Abstract
Background
Extensive studies using the model system Arabidopsis thaliana to elucidate plant defense signaling and pathway networks indicate that salicylic acid (SA) is the key hormone triggering the plant defense response against biotrophic and hemi-biotrophic pathogens, while jasmonic acid (JA) and derivatives are critical to the defense response against necrotrophic pathogens. Several reports demonstrate that SA limits nematode reproduction.
Results
Here we translate knowledge gained from studies using Arabidopsis to soybean. The ability of thirty-one Arabidopsis genes encoding important components of SA and JA synthesis and signaling in conferring resistance to soybean cyst nematode (SCN: Heterodera glycines) are investigated. We demonstrate that overexpression of three of thirty-one Arabidoposis genes in transgenic soybean roots of composite plants decreased the number of cysts formed by SCN to less than 50% of those found on control roots, namely AtNPR1(33%), AtTGA2 (38%), and AtPR-5 (38%). Three additional Arabidopsis genes decreased the number of SCN cysts by 40% or more: AtACBP3 (53% of the control value), AtACD2 (55%), and AtCM-3 (57%). Other genes having less or no effect included AtEDS5 (77%), AtNDR1 (82%), AtEDS1 (107%), and AtPR-1 (80%), as compared to control. Overexpression of AtDND1 greatly increased susceptibility as indicated by a large increase in the number of SCN cysts (175% of control).
Conclusions
Knowledge of the pathogen defense system gained from studies of the model system, Arabidopsis, can be directly translated to soybean through direct overexpression of Arabidopsis genes. When the genes, AtNPR1, AtGA2, and AtPR-5, encoding specific components involved in SA regulation, synthesis, and signaling, are overexpressed in soybean roots, resistance to SCN is enhanced. This demonstrates functional compatibility of some Arabidopsis genes with soybean and identifies genes that may be used to engineer resistance to nematodes.
Keywords: Arabidopsis, Composite plants, Gene overexpression, Jasmonic acid, Resistance, Salicylic acid, Soybean, Soybean cyst nematode, Transgenic roots
Background
Plant parasitic nematodes cause billions of dollars in losses each year worldwide [1-3]. The root-knot nematode, genus Meloidogyne, attacks over 3000 species of plants [4,5], while the soybean cyst nematode (Heterodera glycines) has a much narrower host range and is responsible for almost one billion dollars per year in losses in the US [2,3]. Although some soybean genotypes are resistant to specific populations of SCN, no soybean genotype is known to be resistant to all SCN populations. Several genes conferring partial Resistance to Heteroderaglycines (Rhg) have been mapped, and, recently, genes at the rhg1 and Rhg4 loci have been elucidated [6-9].The defense response of soybean to SCN has been examined at the physiological, genetic, and molecular level, and several reports indicate that salicylic acid (SA), jasmonic acid (JA), ethylene (ET), and indole acetic acid (IAA) play a role in resistance and susceptibility of plants to nematodes [8,10-15]. However, the roles of defense-related hormones and specific components of their synthesis and signaling pathways in providing resistance in plants to nematodes are unknown.
The plant defense response is complex. Plants launch a myriad of local and systemic defense responses to protect themselves from invasion by pests and pathogens. Several hormones are involved in inducing the defense response and regulating defense response networks, including SA, JA, ET, and IAA [11,12,16-18]. Plants react to pathogen-associated or microbe–associated molecular patterns (PAMPs/MAMPs) which are sensed by plants through leucine-rich repeat (LRR) receptors. PAMPs signal stomatal closure and stimulate innate immunity in plants. In general, SA activates the defense response to biotrophic and hemi-biotrophic pathogens, induces systemic acquired resistance (SAR), and triggers the expression of SAR-associated pathogenesis related genes PR-1, PR-5, and others [12,18]. The role of specific components of SA regulation, synthesis, and signaling in defending plants against parasitic nematodes is not well understood. However, SA does play a role in decreasing susceptibility to root-knot nematode (RKN) in cow pea [19] and tomato [20,21], and to the cyst nematode, Heterodera schachtii, in Arabidopsis[10]. Likewise, JA also plays a role in resistance of plants to nematodes. Foliar spraying of tomato with JA reduced galling and the final population of RKN (M. incognita); [22-26], as did pre-treatment of tomato seeds with JA [21], indicating a role for JA in plant resistance to nematodes.
Little is known of the role of specific components of SA regulation and signaling in the interaction of soybean with the soybean cyst nematode (SCN; Heterodera glycines), the major pest of soybean in the US. Although soybean genes conferring resistance to SCN have been identified, these do not provide resistance to all SCN populations. Resistance in soybean to SCN is multigenic, and several major loci for resistance have been identified [15,27-31]. For example, in soybean cv Peking, several genes (rhg1, rhg2, rhg3, and Rhg4) have been reported that confer resistance to SCN race 1 [15,32], yet none of these genes confers complete resistance to all SCN populations. Therefore, we are applying to soybean a portion of the vast knowledge that has been gained from studies on the model plant Arabidopsis and its large array of mutants on the role of SA and JA in the plant defense response to identify important components that may be useful in decreasing susceptibility of plants to nematodes, and especially of soybean to SCN.
Arabidopsis has been used widely as a model system to study plant defense pathways, usually with bacterial and fungal pathogens [11,12,17,33-39]. Much attention has been focused on SA production and signaling pathways using Arabidopsis mutants infected with bacterial and fungal pathogens (Figure 1) [12,16-18,35-44]. For example, when a virulent strain of the biotrophic pathogen, Pseudomonas syringae, attacks Arabidopsis, the AvrRPS4 effector protein of P. syringae secreted by the type III secretion system is detected by the plant receptor RPS4, a Toll-interleukin-nucleotide binding-leucine-rich repeat (TIR-NB-LRR) that mediates the induction of antimicrobial defenses to provide disease resistance. The nucleo-cytoplasmic protein ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1), which is a lipase-like protein, connects RPS4 with downstream defense pathways and regulates the accumulation of SA [45,46]. EDS1 is essential for production of the hypersensitive response and mobilization of defense pathways [47-49]. EDS1 can dimerize and interact with another lipase-like protein, phytoalexin deficient 4 (PAD4) [48,49]. Both EDS1 and PAD4 are required for the accumulation of SA and they are involved in ROS signaling [46,50]. PAD4 protein is required for amplification of weak signals resulting from pathogen infection. Another important component of the defense response is EDS5, a multi-drug transporter member of the MATE family of transporters [50].
Figure 1.
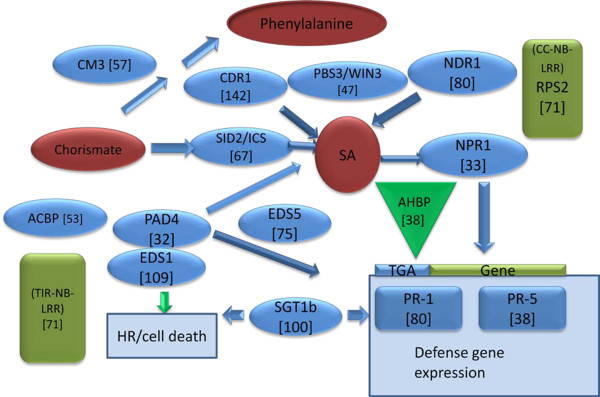
Representation of some components involved in regulation and biosynthesis of salicylic acid and associated signaling. SCN Female Indices (FI) of the genes examined are provided in brackets. Control Female Index = 100.
It is postulated that SA can be synthesized through two different pathways in Arabidopsis[51,52]. One pathway involves the enzyme isochorismate synthase (ICS; EC 5.4.4.2), which catalyzes the conversion of chorismate to isochorismate. The enzyme chorismate mutase (CM; EC 5.4.99.5) catalyzes the competing chemical reaction and converts chorismate to prephenate. This would divert chorismate to produce other compounds, such as phenylalanine and tyrosine. The Arabidopsis sid2-2 (SA INDUCTION-DEFICIENT 2) mutation has been mapped to the locus encompassing the ICS (SID1) gene [52]. Upon synthesis, SA can bind directly with NPR1, which is encoded by AtNPR1 (NONEXPRESSOR OF PR1), also known as NIM1 (NON-INDUCIBLE IMMUNITY 1). NPR1 is an SA receptor that is a transcriptional regulator of genes involved in the SA-dependent defense response [53], including the SA marker gene PR-1 (PATHOGENESIS RELATED 1). NPR1 interacts with transcription factor TGA2 family members, including AHBP-1b, and the complex binds to SA-responsive promoter elements of PR-1 and other SA-dependent defense genes to regulate expression [54].
SA can also be regulated independently of EDS1 and PAD4. NDR1 (NON-RACE SPECIFIC DISEASE RESISTANCE 1) is a positive regulator of SA that works independently of EDS1 and PAD4 [55] NDR1 is an integrin-like protein that can associate with RIN4, while RIN4 can associate with RPM1 and RPS2 [56]. NDR1 may play a role in electrolyte release upon infection of Arabidopsis by P. syringae, while RIN4 regulates stomatal apertures in conjunction with H + −ATPases of the plasma membrane of Arabidopsis during pathogen attack [57].
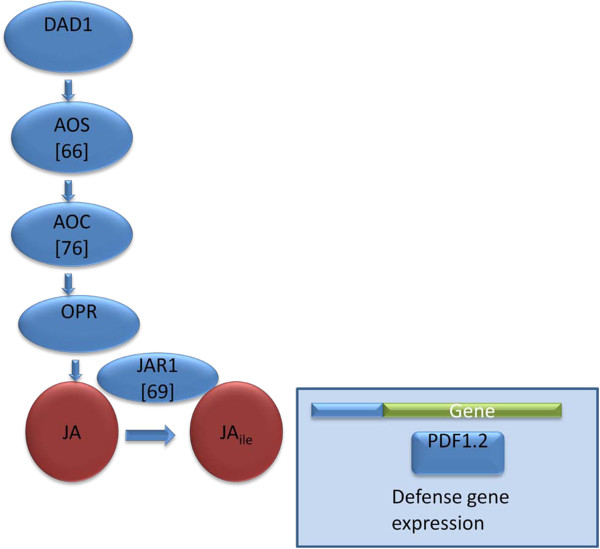
JA, JAile, and related lipid-derived compounds also act as signals in the plant defense response and are associated with resistance to necrotrophic pathogens [16,58,59]. The pathway leading to JA and JAile synthesis and some components related to JA signaling and control are depicted in Figure 2. Allene oxide synthase (AOS) and allene oxide cyclase (AOC) are two enzymes important to JA synthesis. JAR1 encodes an ATP-dependent JA-amido synthase that conjugates isoleucine with JA to form JAile, which plays an essential role in JA signaling.
Figure 2.
Representation of some components involved in regulation and biosynthesis of jasmonic acid and associated signaling. The SCN FI is provided in brackets.
In this paper, we examine the role of some components of the plant defense response in conferring resistance in soybean to SCN. We show that specific Arabidopsis genes, namely AtNPR1, AtTGA2, AtICS1, and AtPR5 which encode components of SA regulation, biosynthesis, and downstream effectors, can decrease susceptibility of soybean to SCN when expressed in transgenic soybean roots. We also demonstrate that expression of Arabidopsis genes encoding AOS, AOC, and JAR1, which are involved in the synthesis and modification of JA, only modestly decrease susceptibility of soybean roots to SCN. These results indicated that some Arabidopsis genes can be directly used in soybean, thus directly applying knowledge of the defense response gained from studies using Arabidopsis as a model system to soybean to decrease susceptibility to nematodes.
Results
Expression of Arabidopsis genes in soybean roots
SA and JA are well known regulators of the plant defense response as described through studies of the model plant, Arabidopsis. To determine if some of these Arabidopsis genes could be directly used in soybean to translate knowledge from Arabidopsis to soybean, we selected and cloned genes encoding numerous components of SA and JA synthesis, regulation, and signaling from the literature describing the defense response of Arabidopsis to pathogens (Table 1). To broaden the scope of our study, we also selected several Arabidopsis genes less well defined in function or that represented other portions of the plant defense response less dependent upon SA and JA. Open reading frames (ORFs) of thirty-one Arabidopsis genes were cloned into the gene expression vector pRAP15 [8,60,61] using the primers listed in Additional file 1: Table S1. The inserted ORFs were sequenced to confirm their identity and to ensure their sequence was conserved. The vector with insert was transformed by Agrobacterium rhizogenes K599 into cells at the base of the cut stem of soybean seedlings. Approximately 35 days after transformation, untransformed roots were removed from the composite plants, and the transformed roots were inoculated with SCN.
Table 1.
The effect of expression of thirty-one Arabidopsis genes on the number of SCN cysts at 35 dai was determined The number of plants (n), Standard Error of the Mean (SEM) and Female Index (FI) are provided. The control FI = 100
| Phytozome ID | Gene |
Gene |
SEM |
pRAP15 |
SEM |
FI |
P-value |
|---|---|---|---|---|---|---|---|
| n | n | (% of control) | |||||
| AT1G64280.1 |
NPR1 |
12 |
16 |
10 |
26 |
33 |
0.003 |
| AT5G06950.1 |
TGA2 |
9 |
13 |
27 |
13 |
38 |
<0.0001 |
| AT1G75040.1 |
PR-5 |
10 |
13 |
10 |
26 |
38 |
0.001 |
| AT4G24230.6 |
ACBP3 |
12 |
11 |
15 |
21 |
53 |
0.03 |
| At4G37000.1 |
ACD2 |
11 |
10 |
15 |
21 |
55 |
0.04 |
| AT1G69370.1 |
CM3 |
11 |
22 |
10 |
26 |
57 |
0.03 |
| AT5G42650.1 |
AOS |
12 |
11 |
15 |
21 |
66 |
0.11 |
| AT5G50260.1 |
CEP1 |
12 |
11 |
15 |
21 |
66 |
0.11 |
| AT1G74710.2 |
ICS1 |
12 |
13 |
28 |
10 |
67 |
0.002 |
| AT2G46370.4 |
JAR1 |
12 |
7 |
24 |
6 |
69 |
0.07 |
| AT3G03600.1 |
RPS2 |
9 |
16 |
15 |
21 |
71 |
0.22 |
| AT5G48485.1 |
DIR1 |
10 |
14 |
15 |
21 |
72 |
0.22 |
| AT4G39030.1 |
EDS5 |
13 |
16 |
11 |
26 |
75 |
0.39 |
| AT3G25760.1 |
AOC |
9 |
17 |
28 |
10 |
76 |
0.06 |
| AT3G26830.1 |
PAD3 |
14 |
4 |
17 |
9 |
79 |
0.28 |
| AT5G54250.1 |
DND2 |
11 |
15 |
15 |
21 |
79 |
0.35 |
| AT3G20600.1 |
NDR1 |
10 |
13 |
11 |
26 |
80 |
0.46 |
| AT2G14610.1 |
PR-1 |
8 |
23 |
28 |
10 |
80 |
0.20 |
| AT5G13160.1 |
PBS1 |
11 |
19 |
27 |
13 |
81 |
0.20 |
| AT1G02170.1 |
LOL3 |
11 |
13 |
28 |
10 |
83 |
0.10 |
| AT3G25070.1 |
RIN4 |
10 |
27 |
10 |
19 |
89 |
0.53 |
| AT4G20380.8 |
LSD1 |
7 |
31 |
28 |
10 |
95 |
0.78 |
| AT4G11260.1 |
SGT1b |
16 |
7 |
17 |
9 |
100 |
0.99 |
| AT5G64930.1 |
CPR5 |
12 |
7 |
17 |
9 |
102 |
0.97 |
| AT1G12560.1 |
ExPA7 |
12 |
5 |
14 |
6 |
108 |
0.72 |
| AT3G48090.1 |
EDS1 |
11 |
22 |
11 |
26 |
109 |
0.80 |
| AT2G17265.1 |
DMR1 |
11 |
20 |
15 |
21 |
110 |
0.67 |
| AT1G05180.1 |
AXR1 |
15 |
17 |
17 |
9 |
115 |
0.67 |
| AT4G21610.1 |
MC2 |
10 |
27 |
28 |
10 |
135 |
0.06 |
| AT5G33340.1 |
CDR1 |
12 |
23 |
11 |
26 |
142 |
0.23 |
| AT5G15410.1 | DND1 | 13 | 16 | 17 | 9 | 175 | 0.047 |
The effect of expression of thirty-one genes on the number of SCN cysts at 35 dai (days after inoculation) was determined (Table 1). Six genes decreased the number of cysts more than 40%, thus conferring partial resistance to SCN. Three of these genes, AtNPR1, AtTGA2, and AtPR-5, decreased the number of cysts more than 60%, while three others, AtACBP3, AtACD2, and AtCM3 decreased the number of cysts 40%. One Arabidopsis gene, AtDND1, increased the number of cysts of SCN to 175% of the control, thus making the soybean roots more susceptible to SCN (Table 1).
RNA was extracted from a subset of transformed roots for genes listed in Additional file 2: Table S2 to check for expression of the Arabidopsis gene by PCR. The amplicons were separated and visualized by gel electrophoresis and staining (Figure 3) to confirm that the ORFs were expressed in the composite root. All roots tested expressed the transcript.
Figure 3.
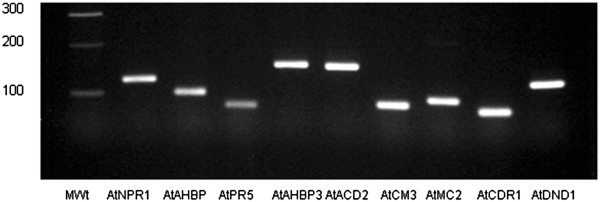
Expression of transcripts from each gene in transformed roots. RNA was converted to cDNA and used as template for PCR amplification of a fragment of each gene. Agarose gel containing amplicons representing a portion of AtNPR1, AtTGA2, AtPR-5, AtACBP3, AtACD2, AtCM-3, AtMC2, AtCDR1, AtDND1, respectively. Molecular weight markers (MWt) are shown in the first lane. Each lane represents a transgenic root from an individual plant. The cDNA from RNA extracted from wild type (WT) roots did not produce an amplicon for any of these genes. However, cDNA from the wild type was present, and an amplicon was produced by PCR when the cDNA was used as template with primers for a soybean control gene. RNA was extracted from three roots, individually, and independently made into cDNA. Each was examined by PCR and visualized as above. All samples from transgenic roots produced amplicons according to the appropriate Arabidopsis gene.
In addition, the abundance of transcript of two genes providing the most protection against SCN, AtNPR1 and AtTGA2, was determined by qRT-PCR using gene specific primers (Additional file 2: Table S2). Transcript number was calculated using the sigmoidal method [62]. The number of transcripts of AtNPR1 was 40,500 molecules and for AtTGA2 was 60,500 molecules in transformed roots, while no transcripts of either gene were detectable in the control roots. In all samples, the expression level was similar for the housekeeping gene encoding ubiquitin-3 (Figure 4).
Figure 4.
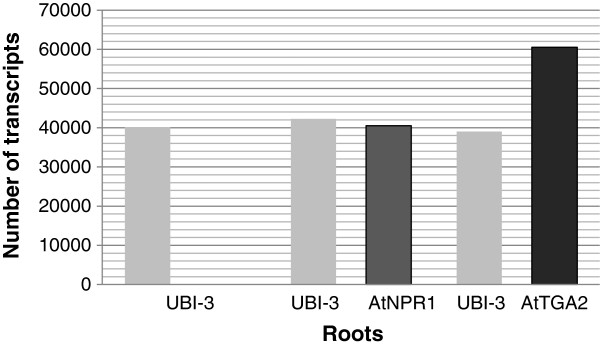
Number of transcripts of AtNPR1 and AtTGA2 in transformed roots were determined in roots transformed with AtNPR1 and AtTGA2. No transcripts of either gene were detectable in the control roots. In all samples, the expression level was similar for the soybean housekeeping gene encoding ubiquitin-3 (GmUBI-3).
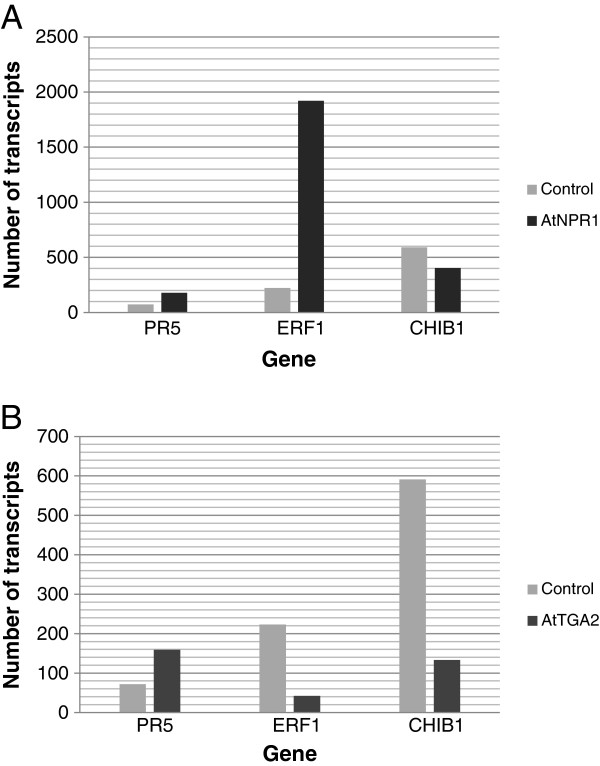
The number of transcripts of three defense-related genes, GmPR5, GmCHIB1, and GmERF1was also determined using qRT-PCR (Figure 5). In roots overexpressing AtNPR1, there were 178 transcripts of GmPR5, while in roots overexpressing AtTGA2, there were 159 transcripts. In control roots, there were only 38 transcripts of GmPR5. Thus, GmPR5 was elevated approximately 4-fold in roots overexpressing AtTGA2. Transcripts of GmCHIB1 were also elevated in these roots. There were 403 transcripts of GmCHIB1 in roots overexpressing AtNPR1 and 133 transcripts in roots overexpressing AtTGA2. There were only 53 transcripts of GmCHIB1 in control roots. This represents an increase in expression of GmCHIB1 by approximately 8- and 2.5-fold in soybean roots overexpressing AtNPR1 and AtTGA2, respectively. In contrasts, the number of transcripts of GmERF1 decreased in soybean roots overexpressing AtNPR1. Only 42 transcripts of GmERF1 were present, whereas control plants contained 1921 transcripts. Similarly, roots overexpressing AtTGA2 contained fewer GmEFR1 transcripts than in control roots with only 75 transcripts present. Thus, GmERF1 expression was greatly decreased in both sets of transgenic roots.
Figure 5.
The number of transcripts of three defense-related genes, GmPR5 , GmCHIB1 , and GmERF1 was determined in roots transformed with (A) AtNPR1 and (B) AtTGA2 using qRT-PCR.
SA-related genes
Activation of the defense response in plants is initiated through several parallel signaling pathways. In gene-for-gene resistance, host resistance (R) proteins indirectly recognize pathogen effectors to initiate resistance [63]. The coiled-coiled-nucleotide-binding site-leucine-rich repeat (CC-NB-LRR) and the TIR-NB-LRR classes of proteins are two major sub-groups of R protein [64]. In this study, we selected the Arabidopsis protein NON-RACE-SPECIFIC DISEASE RESISTANCE 1 (NDR1) as a representative CC-NB-LRR R-protein, because of its known role in activating the SA-mediated defense response in Arabidopsis. RPS2, encoded by AtRPS2, was selected as a representative of the TIR-NB-LRR class of proteins. Both AtNDR1 and AtRPS2 were overexpressed in transgenic soybean roots to determine their effect on SCN growth and maturation as measured by the female index (FI), which expresses the number of mature SCN females 35 days after root inoculation as a percent of the control value. Overexpression of AtNDR1 slightly inhibited SCN development (FI = 80), while AtRPS2 had a slightly greater inhibitory effect (FI = 71), but the effect was not statistically significant (P > 0.05) (Table 1) for either gene.
Arabidopsis NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (AtNPR1) is downstream of the R proteins NDR1 and RPS2. NPR1 is a key regulator of SAR and plays a critical role as a SA signal transducer in Arabidopsis[38,44]. When AtNPR1 was overexpressed, the FI decreased to 33% of the control. AtNPR1 had the lowest FI value among the Arabidopsis genes tested in this study (Table 1).
Alignment of the 593 aa of AtNPR1 with its closest soybean counterpart, the product of Glyma09g07440.1, indicates conservation of only 273 aa, although there are also many conservative substitutions (Figure 6). There are five soybean genes encoding proteins closely related to AtNPR1. The protein encoded by Glyma09g07440.1 is most closely related to the protein encoded by Glyma09g02430.1, and these are closely related to the proteins encoded by Glyma15g13320.1, Glyma14g03510.1, and Glyma02g45260.1. All five soybean putative NPR1 proteins contain a BTB/POZ domain, ankyrin repeats (domain CLO465), a NPR1/NIM1-like defense protein C terminal, and a domain of unknown function (DUF3420), as does AtNPR1.
Figure 6.
Alignment of the Arabidopsis and soybean protein sequences of NPR1 using Clustal 2.1, showing the BTB/POZ domain (underlined), ankyrin repeats (domain CLO465; underlined and bold)), and NPR1/NIM1-like defense protein C terminal (bold). (*) = identical aa; (:) = highly conserved aa substitution; (.) = conserved substitution.
Expression of AtTGA2, encoding the TGA-type basic leucine zipper bZip transcription factor AHBP-1B, in soybean roots decreased the FI of SCN to 38% of the control (Table 1). There are numerous soybean homologs of AtTGA2. The four most highly conserved are Glyma13g26280.1, Glyma15g37220.1, Glyma20g39050.2, and Glyma10g44270.1. The amino acid sequence of AtTGA2 is closely related to Glyma20g24766.1 (5e-56; Figure 7). The alternative transcript, Glyma20g24766.2, is exactly the same, except it lacks the 5′ leader sequence. The Glyma20g24766.1 transcript appears to contain a chloroplast transit sequence as predicted using ChloroP1.1, Technical University of Denmark, http://www.cbs.dtu.dk/services/ChloroP/, while Glyma20g24766.2 does not contain this 5′ transit sequence. We used the alternate sequence Glyma20g39050.2 that lacks a 5′ leader sequence that is also missing in AtTGA2. The AtTGA2 and GmTGA2 protein sequences are highly conserved in the bZIP domain [65-68], with only two aa differences in the 39 aa domain. This region is highly conserved with only eight aa substitutions in the 65 aa region. A second, domain, DOG1 [69] is found toward the carboxy terminus and is involved in the control of seed dormancy [54]. The DOG1 domain is less conserved between these Arabidopsis and soybean proteins.
Figure 7.
Multiple sequence alignment of the Arabidopsis and soybean protein sequences of TGA2-1B using Clustal 2.1, showing the bZIP domain (bold) and the DOG1 domain (underlined). (*) = identical aa; (:) = highly conserved aa substitution; (.) = conserved substitution.
In Arabidopsis, SA interacts with the receptor NPR1, which then interacts with the transcription factor TGA2 to modulate the transcription of some genes, including PR-1 and PR-5. Thus, we examined the effect of overexpression of AtPR-1 and AtPR-5 on the FI of SCN. Overexpression of AtPR-1 did not have a statistically significant effect (FI = 80; P = 0.2) on the number of SCN cysts. In contrast, when AtPR-5 was overexpressed in soybean roots, the FI was decreased to 38% of the control (Table 1), while overexpression of AtPR-1 had only a mild effect on the FI, which was 80% of the control. In soybean there is a large family of over 15 Gm-PR-5 genes with Glyma14g08380 and Glyma17g36680 being the most closely related to the Arabidopsis PR-5 gene, AT1G75040 (Figure 8). The sequences of the proteins encoded by the Arabidopsis and two most closely related soybean genes are highly conserved.
Figure 8.

Multiple sequence alignment of the Arabidopsis and soybean protein sequences of PR-5 using Clustal 2.1. (*) = identical aa; (:) = highly conserved aa substitution; (.) = conserved substitution.
Three well-studied genes involved in SA are AtPAD4, AtEDS1, and AtEDS5. Previously, we demonstrated that expression of AtPAD4 greatly decreased the development of female SCN to 32% of the control [60]. Here, we examined the effect of overexpression of AtEDS5 and AtEDS1, neither of which significantly affected SCN development, with FI values of 77 and 109, respectively.
SA can be synthesized through a shorter pathway involving ICS, or through a longer pathway through phenylalanine using CM. Therefore, we expressed AtICS1 and AtCM-3 in soybean roots to determine their effects on SCN maturation. Overexpression of AtICS1 in roots had a modest inhibitory effect (FI = 67, P = 0.002). Because AtICS1 did not strongly affect the FI, we anticipated that expression of CM would have minimal inhibitory effect on the FI of SCN or, perhaps, increase susceptibility, because CM competes with ICS for the common substrate chorismate. However, expression of AtCM-3 also significantly inhibited SCN growth (FI = 57, P = 0.03).
WIN3, encoded by HOPW1-1-INTERACTING 3 (WIN3), is involved in regulating SA and disease resistance [70-72], though the mechanism is unclear [73]. Overexpression of AtWIN3 decreased the FI of SCN to 47% of the control.
Arabidopsis ACBP3 is an acyl-coenzyme A (CoA)-binding protein [74]. Transgenic Arabidopsis overexpressing AtACBP3 displayed constitutive expression of the pathogenesis-related genes PR-1 (unknown function), PR-2 (β-1,3-glucanase), and PR-5 (osmotin), and resistance to P. syringae DC3000 was dependent upon the NPR1 mediated signaling pathway [75]. Overexpression of AtACBP3 in soybean roots resulted in a decrease of the FI of SCN to 53% of the control.
Overexpression of AtCPR5 (CONSTITUTIVE EXPRESSOR OF PATHOGEN RELATED GENES 5) in soybean roots had little effect on the female index of SCN. CPR5 mutants constitutively express PR genes at a high level [76,77]; display defects in cell division, endoreduplication, and cell wall production [78,79]; and are of reduced stature and exhibit the formation of spontaneous lesions [79,80].
JA-related genes
JA and related compounds are important in defense responses, especially the response to necrotrophic pathogens [81,82]. JA and JAile are synthesized through a series of enzymatic steps (Figure 2), including the enzymes allene oxide synthase (AOS (DDE2); EC 4.2.1.92); allene oxide cyclase (AOC; EC 5.3.99.6); and jasmonic acid-amido synthetase (JAR1; EC 6.3.2.-.). JAR1 conjugates JA with isoleucine to form JA-Ile, which is considered to be one of the active forms of JA [74-77]. Overexpression of the Arabidopsis genes AtAOS, AtAOC, and AtJAR1 did not influence the FI of SCN in a statistically significant manner (66% (P = 0.11), 76% (P = 0.06), and 69% (P = 0.07) of the control, respectively; Table 1). These data do not suggest overexpression of [83] these genes will improve resistance in soybean to SCN.
Other Arabidopsis genes
Overexpression of AtRIN4 genes in soybean roots had little effect on the female index of SCN (Table 1). RIN4 is a negative regulator of innate immunity in plants [57]. It regulates stomatal closure. It appears to be peripheral to the defense response of soybean roots to nematode attack, as it did not significantly alter the FI of SCN.
In Arabidopsis, the chloroplast protein ACCELERATED CELL DEATH 2 (ACD2) modulates the amount of cell death that occurs in Arabidopsis leaves infected with P. syringae[84]. When the AtACD2 gene was overexpressed in soybean roots, the FI of SCN was reduced to 55% of the control (Table 1).
Cysteine endopeptidases containing a C-terminal endoplasmic reticulum retention signal, KDEL, are involved plant cell death [85]. Overexpression of the cysteine endopeptidase encoded by AtCEP1 reduced the FI of SCN to 66% of the control which was not statistically significant (Table 1).
Arabidopsis genes that increased susceptibility when overexpressed
The AtDND1 gene AT5G15410.1 encodes the cyclic nucleotide-gated cation channel protein DEFENSE NO DEATH 1 (DND1), and is involved in production of the hypersensitive response [86]. The Arabidopsis dnd1 mutant produces elevated amounts of SA. Overproduction of AtDND1 in soybean roots did not provide resistance to SCN; rather, it enhanced susceptibility. The FI of transgenic soybean roots containing AtDND1 was 175% of the control, the largest increase in susceptibility of the genes tested here (Table 1). The protein sequence of DND1 is highly conserved between Arabidopsis and soybean (Glyma18g49890.1) as indicated in Figure 9. It contains a cyclic nucleotide-binding domain as indicated by a significant (e-value = 5.8) Pfam-A match.
Figure 9.
Multiple sequence alignment of the Arabidopsis and soybean protein sequences of DND1 using Clustal 2.1. (*) = identical aa; (:) = highly conserved aa substitution; (.) = conserved substitution.
Overexpression of two other Arabidopsis genes did not alter susceptibility of soybean to SCN at a statistically significant level. The first gene, CONSTITUTIVE DISEASE RESISTANCE 1 (AtCDR1), encodes an aspartic protease [87]. When AtCDR1 was overexpressed in soybean roots, the FI was 142% of the control (P = 0.23) (Table 1). The second gene AtMC2 (LOL2 (LSD1-LIKE)) encodes the positive regulator of cell death during the hypersensitive response and is a conserved paralog of LSD1 [88,89]. LSD1 is a negative regulator of plant programmed cell death. Overexpression of AtMC2 in soybean roots yielded a FI of 135% (P = 0.06) (Table 1).
Discussion
Resistance to SCN is a multigenic trait and several genetic loci have been mapped [15,27-29,90-93]. Recently, the identity of genes residing at the rhg1 and Rhg4 loci have been reported [6-9] which confer some resistance to SCN. However, none of these loci alone provides full resistance to any one SCN population. For example, in a cross between soybean cv Essex and Forrest, rhg1 and Rhg4 accounted for about 65% of the variation in resistance found in the resultant inbred population to SCN [94]. Other soybean genes have been identified that confer partial resistance to SCN when overexpressed in roots [8,9,60,61].
An option to developing resistance to nematodes is to use defense-related genes that have been described in the literature. Much of the literature describing work with the defense response of Arabidopsis is concerned with elucidating defense response signaling, regulation, and pathways important to bacterial and fungal pathogens that attack the leaf of the plant. Although this research may be applicable to resistance of plants to nematodes and to agronomic crops such as soybean, little published work has yet translated the knowledge gained from these important studies in Arabidopsis to soybean and other important crops. Direct translation of research in Arabidopsis, includes transforming Arabidopsis genes directly into crop plants to determine if they have a positive or negative effect in that crop on disease resistance. Here we have shown that some Arabidopsis genes, when overexpressed in soybean roots, are compatible and confer resistance to SCN.
SA plays an important role in the plant defense response to pathogens. SA regulates SAR, local disease resistance, host cell death, and expression of genes involved in the defense response [44]. In tomato, SA is important to resistance to three RKN species [21]. Transgenic tomato expressing NahG, encoding salicylate hydrolase which degrades SA, was less resistant to RKN. However, resistance to RKN induced in tomato through the application of cell suspensions of the biocontrol bacterium Pseudomonas aeruginosa is independent of the accumulation of SA [95]. Thus, it may be that SA plays a role in providing resistance to RKN in tomato, but there may be other SA-independent mechanisms that also confer resistance. Uehara et al. [96] showed that inhibition of the SA signaling pathway in tomato harboring the Hero A gene increased susceptibility to Globodera rostochiensis. A protective effect against gall eelworm was seen in tomatoes when seeds were soaked in SA [14]. These and other reports show a strong link between SA and nematode resistance.
Examination of Arabidopsis mutants has played a key role in our understanding of the defense response and is the subject of many reviews [17,18,35,36,38,39]. It is postulated that SA can be synthesized through two different biochemical pathways [52,97]. In the first pathway, chorismate is converted to isochorismate via the action of ICS; then, SA is produced from isochorismate by isochorismate pyruvate lyase. Examination of ICS1 mutants, sid2-1 and sid2-2, of Arabidopsis indicate that loss of ICS1 activity dramatically decreases SA levels [48]. Most SA synthesized and relevant to plant defense in Arabidopsis appears to be made through this pathway. The sid2 mutant does not accumulate SA upon inoculation with P. syringae, and PR-1 expression is reduced greatly. However, PR-2 and PR-5 are expressed [90]. The second possible pathway diverts chorismate via CM to phenylalanine, which is converted to cinnamic acid by phenylalanine ammonia lyase (PAL) and progresses through a series of reactions to form SA. Previously, we demonstrated that overexpression in transgenic soybean roots of two different soybean genes encoding PAL did not greatly affect SCN maturation, with FI values of 94 and 111% [8]. However, here we show that overexpression of CM and ICS, representatives of the two different pathways, decrease the FI to 57% and 67% of the control, respectively. However, overexpression of PAL, CM, or ICS alone does not confer resistance to the level provided by overexpression of several other SA-related genes individually.
Genes decreasing susceptibility of soybean to SCN
If the SA-related defense response is a major factor in soybean resistance to SCN, then components regulated by SA may confer resistance to SCN when overexpressed. Our results show that several Arabidopsis genes involved in SA regulation, synthesis, and signaling conferred resistance to SCN when overexpressed in soybean roots. The Arabidopsis genes AtNPR1, AtTGA2, AtPR-5, and several others related to SA strongly decreased the FI of SCN in transgenic soybean roots (Figure 1). NPR1 is a master regulator of the SA-related defense response, and it is a receptor for SA [51]. NPR1 binds SA and interacts with TGA transcription factors, such as the transcription factor AHBP-1b/TGA2, perhaps through its ankyrin domain [54]. NPR1 and TGA transcription factors work downstream of SA and are important to the expression of the genes encoding PR-1, PR-5, and others [98-102]. Expression of these genes is completely abolished in Arabidopsis plants carrying the npr1 mutation [83]. Recently, Pant et al. [103] demonstrated that a Gm ortholog, Glyma09g02430, of Arabidopsis NPR1 reduced SCN cysts to approximately 30% of the control, in agreement with our data for overexpression of AtNPR (FI = 33%). There are reports in numerous plants indicating that overexpression of NPR1 results in defense against fungal and bacterial pathogens. Overexpression of AtNPR1 in Arabidopsis conferred resistance to P. syringae and Peronospora parasitica[104]. Overexpression of AtNPR1 in rice conferred resistance to the rice bacterial blight pathogen Xanthomonas oryzae[105]. Overexpression of AtNPR1 in wheat conferred resistance to fusarium head blight, caused by Fusarium graminearum. The apple MpNPR1 gene confers resistance to two fungal pathogens of apple, Venturia inaequalis and Gymnosporangium juniper-virginianae[106,107] complemented Arabidopsis npr1-1 mutants with soybean homologs GmNPR1-1 and NPR1-2, and PR-1 was induced in the transformed plants after infection with P. syringae and after treatment with the SAR inducer, 2,6-dichloroisonicotinic acid.
NPR1 interacts with the transcription factor TGA2 to modulate expression of some plant defense genes, such as PR-1 and PR5 [108]. When we overexpressed AtTGA2, the FI of SCN was decreased to 38, showing that TGA2 can also confer resistance to SCN. This is further supported by our data showing the reduction of the FI to 38% of the control due to PR-5 overexpression. PR-5 is a thaumatin-like protein involved in the defense response [109], perhaps creating transmembrane pores to disrupt the membranes of pathogens [110]. Some PR-5 proteins exhibit anti-fungal activity [109,111]. When Prunus domestica PR-5 was overexpressed in transgenic Arabidopsis, the plants displayed more resistance to the fungal pathogen Alternaria brassicola[112].
PAD4 is found upstream of SA production ([113]; Figure 1). PAD4 is a lipase like protein [48] that can form molecular complexes with EDS1 to modulate SA defense signaling [114]. Ectopic expression of PAD4 reduces feeding time of green peach aphids on transgenic Arabidopsis plants. The aphid spends more time actively feeding on pad4 mutants [115]. Previously, we showed that AtPAD4 conferred resistance to SCN and RKN [60]. EDS1 is also upstream of SA, and it can interact with PAD4 [114,116,117]. Recently, Pant et al. [103] demonstrated that GmEDS1 (Glyma06g19890) had a great effect on SCN and reduced SCN cysts by approximately 80%. This is in contrast to our data indicating that overexpression of AtEDS1 did not decrease the FI of SCN. GmEDS1 is composed of 620 aa and AtEDS1 is composed of 623 aa (Figure 10). The amino acid sequences of GmEDS1 and AtEDS1 have 239 aa in common. Furthermore, they have another 140 aa that are closely related substitutions. Apparently, this conservation is not enough for AtEDS1 to provide resistance to SCN as did GmEDS1.
Figure 10.
Multiple sequence alignment of the Arabidopsis and soybean protein sequences of EDS1 using Clustal 2.1. (*) = identical aa; (:) = highly conserved aa substitution; (.) = conserved substitution.
The EDS5 gene, also found upstream of SA, encodes a membrane protein with homology to multidrug and toxin extrusion (MATE) transporters [50]. The eds5 mutant accumulates very little SA and exhibits a reduction in PR-1 transcripts when infected with nematodes [19,118]. We show that overexpression of AtEDS5 reduced the number of mature female cysts only modestly, to 75% of the control.
ACBP3 is one of six acyl-coenzyme A (CoA)-binding proteins in Arabidopsis[66]. ACBP binds to acyl-CoA esters and protects acyl-CoAs from degradation [119]. Bovine ACBP overexpression in yeast leads to an increase in the acyl-CoA pool size [120]. Overexpression of AtACBP6 increased freezing tolerance in Arabidopsis [121]. These plants also showed a decrease in phosphatidyl choline and an increase in phosphatidic acid. Infection of Arabidopsis by either Botrytis cinerea or P. syringae pv tomato DC3000 induces the expression of AtACBP3, as does treatment with the fungal elicitor arachidonic acid [75]. The authors also showed that resistance to P. syringe was conferred by ACBP3 overexpression in an NPR1-dependent manner and that PR-1, PR-2, and PR-5 were constitutively expressed. When we overexpressed AtACBP in transgenic soybean roots, the number of SCN cysts decreased to 53% of the control at 35 dai.
The ACD2 gene in Arabidopsis encodes red chlorophyll reductase [122], which catalyzes the degradation of the porphyrin portion of chlorophyll [123]. ACD2 modulates cell death in Arabidopsis infected with P. syringae. It is localized to the chloroplast. Upon infection by P. syringae, localization of the protein changes, and it is localized in chloroplasts, mitochondria, and to a lesser degree the cytosol [84]. The accumulation of chlorophyll breakdown products may trigger cell death [122]. When we overexpressed the AtACD2 gene in soybean roots, the FI of SCN was reduced to 55% of the control.
Jasmonic acid
JA and ethylene are also important in the plant defense response, especially the response to necrotrophic pathogens [58]. JA and ethylene interact antagonistically with SA. Mechanical damage and wounding activates JA synthesis in Arabidopsis, potato, tomato, and other plants [58,124,125]. The role of JA in defense against nematodes is being examined by several laboratories. There are recent reports that JA is involved in defense in rice plants against nematodes [126,127]. Exogenous application of methyl-JA on rice shoots reduced galls by 63% per plant. In contrast, Bhattarai et al. [128] showed that JA is not required for resistance in tomato to RKN. They used nearly isogenic tomato cultivars resistant and susceptible to RKN to study gene expression using microarrays. The tomato jai1 mutant, altered in JA signaling, reduced the susceptibility of tomato to RKN. Furthermore, they showed that auxin-related genes were differentially expressed in compatible and incompatible interactions with RKN. Neither foliar spray nor soil-drenching of tomato plants with SA, JA, or methyl-JA affected galling of roots by RKN [129]. We show that overexpression of three genes involved in JA/JAile production, AtAOS, AtAOC, and AtJAR1reduced the FI of SCN to 66% (P = 0.06) , 75% (P = 0.6) and 69% (P = 0.06) of the control. Although, the data are at the borderline of significance, the trend suggests that JA/ JAile may provide some degree of resistance to SCN in soybean. Further work is needed in this area, as the effects of SA and JA in roots has not been explored. Nor has SA-JA antagonism been documented in root systems. Perhaps, SA and JA interactions are not completely antagonistic at all times in all tissues. Or perhaps, exogenous application of plant hormones can provide resistance to nematodes, but the level of the plant hormone necessary to achieve resistance is not normally achieved during nematode attack.
Genes increasing susceptibility of soybean to SCN
Overexpression of AtDND1 resulted in the greatest increase in susceptibility of soybean roots to SCN of all genes tested here. DND1 is a known negative regulator of plant immunity [130-132]. Its promoter is the target of the transcriptional co-repressor, Topless-related 1 (TPR1), which may function through repression of negative regulators to activate R protein-mediated immunity responses [133]. When AtDND1 was overexpressed in soybean roots, it decreased resistance to SCN as reflected by the female Index of 175% as compared to the control.
Arabidopsis defense genes not impacting SCN susceptibility
Overexpression of AtCDR1, which encodes an apoplastic aspartic protease, resulted in a non-significant increase in susceptibility to SCN. This result contrasts with those of previous studies on plant resistance to bacterial and fungal pathogens. Overexpression of CDR1 in T-DNA activation tagging studies yielded dwarf Arabidopsis plants and increased resistance to P. syringae[134]. Antisense CDR1 Arabidopsis plants were compromised in resistance to P. syringae. A rice aspartic protease, encoded by OsCDR1, was identified by Presad et al. [135]. When they overexpressed OsCDR1 in Arabidopsis, the plants were more resistant to the necrotrophic fungal pathogen Alternaria brassicicola. When the gene was overexpressed in rice plants, the plants were more resistant to Xanthomonas oryzae, the rice blast fungus, and to Magnaporthe oryzae, which causes bacterial blight.
Conclusions
Expression of several Arabidopsis genes provided protection to soybean against SCN. In fact, several genes provided increases in resistance comparable to or better than that provided by Rhg1 and Rhg4, two naturally occurring resistance gene loci in soybean. However, not all Arabidopsis genes provided resistance. In fact, overexpression of AtEDS1 did not increase resistance, although a GmEDS1 ortholog was recently reported as providing resistance. These data indicate that some Arabidopsis genes can be used directly in soybean to confer resistance, especially genes associated with SA regulation, signaling, and synthesis, but not all Arabidopsis orthologs will provide the same results as the orthologous soybean gene. These and similar studies may provide useful insights into protein conservation and function, and several of these Arabidopsis genes may prove useful in engineering plants with broad resistance to nematodes.
Methods
Bioinformatics
Thirty-one genes were selected from published studies defining Arabidopsis mutants displaying phenotypes affecting SA and JA production, regulation, and signaling. The DNA sequence of the gene was obtained from The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/). The DNA sequences of soybean genes used in multiple sequence alignments with Arabidopsis genes were obtained at Phytozome.net (Joint Genome Institute, U.S.D.O.E., Center for Integrative Genomics, U.C. Berkeley) using the Glycine max genome [136]. Primers for PCR amplification of the open reading frame of each gene were designed using Primer 3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) or OligoAnalyzer 3.1 (Integrated DNA Technologies, Coralville, IA.) Multiple sequence alignments were made using CLUSTAL 2.1 (http://www.genome.jp/tools/clustalw/). Protein domains were identified using Pfam (http://pfam.sanger.ac.uk/search).
Amplification and cloning of ORFs
The open reading frames (ORFs) of Arabidopsis target genes (Additional file 1: Table S1) were amplified by PCR and cloned into pRAP15 using the Gateway® system (Invitrogen, Carlsbad, CA) as described previously [8,60]. Templates for PCR were from cDNA libraries derived from Arabidopsis RNA. Arabidopsis cDNA was constructed from RNA extracted from A. thaliana (Columbia) whole plants and converted into cDNA as described by [8,60]. ORFs were PCR amplified using gene-specific PCR primers that contained CACC at the 5′end of the forward primer for directional cloning using the Gateway® (Invitrogen) system (Additional file 1: Table S1).
The PCR-amplified ORFs were cloned into pENTR using a pENTR™ Directional TOPO® Cloning Kit (Invitrogen) and transformed into Escherichia coli using One Shot® Mach1™ T-1 chemically competent cells (Invitrogen). Transformed colonies were selected using 50 μg mL−1 kanamycin. Each cloned ORF was DNA sequenced using the vector-specific primers M13-F and M13-R to confirm identity and integrity (Additional file 3: Table S3). Then, each ORF was directionally cloned into pRAP15, a gene expression vector [8,9], at the attR1 and attR2 sites using Invitrogen’s Gateway® technology and LR Clonase™ II Enzyme Mix (Invitrogen). The Clonase II reaction product was used to transform E. coli cells, and transformed colonies were selected on 10 μg mL−1 tetracycline plates. Presence of the insert in the correct orientation downstream from the FMV promoter was confirmed by PCR using the FMV-specific primer FMV-F (Additional file 3: Table S3) and the A. thaliana gene-specific reverse primer. The pRAP15 vector bearing each ORF was used to transform competent Agrobacterium rhizogenes ‘K599’ cells using the freeze-thaw method [137] with selection on 5 μg mL−1 tetracycline plates. Presence of the ORF in the pRAP15 vector was confirmed as described above. Expression of the ORF was controlled by the Figwort Mosaic Virus (FMV) promoter. The pRAP15 vector contains the gene encoding enhanced green fluorescent protein gene (eGFP) [138] regulated by the rolD promoter to provide strong eGFP expression in the root for identification of transformed roots. Presence of the gene encoding eGFP was confirmed by PCR using eGFP-F and eGFP-R primers (Additional file 3: Table S3), and eGFP was confirmed visually in transgenic roots. Presence of the A. rhizogenes Ri plasmid was confirmed by PCR using Ri-F and Ri-R primers (Additional file 3: Table S3).
Formation and confirmation of composite soybean plants
Composite soybean plants consisting of untransformed shoots and transformed roots were produced as described previously [139,140]A. rhizogenes clones containing each ORF were grown as described previously [8]. Transformed control roots were produced using A. rhizogenes containing empty pRAP15 with no ORF. Briefly, one hundred soybean cv. Williams 82 PI518671 plants were grown in Promix in the greenhouse for 5–7 days. The plantlets were cut at the soil line and transformed with A. rhizogenes grown to an OD600 of 0.5. The stems were rinsed, and the plantlets were planted in the greenhouse and grown for four to five weeks. The plantlets were gently removed from the Promix, and non-transformed roots were excised. Transformed roots were retained after being recognized by fluorescence of eGFP using a Dark Reader Spot lamp (Clare Chemical Research, Dolores, CO). Plants were replanted in Promix and grown an additional two weeks. The non-transformed roots were removed a second time and approximately 12 to 20 healthy plants with only transformed roots were planted in sand and inoculated with SCN.
The presence of Arabidopsis genes in soybean transgenic roots was confirmed by PCR. Briefly, transgenic soybean roots from each construct were harvested, flash-frozen in liquid nitrogen in 2 ml microfuge tubes, and stored at -80C. After grinding 100 mg root tissue with a mortar and pestle in liquid nitrogen, total RNA was extracted using the RNAeasy Plant Mini Kit (Qiagen, Valencia, CA). Extracted samples of RNA were treated with TURBO™ DNAse I (Ambion, Carlsbad, CA) to remove residual genomic DNA. RNA was tested for genomic DNA contamination by PCR amplification using soybean primers for the soybean gene AW31036. No amplification products were produced when the RNA samples were used as template, but an amplification product was produced when genomic DNA served as template. One milligram of each RNA was converted to cDNA using the Superscript III First-Strand Syntheses System for RTPCR (Invitrogen, Carlsbad, CA), using oligo (dT)12–18 to prime the first strand of cDNA. To test the presence of the Arabidopsis constructs in the transgenic soybean roots, each cDNA served as template for amplification using gene-specific primers in a PCR reaction with Taq DNA polymerase (Invitrogen). Ten microliters of each reaction was electrophoresed on a 2% SB agarose gel for one hour at 150 volts. A 1Kb Plus ladder (Invitrogen) was included to estimate the size of the amplicons. The gel was photographed using a UV light box with an EOS Rebel T3i camera (Canon, Arlington, VA) with a HD UV filter (Canon). Images created with EOS imaging software (Canon) were annotated in Adobe Imaging software (Adobe, San Jose, CA). The nine gene-specific primers were tested by PCR to confirm that Arabidopsis cDNA was the source of the amplicon and not soybean DNA. PCR containing Arabidopsis cDNA produced amplicons, and only the positive control soybean-derived primer pairs gave amplification products when soybean DNA was used, confirming that the primers were specific to only the Arabidopsis genes within the soybean roots.
Preparation of nematodes
SCN line NL1-RHg was maintained on susceptible Glycine max cv. ‘Essex” as described previously [141]. Roots were washed to dislodge SCN cysts, which were captured between nested 850-μm and 250-μm sieves. Cysts were purified by sucrose flotation [142] and crushed against a 7.6-cm diameter 250-μm sieve with a rubber stopper partially submerged in water to release the eggs. Eggs captured in a tray below the sieve were poured through a 61-μm sieve and collected on a 25-μm sieve. Eggs were cleaned by soaking in 0.5% sodium hypochlorite for 1.5 minutes, and then rinsed in sterile deionized distilled water. The eggs were poured into a small tray and hatched in a solution of 3 mM ZnSO4 on a rotary shaker at 26°C and 25 rpm. After four days, the hatching solution was passed through a 30-μm mesh nylon cloth (Spectrum Labs Inc, Rancho Dominguez, CA), which retained the unhatched eggs and liberated the J2 stage SCN in the solution collected below the cloth. To concentrate the J2s, 200 mL of the solution was placed in a 1 L glass beaker and placed on a rotary shaker at 100 rpm. J2s were collected from the center bottom of the beaker with a Pasteur pipette. Three 5-μL aliquots of the J2 solution were examined under the microscope to determine the concentration and viability of the J2s. The solution was diluted to a concentration of 1000 J2 mL−1 with sterile water.
Nematode assay
Twelve transformed composite plants for each construct tested were inoculated with 2000 J2 nematodes per plant. Two holes 4-cm deep were made in the sand on either side of each plant. One mL of a 1000 J2 mL−1 suspension was added to each hole and covered with sand. At 35 days after inoculation (dai), the cysts were collected from the roots of each plant between nested 850-μm and 250-μm sieves and rinsed onto lined filter paper in a Buchner funnel under vacuum [143]. Cysts were counted under a dissection microscope. Plant roots transformed with empty vector were used as the positive control for the female index as described below.
PCR assays
Expression of ORFs in transformed soybean roots was confirmed by RT-PCR as described previously [60]. Three individual soybean roots were harvested per construct. RNA was extracted using a Qiagen RNeasy Mini Kit according to the manufacturer’s instructions. Contaminating DNA was removed by DNase digestion using a TURBO™ DNase kit (Ambion) according to the manufacturer’s instructions. RNA was tested as template by PCR to confirm that no contaminating DNA was present. The RNA was converted into cDNA using the Superscript III First-Strand Synthesis System for RT-PCR (Invitrogen) according to manufacturer’s instructions. Soybean roots transformed with pRAP15 served as controls. Primers were designed to produce an amplicon between 100 and 200 bp (Additional file 2: Table S2). The gene encoding rs-21 (Glyma09g00210.1) served as a positive control [144]. Reactions containing no RNA were used as negative controls.
RT-PCR reactions were conducted in triplicate for each root sample using the Brilliant II SYBR® Green QPCR Master Mix Kit (Agilent Technologies) according to the manufacturer’s instructions. Primer sequences for RT-PCR are provided in Additional file 2: Table S2. Genomic DNA (gDNA) was isolated from individual roots using the DNeasy Plant Mini kit (Qiagene, USA). Three independently transformed roots were examined for each gene transformation. The gDNA served as template in PCR containing primers specific to the Arabidopsis gene.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Three individual roots (100 mg each) were collected that were transformed with either AtNPR1, AtTGA2, or the empty pRAP15 vector as control, respectively. Each root represented an independent transformation event. RNA was extracted from each root using an Ultra Clean Plant RNA Isolation Kit (MOBIO, Carlsbad, CA). Genomic DNA was removed using DNase I. Single-stranded cDNA was synthesize from the RNA using SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA) and oligo dT primers, according to the manufacturer’s instructions. All qRT-PCR primer pairs were designed to flank a region that contains one intron to ensure that product was amplified from cDNA. Primers (Additional file 4: Table S4) were specific to the flanking region of the Arabidopsis AtNPR1 and AtTGA2 genes, yielding amplicons of approximately 150 bp. The soybean ubiquitin-3 (GmUBI-3) gene, GenBank accession D28123, served as a positive qRT-PCR control to demonstrate that soybean RNA was present in all samples. Expression levels of the defense-related soybean genes ERF1 (Glyma20g34570), encoding the ethylene response factor 1; CHIB1 (Glyma10g27870), encoding a basic chitinase protein; and PR-5 (Glyma05g38110), encoding an osmotin-like protein also were determined by qRT-PCR. Other controls for qRT-PCR included reactions containing no template and qRT-PCR reactions containing no reverse transcriptase. qRT-PCR was performed on three biological replicates with each reaction replicated three times. The Stratagene Mx3000P Real-Time PCR system (Stratagene, La Jolla, CA) was used to determine transcript abundance as described by the manufacturer. SYBR Green was used to measure DNA accumulation during the reaction. The Ct (cycle at which there is the first clearly detectable increases in fluorescence) values were calculated using software supplied with the Stratagene Mx3000P Real-Time PCR system. The dissociation curve of amplified products was used to demonstrate the production of only one product per reaction. To further ensure that only one product was formed in each reaction, the PCR products were analyzed on 0.8% agarose gels and visualized under UV light. Absolute quantification of transcript levels was performed according to the sigmoidal model described by (Rutledge and Stewart, 2008) [62].
Statistical analysis
Outliers in the female count data were removed using Grubbs’ test [145] at the GraphPad QuickCalcs Web site (http://graphpad.com/quickcalcs/grubbs1/). Normality of the data was checked using the Shapiro-Wilk test ([146]; online version implemented by S. Dittami, http://scistatcalc.blogspot.com/2013/10/shapiro-wilk-testcalculator.html). Means were compared using Welch’s unpaired t test for unequal variance [147,148] at the GraphPad QuickCalcs Web site (http://graphpad.com/quickcalcs/ttest1/). The female index (FI) was calculated as follows: FI = (Ng/Nc) X 100, where Ng = mean number of females for the gene of interest and Nc = mean number of females for the empty pRAP15 control.
The female index was calculated as described below from 7–16 experimental and 10 or more control plants [8,60,61,139,140,149-153]. Experiments on Arabidopsis genes overexpressed in roots of soybean composite plants were conducted according to published procedures, such as those of Golden et al. [153], Riggs and Schmidtt [149,150] Kim et al. [151] and Niblack et al. [152]. In the experiments of Golden et al. [153], the labs that originally developed and modified the FI, the FI is typically calculated from a total of 3–10 experimental and 3–10 control plants, each individual plant serving as a replicate. Experimental replicates may or may not be performed. All of the experiments presented here exceed these published studies in that regard and are conducted with similar plant numbers to recently published studies [8,9,60,61,139,140,154]. In the present analysis, the number of experimental plants met or exceeded that in investigations testing SCN infection in genetically engineered soybean [155-158]. Herein, we also report standard error of the mean (SEM) for experimental and control groups.
Availability of supporting data
The data supporting the results of this article are included within the article.
Abbreviations
AOC: Allene oxide cyclase; AOS: Allene oxide synthase; CM: Chorismate mutase; dai: days after inoculation; ET: Ethylene; FI: Female index; gDNA: genomic DNA; IAA: Indole acetic acid; ICS: Isochorismate mutase; JA: Jasmonic acid; LOX: Lipoxygenase; OPR3: 12-oxophytodienoic acid reductase; ORFs: Open reading frames; PAL: Phenylalanine ammonia lyase: PAMP, Pathogen-associated molecular patterns; PCR: Polymerase chain reaction; PR: Pathogenesis-related; RKN: Root-knot nematode; SA: Salicylic acid; SAR: Systemic acquired resistance; SCN: Soybean cyst nematode.
Competing interests
The authors have no competing interests.
Authors’ contributions
BM conceived of and designed the experiments, analyzed the data, drafted the manuscript; HB grew the plants, transformed the plant roots, and trimmed the roots; MM inoculated roots with nematodes, harvested and counted nematodes; SK trimmed the roots, harvested and counted the nematodes, and cloned genes; EB cloned genes, transformed the plant roots, and trimmed the roots; RY cloned the genes, trimmed the roots, harvested and counted the nematodes. All authors read and approved the final manuscript.
Supplementary Material
Primers used to PCR amplify ORFs for cloning into pRAP15.
Primers used in RT-PCR assays.
Primers used to confirm clone identity.
Primers used for qRT-PCR.
Contributor Information
Benjamin F Matthews, Email: ben.matthews@ars.usda.gov.
Hunter Beard, Email: hunter.beard@ars.usda.gov.
Eric Brewer, Email: eric.brewer@ars.usda.gov.
Sara Kabir, Email: sara_kabir1@hotmail.com.
Margaret H MacDonald, Email: margaret.macdonald@ars.usda.gov.
Reham M Youssef, Email: reham.youssef@ars.usda.gov.
Acknowledgements
The authors thank Dr. Leslie Wanner for critical review of the manuscript, Patrick Elias for DNA sequencing, Dr. Hua Lu for discussions, and Andrea Maldonado for cloning genes. Financial support from United Soybean Board no.1254 is gratefully acknowledged.
Mention of trade name, proprietary product or vendor does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture or imply its approval to the exclusion of other products or vendors that also may be suitable. The authors have no conflict of interest.
References
- Koenning S, Overstreet C, Noling J, Donald P, Becker J, Fortnum B. Survey of crop losses in response to phytoparasitic nematodes in the United States for 1994. J Nematol. 1999;31(4S):587. [PMC free article] [PubMed] [Google Scholar]
- Wrather JA. Soybean disease loss estimates for the United States, 1996–2010. http://aes.missouri.edu/delta/research/soyloss.stm; 2010. [DOI] [PubMed]
- Wrather JA, Koenning SR. Estimates of disease effects on soybean yields in the United States 2003 to 2005. J Nematol. 2006;38(2):173–180. [PMC free article] [PubMed] [Google Scholar]
- Sasser JNF, Freckman DW. In: Vistas on Nematology. Veek JS, Dickson DW, editor. Hyattsville: Society of Nematologists; 1987. A world perspective on nematology: the role of the society; pp. 7–14. [Google Scholar]
- Sasser JN, Hartman KM, Carter CE. North Carolina State University and US Agency for International Development. Raleigh, NC, USA; 1987. Summary of preliminary crop germplasm evaluation of resistance to root-knot nematodes; pp. 1–88. [Google Scholar]
- Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, Diers BW, Jiang J, Hudson ME, Bent AF. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science. 2012;338(6111):1206–1209. doi: 10.1126/science.1228746. [DOI] [PubMed] [Google Scholar]
- Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, Yang C, Jamai A, El-Mellouki T, Juvale PS, Hill J, Baum TH, Cianzio S, Whitham SA, Korkin D, Mitchum MG, Meksem K. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature. 2012;492(7428):256–260. doi: 10.1038/nature11651. [DOI] [PubMed] [Google Scholar]
- Matthews BF, Beard H, MacDonald MH, Kabir S, Youssef RM, Hosseini P, Brewer E. Engineered resistance and hypersusceptibility through functional metabolic studies of 100 genes in soybean to its major pathogen, the soybean cyst nematode. Planta. 2013;237(5):1337–1357. doi: 10.1007/s00425-013-1840-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsye PD, Kumar R, Hosseini P, Jones CM, Tremblay A, Alkharouf NW, Matthews BF, Klink VP. Mapping cell fate decisions that occur during soybean defense responses. Plant Mol Biol. 2011;77(4–5):513–528. doi: 10.1007/s11103-011-9828-3. [DOI] [PubMed] [Google Scholar]
- Wubben MJ, Jin J, Baum TJ. Cyst nematode parasitism of Arabidopsis thaliana is inhibited by salicylic acid (SA) and elicits uncoupled SA-independent pathogenesis-related gene expression in roots. Mol Plant Microbe Interact. 2008;21(4):424–432. doi: 10.1094/MPMI-21-4-0424. [DOI] [PubMed] [Google Scholar]
- Bari R, Jones JD. Role of plant hormones in plant defence responses. Plant Mol Biol. 2009;69(4):473–488. doi: 10.1007/s11103-008-9435-0. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Molinari S, Loffredo E. The role of salicylic acid in defense response of tomato to root-knot nematodes. Physiol Mol Plant Pathol. 2006;68(1):69–78. [Google Scholar]
- Zinovieva S, Vasyukova N, Udalova ZV, Gerasimova N, Ozeretskovskaya O. Involvement of salicylic acid in induction of nematode resistance in plants. Biol Bull. 2011;38(5):453–458. doi: 10.1134/S1062359011050177. [DOI] [PubMed] [Google Scholar]
- Matson A, Williams L. Evidence of a fourth gene for resistance to the soybean cyst nematode. Crop Science. 1965;5:477. doi: 10.2135/cropsci1965.0011183X000500050032x. [DOI] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002;5(4):325–331. doi: 10.1016/S1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis —2001 status. Curr Opin Plant Biol. 2001;4(4):301–308. doi: 10.1016/S1369-5266(00)00177-1. [DOI] [PubMed] [Google Scholar]
- Nandi B, Kundu K, Banerjee N, Babu SPS. Salicylic acid-induced suppression of Meloidogyne incognita infestation of okra and cowpea. Nematology. 2003;5(5):747–752. doi: 10.1163/156854103322746922. [DOI] [Google Scholar]
- Vasyukova N, Zinov'eva S, Udalova ZV, Panina YS, Ozeretskovskaya O, Sonin M. The role of salicylic acid in systemic resistance of tomato to nematodes. Doklady Biological Sciences. 2003;391:343–345. doi: 10.1023/A:1025158702692. [DOI] [PubMed] [Google Scholar]
- Branch C, Hwang CF, Navarre DA, Williamson VM. Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Mol Plant Microbe Interact. 2004;17(4):351–356. doi: 10.1094/MPMI.2004.17.4.351. [DOI] [PubMed] [Google Scholar]
- Cooper WR, Jia L, Goggin L. Effects of jasmonate-induced defenses on root-knot nematode infection of resistant and susceptible tomato cultivars. J Chem Ecol. 2005;31(9):1953–1967. doi: 10.1007/s10886-005-6070-y. [DOI] [PubMed] [Google Scholar]
- Annigeri S, Shakil N, Kumar J, Singh K. Effect of Jasmonate (Jasmonic Acid) foliar spray on resistance in tomato infected with Root-knot nematode, Meloidogyne incognita. Ann Plant Prot Sci. 2011;19(2):446–450. [Google Scholar]
- Zhu H, Hu F, Wang R, Zhou X, Sze S-H, Liou LW, Barefoot A, Dickman M, Zhang X. Arabidopsis Argonaute10 Specifically Sequesters miR166/165 to Regulate Shoot Apical Meristem Development. Cell. 2011;145(2):242–256. doi: 10.1016/j.cell.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T, Tomitaka Y, Abe H, Tsuda S, Futai K, Mizukubo T. Expression profile of jasmonic acid-induced genes and the induced resistance against the root-knot nematode (Meloidogyne incognita) in tomato plants (Solanum lycopersicum) after foliar treatment with methyl jasmonate. J Plant Physiol. 2011;168(10):1084–1097. doi: 10.1016/j.jplph.2010.12.002. [DOI] [PubMed] [Google Scholar]
- Vasyukova N, Zinovieva S, Udalova ZV, Gerasimova N, Ozeretskovskaya O, Sonin M. Jasmonic acid and tomato resistance to the root-knot nematode Meloidogyne incognita. Doklady Biological Sciences. 2009;428:448–450. doi: 10.1134/S0012496609050160. [DOI] [PubMed] [Google Scholar]
- Concibido VC, Denny RL, Boutin SR, Hautea R, Orf JH, Young ND. DNA marker analysis of loci underlying resistance to Soybean Cyst Nematode (Heterodera glycines Ichinohe) Crop Sci. 1994;34(1):240–246. doi: 10.2135/cropsci1994.0011183X003400010044x. [DOI] [Google Scholar]
- Concibido VC, Diers BW, Arelli PR. A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci. 2004;44(4):1121–1131. doi: 10.2135/cropsci2004.1121. [DOI] [Google Scholar]
- Diers B, Arelli P. Management of Parasitic Nematodes of Soybean Through Genetic Resistance. Proceedings of World Soybean Research Conference, 6th (Kauffman HE), Chicago, IL, USA: 1999. 1999. pp. 4–7.
- Webb D, Baltazar B, Rao-Arelli A, Schupp J, Clayton K, Keim P, Beavis W. Genetic mapping of soybean cyst nematode race-3 resistance loci in the soybean PI 437.654. Theor Appl Genet. 1995;91(4):574–581. doi: 10.1007/BF00223282. [DOI] [PubMed] [Google Scholar]
- Schuster I, Abdelnoor R, Marin S, Carvalho V, Kiihl R, Silva J, Sediyama C, Barros E, Moreira M. Identification of a new major QTL associated with resistance to soybean cyst nematode (Heterodera glycines) Theor Appl Genet. 2001;102(1):91–96. doi: 10.1007/s001220051622. [DOI] [Google Scholar]
- Caldwell BE, Brim CA, Ross JP. Inheritance of resistance of soybeans to the Cyst Nematode, Heterodera Glycines1. Agron J. 1960;52(11):635–636. doi: 10.2134/agronj1960.00021962005200110007x. [DOI] [Google Scholar]
- Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- Cao H, Li X, Dong X. Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc Natl Acad Sci. 1998;95(11):6531–6536. doi: 10.1073/pnas.95.11.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DMA, Shah J, Klessig DF. Salicylic acid and disease resistance in plants. Crit Rev Plant Sci. 1999;18(4):547–575. doi: 10.1080/07352689991309397. [DOI] [Google Scholar]
- Dong X. Genetic dissection of systemic acquired resistance. Curr Opin Plant Biol. 2001;4(4):309–314. doi: 10.1016/S1369-5266(00)00178-3. [DOI] [PubMed] [Google Scholar]
- Feys BJ, Parker JE. Interplay of signaling pathways in plant disease resistance. Trends Genet. 2000;16(10):449–455. doi: 10.1016/S0168-9525(00)02107-7. [DOI] [PubMed] [Google Scholar]
- Loake G, Grant M. Salicylic acid in plant defence–the players and protagonists. Curr Opin Plant Biol. 2007;10(5):466–472. doi: 10.1016/j.pbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Thomma BP, Penninckx IA, Cammue B, Broekaert WF. The complexity of disease signaling in Arabidopsis. Curr Opin Immunol. 2001;13(1):63–68. doi: 10.1016/S0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E. A central role of salicylic acid in plant disease resistance. Science. 1994;266(5188):1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, Broekaert WF. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci. 1998;95(25):15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Métraux JP, Zhu T, Katagiri F. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 2003;34(2):217–228. doi: 10.1046/j.1365-313X.2003.01717.x. [DOI] [PubMed] [Google Scholar]
- Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 2005;10(2):71–78. doi: 10.1016/j.tplants.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DMA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. Salicylic acid–independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell Online. 2006;18(4):1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich K, Wirthmueller L, Tasset C, Pouzet C, Deslandes L, Parker JE. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment–specific immune responses. Science. 2011;334(6061):1401–1404. doi: 10.1126/science.1211641. [DOI] [PubMed] [Google Scholar]
- Straus MR, Rietz S, Ver Loren van Themaat E, Bartsch M, Parker JE. Salicylic acid antagonism of EDS1‒driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J. 2010;62(4):628–640. doi: 10.1111/j.1365-313X.2010.04178.x. [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci. 1999;96(23):13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman M-A, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20(19):5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux J-P. EDS5, an essential component of salicylic acid–dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell Online. 2002;14(1):275–286. doi: 10.1105/tpc.010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberne MC, Muljono RAB, Verpoorte R. Salicylic acid biosynthesis. New Compr Biochem. 1999;33:295–312. [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414(6863):562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, De Luca V, Després C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012;1(6):639–647. doi: 10.1016/j.celrep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fan W, Kinkema M, Li X, Dong X. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc Natl Acad Sci. 1999;96(11):6523–6528. doi: 10.1073/pnas.96.11.6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci. 1998;95(17):10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knepper C, Savory EA, Day B. Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol. 2011;156(1):286–300. doi: 10.1104/pp.110.169656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G. RIN4 functions with plasma membrane H + −ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009;7(6):e1000139. doi: 10.1371/journal.pbio.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink J. Regulation of gene expression by jasmonate hormones. Phytochemistry. 2009;70:1560–1570. doi: 10.1016/j.phytochem.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Halim V, Vess A, Scheel D, Rosahl S. The role of salicylic acid and jasmonic acid in pathogen defence. Plant Biol. 2006;8(3):307–313. doi: 10.1055/s-2006-924025. [DOI] [PubMed] [Google Scholar]
- Youssef RM, MacDonald MH, Brewer EP, Bauchan GR, Kim KH, Matthews BF. Ectopic expression of AtPAD4 broadens resistance of soybean to soybean cyst and root-knot nematodes. BMC Plant Biol. 2013;13:67. doi: 10.1186/1471-2229-13-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef RM, Matthews BF. Expression of Arabidopsis genes AtNPR1 and AtTGA2 in transgenic soybean roots of composite plants confers resistance to root-knot nematode (Meloidogyne incognita) Int J Curr Biotechnol Volume. 2013;1(10):15–25. [Google Scholar]
- Rutledge RG, Stewart D. A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol. 2008;8(1):47. doi: 10.1186/1472-6750-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knepper C, Day B. From perception to activation: the molecular-genetic and biochemical landscape of disease resistance signaling in plants. The Arabidopsis Book/American Society of Plant Biologists. 2010;8:1–17. doi: 10.1199/tab.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken F, Tameling W. To nibble at plant resistance proteins. Science. 2009;324(5928):744. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- Ellenberger T. Getting a grip on DNA recognition: structures of the basic region leucine zipper, and the basic region helix-loop-helix DNA-binding domains. Curr Opin Struct Biol. 1994;4(1):12–21. doi: 10.1016/S0959-440X(94)90054-X. [DOI] [Google Scholar]
- Anthony-Cahill SJ, Benfield PA, Fairman R, Wasserman ZR, Brenner SL, Stafford W, Altenbach C, Hubbell WL, DeGrado WF. Molecular characterization of helix-loop-helix peptides. Science. 1992;255(5047):979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Hurst HC. Transcription factors 1: bZIP proteins. Protein Profile. 1994;2(2):101–168. [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci. 2006;103(45):17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeeswaran G, Raina S, Acharya BR, Maqbool SB, Mosher SL, Appel HM, Schultz JC, Klessig DF, Raina R. Arabidopsis GH3‒LIKE DEFENSE GENE 1 is required for accumulation of salicylic acid, activation of defense responses and resistance to Pseudomonas syringae. Plant J. 2007;51(2):234–246. doi: 10.1111/j.1365-313X.2007.03130.x. [DOI] [PubMed] [Google Scholar]
- Lee MW, Lu H, Jung HW, Greenberg JT. A key role for the Arabidopsis WIN3 protein in disease resistance triggered by Pseudomonas syringae that secrete AvrRpt2. Mol Plant Microbe Interact. 2007;20(10):1192–1200. doi: 10.1094/MPMI-20-10-1192. [DOI] [PubMed] [Google Scholar]
- Nobuta K, Venu R, Lu C, Beló A, Vemaraju K, Kulkarni K, Wang W, Pillay M, Green PJ, Wang G-l. An expression atlas of rice mRNAs and small RNAs. Nat Biotechnol. 2007;25(4):473–477. doi: 10.1038/nbt1291. [DOI] [PubMed] [Google Scholar]
- Wang N, Qian W, Suppanz I, Wei L, Mao B, Long Y, Meng J, Müller AE, Jung C. Flowering time variation in oilseed rape (Brassica napus L.) is associated with allelic variation in the FRIGIDA homologue BnaA. FRI. a. J Exp Bot. 2011;62(15):5641–5658. doi: 10.1093/jxb/err249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K-C, Li H-Y, Xiao S, Tse M-H, Chye M-L. Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta. 2006;223(5):871–881. doi: 10.1007/s00425-005-0139-2. [DOI] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen Q-F, Chan S-W, Zheng S-X, Ma J, Wang M, Welti R, Chye M-L. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell Online. 2010;22(5):1463–1482. doi: 10.1105/tpc.110.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell Online. 1994;6(12):1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Verbsky ML, Robertson TL, Larkin JC, Kunkel BN. Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in CPR5. Mol Plant Microbe Interact. 1998;11(12):1196–1206. doi: 10.1094/MPMI.1998.11.12.1196. [DOI] [Google Scholar]
- Kirik V, Bouyer D, Schöbinger U, Bechtold N, Herzog M, Bonneville J-M, Hülskamp M. CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr Biol. 2001;11(23):1891–1895. doi: 10.1016/S0960-9822(01)00590-5. [DOI] [PubMed] [Google Scholar]
- Brininstool G, Kasili R, Simmons LA, Kirik V, Hülskamp M, Larkin JC. Constitutive expressor of pathogenesis-related Genes5 affects cell wall biogenesis and trichome development. BMC Plant Biol. 2008;8(1):58. doi: 10.1186/1471-2229-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H-C, Anderson L, Sturre MJ, Hille J, Dijkwel PP. Arabidopsis CPR5 is a senescence-regulatory gene with pleiotropic functions as predicted by the evolutionary theory of senescence. J Exp Bot. 2007;58(14):3885–3894. doi: 10.1093/jxb/erm237. [DOI] [PubMed] [Google Scholar]
- Staswick PE. JAZing up jasmonate signaling. Trends Plant Sci. 2008;13(2):66–71. doi: 10.1016/j.tplants.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci. 1992;89(15):6837–6840. doi: 10.1073/pnas.89.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell Online. 1994;6(11):1583–1592. doi: 10.1105/tpc.6.11.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N, Greenberg JT. Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell Online. 2006;18(2):397–411. doi: 10.1105/tpc.105.036251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Schmid M, Hierl G, Terneus K, Tan L, Lottspeich F, Kieliszewski MJ, Gietl C. KDEL-tailed cysteine endopeptidases involved in programmed cell death, intercalation of new cells, and dismantling of extensin scaffolds. Am J Bot. 2008;95(9):1049–1062. doi: 10.3732/ajb.2007404. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Fengler KA, Yu I-c, Lippok B, Smith RK, Bent AF. The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc Natl Acad Sci. 2000;97(16):9323–9328. doi: 10.1073/pnas.150005697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões I, Faro R, Bur D, Faro C. Characterization of recombinant CDR1, an Arabidopsis aspartic proteinase involved in disease resistance. J Biol Chem. 2007;282(43):31358–31365. doi: 10.1074/jbc.M702477200. [DOI] [PubMed] [Google Scholar]
- Coll N, Epple P, Dangl J. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18(8):1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Zhang Y. Regulating the reapers: activating metacaspases for programmed cell death. Trends Plant Sci. 2012;17(8):487–494. doi: 10.1016/j.tplants.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Glover K, Wang D, Arelli P, Carlson S, Cianzio S, Diers B. Near isogenic lines confirm a soybean cyst nematode resistance gene from PI 88788 on linkage group J. Crop Sci. 2004;44(3):936–941. doi: 10.2135/cropsci2004.0936. [DOI] [Google Scholar]
- Guo B, Sleper D, Lu P, Shannon J, Nguyen H, Arelli P. QTLs associated with resistance to soybean cyst nematode in soybean: meta-analysis of QTL locations. Crop Sci. 2006;46(2):595–602. doi: 10.2135/cropsci2005.04-0036-2. [DOI] [Google Scholar]
- Kopisch-Obuch F, McBroom R, Diers B. Association between soybean cyst nematode resistance loci and yield in soybean. Crop Sci. 2005;45(3):956–965. doi: 10.2135/cropsci2004.0441. [DOI] [Google Scholar]
- Wang D, Diers B, Arelli P, Shoemaker R. Loci underlying resistance to race 3 of soybean cyst nematode in Glycine soja plant introduction 468916. Theor Appl Genet. 2001;103(4):561–566. doi: 10.1007/PL00002910. [DOI] [Google Scholar]
- Meksem K, Pantazopoulos P, Njiti V, Hyten L, Arelli P, Lightfoot D. 'Forrest' resistance to the soybean cyst nematode is bigenic: saturation mapping of the Rhg1and Rhg4 loci. Theoretical and Applied Genetics. 2001;103(5):710–717. doi: 10.1007/s001220100597. [DOI] [Google Scholar]
- Siddiqui I, Shaukat S. Systemic resistance in tomato induced by biocontrol bacteria against the root‒knot nematode, Meloidogyne javanica is independent of salicylic acid production. J Phytopathol. 2004;152(1):48–54. doi: 10.1046/j.1439-0434.2003.00800.x. [DOI] [Google Scholar]
- Uehara T, Sugiyama S, Matsuura H, Arie T, Masuta C. Resistant and susceptible responses in tomato to cyst nematode are differentially regulated by salicylic acid. Plant Cell Physiol. 2010;51(9):1524–1536. doi: 10.1093/pcp/pcq109. [DOI] [PubMed] [Google Scholar]
- Nawrath C, Métraux J-P. Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell Online. 1999;11(8):1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7(5):547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol. 2007;10(4):366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Van Loon L. NPR1: the spider in the web of induced resistance signaling pathways. Curr Opin Plant Biol. 2004;7(4):456–464. doi: 10.1016/j.pbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. Plant Cell Online. 1992;4(6):645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Métraux J-P, Ryals JA. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell Online. 1991;3(10):1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant SRMPD, McNeece BT, Sharma K, Krishnavajhala A, Lawrence GW, Klink VP. Syntaxin 31 functions in Glycine max resistance to the plant parasitic nematode Heterodera glycines. Plant Mol Biol. 2014. doi:10.1007/s11103-014-0172-2. [DOI] [PubMed]
- Dao D, Frank D, Qian N, O’Keefe D, Vosatka RJ, Walsh CP, Tycko B. IMPT1, an imprinted gene similar to polyspecific transporter and multi-drug resistance genes. Hum Mol Genet. 1998;7(4):597–608. doi: 10.1093/hmg/7.4.597. [DOI] [PubMed] [Google Scholar]
- Chern MS, Fitzgerald HA, Yadav RC, Canlas PE, Dong X, Ronald PC. Evidence for a disease‒resistance pathway in rice similar to the NPR1‒mediated signaling pathway in Arabidopsis. Plant J. 2001;27(2):101–113. doi: 10.1046/j.1365-313x.2001.01070.x. [DOI] [PubMed] [Google Scholar]
- Malnoy M, Jin Q, Borejsza-Wysocka E, He S, Aldwinckle H. Overexpression of the apple MpNPR1 gene confers increased disease resistance in Malus × domestica. Mol Plant Microbe Interact. 2007;20(12):1568–1580. doi: 10.1094/MPMI-20-12-1568. [DOI] [PubMed] [Google Scholar]
- Sandhu D, Tasma IM, Frasch R, Bhattacharyya M. Systemic acquired resistance in soybean is regulated by two proteins, orthologous to Arabidopsis NPR1. BMC Plant Biol. 2009;9(1):105. doi: 10.1186/1471-2229-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, Le Su E, Rochon A, Shearer HL, Murmu J, Chu JY, Fobert PR, Després C. The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell Online. 2009;21(11):3700–3713. doi: 10.1105/tpc.109.069971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Reddy A. Cloning and expression of a PR5-like protein from Arabidopsis: inhibition of fungal growth by bacterially expressed protein. Plant Mol Biol. 1997;34(6):949–959. doi: 10.1023/A:1005893119263. [DOI] [PubMed] [Google Scholar]
- Abad LR, D’Urzo MP, Liu D, Narasimhan ML, Reuveni M, Zhu JK, Niu X, Singh NK, Hasegawa PM, Bressan RA. Antifungal activity of tobacco osmotin has specificity and involves plasma membrane permeabilization. Plant Sci. 1996;118(1):11–23. doi: 10.1016/0168-9452(96)04420-2. [DOI] [Google Scholar]
- Vigers GP, Anderson LJ, Caffes P, Brandhuber BJ. Crystal structure of the type-I interleukin-1 receptor complexed with interleukin-1β. Nature. 1997;386(6621):190–194. doi: 10.1038/386190a0. [DOI] [PubMed] [Google Scholar]
- El-Kereamy A, El-Sharkawy I, Ramamoorthy R, Taheri A, Errampalli D, Kumar P, Jayasankar S. Prunus domestica pathogenesis-related protein-5 activates the defense response pathway and enhances the resistance to fungal infection. PLoS One. 2011;6(3):e17973. doi: 10.1371/journal.pone.0017973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell Online. 1998;10(6):1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietz S, Stamm A, Malonek S, Wagner S, Becker D, Medina‒Escobar N, Corina Vlot A, Feys BJ, Niefind K, Parker JE. Different roles of Enhanced Disease Susceptibility1 (EDS1) bound to and dissociated from Phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 2011;191(1):107–119. doi: 10.1111/j.1469-8137.2011.03675.x. [DOI] [PubMed] [Google Scholar]
- Pegadaraju V, Louis J, Singh V, Reese JC, Bautor J, Feys BJ, Cook G, Parker JE, Shah J. Phloem‒based resistance to green peach aphid is controlled by Arabidopsis PHYTOALEXIN DEFICIENT4 without its signaling partner ENHANCED DISEASE SUSCEPTIBILITY1. Plant J. 2007;52(2):332–341. doi: 10.1111/j.1365-313X.2007.03241.x. [DOI] [PubMed] [Google Scholar]
- Rusterucci C, Aviv DH, Holt BF 3rd, Dangl JL, Parker JE. The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell. 2001;13(10):2211–2224. doi: 10.1105/tpc.13.10.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A, Ménard R, Heitz T, Shen WH. Chromatin modification and remodelling: a regulatory landscape for the control of Arabidopsis defence responses upon pathogen attack. Cell Microbiol. 2012;14(6):829–839. doi: 10.1111/j.1462-5822.2012.01785.x. [DOI] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM. Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell Online. 1997;9(3):305–316. doi: 10.1105/tpc.9.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeseth NJ, Pacovsky RS, Newman T, Ohlrogge JB. Characterization of an acyl-CoA-binding protein from Arabidopsis thaliana. Arch Biochem Biophys. 1996;331(1):55–62. doi: 10.1006/abbi.1996.0282. [DOI] [PubMed] [Google Scholar]
- Mandrup S, Jepsen R, Skott H, Rosendal J, Hojrup P, Kristiansen K, Knudsen J. Effect of heterologous expression of acyl-CoA-binding protein on acyl-CoA level and composition in yeast. Biochem J. 1993;290:369–374. doi: 10.1042/bj2900369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q-F, Xiao S, Chye M-L. Overexpression of the Arabidopsis 10-kilodalton acyl-coenzyme A-binding protein ACBP6 enhances freezing tolerance. Plant Physiol. 2008;148(1):304–315. doi: 10.1104/pp.108.123331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach JM, Castillo AR, Hoogstraten R, Greenberg JT. The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc Natl Acad Sci. 2001;98(2):771–776. doi: 10.1073/pnas.98.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich KL, Bovet L, Hunziker PE, Donnison IS, Hörtensteiner S. Molecular cloning, functional expression and characterisation of RCC reductase involved in chlorophyll catabolism. Plant J. 2000;21(2):189–198. doi: 10.1046/j.1365-313x.2000.00667.x. [DOI] [PubMed] [Google Scholar]
- Pena-Cortés H, Albrecht T, Prat S, Weiler EW, Willmitzer L. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta. 1993;191(1):123–128. [Google Scholar]
- Dammann C, Rojo E, Sánchez‒Serrano JJ. Abscisic acid and jasmonic acid activate wound‒inducible genes in potato through separate, organ‒specific signal transduction pathways. Plant J. 1997;11(4):773–782. doi: 10.1046/j.1365-313X.1997.11040773.x. [DOI] [PubMed] [Google Scholar]
- Nahar K, Kyndt T, De Vleesschauwer D, Höfte M, Gheysen G. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiol. 2011;157(1):305–316. doi: 10.1104/pp.111.177576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyndt T, Nahar K, Haegeman A, De Vleesschauwer D, Höfte M, Gheysen G. Comparing systemic defence‒related gene expression changes upon migratory and sedentary nematode attack in rice. Plant Biol. 2012;14(s1):73–82. doi: 10.1111/j.1438-8677.2011.00524.x. [DOI] [PubMed] [Google Scholar]
- Bhattarai KK, Xie Q-G, Mantelin S, Bishnoi U, Girke T, Navarre DA, Kaloshian I. Tomato susceptibility to root-knot nematodes requires an intact jasmonic acid signaling pathway. Mol Plant Microbe Interact. 2008;21(9):1205–1214. doi: 10.1094/MPMI-21-9-1205. [DOI] [PubMed] [Google Scholar]
- Oka Y, Cohen Y, Spiegel Y. Local and systemic induced resistance to the root-knot nematode in tomato by DL-β-amino-n-butyric acid. Phytopathology. 1999;89(12):1138–1143. doi: 10.1094/PHYTO.1999.89.12.1138. [DOI] [PubMed] [Google Scholar]
- Yu I-c, Parker J, Bent AF. Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci. 1998;95(13):7819–7824. doi: 10.1073/pnas.95.13.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagué C, Lin B, Alcon C, Flottes G, Malmström S, Köhler C, Neuhaus G, Pelletier G, Gaymard F, Roby D. HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide–gated channel ion channel family. Plant Cell Online. 2003;15(2):365–379. doi: 10.1105/tpc.006999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowski GI, Smith RK Jr, Yu I-c, Ham JH, Sharma SB, Klessig DF, Fengler KA, Bent AF. Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol Plant Microbe Interact. 2004;17(5):511–520. doi: 10.1094/MPMI.2004.17.5.511. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci. 2010;107(31):13960–13965. doi: 10.1073/pnas.1002828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, Patel K, Dixon RA, Lamb C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004;23(4):980–988. doi: 10.1038/sj.emboj.7600086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BD, Creissen G, Lamb C, Chattoo BB. Overexpression of rice (Oryza sativa L.) OsCDR1 leads to constitutive activation of defense responses in rice and Arabidopsis. Mol Plant Microbe Interact. 2009;22(12):1635–1644. doi: 10.1094/MPMI-22-12-1635. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Hofgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988;16(20):9877–9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci. 1997;94(6):2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Kim K-H, Martins V, MacDonald MH, Beard HS, Alkharouf NW, Lee S-K, Park S-C, Matthews BF. A correlation between host-mediated expression of parasite genes as tandem inverted repeats and abrogation of development of female Heterodera glycines cyst formation during infection of Glycine max. Planta. 2009;230(1):53–71. doi: 10.1007/s00425-009-0926-2. [DOI] [PubMed] [Google Scholar]
- Ibrahim HM, Alkharouf NW, Meyer SL, Aly MA, Gamal El-Din AEKY, Hussein EH, Matthews BF. Post-transcriptional gene silencing of root-knot nematode in transformed soybean roots. Exp Parasitol. 2011;127(1):90–99. doi: 10.1016/j.exppara.2010.06.037. [DOI] [PubMed] [Google Scholar]
- Klink VP, Overall CC, Alkharouf NW, MacDonald MH, Matthews BF. A time-course comparative microarray analysis of an incompatible and compatible response by Glycine max (soybean) to Heterodera glycines (soybean cyst nematode) infection. Planta. 2007;226(6):1423–1447. doi: 10.1007/s00425-007-0581-4. [DOI] [PubMed] [Google Scholar]
- Jenkins W. A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Dis Rep. 1964;48(9):692. [Google Scholar]
- Krusberg L, Sardanelli S, Meyer S, Crowley P. A method for recovery and counting of nematode cysts. J Nematol. 1994;26(4S):599. [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Alkharouf N, MacDonald M, Matthews B. Laser capture microdissection (LCM) and expression analyses of Glycine max (soybean) syncytium containing root regions formed by the plant pathogen Heterodera glycines (soybean cyst nematode) Plant Mol Biol. 2005;59(6):965–979. doi: 10.1007/s11103-005-2416-7. [DOI] [PubMed] [Google Scholar]
- Grubbs FE. Procedures for detecting outlying observations in samples. Technometrics. 1969;11(1):1–21. doi: 10.1080/00401706.1969.10490657. [DOI] [Google Scholar]
- Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52(3/4):591–611. doi: 10.2307/2333709. [DOI] [Google Scholar]
- Welch B. On the comparison of several mean values: an alternative approach. Biometrika. 1951;38(3/4):330–336. doi: 10.2307/2332579. [DOI] [Google Scholar]
- Welch BL. The generalization ofstudent’s’ problem when several different population variances are involved. Biometrika. 1947;34(1/2):28–35. doi: 10.2307/2332510. [DOI] [PubMed] [Google Scholar]
- Riggs R, Schmitt D. Complete characterization of the race scheme for Heterodera glycines. J Nematol. 1988;20(3):392. [PMC free article] [PubMed] [Google Scholar]
- Riggs R, Schmitt D. Optimization of the Heterodera glycines race test procedure. J Nematol. 1991;23(2):149. [PMC free article] [PubMed] [Google Scholar]
- Kim D, Riggs R, Mauromoustakos A. Variation in resistance of soybean lines to races of Heterodera glycines. J Nematol. 1998;30(2):184. [PMC free article] [PubMed] [Google Scholar]
- Niblack T, Arelli P, Noel G, Opperman C, Orf J, Schmitt D, Shannon J, Tylka G. A revised classification scheme for genetically diverse populations of Heterodera glycines. J Nematol. 2002;34(4):279–288. [PMC free article] [PubMed] [Google Scholar]
- Golden AM, Epps JM, Riggs RD, Duclos LA, Fox JA, Bernard RL. Terminology and identity of infraspecific forms of the soybean cyst nematode (Heterodera glycines) Plant Dis Rep. 1970;54:544–546. [Google Scholar]
- Maldonado A, Youssef R, MacDonald M, Brewer E, Beard E, Matthews B. Modification of the expression of two NPR1 suppressors, SNC1 and SNI1, in soybean (Glycine max) confers partial resistance to the soybean cyst nematode (Heterodera glycines) Funct Plant Biol. 2014. http://dx.doi.org/10.1071/FP13323. [DOI] [PubMed]
- Steeves RM, Todd TC, Essig JS, Trick HN. Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Funct Plant Biol. 2006;33(11):991–999. doi: 10.1071/FP06130. [DOI] [PubMed] [Google Scholar]
- McLean MD, Hoover GJ, Bancroft B, Makhmoudova A, Clark SM, Welacky T, Simmonds DH, Shelp BJ. Identification of the full-length Hs1pro-1 coding sequence and preliminary evaluation of soybean cyst nematode resistance in soybean transformed with Hs1pro-1 cDNA. Botany. 2007;85(4):437–441. [Google Scholar]
- Li J, Todd TC, Oakley TR, Lee J, Trick HN. Host-derived suppression of nematode reproductive and fitness genes decreases fecundity of Heterodera glycines Ichinohe. Planta. 2010;232(3):775–785. doi: 10.1007/s00425-010-1209-7. [DOI] [PubMed] [Google Scholar]
- Melito S, Heuberger AL, Cook D, Diers BW, MacGuidwin AE, Bent AF. A nematode demographics assay in transgenic roots reveals no significant impacts of the Rhg1 locus LRR-Kinase on soybean cyst nematode resistance. BMC Plant Biol. 2010;10(1):104. doi: 10.1186/1471-2229-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used to PCR amplify ORFs for cloning into pRAP15.
Primers used in RT-PCR assays.
Primers used to confirm clone identity.
Primers used for qRT-PCR.