Abstract
Priming of defense increases the responsiveness of the plant immune system and can provide broad-spectrum protection against disease. Recent evidence suggests that priming of defense can be inherited epigenetically to following generations. However, the mechanisms of long-lasting defense priming within one generation remains poorly understood. Here, we have investigated the mechanistic basis of long-lasting induced resistance after treatment with β -aminobutyric acid (BABA), an agent that mimics biologically induced resistance phenomena. BABA-induced resistance (BABA-IR) is based on priming of salicylic acid (SA)-dependent and SA-independent defenses. BABA-IR could be detected up to 28 days after treatment of wild-type Arabidopsis. This long-lasting component of the induced resistance response requires the regulatory protein NPR1 and is associated with priming of SA-inducible genes. In contrast, NPR1-independent resistance by BABA was transient and had disappeared by 14 days after treatment. Chromatin immunoprecipitation (ChIP) assays revealed no increased acetylation of histone H3K9 at promoters regions of priming-responsive genes, indicating that this post-translational histone modification is not critical for long-term transcriptional priming. Interestingly, the kyp-6 mutant, which is affected in methyltransferase activity of H3K9, was blocked in long-lasting BABA-IR, indicating a critical requirement of this post-translational histone modification in long-lasting BABA-IR. Considering that KYP suppresses gene transcription through methylation of H3K9 and CpHpG DNA methylation, we propose that KYP enables long-term defense gene priming by silencing suppressor genes of SA/NPR1-dependent genes.
Keywords: priming, induced defense, Arabidopsis, NPR1, KYP
INTRODUCTION
Plants can resist pathogen attack by increasing the responsiveness of their immune system. This phenomenon typically occurs after perception of stress-indicating signals and is known as priming of defense. Priming provides non-specific protection against a wide range of biotic and abiotic stresses (Conrath et al., 2006; Pastor et al., 2012; Tanou et al., 2012), which is associated with relatively minor costs on growth and reproduction (Van Hulten et al., 2006). Induction of defense priming results in a faster and stronger expression of basal immune responses upon pathogen attack (Conrath et al., 2006, 2011), and can render plants immune if the augmented defense reaction precedes immune-suppression by pathogen (Ahmad et al., 2010). In most cases, however, defense priming slows down pathogen colonization and reduces disease. Research over the past decades has identified various chemicals that can mimic biologically induced priming responses. These chemicals are often plant-derived signaling metabolites, such as salicylic acid (SA; Shirasu et al., 1997), jasmonic acid (JA; Kauss et al., 1994; Conrath et al., 2002), azelaic acid (Jung et al., 2009), or herbivore-induced volatiles (Ton et al., 2007; Frost et al., 2008). There are also xenobiotic chemicals that can trigger defense priming in plants. Amongst these, the non-protein amino acid β-aminobutyric acid (BABA; Zimmerli et al., 2000; Ton et al., 2005) and benzo-thiadiazole-7-carbothioic acid S-methyl ester (BTH; Kohler et al., 2002) have emerged as popular agents to study the mechanistic basis of defense priming in plants (Conrath, 2011).
BABA-induced resistance (BABA-IR) mimics component of defense priming that are active during pathogen-induced systemic acquired resistance (SAR) and rhizobacteria-induced systemic resistance (ISR; Van der Ent et al., 2009). Consequently, it provides protection against an exceptionally broad range of pathogens and insects. The signaling pathways controlling BABA-IR against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 relies on production of the plant hormone salicylic acid (SA) and a functional non-expressor of PR GENES (NPR1) protein (Zimmerli et al., 2000). However, BABA-IR against the oomycete pathogen Hyaloperonospora arabidopsidis and the necrotrophic fungi Alternaria brassicicola and Plectosphaerella cucumerina can function independently from NPR1, but requires components of the abcisic acid (ABA) signaling pathway (Ton and Mauch-Mani, 2004; Ton et al., 2005). Both pathways operate independently from each other and provide different mechanisms of defense priming (Ton et al., 2005). The NPR1-independent pathway primes cell wall defense, which leads to augmented deposition of callose-rich papillae after pathogen attack. On the other hand, the NPR1-dependent pathway controls priming of SA-dependent genes, which is marked by enhanced transcription of NPR1-dependent transcription factor (TF) genes that control SA-dependent gene induction (Van der Ent et al., 2009). The latter finding suggested that greater abundance of defense regulatory TFs contributes to transcriptional priming of SA-inducible defense genes. However, TFs have limited turn-over times and their enhanced accumulation after application of a single priming stimulus is not a satisfactory explanation for a long-lasting induced resistance response.
Epigenetic mechanisms, such as histone modifications or DNA methylation, have emerged as important regulatory mechanisms in plant immunity (Alvarez et al., 2010). There is ample evidence that post-translational modifications of histone proteins are influences by JA-, SA-, and ABA-dependent signaling pathways (Devoto et al., 2002; Mosher et al., 2006; Walley et al., 2008; Cho et al., 2012). Furthermore, exposure to disease, herbivores and abiotic stresses can have profound impacts on patterns of symmetric and asymmetric DNA methylation (Pavet et al., 2006; Boyko et al., 2010; Verhoeven et al., 2010; Dowen et al., 2012). It is, therefore, not surprising that priming of defense has been associated with epigenetic regulatory mechanisms (Conrath, 2011; Pastor et al., 2012). First evidence for an epigenetic basis of defense priming came from Jaskiewicz et al. (2011), who demonstrated that infection of Arabidopsis by Pseudomonas syringae pv. maculicola primes stress-inducible expression of transcription factor genes via NPR1-dependent modifications of histone H3 at their promoter regions. Furthermore, López et al. (2011) demonstrated that mutants blocked in RNA-directed DNA methylation are primed to activate SA-inducible defense genes, which was associated with H3 modifications marking a facilitated state of gene transcription: acetylation at lysine residue 9 (H3K9ac) and triple-methylation at lysine 4. Hence, defense priming is often associated with post-translational histone modifications at promoter regions of primed defense genes.
Recently, three independent research groups provided evidence that priming of defense can be inherited epigenetically from isogenic plants that had been treated with pathogens, herbivores, or BABA (Luna et al., 2012; Rasmann et al., 2012; Slaughter et al., 2012). Although these studies demonstrated an epigenetic component of defense priming, the extent by which epigenetic regulation contributes to long-lasting defense priming within one plant generation remains unknown. In this study, we have investigated the mechanisms controlling durable maintenance of defense priming in individual plants after treatment with the chemical agent BABA. We show that only the NPR1-dependent component of BABA-IR is long-lasting in Arabidopsis, which is associated with priming of SA-inducible defense genes. Furthermore, we provide evidence that this long-lasting defense resistance requires the histone methyltransferase KYP.
MATERIALS AND METHODS
PLANT MATERIAL, GROWTH CONDITIONS, AND EXPERIMENTAL DESIGN
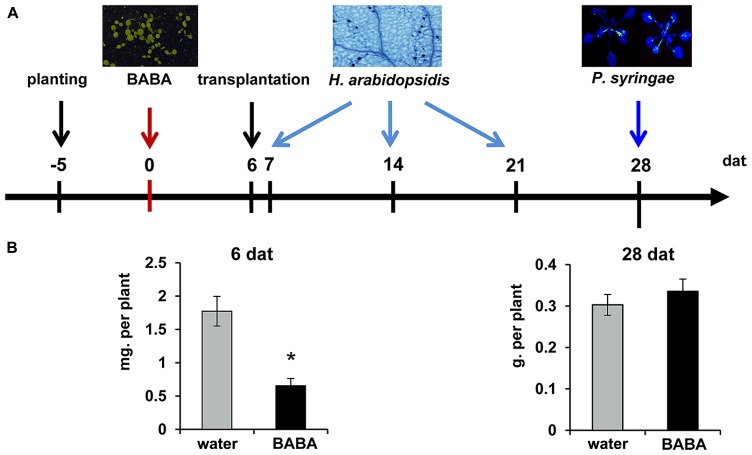
Arabidopsis thaliana (Col-0), npr1-1 (Despres et al., 2003), and kyp-6 (Alonso et al., 2003; Chan et al., 2006) were cultivated in a growth chamber with a 8-h day (150 μE m-2 s-1 at 20°C) and 16-h night (18°C) under at 65% relative humidity. Seeds were planted in 60-ml pots containing a 50% (v/v) sand/M3 mixture and kept at 4°C in the dark for 2 days to break dormancy. Five day-old seedlings were soil-drenched with water or BABA solution (Sigma-Aldrich; Cat.: A4, 420–7) to a final concentration of 40 mg/L in the soil. Six days after treatment, seedlings were transplanted to BABA-free soil (Figure 1A). At different time-points after treatment, plants were examined for differences in fresh weight (7 and 28 days), inoculated with H. arabidopsidis (7, 14, and 21 days), inoculated with Pseudomonas syringae pv. tomato DC3000 luxCDABE (Pst-luxCDABE; Fan et al., 2008; 28 days), or examined for priming of salicylic acid (SA)-inducible gene expression and chromatin modification (28 days).
FIGURE 1.
Experimental design to determine durability of induced resistance. (A) Five-day-old seedlings were soil-drenched with water or BABA (40 mg/L) and transplanted to BABA-free soil at 6 days after induction treatment (dat). Plants were examined for shoot fresh weight at 6 and 28 days after treatment (dat), or inoculated with either Hyaloperonospora arabidopsidis WACO9 (7, 14, 21 dat), or Pseudomonas syringae pv. tomato DC3000 (Pst-luxCDABE; 28 dat). (B) Mean shoot fresh weights of water- and BABA-treated plants at 6 and 28 dat. Error bars represent the SEM. Asterisk indicates a statistically significant difference between water- and BABA-treated plants (Student’s t-test; α < 0.05; n = 15).
GROWTH ANALYSIS AND INDUCED RESISTANCE BIOASSAYS
Plant growth was analyzed by measuring shoot fresh weights at 6 and 28 days after induction treatment (seedlings were excised from the roots, including hypocotyls). Inoculation and determination of pathogen colonization were performed as described previously (Luna et al., 2012). Colonization by H. arabidopsidis was scored at 6 days after inoculation. Colonization by Pst-luxCDABE (Fan et al., 2008) was quantified at 3 days after inoculation.
GENE EXPRESSION ANALYSIS
Water- and BABA-treated plants (n = 30) were sprayed with 0.5 mM SA (Sodium salicylate; Sigma-Aldrich; Cat.: S3007). At 0, 4, 8, and 24 h after application of SA, at least three biologically replicated samples containing pooled material from individual pots, were snap frozen in liquid nitrogen. RNA extraction, cDNA synthesis, and reverse-transcriptase quantitative PCR (RT-qPCR) with gene-specific primers were entirely performed as described before (Luna et al., 2012). Fold induction values were normalized to average 2ΔCt values relative to time-point 0 h before SA application of control-treated plants.
CHROMATIN IMMUNOPRECIPITATION
Chromatin immunoprecipitation (ChIP) assays were carried out as described in the manufacturer’s protocol (EpiQuik Plant ChIP kit; Epigentek, Brooklyn, NY, USA), using mature leaves from 5-week-old plants. For each experiment, at least three biologically replicated samples were collected, each consisting of rosettes from five to seven plants. Chromatin samples were immunoprecipitated using antibodies against acetyl-histone H3K9 (Millipore 07-352). Before and after immunoprecipitation, DNA abundance in chromatin extracts was analyzed by quantitative PCR, using the ABI PRISM® 7900 HT sequence detection system. Two technically replicated reactions per sample were performed in a final reaction volume of 25 μl, containing Jump Start SYBR Green (Sigma-S4438). Sequence-specific primers were used to amplify promoter DNA from PR1, WRKY6, WRKY29, WRKY53 (Jaskiewicz et al., 2011), and WRKY70 (Fw: AATTAGATTCAAGTCCACAACCAA Rv: ATCAAGAAATTGTCATCCAACAC). Results were normalized to DNA amounts in the input control, as described by Haring et al. (2007) with modifications. To prevent possible bias to inaccurate estimations of input DNA, two independent DNA extractions were performed from each chromatin extract. Only if input values differed less than 0.25 Ct values, samples were considered reliable for further analysis and Ct input values were averaged for normalization of immunoprecipitated DNA.
STATISTICS
Average shoot fresh weights and % bacterial bioluminescence were based on at least 15 individual plants per treatment and were analyzed for statistical differences by Student’s t-tests (α = 0.05; SPSS, v19.0). H. arabidopsidis class distributions were based on 50–100 leaves and differences between treatments were analyzed for statistical significance by χ2 contingency tests using SPSS, v19.0. Average fold-change values of gene expression and H3K9ac levels were based on three biological replicates per treatment and statistical differences were determined by Student’s t-tests (α = 0.05; SPSS, v19.0). Each experiment was repeated twice from the onset.
RESULTS
NO LONG-LASTING IMPACTS OF BABA ON PLANT DEVELOPMENT
Induction of disease resistance by BABA can reduce growth of Arabidopsis (Van Hulten et al., 2006; Wu et al., 2010). To examine the long-lasting impact of BABA on plant development, 5-day-old seedlings were soil-drenched with 40 mg/L BABA. Seedlings were kept in BABA-drenched soil for 6 days, after which they were transferred to un-treated soil in order to exclude ongoing induction by excess amounts of soil-based BABA (Figure 1A). Fresh weight analysis of shoots at 6 days after treatment revealed a statistically significant reduction of plant growth in BABA-treated plants (Figure 1B). However, no growth differences were apparent by 28 days after treatment, indicating that BABA-treated plants can fully recover from the induction treatment.
INDUCED RESISTANCE BY BABA LASTS UP TO 4 WEEKS AFTER TREATMENT
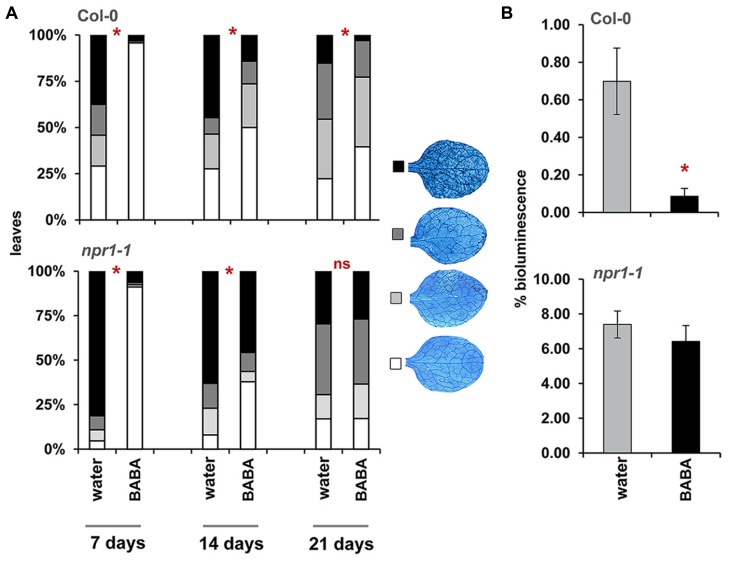
To determine durability of BABA-IR, plants were infected with H. arabidopsidis at 7, 14, and 21 days after BABA application (Figure 2A), after which colonization was microscopically analyzed at 6 days after inoculation. Because of age-related resistance against H. arabidopsidis at later developmental stages, bioluminescent Pseudomonas syringae pv. tomato DC3000 (Pst-luxCDABE) was used to determine disease resistance at 28 days after induction treatment. BABA-treated plants express nearly complete levels of resistance against H. arabidopsidis when inoculated at 7 days after induction treatment, which declined when plants had been inoculated at later time points. Nevertheless, statistically significant levels of induced resistance were still detectable by 21 days after treatment (Figure 2A). Moreover, when plants had been infected Pst-luxCDABE at 28 days after induction treatment, BABA-treated plants still allowed lower levels of leaf colonization by these bacteria (Figure 2B). Hence, BABA-IR declines during the first 2 weeks after treatment, but remains stable during following weeks. Slaughter et al. (2012) demonstrated that plant-endogenous BABA levels at 3 weeks after soil-drench application (40 mg/L) are 10-fold lower than the threshold level required for induced resistance. Accordingly, we conclude that long-lasting resistance by BABA is not due to lingering traces of BABA in the tissue, but rather due to long-lasting physiological changes in the plant.
FIGURE 2.
Durability of BABA-IR in Arabidopsis. (A) Levels of leaf colonization by H. arabidopsidis in wild-type (Col-0) and npr1-1 plants infected at 7, 14, and 21 days after treatment with 40 mg/L BABA (dat). Shown are % of leaves assigned to four different pathogen colonization classes, based on scoring of 50–100 trypan-blue stained leaves at 6 days after inoculation with 105 spores mL-1. Asterisks indicate statistically significant differences between treatments (χ2 contingency test; α < 0.05). (B) Bacterial colonization by Pst-luxCDABE in wild-type (Col-0) and npr1-1 plants at 28 dat (40 mg/L BABA). Shown are average values (±SEM) of relative bioluminescence per plant at 3 days after inoculation. Asterisk indicates a statistically significant difference between water- and BABA-treated plants (Student’s t-test; α < 0.05). ns = no significant.
LONG-LASTING BABA-IR REQUIRES NPR1
The npr1-1 mutant is blocked in SA-dependent defense (Cao et al., 1994). Consequently, this mutant is only capable of expressing the SA-independent component of BABA-IR (Zimmerli et al., 2000). To examine which component is responsible for long-lasting disease protection, we measured durability of BABA-IR in npr1-1 plants. As observed in wild-type plants, npr1-1 expressed relatively high levels of BABA-IR when the plants had been inoculated at 7 days after priming treatment, which declined as time progressed (Figure 2A). However, unlike wild-type plants, BABA-treated npr1-1 failed to express induced resistance to H. arabidopsidis and Pst-luxCDABE when inoculated at 21 and 28 days after induction treatment, respectively (Figure 2). These results indicate that long-lasting BABA-IR is regulated by the NPR1-dependent pathway.
LONG-LASTING PRIMING BY BABA IS ASSOCIATED WITH PRIMING OF SA-INDUCIBLE GENES
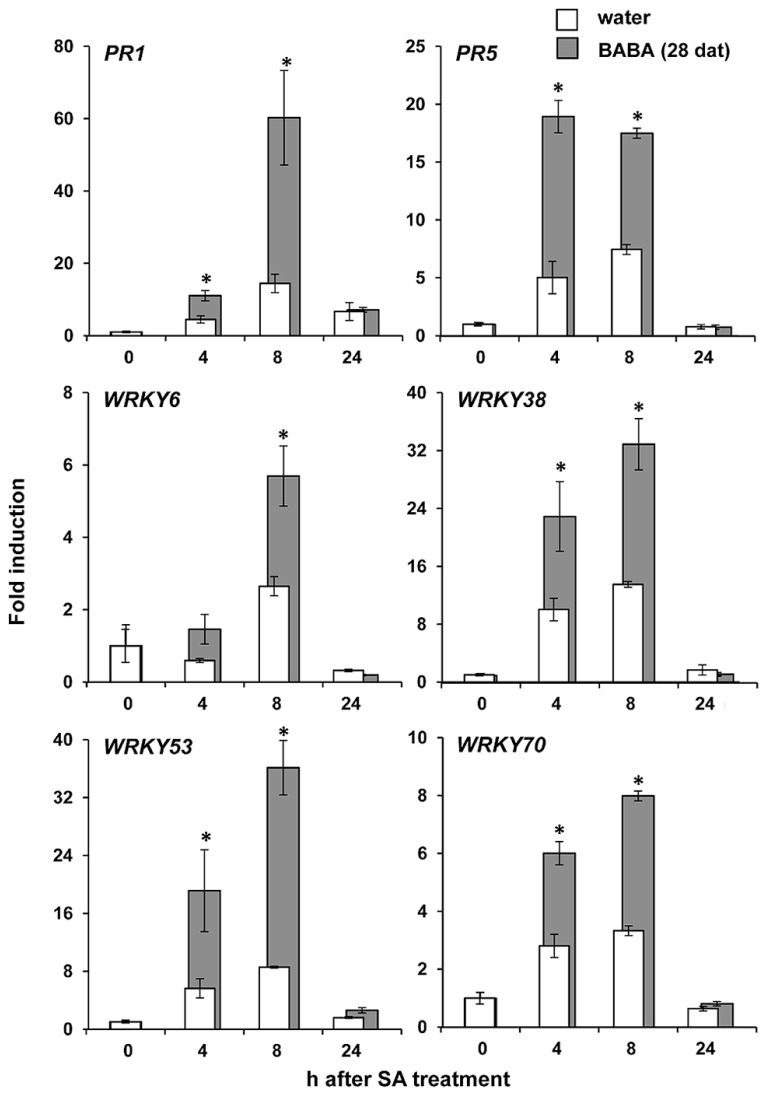
NPR1 regulates priming of SA-dependent defense (Kohler et al., 2002; Van der Ent et al., 2009). Since our experiments revealed that NPR1 is necessary for long-lasting resistance by BABA, we investigated whether the resistance is associated with transcriptional priming of SA responsive genes (PR1, PR5, WRKY70, WRKY6, WRKY53, and WRKY38). At 28 days after BABA application, leaves were sprayed with 0.5 mM SA and harvested at different time-points after treatment for RT-qPCR analysis of defense gene expression. As is shown in Figure 3, basal transcription levels of all genes before SA application were similar in BABA- and control-treated plants. However, after application of SA, all genes tested showed faster and stronger transcriptional induction in BABA-treated plants compared to control plants (Figure 3). Hence, long-lasting induced resistance is not based on enhanced transcription of SA-dependent defense genes, but rather on a transcriptional priming of these genes.
FIGURE 3.
Long-lasting priming of SA-dependent gene transcription. Levels of gene transcription were determined by RT-qPCR analysis of shoot tissues from water- and BABA-treated plants at 28 days after treatment (dat) with 40 mg/L BABA, and different hours (h) after spraying of the leaves with 0.5 mM SA. Data represent average fold-change values (±SEM) relative to expression levels before SA application. Asterisks indicate statistically different levels of expression between control- and BABA-treated plants (Student’s t-test; α < 0.05; n = 3).
SA-DEPENDENT GENE PRIMING IS NOT MARKED BY INCREASED ACETYLATION OF H3K9
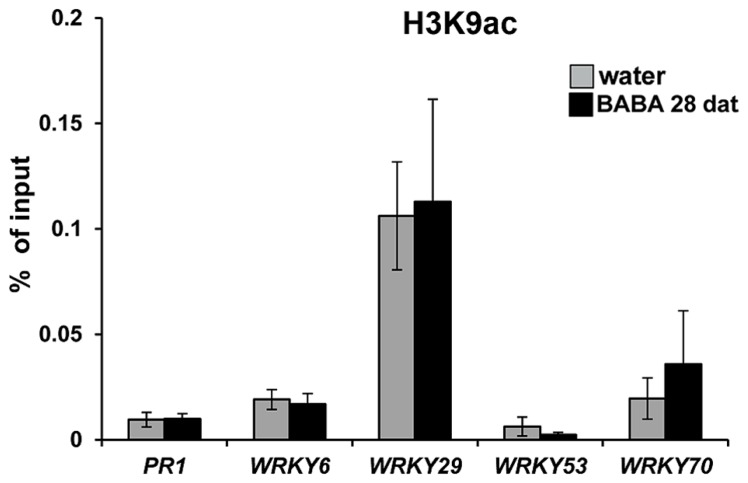
Chromatin remodeling is an epigenetic mechanism that can provide long-lasting changes in the plant’s transcriptional capacity (Berger, 2007). Post-translational modifications at the lysine residue 9 of histone 3 (H3K9) have been shown to regulate gene transcription (Li et al., 2007). Acetylation of H3K9 correlates with increased transcriptional capacity, whereas methylation of this residue correlates with gene silencing (Zhou et al., 2010). Previously, we demonstrated that transgenerational priming of SA-dependent defense in progeny from diseased Arabidopsis is associated with enrichment of H3K9ac at the promoter regions of the primed genes (Luna et al., 2012). This post-translational histone modification has also been associated to short-term defense gene priming after treatment with BTH and BABA (Jaskiewicz et al., 2011; Po-Wen et al., 2013). Based on these observations we assessed levels of H3K9 acetylation in promoters of defense genes displaying long-lasting priming by BABA (Figure 3), using similar primer pairs and H3K9ac antibody as described before (Jaskiewicz et al., 2011; Luna et al., 2012). Unexpectedly, these ChIP analyses revealed that BABA did not have long-term impacts on H3K9ac levels at the promoter of PR1, nor did it consistently affect H3K9ac levels at promoters of WRKY genes (Figure 4). These results indicate that H3K9ac is not a cis-acting requirement for long-lasting defense gene priming by BABA.
FIGURE 4.
Long-lasting defense gene priming is not marked by increased H3K9ac. Levels of H3K9ac at the promoters of the SA-inducible PR1, WRKY6, WRKY29, WRKY53, and WRKY70 genes in water and BABA treated wild-type plants at 28 dat. Chromatin-immunoprecipitation (ChIP) was quantified by qPCR and expressed relative to DNA amounts in chromatin extracts before immunoprecipitation (input). Data represent mean values (±SEM) from three biologically replicated samples.
KYP REGULATES LONG-LASTING DEFENSE GENE PRIMING
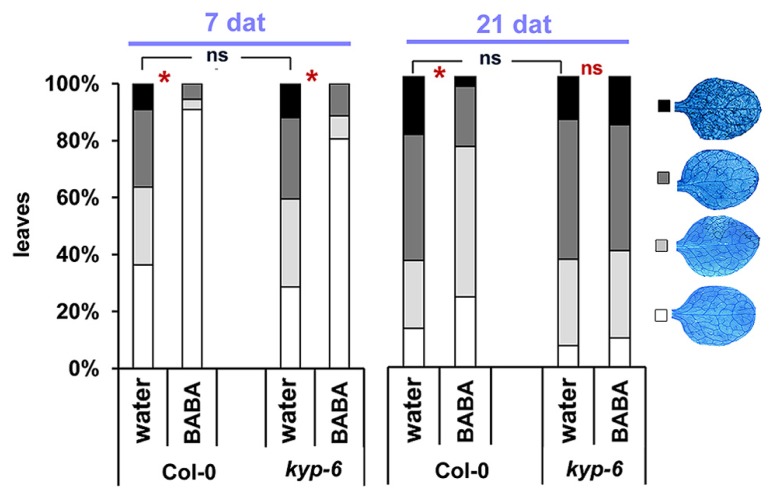
SUVH4/KRYPTONITE (KYP) is a histone methyltransferase that methylates H3K9 residues and its activity results in gene silencing through the interaction with CHROMOMETHYLASE3 (CMT3) DNA methyltransferase (Jackson et al., 2002). Loss of KYP results in decrease H3K9me2 and DNA methylation levels at CpHpG context (Chan et al., 2006). Previously, in an attempt to decipher the epigenetic mechanisms controlling transgenerational immune priming, we identified KYP as a key regulator of this phenomenon (Luna and Ton, 2012). In order to assess the role of this enzyme in long-lasting priming by BABA, we measured levels of BABA-IR against H. arabidopsidis at 7 and 21 days after treatment in the kyp-6 mutant (Jackson et al., 2002, 2004; Chan et al., 2006; Henderson and Jacobsen, 2008). This mutant displayed wild-type levels of basal resistance against H. arabidopsidis and showed significant levels of BABA-IR resistance to H. arabidopsidis when inoculated at 7 days after induction treatment (Figure 5). However, this mutant had lost its ability to express BABA-IR at 21 days after treatment (Figure 5), indicating that KYP acts as a positive regulator of long-term maintenance of BABA-IR, possibly by repressing negative regulatory genes of defense priming.
FIGURE 5.
KYP acts as a positive regulator of long-term maintenance of BABA-IR. BABA-IR against H. arabidopsidis in wild-type and kyp-6 plants at 7 and 21 days after induction treatment with 40 mg/L BABA (dat). For details see legend of Figure 2. Asterisks indicate statistically significant differences between treatments (χ2 contingency test; α < 0.05). ns = no significant.
DISCUSSION
We have investigated the durability of induced resistance after seedling treatment with the priming agent BABA. The response of plants to this non-protein amino acid mimics different biologically induced resistance responses in plants (Ton et al., 2005; Van der Ent et al., 2009). Consequently, this agent is effective against an exceptionally wide range of diseases (Jakab et al., 2001; Cohen, 2002). BABA primes the plant immune system via at least two independent signaling pathways, which differ in their dependency for NPR1 (Zimmerli et al., 2000). The NPR1-independent pathway primes pathogen-inducible expression of cell wall defense, whereas the NPR1-dependent pathway primes SA-inducible genes (Ton et al., 2005; Van der Ent et al., 2009). In this study, we showed that NPR1-independent resistance by BABA is transient and disappears within 2 weeks after application (Figure 6A). On the other hand, NPR1-dependent BABA-IR is long-lasting and remains up to 28 days after treatment (Figure 6A). Consistent with the idea that long-lasting BABA-IR involves epigenetic regulation, Slaughter et al. (2012) demonstrated increased defense phenotypes in progeny from isogenic Arabidopsis lines upon treatment with BABA. These phenotypes included reduced disease susceptibility to H. arabidopsidis and a sensitization to priming treatment by BABA, i.e., plants were “primed to be primed.” Interestingly, transgenerational effects by BABA were largely blocked in the impaired in BABA-induced sterility1 (ibs1) mutant, which had previously been reported to be affected in priming of NPR1-dependent defense by BABA (Ton et al., 2005). Furthermore, we recently demonstrated epigenetic inheritance of NPR1-dependent resistance from Pst-luxCDABE-infected Arabidopsis, which remained stable up to two generations. We, therefore, conclude that long-lasting NPR1-dependent resistance within the same generation has an epigenetic component, which can, at least partially, be transmitted to following generations.
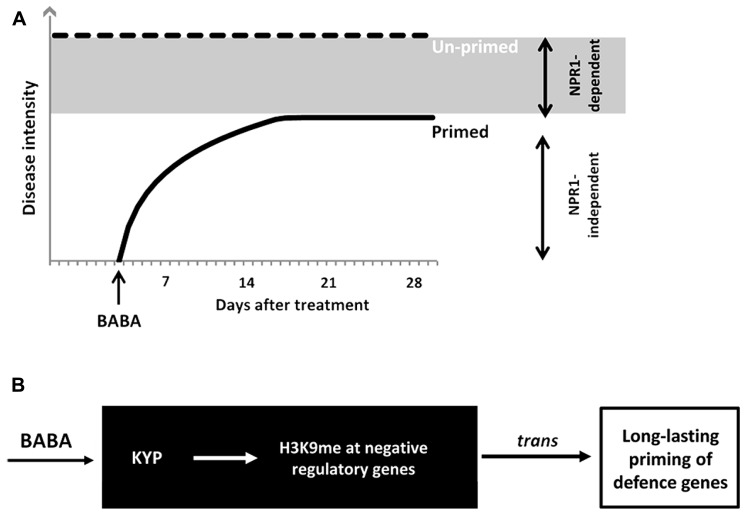
FIGURE 6.
Model of durability of BABA-IR. (A) BABA primes SA-inducible defense genes in a NPR1-dependent and -independent manner. BABA-IR lasts up to 28 days in wild-type plants, but only up to 14 days in npr1-1 plants. Hence, long-lasting disease protection by BABA acts through a NPR1-dependent signaling pathway. (B) KYP-mediated control of long-lasting priming by BABA. The kyp-6 mutant is blocked in long-lasting induced resistance by BABA, but expressed wild-type levels of disease susceptibility. This indicates that KYP stimulates long-lasting defense gene priming by BABA. Since KYP mediates transcriptional silencing through H3K9 methylation and CpHpG DNA methylation, we propose that KYP promotes long-lasting defense priming through silencing of trans-acting genes that encode for negative regulators of defense priming.
Previous studies have demonstrated a clear correlation between H3K9 acetylation and transcriptional priming at defense gene promoters (Jaskiewicz et al., 2011; López et al., 2011; Luna et al., 2012; Po-Wen et al., 2013). Using similar gene primers as described earlier (Luna et al., 2012), our ChIP experiments showed that long-lasting priming of SA-inducible defense genes within one generation can occur independently from increased H3K9 acetylation (Figure 4). Although we cannot exclude the possibility that flanking promoter regions of these genes did show enhanced H3K9 acetylation, our findings indicate that H3K9 acetylation at the regions analyzed is not a strict requirement for long-lasting priming of SA-inducible genes. The difference between our current H3K9ac results and those reported in progenies from Pst-luxCDABE-infected plants (Luna et al., 2012) suggests that the within-generational effects by BABA and transgenerational effects by pathogens are based on different epigenetic mechanisms. This is further supported by the fact that transgenerational resistance by BABA is lost after one stress-free generation (Slaughter et al., 2012), whereas transgenerational resistance by Pst-luxCDABE is maintained over 1 stress-free generation (Luna et al., 2012).
Interestingly, the kyp-6 mutant failed to retain long-lasting induced resistance by BABA (Figure 5), suggesting that KYP enables long-lasting defense priming by BABA. Although the possibility that the kyp-6 mutant harbors an independent second mutation disrupting long-lasting BABA-IR cannot be excluded, the phenotype of the kyp-6 mutant strongly suggests that H3K9 methyltransferase activity by the KYP protein is necessary for long-term maintenance of SA-dependent defense priming in BABA-treated plants. Since KYP-dependent H3K9 di-methylation and associated CpHpG DNA methylation are repressive mechanisms of gene expression (Kass et al., 1997; Zhou et al., 2010), we propose that KYP maintains BABA-IR by silencing repressive regulatory genes of SA-dependent defense genes, thereby priming their responsiveness to pathogen infection (Figure 6B). Previously, we proposed that disease-induced repression of RNA-directed DNA methylation is responsible for transmission of defense priming (Luna et al., 2012). Further research is required to resolve the interaction between KYP and components of RNA-directed DNA methylation with regards to long-lasting BABA-IR.
There is no evidence to support that post-translational histone modifications themselves can be transmitted through meiosis in plants. It is, therefore, plausible that transgenerational transmission of BABA-IR is determined by differentially methylated DNA regions (DMRs) that can faithfully be transmitted through meiosis. Indeed, we recently reported that transgenerational resistance in progeny from PstDC30000-infected Arabidopsis is most likely transmitted through a reduction in non-CpG DNA methylation (Luna and Ton, 2012; Luna et al., 2012). Interestingly, however, Slaughter et al. (2012) did not detect consistent changes in DNA methylation at priming-responsive defense genes in progenies of BABA-treated plants, indicating that transgenerational priming of defense genes by BABA is regulated by trans-acting DMRs. Future research is required to decipher the complex interplay between small RNAs, DNA (hypo)methylation, and post-translational histone modifications (Bond and Baulcombe, 2014).
Safeguarding food security represents an urgent challenge in this century, which is further aggravated by climate change that can render agricultural lands less suitable for crop production. Consequently, there is a pressing need to improve the efficiency of sustainable food production, including intensification of durable crop protection strategies (Royal-Society, 2009). Although usage of modern fungicides poses relatively little direct risks on food safety and soil ecology, repeated applications of fungicides demand considerable energy consumption. Integration of long-lasting induced resistance in existing disease management schemes would allow fewer energy costs to reach similar levels of disease protection. Worrall et al. (2012) recently reported that seed treatment of tomato with BABA provides long-lasting protection against powdery mildew. Considering that some crops are cultivated hydroponically, seedling application of BABA would provide another means of achieving long-lasting induced resistance against disease. Our present study has uncovered chromatin remodeling as an important regulatory mechanism of long-lasting induced resistance. Future research is required to identify the trans-acting genes that become targeted by chromatin remodeling after priming treatment, which will help to optimize the efficiency of durable induced resistance in plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Victor Flors, Dr. Mike Roberts and the two referees for fruitful discussions and helpful feedback on earlier versions of the manuscript. The research was supported by a consolidator grant from the European Research Council (ERC; no. 309944-Prime-A-Plant) to Jurriaan Ton, a Research Leadership Award from the Leverhulme Trust (no. RL-2012-042) to Jurriaan Ton and a grant from the Felix Thornley Cobbold Agricultural Trust to Jurriaan Ton and Estrella Luna.
REFERENCES
- Ahmad S., Gordon-Weeks R., Pickett J., Ton J. (2010). Natural variation in priming of basal resistance: from evolutionary origin to agricultural exploitation. Mol. Plant Pathol. 11 817–827 10.1111/j.1364-3703.2010.00645.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Alvarez M. E., Nota F., Cambiagno D. A. (2010). Epigenetic control of plant immunity. Mol. Plant Pathol. 11 563–576 10.1111/j.1364-3703.2010.00621.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447 407–412 10.1038/nature05915 [DOI] [PubMed] [Google Scholar]
- Bond D. M., Baulcombe D. C. (2014). Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 24 100–107 10.1016/j.tcb.2013.08.001 [DOI] [PubMed] [Google Scholar]
- Boyko A., Blevins T., Yao Y., Golubov A., Bilichak A., Ilnytskyy Y., et al. (2010). Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 5:e9514 10.1371/journal.pone.0009514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Bowling S. A., Gordon A. S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592 10.1105/tpc.6.11.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. W. L., Henderson I. R., Zhang X., Shah G., Chien J. S. C., Jacobsen S. E. (2006). RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in Arabidopsis. PLoS Genet. 2:e83 10.1371/journal.pgen.0020083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J.-N., Ryu J.-Y., Jeong Y.-M., Park J., Song J.-J., Amasino R. M., et al. (2012). Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 22 736–748 10.1016/j.devcel.2012.01.024 [DOI] [PubMed] [Google Scholar]
- Cohen Y. R. (2002). β-aminobutyric acid-induced resistance against plant pathogens. Plant Dis. 86 448–457 10.1094/PDIS.2002.86.5.448 [DOI] [PubMed] [Google Scholar]
- Conrath U. (2011). Molecular aspects of defence priming. Trends Plant Sci. 16 524–531 10.1016/j.tplants.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Conrath U., Beckers G. J., Flors V., Garcia-Agustin P., Jakab G., Mauch F., et al. (2006). Priming: getting ready for battle. Mol. Plant Microbe Interact. 19 1062–1071 10.1094/MPMI-19-1062 [DOI] [PubMed] [Google Scholar]
- Conrath U., Pieterse C. M. J., Mauch-Mani B. (2002). Priming in plant–pathogen interactions. Trends Plant Sci. 7 210–216 10.1016/S1360-1385(02)02244-6 [DOI] [PubMed] [Google Scholar]
- Despres C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., et al. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15 2181–2191 10.1105/tpc.012849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., Patrick E., et al. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin–ligase complex in Arabidopsis. Plant J. 32 457–466 10.1046/j.1365-313X.2002.01432.x [DOI] [PubMed] [Google Scholar]
- Dowen R. H., Pelizzola M., Schmitz R. J., Lister R., Dowen J. M., Nery J. R., et al. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. U.S.A. 109 E2183–E2191 10.1073/pnas.1209329109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Crooks C., Lamb C. (2008). High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 53 393–399 10.1111/j.1365-313X.2007.03303.x [DOI] [PubMed] [Google Scholar]
- Frost C. J., Mescher M. C., Carlson J. E, De Moraes C. M. (2008). Plant defense priming against herbivores: getting ready for a different battle. Plant Physiol. 146 818–824 10.1104/pp.107.113027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M., Offermann S., Danker T., Horst I., Peterhansel C., Stam M. (2007). Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3 11 10.1186/1746-4811-3-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I. R., Jacobsen S. E. (2008). Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 22 1597–1606 10.1101/gad.1667808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J., Johnson L., Jasencakova Z., Zhang X., Perezburgos L., Singh P., et al. (2004). Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112 308–315 10.1007/s00412-004-0275-7 [DOI] [PubMed] [Google Scholar]
- Jackson J. P., Lindroth A. M., Cao X., Jacobsen S. E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416 556–560 10.1038/nature731 [DOI] [PubMed] [Google Scholar]
- Jakab G., Cottier V., Toquin V., Rigoli G., Zimmerli L., Métraux J.-P., et al. (2001). β-Aminobutyric acid-induced resistance in plants. Eur. J. Plant Pathol. 107 29–37 10.1023/A:1008730721037 [DOI] [Google Scholar]
- Jaskiewicz M., Conrath U., Peterhansel C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12 50–55 10.1038/embor.2010.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H. W., Tschaplinski T. J., Wang L., Glazebrook J., Greenberg J. T. (2009). Priming in systemic plant immunity. Science 324 89–91 10.1126/science.1170025 [DOI] [PubMed] [Google Scholar]
- Kass S. U., Pruss D., Wolffe A. P. (1997). How does DNA methylation repress transcription? Trends Genet. 13 444–449 10.1016/S0168-9525(97)01268-7 [DOI] [PubMed] [Google Scholar]
- Kauss H., Jeblick W., Ziegler J., Krabler W. (1994). Pretreatment of parsley (Petroselinum crispum L.) suspension cultures with methyl jasmonate enhances elicitation of activated oxygen species. Plant Physiol. 105 89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Schwindling S., Conrath U. (2002). Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 128 1046–1056 10.1104/pp.010744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J. L. (2007). The role of chromatin during transcription. Cell 128 707–719 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- López A., Ramírez V., García-Andrade J., Flors V., Vera P. (2011). The RNA silencing enzyme RNA polymerase V is required for plant immunity. PLoS Genet. 7:e1002434 10.1371/journal.pgen.1002434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E., Bruce T. J. A., Roberts M. R., Flors V., Ton J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158 844–853 10.1104/pp.111.187468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E., Ton J. (2012). The epigenetic machinery controlling transgenerational systemic acquired resistance. Plant Signal. Behav. 7 615–618 10.4161/psb.20155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R. A., Durrant W. E., Wang D., Song J., Dong X. (2006). A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18 1750–1765 10.1105/tpc.105.039677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor V., Luna E., Mauch-Mani B., Ton J., Flors V. (2012). Primed plants do not forget. Environ. Exp. Bot.. 10.1016/j.envexpbot.2012.02.013 [DOI] [Google Scholar]
- Pavet V., Quintero C., Cecchini N. M., Rosa A. L., Alvarez M. E. (2006). Arabidopsis displays centromeric dna hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae. Mol. Plant Microbe Interact. 19 577–587 10.1094/MPMI-19-0577 [DOI] [PubMed] [Google Scholar]
- Po-Wen C., Singh P., Zimmerli L. (2013). Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol. Plant Pathol. 14 58–70 10.1111/j.1364-3703.2012.00827.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmann S., De Vos M., Casteel C. L., Tian D., Halitschke R., Sun J. Y., et al. (2012). Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol. 158 854–863 10.1104/pp.111.187831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royal-Society. (2009). Reaping the Benefits: Science and the Sustainable Intensification of Global Agriculture. London: The Royal Society [Google Scholar]
- Shirasu K., Nakajima H., Rajasekhar V. K., Dixon R. A., Lamb C. (1997). Salicylic acid potentiates an agonist-dependent gain control that amplifies pathogen signals in the activation of defense mechanisms. Plant Cell 9 261–270 10.1105/tpc.9.2.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter A., Daniel X., Flors V., Luna E., Hohn B., Mauch-Mani B. (2012). Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 158 835–843 10.1104/pp.111.191593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanou G., Fotopoulos V., Molassiotis A. (2012). Priming against environmental challenges and proteomics in plants: update and agricultural perspectives. Front. Plant Sci. 3:216 10.3389/fpls.2012.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J., D’Alessandro M., Jourdie V., Jakab G., Karlen D., Held M., et al. (2007). Priming by airborne signals boosts direct and indirect resistance in maize. Plant J. 49 16–26 10.1111/j.1365-313X.2006.02935.x [DOI] [PubMed] [Google Scholar]
- Ton J., Jakab G., Toquin V., Flors V., Iavicoli A., Maeder M. N., et al. (2005). Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 17 987–999 10.1105/tpc.104.029728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J., Mauch-Mani B. (2004). Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 38 119–130 10.1111/j.1365-313X.2004.02028.x [DOI] [PubMed] [Google Scholar]
- Van der Ent S., Van Hulten M., Pozo M. J., Czechowski T., Udvardi M. K., Pieterse C. M. J. et al. (2009). Priming of plant innate immunity by rhizobacteria and β-aminobutyric acid: differences and similarities in regulation. New Phytol. 183 419–431 10.1111/j.1469-8137.2009.02851.x [DOI] [PubMed] [Google Scholar]
- Van Hulten M., Pelser M., Van Loon L. C., Pieterse C. M., Ton J. (2006). Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103 5602–5607 10.1073/pnas.0510213103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K. J. F., Jansen J. J., Van Dijk P. J., Biere A. (2010). Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 185 1108–1118 10.1111/j.1469-8137.2009.03121.x [DOI] [PubMed] [Google Scholar]
- Walley J. W., Rowe H. C., Xiao Y., Chehab E. W., Kliebenstein D. J., Wagner D., et al. (2008). The chromatin remodeler splayed regulates specific stress signaling pathways. PLoS Pathog. 4:e1000237 10.1371/journal.ppat.1000237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall D., Holroyd G. H., Moore J. P., Glowacz M., Croft P., Taylor J. E., et al. (2012). Treating seeds with activators of plant defence generates long-lasting priming of resistance to pests and pathogens. New Phytol. 193 770–778 10.1111/j.1469-8137.2011.03987.x [DOI] [PubMed] [Google Scholar]
- Wu C.-C., Singh P., Chen M.-C., Zimmerli L. (2010). L-Glutamine inhibits beta-aminobutyric acid-induced stress resistance and priming in Arabidopsis. J. Exp. Bot. 61 995–1002 10.1093/jxb/erp363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wang X., He K., Charron J.-B., Elling A., Deng X. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 72 585–595 10.1007/s11103-009-9594-7 [DOI] [PubMed] [Google Scholar]
- Zimmerli L., Jakab G., Métraux J.-P., Mauch-Mani B. (2000). Potentiation of pathogen-specific defense mechanisms in Arabidopsis by aminobutyric acid. Proc. Natl. Acad. Sci. U.S.A. 97 12920–12925 10.1073/pnas.230416897 [DOI] [PMC free article] [PubMed] [Google Scholar]