlled CIK cells without the infection by the virus grew well (Figure 1).
Figure 1.
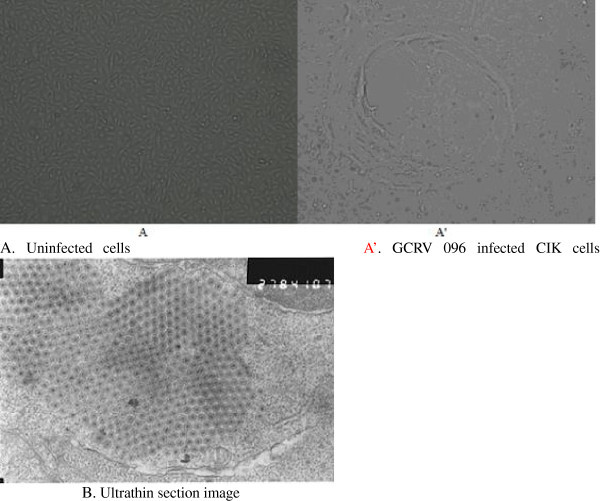
CPE in the CIK cells 3 d after GCRV 096 isolate inoculation (A, A’ 100×) and Crystalline array of viral particles (B 50,000×). Notes: A. The control CIK cells without GCRV096 inoculation. A’. CPE in the CIK cells 3 d after GCRV 096 isolate inoculation. B: Crystalline array of viral particles.
A large number of virus particles without the envelope structure crystalline in CIK cells were detected from transmission electron microscopy ultrathin section of CIK cells infected with GCRV 096 (Figure 1). The shape, size and the arrangement of GCRV 096 were similar to those of grass carp reovirus (Ke et al. 1990).
Detection by RT-PCR
vp4, vp6 and vp7 genes were PCR amplified from GCRV 096, subcloned into a pMD18-T vector and sequenced. The length of vp4, vp6 and vp7 genes in GCRV 096 was 1981 bp, 1258 bp, and 855 bp, respectively (GenBank accession numbers: JN206664, HQ452490 , and JN206665).
Sequence analysis
vp4, vp6 and vp7 genes in the these GCRV isolates contain 184, 447 and 375 informative sites, respectively. Table 2 shows the identity and divergence among GCRV isolates based on vp4, vp6 and vp7 genes, respectively. Based on the data shown in Table 2, it is apparent that genetic distances of vp4, vp6 and vp7 genes among GCRV 096, GCRV 873, GCRV 875, GCRV JX01, GCRV 876 and GCRV 991 or AGCRV and AGVRV PB01-155 were small, and their homologous rates were high. Also, genetic distances among GCRV HZ08, GCRV GD108, GCRV 918, GCRV HuNan794, GCRV HeNan988, GCRV 106, GCRV ZS11, GCRV QC11, GCRV HN12, GCRV HS11, GCRV YX11, GCRV JS12, GCRV QY12, GCRV JX02, and GCRV 097 were small, with elevated homologous rates. Other genetic distances were far and the genetic identities were small.
Table 2.
Identity (above the diagonal) and divergence (under the diagonal) between GCRV isolates based on the vp4 , vp6 , vp7 gene [×1000]
| Based on the vp4 gene | Based on the vp7 gene | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GCRV | 155 | ARV | 096 | 097 | 873 | 108 | 988 | 794 | H08 | 106 | 918 | GCRV | 155 | AVR | 096 | 873 | 875 | 876 | 991 | 108 | H08 | J01 | J02 |
| 155 | 1000 | 599 | 317 | 603 | 299 | 293 | 292 | 286 | 293 | 296 | 155 | 1000 | 203 | 303 | 279 | 287 | 289 | 219 | 214 | 302 | 213 | ||
| ARV | 0 | 599 | 317 | 603 | 299 | 293 | 292 | 286 | 293 | 296 | ARV | 0 | 203 | 303 | 279 | 287 | 289 | 219 | 214 | 302 | 213 | ||
| 096 | 453 | 453 | 295 | 993 | 303 | 299 | 298 | 304 | 298 | 304 | 096 | 1178 | 1178 | 212 | 196 | 197 | 197 | 199 | 208 | 213 | 207 | ||
| 097 | 946 | 946 | 827 | 311 | 981 | 990 | 987 | 994 | 990 | 987 | 873 | 767 | 767 | 920 | 902 | 999 | 1000 | 199 | 197 | 994 | 205 | ||
| 873 | 445 | 445 | 5 | 827 | 298 | 297 | 296 | 299 | 296 | 302 | 875 | 856 | 856 | 910 | 100 | 901 | 902 | 222 | 208 | 899 | 206 | ||
| 108 | 967 | 967 | 886 | 20 | 872 | 970 | 970 | 963 | 971 | 968 | 876 | 776 | 776 | 855 | 1 | 102 | 999 | 204 | 200 | 993 | 197 | ||
| 988 | 971 | 971 | 891 | 10 | 882 | 31 | 997 | 986 | 998 | 986 | 991 | 782 | 782 | 848 | 0 | 100 | 1 | 204 | 200 | 995 | 195 | ||
| 794 | 977 | 977 | 896 | 13 | 888 | 31 | 3 | 985 | 998 | 985 | 108 | 1218 | 1218 | 920 | 885 | 891 | 865 | 870 | 986 | 203 | 987 | ||
| H08 | 941 | 941 | 875 | 6 | 863 | 35 | 14 | 14 | 986 | 984 | H08 | 1194 | 1194 | 904 | 863 | 880 | 843 | 848 | 14 | 202 | 998 | ||
| 106 | 973 | 973 | 893 | 10 | 884 | 30 | 2 | 2 | 13 | 987 | J01 | 772 | 772 | 897 | 6 | 103 | 7 | 5 | 890 | 864 | 193 | ||
| 918 | 962 | 962 | 897 | 13 | 888 | 33 | 15 | 15 | 17 | 13 | J02 | 1300 | 1300 | 913 | 858 | 876 | 839 | 844 | 14 | 2 | 859 | ||
| Based on the vp6 gene | |||||||||||||||||||||||
| GCRV | 155 | ARV | 096 | 104 | 873 | 875 | 876 | 991 | 108 | 988 | H12 | H11 | 794 | H08 | J12 | N11 | Q11 | Q12 | Y11 | Z11 | 106 | 918 | |
| 155 | 1000 | 548 | 230 | 548 | 547 | 547 | 548 | 196 | 233 | 241 | 235 | 233 | 204 | 235 | 235 | 235 | 235 | 235 | 233 | 233 | 232 | ||
| ARV | 0 | 548 | 230 | 548 | 547 | 547 | 548 | 196 | 233 | 241 | 235 | 233 | 204 | 235 | 235 | 235 | 235 | 235 | 233 | 233 | 232 | ||
| 096 | 500 | 500 | 220 | 994 | 998 | 998 | 997 | 201 | 242 | 235 | 235 | 241 | 202 | 233 | 233 | 239 | 233 | 233 | 243 | 241 | 241 | ||
| 104 | 936 | 936 | 769 | 238 | 246 | 246 | 246 | 190 | 232 | 229 | 232 | 232 | 198 | 238 | 238 | 228 | 238 | 232 | 232 | 232 | 223 | ||
| 873 | 503 | 503 | 6 | 751 | 999 | 999 | 998 | 204 | 243 | 235 | 235 | 242 | 205 | 232 | 232 | 239 | 232 | 232 | 243 | 242 | 242 | ||
| 875 | 516 | 516 | 3 | 751 | 1 | 1000 | 999 | 211 | 238 | 231 | 231 | 238 | 203 | 233 | 233 | 236 | 233 | 233 | 238 | 238 | 237 | ||
| 876 | 516 | 516 | 3 | 751 | 1 | 0 | 999 | 211 | 238 | 231 | 231 | 238 | 203 | 233 | 233 | 236 | 233 | 233 | 238 | 238 | 237 | ||
| 991 | 514 | 514 | 4 | 755 | 2 | 1 | 1 | 211 | 238 | 231 | 236 | 238 | 200 | 233 | 233 | 236 | 233 | 233 | 238 | 238 | 237 | ||
| 108 | 1427 | 1427 | 1340 | 1527 | 1261 | 1236 | 1236 | 1236 | 207 | 197 | 203 | 207 | 965 | 204 | 204 | 203 | 204 | 197 | 200 | 200 | 199 | ||
| 988 | 834 | 834 | 682 | 939 | 693 | 665 | 665 | 665 | 1157 | 975 | 972 | 998 | 208 | 971 | 971 | 990 | 971 | 973 | 998 | 998 | 995 | ||
| H12 | 844 | 844 | 680 | 959 | 692 | 659 | 659 | 659 | 1214 | 26 | 991 | 975 | 210 | 990 | 990 | 979 | 990 | 992 | 975 | 976 | 973 | ||
| H11 | 850 | 850 | 683 | 962 | 694 | 662 | 662 | 662 | 1211 | 29 | 9 | 973 | 205 | 998 | 998 | 981 | 998 | 999 | 973 | 974 | 971 | ||
| 794 | 838 | 838 | 685 | 931 | 697 | 670 | 670 | 670 | 1149 | 2 | 25 | 28 | 206 | 972 | 972 | 990 | 972 | 974 | 998 | 999 | 996 | ||
| H08 | 1400 | 1400 | 1288 | 1510 | 1210 | 1196 | 1196 | 1196 | 32 | 1113 | 1165 | 1162 | 1106 | 204 | 204 | 200 | 204 | 205 | 206 | 206 | 204 | ||
| J12 | 850 | 850 | 683 | 958 | 694 | 662 | 662 | 662 | 1228 | 29 | 10 | 2 | 29 | 1178 | 1000 | 980 | 1000 | 998 | 972 | 973 | 970 | ||
| N11 | 850 | 850 | 683 | 958 | 694 | 662 | 662 | 662 | 1228 | 29 | 10 | 2 | 29 | 1178 | 0 | 980 | 1000 | 998 | 972 | 973 | 970 | ||
| Q11 | 834 | 834 | 689 | 923 | 701 | 670 | 670 | 670 | 1142 | 10 | 22 | 19 | 10 | 1099 | 20 | 20 | 980 | 982 | 992 | 991 | 988 | ||
| Q12 | 850 | 850 | 683 | 958 | 694 | 662 | 662 | 662 | 1228 | 29 | 10 | 2 | 29 | 1178 | 0 | 20 | 20 | 988 | 972 | 973 | 970 | ||
| Y11 | 854 | 854 | 686 | 958 | 698 | 667 | 667 | 667 | 1219 | 28 | 8 | 1 | 27 | 1170 | 2 | 2 | 19 | 2 | 974 | 975 | 971 | ||
| Z11 | 834 | 834 | 682 | 939 | 693 | 665 | 665 | 665 | 1142 | 2 | 25 | 28 | 2 | 1099 | 29 | 29 | 8 | 29 | 27 | 999 | 996 | ||
| 106 | 838 | 838 | 685 | 935 | 697 | 670 | 670 | 670 | 1149 | 2 | 24 | 27 | 1 | 1106 | 28 | 28 | 9 | 28 | 26 | 1 | 997 | ||
| 918 | 851 | 851 | 692 | 946 | 704 | 677 | 677 | 677 | 1154 | 5 | 28 | 30 | 4 | 1124 | 31 | 31 | 12 | 31 | 29 | 4 | 3 | ||
Simulation results of evolutionary model and phylogenetic relationships of GCRV isolates
Simulation results of the GCRV evolutionary model based on vp4, vp6 and vp7 gene from ModelTest3.7 (Posada and Crandall 1998) are shown in Table 3. The simulation results are used to construct phylogenetic trees.
Table 3.
Simulation results of the evolutionary model
| vp4 gene | vp6 gene | vp7 gene | |||
|---|---|---|---|---|---|
| Model selected: | HKY + G | GTR + G | HKY + G | TVMef + G | K80 + G |
| -lnL = | 10960.6680 | 10947.8525 | 10486.8574 | 10480.6113 | 6139.4121 |
| K = | 5 | 9 | 5 | 8 | 2 |
| AIC = | 21913.7051 | 20977.2227 | |||
| Base frequencies: | |||||
| freqA = | 0.2726 | 0.2724 | 0.2561 | 0.2530 | |
| freqC = | 0.2541 | 0.2476 | 0.2612 | 0.2586 | |
| freqG = | 0.2361 | 0.2401 | 0.2285 | 0.2313 | |
| freqT = | 0.2371 | 0.2400 | 0.2541 | 0.2571 | |
| Substitution model: | |||||
| R(a) [A-C] = | 1.9847 | 1.8471 | |||
| R(b) [A-G] = | 3.8446 | 7.0961 | |||
| R(c) [A-T] = | 1.3223 | 1.3157 | |||
| R(d) [C-G] = | 1.7384 | 1.3664 | |||
| R(e) [C-T] = | 4.4150 | 7.0961 | |||
| R(f) [G-T] = | 1.0000 | 1.0000 | |||
| i/tv ratio = | 1.3563 | 2.5439 | |||
| Proportion of invariable | |||||
| sites = | 0 | 0 | 0 | 0 | 0 |
| Gamma distribution | |||||
| shape parameter = | 4.2112 | 4.3849 | 3.8529 | 3.7611 | 3.8868 |
Topological structures of constructed phylogenetic trees, based on vp4, vp6 and vp7 genes of GCRV in this article are basically coincident. According to evolutionary simulation results, there is the UPGMA tree constructed based on vp4 gene in Figure 2. The results showed that the cluster on the top of the UPGMA tree consisted of GCRV 106, GCRV HeNan988, GCRV HuNan794, GCRV 097, GCRV 918, GCRV GD108 and GCRV HZ08. The second cluster was AGCRV PB01-155 and AGCRV. The third cluster contained GCRV 096 and GCRV 873.
Figure 2.
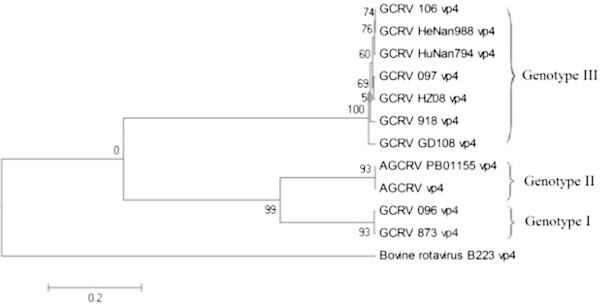
The constructed UPGMA tree based on the vp4 gene (Numbers indicate degree of confidence) was created first in MEGA software and completed with Microsoft Paint program.
In Figure 3, the MP tree was constructed based on vp6 gene. On the MP tree, the cluster on the top consisted of GCRV 106, GCRV HeNan988, GCRV HuNan794, GCRV 918, GCRV ZS11, GCRV QC11, GCRV HN12, GCRV HS11, GCRV YX11, GCRV JS12, GCRV QY12, GCRV GD108 and GCRV HZ08. The second cluster was GCRV 104. The next cluster was AGCRV PB01-155 and AGCRV. The last cluster contained GCRV 096, GCRV 875, GCRV 876, GCRV 991 and GCRV 873.
Figure 3.
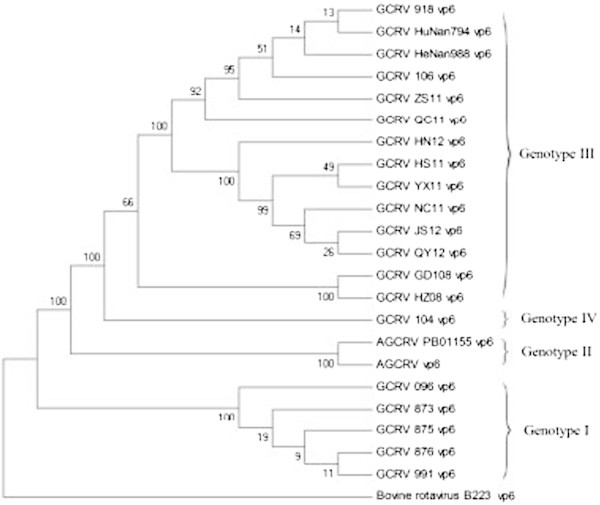
The constructed MP tree based on the vp6 gene (Numbers indicate the degree of confidence) was created first in PAUP software and completed with Microsoft Paint program.
In Figure 4, the UPGMA tree was constructed based on vp7 gene. The cluster on the top of this tree consisted of GCRV 096, GCRV 875, GCRV 876, GCRV JX01, GCRV 991 and GCRV 873. The second cluster was GCRV GD108, GCRV HZ08 and GCRV JX02. The last cluster contained AGCRV PB01-155 and AGCRV. The phylogenetic relationships of GCRV096, GCRV 991, GCRV 876, GCRV 873, GCRV JX01, and GCRV 875 or GCRV HZ08, GCRV JX02, and GCRV GD108 are relatively close.
Figure 4.
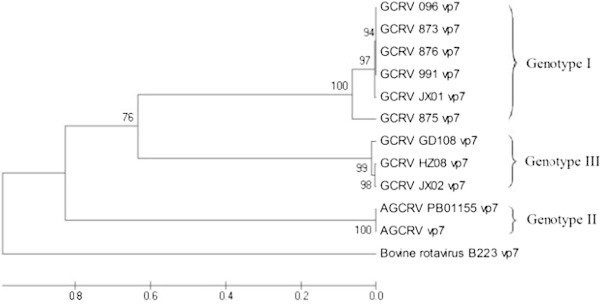
The constructed UPGMA tree based on the vp7 gene (Numbers indicate the degree of confidence) was created first in MEGA software and completed with Microsoft Paint program.
Sequence analysis of vp4, vp6 and vp7 genes in GCRV isolated to the same genotype
By analysing variable sites, we found the ratios of variation sites located on the third condon and transitions were respectively 71.6% and 82.1% in vp4 gene; 57.1% and 80.0% in vp6 gene; 77.3% and 89.3% in vp7 gene.
Discussion
Amongst all aquareovirus isolates, GCRV is one of the most pathogenic agents (Fang et al. 2002). GCRV can cause fatal epidemics of haemorrhagic disease in grass carp, and affects approximately 85% of fingerling and yearling populations (Jiang and Ahne 1989). Many GCRV isolates have been isolated in recent years, and various of them have been reported to exhibit distinctive differences in virulence (Fang et al. 2002). Moreover, new GCRV isolates were found constantly. In this study, GCRV 096 is a new GCRV isolate similar to GCRV 873, GCRV 875, GCRV 876, GCRV 991 and GCRV JX01.
In order to analyse the difference among GCRV isolates as well as their evolutionary relatiohship, it is necessary to genotyping. Currently, uniform criteria in place for virus genotyping are still unavailable. In hepatitis C virus, a more than 30% nucleotide sequence divergence between genotypes is generally considered standard (Simmonds 2004). The genetic heterogeneity among genotypes of hepatitis E virus has been shown to be more than 20% (Schlauder and Mushahwar 2001). In GCRV, relatively conservative vp4, vp6 and vp7 gene encode major outer capsid proteins and consist of many variable sites (Rangel et al. 1999). So, vp4, vp6 and vp7 gene could be used for GCRV genotyping.
The genetic distances among GCRV 096, GCRV JX01, GCRV 873, GCRV 875, GCRV 876 and GCRV 991 were small with high homologous rates. Furthermore, these isolates clustered together into one cluster on constructed phylogenetic trees. These results present that GCRV 096, GCRV JX01, GCRV 873, GCRV 875, GCRV 876 and GCRV 991 are attributed to the same genotype, i.e. genotype І. Genetic distances between AGCRV PB01-155 and AGVRV were small and their homologous rates were also high. On phylogenetic trees, AGCRV and AGCRV PB01-155 separately clustered into one cluster. These results indicate that AGCRV and AGCRV PB01-155 are attributed to a new genotype, i.e. genotype II. Genetic distances among GCRV HZ08, GCRV GD108, GCRV 918, GCRV HuNan794, GCRV HeNan988, GCRV 106, GCRV ZS11, GCRV QC11, GCRV HN12, GCRV HS11, GCRV YX11, GCRV JS12, GCRV QY12, GCRV JX02, and GCRV 097 were extremely small with especially high homologous rates. Furthermore, these isolates clustered together into one cluster on phylogenetic trees. GCRV HZ08, GCRV GD108, GCRV 918, GCRV HuNan794, GCRV HeNan988, GCRV 106, GCRV ZS11, GCRV QC11, GCRV HN12, GCRV HS11, GCRV YX11, GCRV JS12, GCRV QY12, GCRV JX02, and GCRV 097 were attributed to another new genotype, i.e. genotype III. In contrast, genetic distances between GCRV 104 and other GCRV isolates were large, and their homologous rates were small. On the phylogenetic tree (Figure 4), GCRV 104 separately clustered into one cluster. GCRV 104 is attributed to a new genotype, i.e. genotype IV.
The genotyping results obtained are consistent with previous research conclusions. The study of Wang indicated there were different genotypes of GCRV in China (Wang et al. 2012a). The biological characteristics of GCRV isolates belonging to the same genotype indicated they were analogous. For example, in an artificial infection test, GCRV HZ08 and GCRV GD108 can cause mortality of 60–80% of the yearly grass carp (approx. 10 cm in length), without obvious CPE in CIK cells (Ye et al. 2012; Zhang et al. 2010b). However, American grass carp reovirus (AGCRV) is not strongly connected with infectious disease in fish, although it is commonly detected by cell culture during routine inspections of healthy fish (Goodwin et al. 2010). GCRV 873, GCRV 096, GCRV 875, GCRV 876, GCRV 991 and GCRV JX01 can arouse significant CPE in CIK cells (Zhang et al. 2010a; Wang et al. 2012b). Furthermore, other characteristics of these two isolates were also similar. The genomic segments pattern of GCRV 875 was found to be similar to that of GCRV 873 (Fang et al. 2002). Polyacrylamide gel electrophoresis atlases of GCRV 873, GCRV 875, GCRV 876 and GCRV 991 were also the same (Fang et al. 2002).
The comparative analysis of the geographic location (Table 1) of collected GCRV isolates together with the difference between GCRV isolates and GCRV genotyping indicated there was no obvious relationship between the evolution of GCRV and geographical distribution of GCRV. In the same genotype, the ratios of variation sites on the third condon and the transitions in vp4, vp6 and vp7 gene were high.
Hemorragic disease of grass carp outbreaks seriously in China. Many isolates of grass carp reovirus have been discovered while new isolates are being isolated constantly. The systematic difference comparison of the different GCRV isolates has not been reported. In this study, we have verified the diference among various GCRV genotypes. GCRV genotyping has important significance to diagnosis and treatment in hemorrhagic disease of grass carp, especially to vaccine development. Comparison of different GCRV isolates and genotyping are helpful to further our understanding in GCRV genetic variation and evolution and the development of more effective preventative strategies against GCRV.
This study provides a foundation for revealing differences among GCRV isolates. Simultaneously, it is significant for the further research on genetically engineered vaccines against grass carp haemorrhagic disease and grass carp breeding for disease resistance.
Acknowledgements
The study was supported by the National 973 Plan Project in China (No. 2009CB118704). The authors would like to thank Dr. JY Shen for her help.
Abbreviations
- bp
Base pairs
- CIK
Grass carp kidney cell line
- CPE
Cytopathic effect
- dNTPs
Deoxynucleotide triphosphates
- FBS
Fetal bovine serum
- GCRV
Grass carp reovirus
- MgCl2
Magnesim chloride
- min
Minute
- ML
Maximum likelihood
- MP
Maximum parsimony
- PBS
Phosphate-buffered saline
- PCR
Polymerase chain reaction
- RT-PCR
Reverse transcription-PCR
- TBR
Tree-bisection-recorrnection.
Footnotes
Competing interests
The authors declare that they have no competing interests. The authors alone are responsible for the content and writing of the paper.
Authors’ contributions
YXY, JJC and WZH conceived and designed the experiments described in this work and wrote the manuscript. YXY, WY and XLF performed and analyzed the data. JJC and WZH supervised the work and analyzed the results. All the authors read and approved the final manuscript.
Contributor Information
Xiu-ying Yan, Email: yanxiuying1201@126.com.
Ya Wang, Email: yawang_1688@126.com.
Ling-fang Xiong, Email: xlf530@126.com.
Ji-chang Jian, Email: jianjc@gmail.com.
Zao-he Wu, Email: wuzaohe@163.com.
References
- Chen YS, Jiang YL. Morphology and physical and chemical properties of grass carp hemorrhage virus. KeXue Tong-Bao. 1983;18:1138–1140. [Google Scholar]
- Zuo WG, Qian HX, Xu YF, Du SY, Yang XL. A cell line derived from the kidney of grass carp (ctenopharyngodon idellus) J Fish China. 1986;10(1):11–17. [Google Scholar]
- Lu YA, Lannan CN, Rohovec JS, Fryer JL. Fish cell lines: establishment and characterization of three new cell lines from grass carp (Ctenopharyngodon idella) Vitro Cell Dev Biol. 1990;26(3 Pt 1):275–279. doi: 10.1007/BF02624457. [DOI] [PubMed] [Google Scholar]
- Fang Q, Xiao TY, Li L, Zou GP, Zhang HY, Wang YP. Infection Characterizations of four Grass Carp Reovirus (GCRV) strains. Virol Sin. 2002;17:182–184. [Google Scholar]
- Chi YY, Tian YY, Ye X, Deng GC, Li J, Wang HJ. Molecular properties of grass carp reovirus in southern China and establishment of duplex PCR detection method. Bing Du Xue Bao. 2011;27(4):358–365. [PubMed] [Google Scholar]
- Ye X, Tian YY, Deng GC, Chi YY, Jiang XY. Complete genomic sequence of a reovirus isolated from grass carp in China. Virus Res. 2012;163(1):275–283. doi: 10.1016/j.virusres.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Zeng WW, Wang Q, Wang YY, Xu DH, Wu SQ. A one-step molecular biology method for simple and rapid detection of grass carpCtenopharyngodon idella reovirus (GCRV) HZ08 strain. J Fish Biol. 2013;82(5):1545–1555. doi: 10.1111/jfb.12088. [DOI] [PubMed] [Google Scholar]
- Mohd Jaafar F, Goodwin AE, Belhouchet M, Merry G, Fang Q, Cantaloube JF, Biagini P, de Micco P, Mertens PPC, Attoui H. Complete characterisation of the American grass carp reovirus genome (genus Aquareovirus: family Reoviridae) reveals an evolutionary link between aquareoviruses and coltiviruses. Virology. 2008;373(2):310–321. doi: 10.1016/j.virol.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Zhang LL, Luo Q, Fang Q, Wang YP. An improved RT-PCR assay for rapid and sensitive detection of grass carp reovirus. J Virol Methods. 2010a;169(1):28–33. doi: 10.1016/j.jviromet.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Rangel AA, Rockemann DD, Hetrick FM, Samal SK. Identification of grass carp haemorrhage virus as a new genogroup of aquareovirus. J Gen Virol. 1999;80(9):2399–2402. doi: 10.1099/0022-1317-80-9-2399. [DOI] [PubMed] [Google Scholar]
- Fang Q, Attoui H, Cantaloube JF, Biagini P, Zhu ZY, De Micco P, De Lamballerie X. Sequence of genome segments 1, 2, and 3 of the grass carp reovirus (Genus Aquareovirus, Family Reoviridae) Biochem Biophys Res Commun. 2000;274(3):762–766. doi: 10.1006/bbrc.2000.3215. [DOI] [PubMed] [Google Scholar]
- Su JG, Huang T, Dong J, Heng JF, Zhang RF, Peng LM. Molecular cloning and immune responsive expression of MDA5 gene, a pivotal member of the RLR gene family from grass carp Ctenopharyngodon idella. Fish Shellfish Immun. 2010;28(4):712–718. doi: 10.1016/j.fsi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Attoui H, Fang Q, Mohd Jaafar F, Cantaloube JF, Biagini P, de Micco P, de Lamballerie X. Common evolutionary origin of aquareoviruses and orthoreoviruses revealed by genome characterization of Golden shiner reovirus, Grass carp reovirus, Striped bass reovirus and golden ide reovirus (genus Aquareovirus, family Reoviridae) J Gen Virol. 2002;83(8):1941–1951. doi: 10.1099/0022-1317-83-8-1941. [DOI] [PubMed] [Google Scholar]
- Fan C, Shao L, Fang Q. Characterization of the nonstructural protein NS80 of grass carp reovirus. Arch Virol. 2010;155(11):1755–1763. doi: 10.1007/s00705-010-0753-6. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wang Q, Shi CB, Zeng WW, Liu Y, Wu SQ. Molecular analysis of grass carp reovirus HZ08 genome segments 1-3 and 5-6. Virus Genes. 2010b;41(1):102–104. doi: 10.1007/s11262-010-0489-0. [DOI] [PubMed] [Google Scholar]
- Ma J, Wang W, Zeng L, Fan Y, Xu J, Zhou Y. Inhibition of the replication of grass carp reovirus in CIK cells with plasmid-transcribed shRNAs. J Virol Methods. 2011;175(2):182–187. doi: 10.1016/j.jviromet.2011.05.008. [DOI] [PubMed] [Google Scholar]
- Fang Q, Ke LH, Cai YQ. Growth characteristics and high titer culture of grass carp hemorrhage virus (GCHV)-873 in vitro. Virol Sinion. 1989;3:314–319. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15(2):174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP: Phylogenetic analysis using parsimony and other methods Associates. 1998. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ke LH, Fang Q, Cai YQ. Characteristics of a novel isolate of grass carp hemorrhagic virus. ACTA Hydrobiol Sin. 1990;14(2):153–159. [Google Scholar]
- Jiang Y, Ahne W. Some properties of the etiological agent of the hemorrhagic disease of grass carp and black carp. In: Ahne W, Kurstak E, editors. Viruses of Lower Vertebrates. Berlin: Springer-Verlag; 1989. pp. 227–239. [Google Scholar]
- Simmonds P. Genetic diversity and evolution of hepatitis C virus–15 years on. J Gen Virol. 2004;85(11):3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- Schlauder GG, Mushahwar IK. Genetic heterogeneity of hepatitis E virus. J Med Virol. 2001;65(2):282–292. doi: 10.1002/jmv.2031. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zeng WW, Liu C, Zhang C, Wang YY, Shi CB, Wu SQ. Complete genome sequence of a reovirus isolated from grass carp, indicating different genotype of GCRV in China. J Virol. 2012a;86(22):12466. doi: 10.1128/JVI.02333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin AE, Merry GE, Attoui H. Detection and prevalence of the nonsyncytial American grass carp reovirus Aquareovirus G by quantitative reverse transcriptase polymerase chain reaction. J Aquat Anim Health. 2010;22(1):8–13. doi: 10.1577/H09-025.1. [DOI] [PubMed] [Google Scholar]
- Wang T, Xu D, Lv LQ. Detection of the co-infection of different grass carp reovirus strains using dsRNA sequencing technology. J Shanghai Ocean Univ. 2012;21(5):756–762. [Google Scholar]


