Abstract
Background
Sporotrichosis is a chronic subcutaneous mycosis of humans and animals, which is typically acquired by traumatic inoculation of plant material contaminated with Sporothrix propagules, or via animals, mainly felines. Sporothrix infections notably occur in outbreaks, with large epidemics currently taking place in southeastern Brazil and northeastern China. Pathogenic species include Sporothrix brasiliensis, Sporothrix schenckii s. str., Sporothrix globosa, and Sporothrix luriei, which exhibit differing geographical distribution, virulence, and resistance to antifungals. The phylogenetically remote species Sporothrix mexicana also shows a mild pathogenic potential.
Methods
We assessed a genetically diverse panel of 68 strains. Susceptibility profiles of medically important Sporothrix species were evaluated by measuring the MICs and MFCs for amphotericin B (AMB), fluconazole (FLC), itraconazole (ITC), voriconazole (VRC), posaconazole (PCZ), flucytosine (5FC), and caspofungin (CAS). Haplotype networks were constructed to reveal interspecific divergences within clinical Sporothrix species to evaluate genetically deviant isolates.
Results
ITC and PCZ were moderately effective against S. brasiliensis (MIC90 = 2 and 2 μg/mL, respectively) and S. schenckii (MIC90 = 4 and 2 μg/mL, respectively). PCZ also showed low MICs against the rare species S. mexicana. 5FC, CAS, and FLC showed no antifungal activity against any Sporothrix species. The minimum fungicidal concentration ranged from 2 to >16 μg/mL for AMB against S. brasiliensis and S. schenckii, while the MFC90 was >16 μg/mL for ITC, VRC, and PCZ.
Conclusion
Sporothrix species in general showed high degrees of resistance against antifungals. Evaluating a genetically diverse panel of strains revealed evidence of multidrug resistant phenotypes, underlining the need for molecular identification of etiologic agents to predict therapeutic outcome.
Keywords: Sporothrix schenckii, Sporothrix brasiliensis, Multidrug resistance, MIC, MFC, Intraspecific diversity
Background
Sporotrichosis is a (sub)cutaneous mycosis of man and animals that is highly prevalent in tropical and subtropical areas, and currently reaching epidemic proportions in Brazil [1-3] and China [4,5]. Over the last decade, sporotrichosis has changed from a relatively obscure endemic infection to an epidemic zoonotic health problem in the South and Southeast Regions of Brazil [2]. Infection typically occurs by cutaneous inoculation of the microorganism from environmental sources [6,7] or via zoonotic transmission [3]. Sporotrichosis is a chronic disease that can spread lymphatically and most often presents as a localized lesion with subcutaneous nodules that eventually break through the skin [7,8]. Although rare, hematogenous dissemination can also occur depending on the immune status of the host [9].
Recent studies on molecular phylogeny clarified species boundaries within clinical isolates of Sporothrix and provided strong support for many cryptic groups. The major causative agents of sporotrichosis are several closely related thermodimorphic fungi of the genus Sporothrix [10,11], including Sporothrix brasiliensis, Sporothrix schenckii s. str., Sporothrix globosa, and Sporothrix luriei [2,5]. These species have distinct virulence profiles [12], karyotypes [13] and geographic distributions ranging from regional [1,10] to global [5]. The Sporothrix genus, embedded in the plant-associated order Ophiostomatales, exhibits high genetic diversity [10,11], which is accompanied by a diversity of in vitro responses to the main antifungal agents [14] used in sporotrichosis treatment [7,15]. Such agents include potassium iodide [16] and itraconazole [17] for localized cutaneous and lymphocutaneous forms, and amphotericin B for disseminated cases [6,9,16]. Screening for antifungal susceptibilities during epidemiological surveillance programs could help to uncover putative multidrug resistant Sporothrix strains, and improve our ability to adjust therapeutic regimens and reduce relapse.
Despite the considerable spread of the Brazilian epidemic of sporotrichosis over the last two decades [1-3,10] and the increased occurrence of atypical cases [9], no study of this illness to date has integrated molecular epidemiology and antifungal drug susceptibility. The present study aimed to evaluate the resistance profiles to currently used antifungals in a genetically diverse panel of Sporothrix strains representing the main pathogenic species.
Methods
Fungal isolates
This study included 68 Sporothrix isolates that were obtained from clinical lesions of patients with varying degrees of disease severity (n = 65 human/n = 2 animal) or from environmental sources (n = 1). The isolates were originally received as S. schenckii s.l., and were subsequently identified down to the molecular species level by Rodrigues et al.[1,3,18] and Fernandes et al. [12,19] (Additional file 1: Table S1). Isolates were stored at room temperature in slant cultures on Sabouraud dextrose agar (SDA; Difco Laboratories, Detroit, MI). Ethical approval was provided by Institutional Committee (UNIFESP-0244/11).
Antifungal agents and susceptibility testing
Culture conditions and inoculum preparation were as described by Marimon et al. [14]. Using the methods recommended in the CLSI standard document M38-A2, we evaluated minimum inhibitory concentrations (MIC) for the following seven antifungal agents: amphotericin B (AMB), fluconazole (FLC), itraconazole (ITC), voriconazole (VRC), posaconazole (PCZ), flucytosine (5FC), and caspofungin (CAS). The final drug concentrations ranged from 0.03 to 16 μg/mL for AMB, ITC, VRC, PCZ, and CAS, and from 0.12 to 64 μg/mL for 5FC and FLC. The microplates were incubated at 35°C and read after 72 h. Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258 were used as quality controls for the experiments.
To obtain the minimum fungicidal concentration (MFC), for each serial dilution, 10 μL was taken from each well with no visible growth and spread on Sabouraud dextrose agar (Difco Laboratories, USA). Plates were then incubated at 28°C for 48–72 h. The MFC was defined as the lowest drug concentration that yielded three or fewer colonies, i.e., 95.5 to 99% killing activity [20]. The Mann–Whitney U test was used to determine the distribution of MIC and MFC data. A p value of <0.05 was considered to indicate statistical significance.
Molecular characterization
The isolates were genetically characterized [1,10] based on calmodulin gene sequencing using the primers CL1 (5′-GAR TWC AAG GAG GCC TTC TC) and CL2A (5′-TTT TTG CAT CAT GAG TTG GAC) [21], as well as sequencing part of the rRNA operon [5] using the primers ITS1 (5′-TCC GTA GGT GAA CCT TGC GG) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC) [22]. We choose these loci because they have previously been extensively used for phylogeny and taxonomy of this complex of cryptic species [1-3,5,10,11,23]. To increase the quality of sequence data, sense and antisense sequences were determined with an ABI 3730 DNA Analyser (Applied Biosystems, Inc., Foster City, CA, USA) and thereafter assembled into single sequences via CAP3 using bases with a Phred quality score of ≥30. All sequences were deposited online at GenBank (Additional file 1: Table S1).
Phylogenetic histories were reconstructed using maximum likelihood (ML) and neighbor-joining (NJ) methods with the Mega5 software [24], using 1,000 bootstrap replicates to estimate the confidence values for individual branches [25]. Evolutionary distances were calculated using the Tamura 3-parameter model [26]. The rate of change between sites was modeled with a gamma distribution (shape parameter = 1). Nucleotide (π) and haplotype (Hd) diversities [27] were estimated using DnaSP software version 5.10 [28]. Haplotype networks were used to visualize differences and diversity among S. brasiliensis and S. schenckii sequence data [28]. Gaps and missing data were excluded in the calculations. Median-joining networks [29] for the concatenate dataset (CAL + ITS) were obtained and visualized using the Network 4.610 software, as previously described by Rodrigues et al. [3].
Results
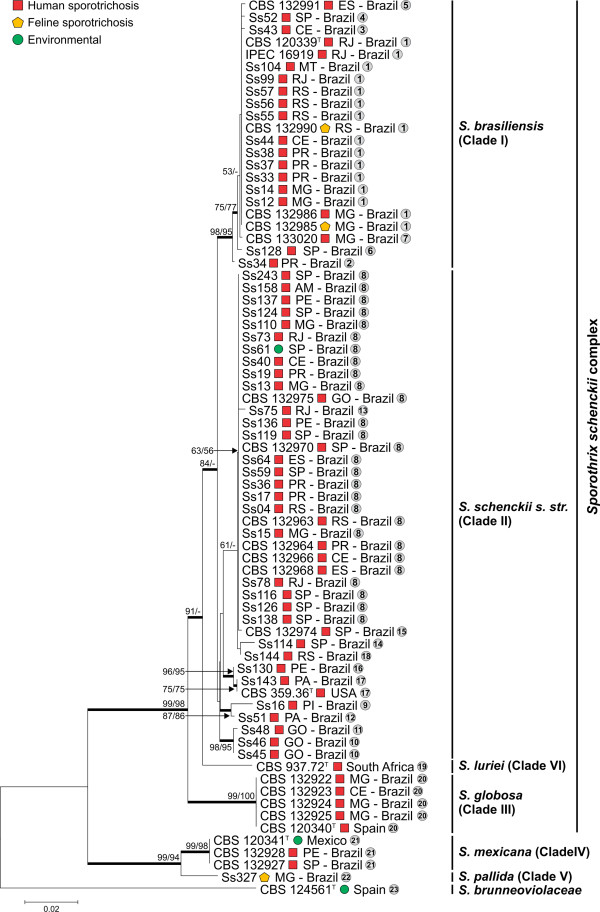
The combined dataset (CAL + ITS) yielded a sequence alignment of 1,397 positions, including 866 invariable characters, 232 variable parsimony-informative sites (16.6%), and 185 variable singletons. The topologies of the ML and NJ trees were congruent (Icong = 3.66; p = 1.68 e-24) [30] and the isolates were distributed among the six major clades (Figure 1), in agreement with previous studies [1-3,10,11].
Figure 1.
Phylogenetic relationships of calmodulin-encoding gene and ITS sequences among clinical and environmental Sporothrix species. The phylogenetic tree were estimated in MEGA5 using the Tamura 3-parameter model. The numbers close to the branches represent indices of support (NJ/ML) based on 1,000 bootstrap replications. The branches with bootstrap support higher than 70% are indicated in bold. Haplotypes numbers were assigned to each isolate and are indicated inside the circles.
Table 1 summarizes the MIC and MFC ranges for the human clinical Sporothrix isolates. Sporothrix brasiliensis exhibited low genetic diversity (π = 0.00194), and comprised seven haplotypes (H1–H7). Of the tested drugs, the triazoles ITC and PCZ showed the best activity against S. brasiliensis strains, with MICs ranging from 0.25 to 4 μg/mL (Figure 2A) and from 0.5 to 2 μg/mL (Figure 2C), respectively. The MICs for ITC were lower in S. brasiliensis compared to in S. schenckii (p = 0.0001) and S. mexicana (p = 0.0031). We detected moderate activity for the polyene AMB against S. brasiliensis strains, with MICs ranging from 1 to 8 μg/mL (Figure 2E), while CAS, 5FC, FLC, and VRC showed poor activity.
Table 1.
Minimum inhibitory concentration (MIC) and minimum fungicidal concentration (MFC) of clinical isolates belonging to the Sporothrix schenckii complex
| Species |
AMB (µg/mL) |
FLC (µg/mL) |
ITC (µg/mL) |
VRC (µg/mL) |
PCZ (µg/mL) |
5FC (µg/mL) |
CAS (µg/mL) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MFC | MIC | MFC | MIC | MFC | MIC | MFC | MIC | MFC | MIC | MFC | MIC | MFC | ||
|
S. brasiliensis (n = 22) |
Range |
1-8 |
2 - >16 |
>64 |
>64 |
0.25 - 4 |
1 - > 16 |
2 - >16 |
16 - >16 |
0.5 - 2 |
1 - >16 |
64 - >64 |
>64 |
>16 |
>16 |
| 50% |
4 |
4 |
>64 |
>64 |
1 |
16 |
16 |
>16 |
1 |
>16 |
>64 |
>64 |
>16 |
>16 |
|
| 90% |
4 |
16 |
>64 |
>64 |
2 |
>16 |
>16 |
>16 |
2 |
>16 |
>64 |
>64 |
>16 |
>16 |
|
|
S. schenckii (n = 39) |
Range |
1 - >16 |
2 - >16 |
>64 |
>64 |
0.25 - >16 |
0.5 - > 16 |
4 - >16 |
4 - >16 |
0.06 - >16 |
1 - >16 |
64 - >64 |
64 - >64 |
2 - >16 |
>16 |
| 50% |
8 |
16 |
>64 |
>64 |
1 |
16 |
16 |
>16 |
1 |
>16 |
64 |
>64 |
>16 |
>16 |
|
| 90% |
>16 |
>16 |
>64 |
>64 |
4 |
>16 |
>16 |
>16 |
2 |
>16 |
>64 |
>64 |
>16 |
>16 |
|
|
S. globosa (n = 4) |
Range |
2 - >16 |
4 - >16 |
>64 |
>64 |
0.5 - >16 |
0.5 - >16 |
2 - 8 |
8 - >16 |
0.5 - 16 |
1 - >16 |
8 - >64 |
>64 |
8 - > 16 |
>16 |
| 50% |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
| 90% |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
|
S. mexicana (n = 3) |
Range |
8 - >16 |
8 - >16 |
>64 |
>64 |
2 - >16 |
2 - >16 |
1 - >16 |
16 - > 16 |
1 - >16 |
>16 |
64 - >64 |
>64 |
8 - >16 |
>16 |
| 50% |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
| 90% |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
| Overall (n = 68) | Range |
1 - >16 |
2 - >16 |
>64 |
>64 |
0.25 - >16 |
0.5 - >16 |
1 - >16 |
4 - >16 |
0.06 - >16 |
1 - >16 |
8 - >64 |
64 - >64 |
2 - >16 |
>16 |
| 50% |
4 |
16 |
>64 |
>64 |
1 |
>16 |
16 |
>16 |
1 |
>16 |
>64 |
>64 |
>16 |
>16 |
|
| 90% | >16 | >16 | >64 | >64 | >16 | >16 | >16 | >16 | 2 | >16 | >64 | >64 | >16 | >16 | |
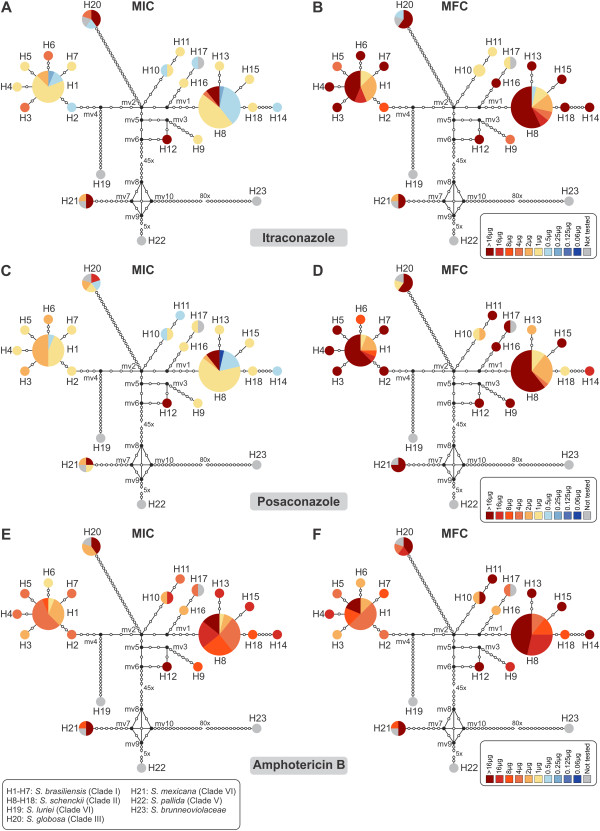
Figure 2.
Genetic diversity and antifungal susceptibility profiles in causative agents of sporotrichosis. Median-joining haplotype network of Sporothrix schenckii complex isolates based on calmodulin and ITS concatenated sequences. The circumference size is proportional to the haplotype frequency. Black dots (median vectors) represent unsampled or extinct haplotypes in the population. Mutational steps are represented by white dots and, in cases of long branches, by values. The charts represent the MIC (A, C, and E) and MFC (B, D, and F) distributions within the population for ITC, PCZ, and AMB.
Sporothrix schenckii represented by the genetically heterogeneous clade II (π = 0.00917) was divided into ten haplotypes (H8–H18) and showed a broad MIC range for the different antifungals. ITC (Figure 2A) and PCZ (Figure 2C) presented good activity against most S. schenckii s. str. strains, with MICs ranging from 0.25 to >16 μg/mL and from 0.06 to >16 μg/mL, respectively. The MICs for PCZ were slightly higher in S. schenckii compared to in S. brasiliensis (p = 0.0076). AMB presented high MICs against S. schenckii isolates, ranging from 1 to >16 μg/mL (Figure 2E), while the remaining compounds—CAS, 5FC, FLC, and VRC—showed very poor activities. Each of the evaluated drugs demonstrated poor in vitro efficacy against the tested S. globosa (H20) and S. mexicana (H21) strains (Figure 2).
As expected, MFCs were at least two-fold higher than the MICs for each drug (Figure 2B, D and F). AMB presented lower MFC values compared to those obtained for the other tested drugs (p = 0.0011; Figure 2F). The MFC for AMB was also lower among S. brasiliensis isolates compared to in S. schenckii (p = 0.0001) and S. mexicana (p = 0.0361). Evaluating the MIC and MFC values together, we identified several multidrug resistant isolates among the S. schenckii s. str. strains. In particular, the deviating haplotype 12 (H12; Figure 2B, D and F), which was represented by a Brazilian strain of human origin (Ss51), showed cross-resistance to all azole drugs and polyenes.
Discussion
Given the high intrinsic antifungal resistance in Sporothrix, studies correlating antifungal susceptibility and genetic diversity among etiological agents of sporotrichosis are overdue. Here we used haplotype networks to estimate genetic diversity among clinical Sporothrix species. This approach has the strength of enabling the recognition of genetically deviating strains in a population, rather than randomly sampled frequent haplotypes with similar genetic background, as represented by the frequent haplotypes H1 and H8 in the S. brasiliensis and S. schenckii populations, respectively. However, haplotype-based strategies are dependent on the molecular marker used to estimate diversity, the set of strains comprising the database as well as the network methods [31]. To date, the partial sequence of CAL and ITS region (ITS1/2 + 5.8 s) has been used to estimate genetic diversity and taxonomy of clinical Sporothrix species [2,5,10,11]. Indeed, due to its highly discriminatory power and polymorphisms we choose these regions to give insights into the genetic diversity in sympatric populations of S. brasiliensis and S. schenckii s. str.
A genetically homogeneous population may be expected to have similar susceptibility profiles among the individuals, but increased fitness during population expansion can result in a diversity of susceptibility profiles. Alternatively, differential responses may emerge over time in ancestral genetically diverse species. Of the phylogenetically related species S. brasiliensis and S. schenckii s. str., the former has a lower genetic diversity [2,3,10,13], and here we found that it also had a correspondingly lower variability of in vitro susceptibility. A larger degree of genetic diversity has been observed among S. schenckii s. str. isolates, which is accompanied by greater variation in antifungal profiles [14] virulence [12] and genomic organization [13]. Moreover, the presence of median vectors (mv1-3, 5, 6) in the haplotype network (Figure 2) may indicate extinct or intermediate unsampled haplotypes within the S. schenckii s. str. populations, possibly suggesting the existence of even more variation. Conversely, the absence of median vectors in the S. brasiliensis population strongly suggests that our analysis detected most of the strain diversity circulating in the human epidemic, thus supporting the low variation of the MIC and MFC values in this species.
Itraconazole is the drug of choice for treating endemic mycoses caused by thermodimorphic fungi, including sporotrichosis. Our findings regarding the in vitro activities of the main antifungal drugs are in agreement with previous studies, indicating that ITC and PCZ were moderately effective against isolates of S. brasiliensis and S. schenckii s. str. [14,32]. Despite the relatively high values found in all studies, several authors have demonstrated the efficacy of ITC in clinical outcome. Barros et al. [17] studied the clinical treatment of a large number of patients in Rio de Janeiro, and reported the success of ITC treatment (50–400 mg/day) in 94.6% patients (n = 610). Although this study did not molecularly identify the phylogenetic species involved, it was most likely S. brasiliensis, as this species is highly prevalent in Rio de Janeiro due to feline sporotrichosis outbreaks [1-3,10]. Such an epidemiological profile could explain the favorable results for ITC. The relatively low MICs found for PCZ also suggest this molecule to be a promising drug in the treatment of sporotrichosis caused by S. schenckii s. str. or S. brasiliensis. Additionally, PCZ also showed low MICs against 2 out of 3 clinical isolates of the rare species S. mexicana, including CBS 132928 (MIC = 1 μg/mL) and CBS 132927 (MIC = 2 μg/mL). Only a few strains of S. mexicana have been described in the literature [1,10], and the species has been reported as tolerant to most commercially available drugs [14,33].
Resistance to amphotericin B has been reported in emerging pathogens, such as Aspergillus terreus, Candida lusitaniae, Fusarium spp., Scedosporium prolificans, and Trichosporon asahii [34,35]. Here we found high MICs for AMB in five isolates of S. schenckii s. str. (Ss17, Ss22, Ss110, and Ss119 from H8, and isolate Ss51 from H12) with MIC and MFC values of >16 μg/mL. The multidrug resistant phenotype found for isolate Ss51 (H12; Figure 2B, D, and F) is particularly noteworthy in relation to its clinical origin, where it showed moderate virulence in a murine model [12]. The high MICs found for the remaining compounds (FLC, VRC, CAS, and 5FC) are in agreement with previous reports [14,32,36].
Fungistatic drugs are capable of inhibiting the cell growth and reproduction of fungi without destroying them. Drug-sensitive pathogens may evolve resistance under the selective pressure imposed by continuous exposure to fungistatic drugs [37]. The molecular mechanism that lies behind the recent emergence of drug-resistant phenotype among Sporothrix species is currently unknown. However, judging from other fungi, the increased and prolonged use of triazoles has raised concerns about resistant infections by Cryptococcus neoformans [38], Candida albicans [39] and Aspergillus fumigatus [40]. Azole resistance may occur through a diversity of mechanisms including the upregulation of multidrug transporter genes that leads to enhanced efflux of azoles and therefore reduce drug accumulation [41]; multiple genetic alterations of the target enzyme that can affect the affinity of the enzyme and therefore prevents azole binding [42], and alteration of metabolism, usually sterol synthesis [43]. More recently, genetic studies correlating antifungal resistance and alteration in chromosome copy number and genomics architecture has delivered an exciting view on the molecular mechanisms governing the increased fitness to the phenotype of azole-resistance [38,44-46].
Candida albicans [39,44,45] and Cryptococcus neoformans [38,46,47] triazole-resistant isolates very often contain an abnormal number of chromosomes. Such genomic plasticity may offer additional copies of drug resistance genes leading chromosomal aneuploidies isolates to overcoming the drug effects [44,45] and allowing rapid adaptive evolution [47]. Although this phenomenon may occur spontaneously [47], drug-resistant aneuploidies arise very frequently within drug-exposed fungal populations and this may support our recent findings on chromosomal polymorphisms in Sporothrix species [13]. The intra-specific polymorphisms in chromosome number and size in S. schenckii s. str. suggested that aneuploidy occur among clinical isolates, and it is tempting to hypothesize that this phenomenon could, in part, be responsible for the differences in drug profiles observed here. Therefore, the high karyotype diversity observed for S. schenckii s. str. [13] is reflected in the genetic diversity found in our haplotype network (Figure 2). However, testing for this pattern requires a larger number of isolates with dissimilar antifungal profiles as well as chromosomal polymorphism.
Fungicidal drugs may be defined as those that lead to a reduction of 99.9% of the initial inocula [48]. It is difficult to make comparisons between published MFCs studies, since most were performed before the introduction of clinical species beyond S. schenckii s. str. to the genus Sporothrix. Two or more of these newer species were likely involved in the studies of McGinnis et al. [49] and Silveira et al. [50]. In all clinical Sporothrix spp. evaluated, we found significant differences between the minimum concentrations needed to inhibit fungal cell growth and the concentration required for colony count reduction. In all studied species, most isolates were able to grow after 72 h in the presence of each drug at the maximum concentration tested, which is in agreement with results obtained by Trilles et al. [36]. The moderately low MICs for triazoles imply a great resistance to killing among isolates of S. brasiliensis, the most pathogenic among Sporothrix species.
Spontaneous cure, as well as relapse, are common features of sporotrichosis. Our present data show that most antifungal agents had only a fungistatic effect against clinical isolates. It remains unclear whether there is a correlation between in vitro MIC/MFC data and clinical outcome in human sporotrichosis. Experimental murine models have shown the efficacy of posaconazole (5 mg/kg) for treating S. brasiliensis and S. schenckii s. str. infections [51]. Voriconazole (40 mg/kg) has been demonstrated to only reduce fungal load in mice infected with S. schenckii s. str., and to have no activity against S. brasiliensis [52]. No studies to date have linked the dissimilar phylogenetic species to in vitro and in vivo data. Our in vitro results for the highly tolerant strain CBS 133020 (=Ss265, haplotype 7), an isolate originated from a disseminated case of S. brasiliensis in an HIV patient [9], shown that despite ITC and AMB MICs of 1 and 4 μg/mL, respectively, and MFCs of >16 and 8 μg/mL, respectively, the patient showed positive clinical outcome after administration of intravenous AMB (including 10 days on L-AMB) [9], indicating a need to monitor MFCs values before and during treatment.
Conclusions
Our study presents antifungal susceptibility profiles using MIC and MFC data, which is particularly noteworthy in light of recent taxonomic changes in medically relevant Sporothrix species. We assessed a genetically diverse set of strains with varying degrees of disease severity (fixed cutaneous, lymphocutaneous, or disseminated), and the antifungal susceptibility assays revealed a broad spectrum for the triazoles (ITC, PCZ, and VRC). This finding may have implications for the choice of antifungal therapy in different epidemiological risk groups, potentially having a substantial impact on clinical outcome. Furthermore, our data indicate the existence of multidrug resistant strains within the S. schenckii complex, underlining the need for continuous susceptibility screening. Further studies should be conducted to connect in vitro and clinical data, as well as to investigate the molecular mechanisms underlying the different resistance profiles among S. schenckii and its relatives.
Abbreviations
ITS: Internal transcribed spacer; CAL: Calmodulin; MICs: Minimum inhibitory concentration; MFC: Minimum fungicidal concentration; AMB: Amphotericin B; FLC: Fluconazole; ITC: Itraconazole; VRC: Voriconazole; PCZ: Posaconazole; 5FC: Flucytosine; CAS: Caspofungin; s. str.: sensu stricto; s.l.: sensu lato; ML: Maximum likelihood; NJ: Neighbor-joining; mv: Median vector.
Competing interests
The authors declare that they have no competing interests. The authors alone are responsible for the content and writing of the paper.
Authors’ contributions
AMR and DCP performed the experiments. AMR, ALC, and ZPC designed the experiments. AMR, GSdH, RSNB, JJCS, MFG, ALC, and ZPC analyzed the data, and drafted the manuscript together. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Strains, species, origin, haplotypes, and GenBank accession numbers (CAL and ITS) for the Sporothrix spp. isolates used in this study and the reference strains.
Contributor Information
Anderson Messias Rodrigues, Email: amrodrigues.amr@gmail.com.
G Sybren de Hoog, Email: s.hoog@cbs.knaw.nl.
Débora de Cássia Pires, Email: dcpires@hotmail.com.
Raimunda Sâmia Nogueira Brihante, Email: brilhante@ufc.br.
José Júlio da Costa Sidrim, Email: sidrim@ufc.br.
Marcos Fabio Gadelha, Email: mfgrocha@gmail.com.
Arnaldo Lopes Colombo, Email: arnaldolcolombo@gmail.com.
Zoilo Pires de Camargo, Email: zpcamargo1@gmail.com.
Acknowledgements
The authors are thankful for funding from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2009/54024-2) and National Council for Scientific and Technological Development (CNPq 472600/2011-7). AMR (FAPESP 2011/07350-1) is a fellow of FAPESP.
References
- Rodrigues AM, de Hoog S, de Camargo ZP. Emergence of pathogenicity in the Sporothrix schenckii complex. Med Mycol. 2013;14:405–412. doi: 10.3109/13693786.2012.719648. [DOI] [PubMed] [Google Scholar]
- Rodrigues AM, de Hoog GS, Zhang Y, Camargo ZP. Emerging sporotrichosis is driven by clonal and recombinant Sporothrix species. Emerg Microbes Infect. 2014;14:e32. doi: 10.1038/emi.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues AM, de Melo TM, de Hoog GS, Schubach TMP, Pereira SA, Fernandes GF, Bezerra LML, Felipe MS, de Camargo ZP. Phylogenetic analysis reveals a high prevalence of Sporothrix brasiliensis in feline sporotrichosis outbreaks. PLoS Negl Trop Dis. 2013;14:e2281. doi: 10.1371/journal.pntd.0002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Li SS, Zhong SX, Liu YY, Yao L, Huo SS. Report of 457 sporotrichosis cases from Jilin province, northeast China, a serious endemic region. J Eur Acad Dermatol Venereol. 2013;14:313–318. doi: 10.1111/j.1468-3083.2011.04389.x. [DOI] [PubMed] [Google Scholar]
- Zhou X, Rodrigues AM, Feng P, Hoog GS. Global ITS diversity in the Sporothrix schenckii complex. Fungal Divers. 2013. pp. 1–13. doi:10.1007/s13225-013-0220-2.
- Kauffman CA, Bustamante B, Chapman SW, Pappas PG. Clinical practice guidelines for the management of sporotrichosis: 2007 update by the Infectious Diseases Society of America. Clin Infect Dis. 2007;14:1255–1265. doi: 10.1086/522765. [DOI] [PubMed] [Google Scholar]
- Kauffman CA. Sporotrichosis. Clin Infect Dis. 1999;14:231–237. doi: 10.1086/520190. [DOI] [PubMed] [Google Scholar]
- Bonifaz A, Vázquez-González D. Diagnosis and treatment of lymphocutaneous sporotrichosis: What are the options? Curr Fungal Infect Rep. 2013;14:252–259. doi: 10.1007/s12281-013-0140-3. [DOI] [Google Scholar]
- Silva-Vergara ML, de Camargo ZP, Silva PF, Abdalla MR, Sgarbieri RN, Rodrigues AM, dos Santos KC, Barata CH, Ferreira-Paim K. Disseminated Sporothrix brasiliensis infection with endocardial and ocular involvement in an HIV-infected patient. Am J Trop Med Hyg. 2012;14:477–480. doi: 10.4269/ajtmh.2012.11-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimon R, Cano J, Gené J, Sutton DA, Kawasaki M, Guarro J. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J Clin Microbiol. 2007;14:3198–3206. doi: 10.1128/JCM.00808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimon R, Gené J, Cano J, Trilles L, Dos Santos Lazéra M, Guarro J. Molecular phylogeny of Sporothrix schenckii. J Clin Microbiol. 2006;14:3251–3256. doi: 10.1128/JCM.00081-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes GF, dos Santos PO, Rodrigues AM, Sasaki AA, Burger E, de Camargo ZP. Characterization of virulence profile, protein secretion and immunogenicity of different Sporothrix schenckii sensu stricto isolates compared with S. globosa and S. brasiliensis species. Virulence. 2013;14:241–249. doi: 10.4161/viru.23112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AA, Fernandes GF, Rodrigues AM, Lima FM, Marini MM, dos Feitosa SL, de Melo Teixeira M, Felipe MSS, da Silveira JF, de Camargo ZP. Chromosomal polymorphism in the Sporothrix schenckii complex. PLoS ONE. 2014;14:e86819. doi: 10.1371/journal.pone.0086819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimon R, Serena C, Gené J, Cano J, Guarro J. In vitro antifungal susceptibilities of five species of Sporothrix. Antimicrob Agents Chemother. 2008;14:732–734. doi: 10.1128/AAC.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman CA, Hajjeh R, Chapman SW, Group MS. Practice guidelines for the management of patients with sporotrichosis. Clin Infect Dis. 2000;14:684–687. doi: 10.1086/313751. [DOI] [PubMed] [Google Scholar]
- Yamada K, Zaitz C, Framil VM, Muramatu LH. Cutaneous sporotrichosis treatment with potassium iodide: a 24 year experience in São Paulo State, Brazil. Revista do Instituto de Medicina Tropical de Sao Paulo. 2011;14:89–93. doi: 10.1590/s0036-46652011000200006. [DOI] [PubMed] [Google Scholar]
- de Lima Barros MB, Schubach AO, de-Vasconcellos Carvalhaes De-Oliveira R, Martins EB, Teixeira JL, Wanke B. Treatment of cutaneous sporotrichosis with Itraconazole—Study of 645 patients. Clin Infect Dis. 2011;14:e200–e206. doi: 10.1093/cid/cir245. [DOI] [PubMed] [Google Scholar]
- Rodrigues AM, de Hoog GS, Camargo ZP. Genotyping species of the Sporothrix schenckii complex by PCR-RFLP of calmodulin. Diagn Microbiol Infect Dis. 2014;14:383–387. doi: 10.1016/j.diagmicrobio.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Fernandes GF, dos Santos PO, Amaral CC, Sasaki AA, Godoy-Martinez P, Camargo ZP. Characteristics of 151 Brazilian Sporothrix schenckii isolates from 5 different geographic regions of Brazil: A forgotten and re-emergent pathogen. Open Mycol J. 2009;14:48–58. doi: 10.2174/1874437000903010048. [DOI] [Google Scholar]
- Espinel-Ingroff A, Chaturvedi V, Fothergill A, Rinaldi MG. Optimal testing conditions for determining MICs and Minimum Fungicidal Concentrations of new and established antifungal agents for uncommon molds: NCCLS collaborative study. J Clin Microbiol. 2002;14:3776–3781. doi: 10.1128/JCM.40.10.3776-3781.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Nirenberg H, Aoki T, Cigelnik E. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience. 2000;14:61–78. doi: 10.1007/BF02464387. [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. In: PCR Protocols: A Guide to Methods and Applications. Innis M, Gelfand D, Shinsky J, White T, editor. New York, USA: Academic Press; 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics; pp. 315–322. [Google Scholar]
- Marimon R, Gené J, Cano J, Guarro J. Sporothrix luriei: a rare fungus from clinical origin. Med Mycol. 2008;14:621–625. doi: 10.1080/13693780801992837. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;14:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolution confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;14:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol. 1992;14:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;14:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;14:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- de Vienne DM, Giraud T, Martin OC. A congruence index for testing topological similarity between trees. Bioinformatics. 2007;14:3119–3124. doi: 10.1093/bioinformatics/btm500. [DOI] [PubMed] [Google Scholar]
- Woolley SM, Posada D, Crandall KA. A comparison of phylogenetic network methods using computer simulation. PLoS ONE. 2008;14:e1913. doi: 10.1371/journal.pone.0001913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DC, Lopes PGM, Spader TB, Mahl CD, Tronco-Alves GR, Lara VM, Santurio JM, Alves SH. Susceptibilities to antifungal agents of Sporothrix albicans, S. brasiliensis and S. luriei of the S. schenckii complex identified in Brazil. J Clin Microbiol. 2011;14:3047–3049. doi: 10.1128/JCM.00255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilhante RS, Malaquias AD, Caetano EP, Castelo-Branco Dde S, Lima RA, Marques FJ, Silva NF, Alencar LP, Monteiro AJ, Camargo ZP, Bandeira Tde J, Rodrigues AM, Cordeiro Rde A, Moreira JL, Sidrim JJ, Rocha MF. In vitro inhibitory effect of miltefosine against strains of Histoplasma capsulatum var. capsulatum and Sporothrix spp. Med Mycol. 2014;14(3):320–325. doi: 10.1093/mmy/myt027. [DOI] [PubMed] [Google Scholar]
- Nucci M, Marr KA. Emerging fungal diseases. Clin Infect Dis. 2005;14:521–526. doi: 10.1086/432060. [DOI] [PubMed] [Google Scholar]
- Colombo AL, Padovan ACB, Chaves GM. Current knowledge of Trichosporon spp. and trichosporonosis. Clin Microbiol Rev. 2011;14:682–700. doi: 10.1128/CMR.00003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trilles L, Fernández-Torres B, dos Santos Lazéra M, Wanke B, de Oliveira Schubach A, de Almeida Paes R, Inza I, Guarro J. In vitro antifungal susceptibilities of Sporothrix schenckii in two growth phases. Antimicrob Agents Chemother. 2005;14:3952–3954. doi: 10.1128/AAC.49.9.3952-3954.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JB. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat Rev Microbiol. 2005;14:547–556. doi: 10.1038/nrmicro1179. [DOI] [PubMed] [Google Scholar]
- Sionov E, Lee H, Chang YC, Kwon-Chung KJ. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 2010;14:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;14:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders E, van der Lee HA, Kuijpers J, Rijs AJ, Varga J, Samson RA, Mellado E, Donders AR, Melchers WJ, Verweij PE. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 2008;14:e219. doi: 10.1371/journal.pmed.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S, Barker KS, Znaidi S, Schneider S, Dierolf F, Dunkel N, Aid M, Boucher G, Rogers PD, Raymond M, Morschhäuser J. Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob Agents Chemother. 2011;14:2212–2223. doi: 10.1128/AAC.01343-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre B, Didmon M, Ryder NS. Multiple amino acid substitutions in lanosterol 14alpha-demethylase contribute to azole resistance in Candida albicans. Microbiology. 1999;14:2715–2725. doi: 10.1099/00221287-145-10-2715. [DOI] [PubMed] [Google Scholar]
- Willger SD, Puttikamonkul S, Kim KH, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA Jr. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;14:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, Ammar R, Torti D, Nislow C, Cowen LE. Genetic and genomic architecture of the evolution of resistance to antifungal drug combinations. PLoS Genet. 2013;14:e1003390. doi: 10.1371/journal.pgen.1003390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison BD, Hashemi J, Bibi M, Pulver R, Bavli D, Nahmias Y, Wellington M, Sapiro G, Berman J. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 2014;14:e1001815. doi: 10.1371/journal.pbio.1001815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Chang YC. Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog. 2012;14:e1003022. doi: 10.1371/journal.ppat.1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Feretzaki M, Li W, Floyd-Averette A, Mieczkowski P, Dietrich FS, Heitman J. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity De Novo in the yeast Cryptococcus neoformans. PLoS Biol. 2013;14:e1001653. doi: 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybill JR, Burgess DS, Hardin TC. Key issues concerning fungistatic versus fungicidal drugs. Eur J Clin Microbiol Infect Dis. 1997;14:42–50. doi: 10.1007/BF01575120. [DOI] [PubMed] [Google Scholar]
- McGinnis MR, Nordoff N, Li RK, Pasarell L, Warnock DW. Sporothrix schenckii sensitivity to voriconazole, itraconazole and amphotericin B. Med Mycol. 2001;14:369–371. doi: 10.1080/mmy.39.4.369.371. [DOI] [PubMed] [Google Scholar]
- Silveira CP, Torres-Rodríguez JM, Alvarado-Ramírez E, Murciano-Gonzalo F, Dolande M, Panizo M, Reviakina V. MICs and minimum fungicidal concentrations of amphotericin B, itraconazole, posaconazole and terbinafine in Sporothrix schenckii. J Med Microbiol. 2009;14:1607–1610. doi: 10.1099/jmm.0.007609-0. [DOI] [PubMed] [Google Scholar]
- Fernández-Silva F, Capilla J, Mayayo E, Guarro J. Efficacy of posaconazole in murine experimental sporotrichosis. Antimicrob Agents Chemother. 2012;14:2273–2277. doi: 10.1128/AAC.05376-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Silva F, Capilla J, Mayayo E, Guarro J. Modest efficacy of voriconazole against murine infections by Sporothrix schenckii and lack of efficacy against Sporothrix brasiliensis. Mycoses. 2014;14:121–124. doi: 10.1111/myc.12112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strains, species, origin, haplotypes, and GenBank accession numbers (CAL and ITS) for the Sporothrix spp. isolates used in this study and the reference strains.