Abstract
Karyotypic diversification is more prominent in Equus species than in other mammals. Here, using next generation sequencing technology, we generated and de novo assembled quality genomes sequences for a male wild horse (Przewalski's horse) and a male domestic horse (Mongolian horse), with about 93-fold and 91-fold coverage, respectively. Portion of Y chromosome from wild horse assemblies (3 M bp) and Mongolian horse (2 M bp) were also sequenced and de novo assembled. We confirmed a Robertsonian translocation event through the wild horse's chromosomes 23 and 24, which contained sequences that were highly homologous with those on the domestic horse's chromosome 5. The four main types of rearrangement, insertion of unknown origin, inserted duplication, inversion, and relocation, are not evenly distributed on all the chromosomes, and some chromosomes, such as the X chromosome, contain more rearrangements than others, and the number of inversions is far less than the number of insertions and relocations in the horse genome. Furthermore, we discovered the percentages of LINE_L1 and LTR_ERV1 are significantly increased in rearrangement regions. The analysis results of the two representative Equus species genomes improved our knowledge of Equus chromosome rearrangement and karyotype evolution.
Horses are recognized as extremely successful domestic animals. Humans in many parts of the world have relied on them for thousands of years1. The genus Equus originated on the North American continent and migrated 2.6 million years ago over the Bering Strait during the Ice Age2. Horses, donkeys, and zebras evolved from the same ancestor. The speciation events were accomplished through acute chromosomal rearrangements, with the rearrangement rate ranging from 2.9 to 22.2 per million years, which is significantly higher than in other mammals3,4,5,6,7,8,9. Equus species possess widely varying diploid chromosome numbers, from 2n = 32 (Mountain zebra) to 66 (Przewalski's horse). Przewalski's horse has a different chromosome number than domestic horses because of a Robertsonian translocation, resulting in one pair of metacentric chromosomes (ECA5) split into two pairs of acrocentric chromosomes10,11,12 (EPR23 and EPR24). Although the offspring produced from a cross between Przewalski's horse and a domestic horse had 65 chromosomes, it was fertile11,13, unlike the mule (2n = 63, offspring of male donkey and female horse) and the hinny (2n = 63, offspring of male horse and female donkey), which are sterile.
Przewalski's horse (“wild horse” hereafter) is the only wild horse species surviving in the world today14. Because of environmental change and human activities, this species dropped to only 12 individuals in the middle of the last century. Today, the number has increased to approximately 2000, located in the field or in zoos, but all of them are descendants of those 12 ancestors15. This event dramatically reduced the genetic variation of the wild horse, which could reduce the ability of the species to adapt to environment change. Severe genetic bottlenecks have also occurred with European bison16, northern elephant seals17 and cheetahs18. Therefore, the wild horse is not only a valuable wildlife resource but also a promising model for the study of population genetics. The Mongolian horse is an ancient horse breed that has been an integral part of the culture of nomadic pastoralists in North Asia. The Mongolian horse has a large population with abundant genetic diversity. This ancient breed has influenced other Northern European horse breeds19. It has acquired many special abilities and attributes, such as endurance and disease resistance, and is well adapted to its harsh conditions—a cold, arid climate and poor grazing opportunities20. Dramatic chromosomal rearrangement in the horse is a notable feature in comparison to other mammals, and this makes the horse an ideal model for studying chromosomal evolution.
In this study, we obtained quality whole-genome sequences of a male wild horse and a male Mongolian horse using next-generation sequencing technology. The genome sequences of the two representative Equus species would improve the genomic maps of the horse. Importantly, based on this, we will focus on karyotypic diversification and explore the genetic mechanisms and evolution rules through analysis of comparative genomics, further uncovering the genetic mechanisms of chromosomal evolution for Equus species.
Results
Genome sequencing and assembly
The wild horse and Mongolian horse genomes were sequenced using the Illumina Hiseq platform. A paired-end library (500 bp) and two mate-paired libraries (3 kb, and 8 kb) were constructed for both the wild horse and Mongolian horse. In total, we generated 231.21 Gb and 224.17 Gb of usable sequences for the wild horse and Mongolian horse. The sequence depth was 93× and 91×, respectively (Supplementary Table S1). The sequencing error rates were 0.000575 and 0.000507 for the wild horse and Mongolian horse, respectively (Supplementary Table S2). After assembly, both the wild horse and Mongolian horse generated the same length of genome sequences (2.38 Gb) (Supplementary Table S3).
We checked 248 core eukaryotic genes21 in our two assemblies, and found the completeness was comparable with that of published genomes sequences assemblies22,23,24,25,26 (Supplementary Table S4). We did not detect any misassemblies27 when comparing our genome assemblies with the wild horse and Mongolian horse sequences available in Genbank. (Supplementary Table S5).
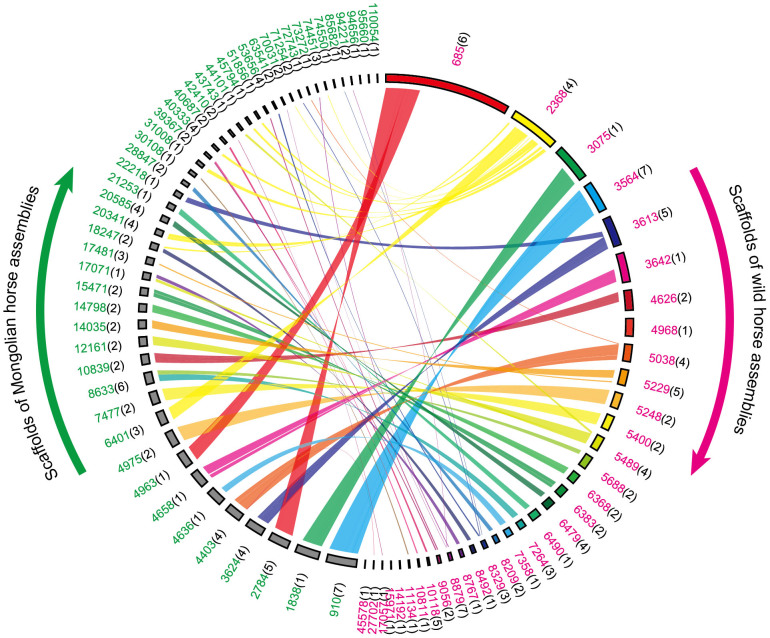
We assembled Y chromosome of wild horse and Mongolian horse (Fig. 1). In previous studies28, 127 markers on horse Y chromosome were reported, and in this study, 87 markers and 103 markers could be detected in wild horse and Mongolian horse assemblies, respectively (Supplementary Table S6, 7). Thus, 34 scaffolds (3,018,288 bp) of wild horse and 48 scaffolds (1,971,029 bp) of Mongolian horse were identified originated from Y chromosome. The length of collinearity regions between wild horse and Mongolian horse was around 1.74 Mbp.
Figure 1. Scaffolds of Y chromosome of wild horse and Mongolian horse.
Thirty-four scaffolds of wild horse and 48 scaffolds of Mongolian horse are shown in this figure, and collinearity regions are linked. Numbers located outside of the brackets are the scaffolds ID of wild horse (carmine) and Mongolian horse (green). Numbers located inside of the brackets represent count of markers detected in the scaffolds.
To improve gene prediction accuracy, eight types of tissue samples (heart, liver, spleen, lung, kidney, brain, spinal cord and muscle) from a female Mongolian horse were used to construct cDNA libraries. The RNA-seq was performed using the 454 FLX+ platform, and 853,978 reads were obtained with an average length of 458 bp (Supplementary Fig. S1). From these transcriptome data, aided by homology-based gene prediction methods, we estimated that the horse genome contained 20,000 to 21,000 protein-coding genes.
Synteny analysis
Robertsonian translocation, which is also called whole-arm translocation or centric-fusion translocation, is a common form of chromosomal rearrangement. Previous studies based on fluorescence in situ hybridization (FISH) results indicate that chromosomes 23 and 24 of the wild horse are homologous with chromosome 5 of the domestic horse. After assembling the wild horse and Mongolian horse genome, we masked out all repetitive sequences and found that EPR23 and 24 of the wild horse and ECA5 of the Mongolian horse could be aligned to the chromosome 5 of the reference genome29. The five probes (LAMC2, LAMB3, VCAM1, UOX and DIA1), which were used in FISH mapping in previous research to confirm that EPR23, 24 is homologous with ECA512, were also identified in both the wild horse and domestic horse genome (Fig. 2).
Figure 2. Synteny analysis.
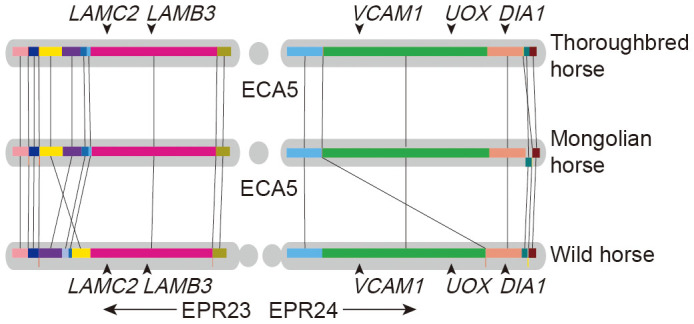
Microsynteny between chromosome 5 of domesticated horses (ECA5) and chromosomes 23 and24 of wild horses (EPR23, EPR24). Locally Collinear Blocks (LCBs) are marked with the same color and connected by straight lines. The probes (LAMC2, LAMB3, VCAM1, UOX, DIA1), which are used for FISH, are also detected in this figure.
To study the relationship between Robertsonian translocation and local rearrangement, we performed whole genome synteny analysis. We compared wild horse genome and Mongolian horse genome to the Thoroughbred horse genome, respectively. Collinearity region between Mongolian horse and Thoroughbred horse (2.25 Gbp) was slightly longer than that between wild horse and Thoroughbred horse (2.23 Gbp). 124 Mbp (5.51%) of wild horse genome and 76 Mbp (3.34%) of Mongolian horse genome could not align to Thoroughbred horse genome. Four types of rearrangement, BRK (insertion of unknown origin), DUP (inserted duplication), INV (inversion), and JMP (relocation), were identified (Supplementary Table S8, 9).
Since artifactual mis-joins of assemblies could be counted as rearrangements, we attempted to estimate the correct rate of these rearrangements breakpoints. We remapped the usable reads to the genomes assemblies of wild horse and Mongolian horse, respectively. Then we checked the number of mapped reads in the breakpoint of each type of rearrangements. If the number was less than three, we considered the assembly was incorrect (Supplementary Fig. S2), otherwise correct (Supplementary Fig. S3). We counted 100 breakpoints for each type of rearrangement, and calculated the correct rate. The correct rates of INV (92%) and JMP (82%) were higher than those for BRK (76%) and DUP (58%) in assemblies of wild horse. In Mongolian horse, the correct rates were similar with those of wild horse (Supplementary Table S10).
The potential rearrangement sites were investigated for potential synapomorphies. We counted the rearrangement events in two situations: (1) Assume genome sequences of Thoroughbred horse and Mongolian horse were consensus, and identify rearrangements in wild horse; (2) Assume genome sequences of Thoroughbred horse and wild horse were consensus, and identify rearrangements in Mongolian horse. We found that rearrangement events in the first situation were dramatically more than that in the second (Supplementary Fig. S4). This result was consistent with phylogeny.
The numbers of rearrangements on each chromosome were counted (Supplementary Table S11). Chromosome 5 does not have a greater number of local rearrangements compared with the other chromosomes, although chromosome 5 had undergone Robertsonian translocation (Fig. 3). We noticed that the number of inversions is far less than that of insertions and relocations in the horse genome. Some chromosomes, including the X chromosome, contain more local rearrangements than others. Local rearrangements in the genome of wild horse are more numerous than in that of the Mongolian horse.
Figure 3. Local rearrangements in the wild horse and Mongolian horse.
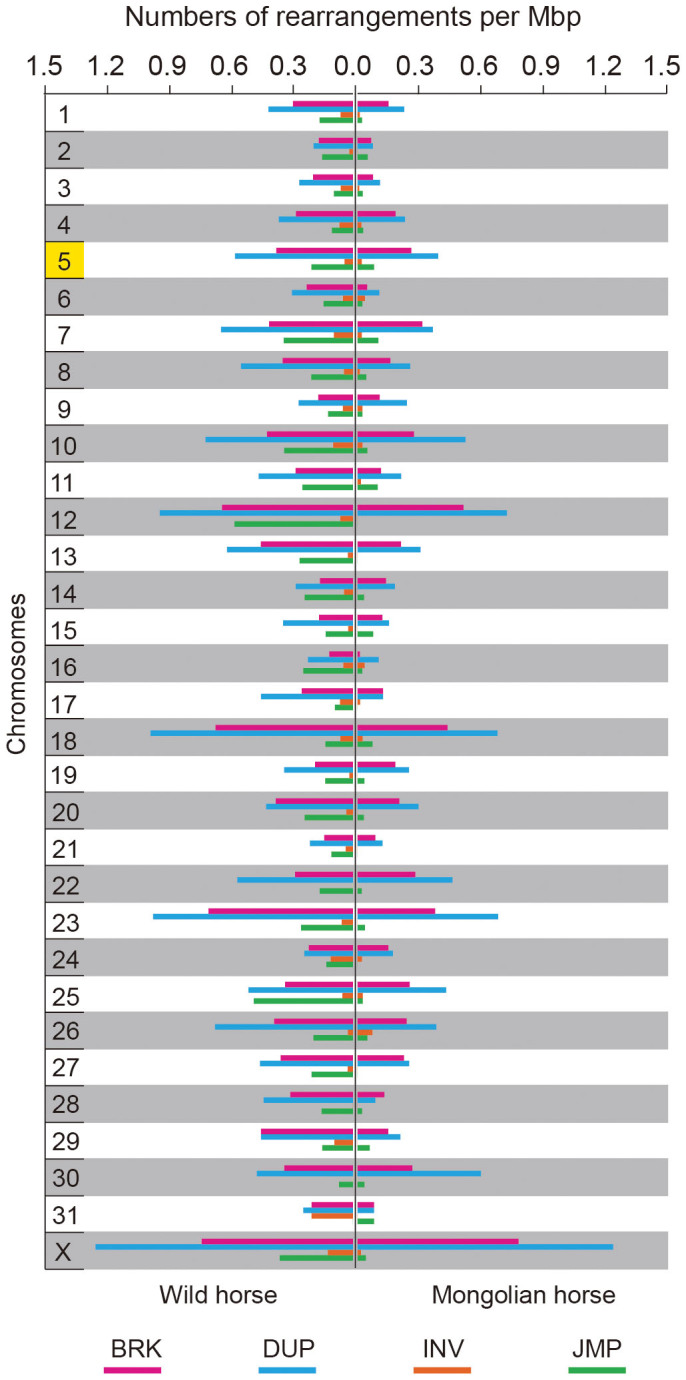
Chromosome 5 of domestic horse had undergone Robertsonian translocation (marked as yellow). Thoroughbred horse genome was used as the reference, so the chromosome undergone Robertsonian translocation was also chromosome 5 for wild horse in this figure. BRK: insertion of unknown origin; DUP: inserted duplication; INV: inversion; JMP: relocation.
Repetitive sequences
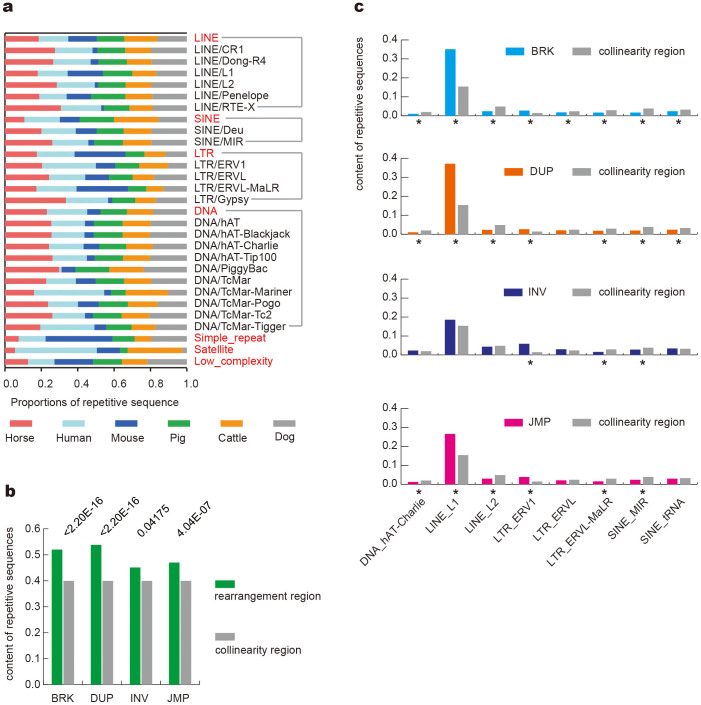
Repetitive sequences comprise approximately 50% of the mammal genomes30 and are associated with syntenic breakpoints and chromosomal fragility31,32,33. Repetitive sequences of six species of mammals (horses29, humans30, mouse34, dogs35, cattle36 and pigs37) were examined in this study (Fig. 4a). Seven common repetitive sequences were identified: short interspersed repeated sequences (SINE), long interspersed repeated sequences (LINE), long terminal repeated (LTR), DNA elements, satellites, simple repeats and low complexity. Broadly, the analysis of these sequences indicated that 41.4% of the horse genome sequences are repetitive sequences, which is comparable to the percentages in humans (46.8%), mouse (42.5%), dogs (40.0%), cattle (47.1%), and pigs (39.1%). LINEs comprise 22.6% of the horse genome, which is more than in humans (19.7%), mouse (19.1%), dogs (19.8%), cattle (21.9%), and pigs (18.4%). SINEs can be found in 7.3% of the horse genome, less than in the human (13.4%), mouse (7.5%), dog (10.6%), cattle (17.0%), and pig (13.0%) genomes (Supplementary Fig. S5).
Figure 4. Analysis of repetitive sequences.
(a) The proportions of repetitive sequences among six species of mammals. Seven common repetitive sequences are marked in red, and the subclasses are marked in black. (b) The content of repetitive sequences is significantly increased in rearrangements regions compared with the collinearity region. The “p-value” is shown on the top. (c) Some repetitive sequences representing content greater than 0.5% of the genome. The content of repetitive sequences significantly increased in BRK/DUP/INV/JMP regions compared with the collinearity region. ‘*' p-value < 0.05.
The distribution of repetitive sequences in each chromosome was also examined. The results indicated that each chromosome contains a similar proportion of repetitive sequences, except the X chromosome, which contains a higher proportion of repetitive sequences than autosomes in the six species.
Using those rearrangement regions of the wild horse genome, we studied the association between rearrangement and repetitive sequences. We found some types of repetitive sequences were significantly increased in the rearrangement regions (Fig. 4b, Supplementary Table S12). This result is consistent with previous findings31,32. Interestingly, the proportions of LINE_L1 and LTR_ERV1 increased, but the proportions of LINE_L2 and several other repetitive sequences decreased (Fig. 4c, Supplementary Table S13 to S16). This result suggests that LINE_L1 and LTR_ERV1 may play a more important role in chromosome rearrangement.
Heterozygosity analysis
We identified 1,280,203 and 2,203,945 heterozygous SNPs (within an individual) in the genomes of the wild horse and Mongolian horse (Supplementary Table S17 to S19). Small indels were also identified in the genomes of the wild horse and Mongolian horse (Supplementary Table S20). The heterozygosity rates were 0.52 × 10−3 and 0.89 × 10−3 in the wild horse and Mongolian horse, respectively. The heterozygosity of the wild horse is considerably lower than that of the Mongolian horse.
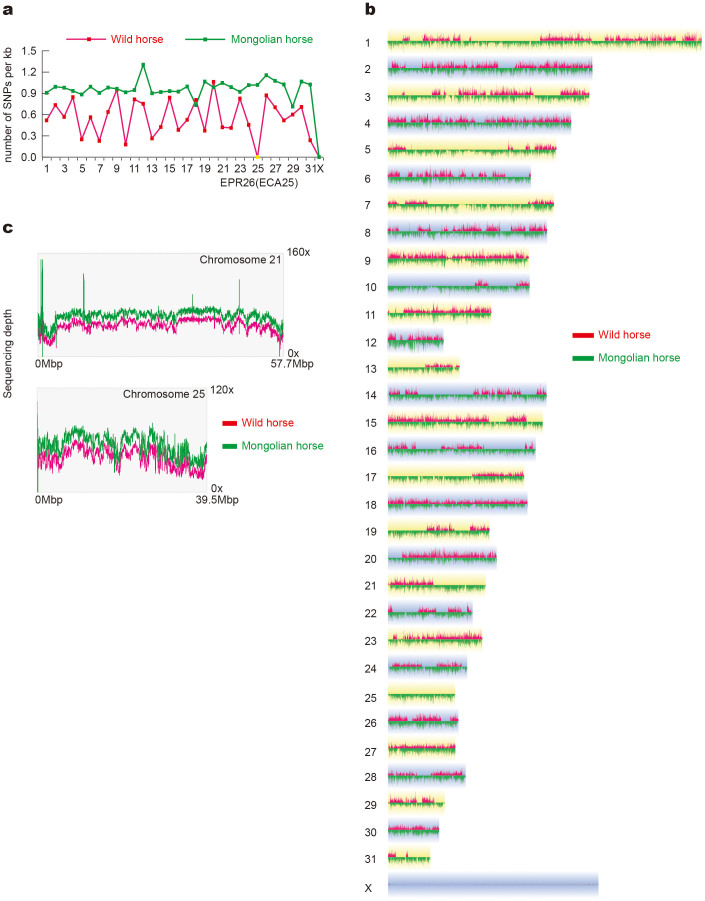
SNPs were not evenly distributed among the wild horse chromosomes but were evenly distributed in the Mongolian horse chromosomes (Fig. 5a). We explored the heterozygosity rates of different regions using sliding windows of 50 kb with a step size of 10 kb. The number of sliding windows with high heterozygosity rate in Mongolian horse genome was considerable greater than that in wild horse genome (Supplementary Fig. S6). Another interesting phenomenon found in the genome of the wild horse was that heterozygous SNPs were completely excluded in many large regions (Fig. 5b). The sequence coverage of those regions in the wild horse was the same as in the Mongolian horse (Fig. 5c, Supplementary Fig. S7).
Figure 5. Effect of genetic bottleneck on genome landscape.
(a) The SNPs distribution of each chromosome in the wild horse and Mongolian horse. For the Mongolian horse, the SNP distribution of each autosome is similar, but for the wild horse, the SNP distribution among the autosomes is different, and there are no SNPs on EPR26 (ECA25 in this figure). (b) Contrast of heterozygous SNPs between the wild horse and Mongolian horse. (c) The sequencing depths of chromosome 21 and 25.
In the wild horse, there is a total length of 1287 M of homozygous regions (there are no SNP in wild horse, and there are more than 0.8SNP/Kbp in Mongolian horse) and 58 homozygous regions were larger than 1 Mbp (Supplementary Table S21). A total of 4508 genes were located in those homozygous regions. Enrichment analysis indicated that these genes were enriched for specific functional categories of olfactory transduction (n = 118, p_value = 8.90e-12), regulation of cell proliferation (n = 11, p_value = 1.70e-02), calcium ion binding (n = 9, p_value = 1.70e-02) and others.
Discussion
In the past decades, researchers have studied chromosomal rearrangement using different conventional methods such as chromosome banding and FISH. However, many local rearrangements are extremely difficult to detect. Here, we sequenced and de novo assembled the homologous chromosomes that had undergone Robertsonian translocation. Our study indicated that Robertsonian translocation did not increase local rearrangements. These findings indicated that Robertsonian translocation and local rearrangements may be caused by different mechanisms. From our results, inversions are rarer than insertions and relocation, suggesting that insertions and relocation may play a more important role in shaping the genome.
Some studies have demonstrated that repetitive sequences are associated with syntenic breakpoints and chromosomal fragility31,32. This study did not reveal significant differences in repetitive sequences among different species and different chromosomes (except the X chromosome). Different strategies of genome sequencing (clone by clone and whole genome shot-gun) may impact the actual content of repetitive sequences in the genomes. Our results suggest that chromosomal local rearrangements are highly associated with repetitive sequences. However, these repetitive sequences did not contribute equally to rearrangement. LINE_L1 and LTR_ERV1 may play a more important role than other repetitive sequences.
In the middle of the last century, the population of the wild horse dropped to only 12 individuals. The genetic bottleneck and inbreeding caused by this event may be the reason for the many more homozygous regions in the wild horse genome. One interesting result is that heterozygous SNPs are completely excluded from chromosome 26 of the wild horse. It was the largest fragment without heterozygous SNPs. As sequencing coverage of EPR26 is similar to other autosomes, we confirmed that there was a pair of chromosome 26 in this individual (Supplementary Fig. S8). Another explanation could be that this pair of chromosome 26 was present because of uniparental isodisomy38,39,40. We also sequenced a short region (~700 bp) in chromosome 26 of several other wild horse samples using the Sanger method and found 6 SNPs, indicating that this region is heterozygous in some other wild horses.
The analysis results of the two representative Equus species improved the genomic maps of the horse. It also revealed the unique aspects of the chromosomal rearrangement and improved our understanding of chromosomal evolution in mammals implicating Equus is thus a promising model to explore the Karyotypic instability. These analysis and discoveries would benefit studies of mammal karyotypic evolution and chromosomal rearrangement, and studies of human disease caused by chromosome aberration.
Methods
Sampling and genome sequencing
Protocols used for this experiment were consistent with those approved by the Institutional Animal Care and Use Committee at Inner Mongolia Agricultural University. For sequencing, a male wild horse was selected from the “YE MA International Group” of Xinjiang, China, and a Mongolian horse was selected from the Xilingol League of Inner Mongolia, China. DNA was extracted from ear tissue and peripheral blood cells. Illumina HiSeq 2000 was used to sequence the genomes of wild horse and Mongolian horse using a shotgun strategy. A pair-end library (500 bp, standard genomic library and sequenced using paired-end reads) and two mate-pair libraries (3 kb, and 8 kb) were constructed for each horse. The length of reads was 101 bp for pair-end library and two mate-pair libraries. Library preparation and sequencing followed the manufacturer's instructions, and sequence reads were collected from the Illumina data processing pipeline.
Data filtering
The following types of reads were filtered out: (1) reads with more than 3 unidentified nucleotides, (2) reads with average phred quality below Q30, and (3) reads with unidentified nucleotides in the first 50 nucleotides.
Genome assembly
The genome sequences of the wild horse and Mongolian horse were assembled with short reads using SOAPdenovo41. We first assembled the short reads of the pair-end library (500 bp) into contigs using sequence overlap information. Then, we used the information of the mate-pair libraries (3 kb and 8 kb) to join the contigs into scaffolds. Finally, “Gapcloser” (http://soap.genomics.org.cn/soapdenovo.html) was used to close the gaps inside the scaffolds.
Genome annotation
For protein-coding gene annotation, ab initio prediction was performed by MAKER42. We generated cDNA data from multiple RNA sources. Using an oligodT-based approach, cDNA libraries were constructed from eight types of tissue samples (heart, liver, spleen, lung, kidney, brain, spinal cord and muscle) from a female Mongolian horse. The library was sequenced using a Roche 454 FLX+ platform.
Estimated sequencing error rate
Data from the X chromosome was used to estimate the sequencing error rate, which is hemizygotic in males. The calculations were not influenced by heterozygous SNPs43. All qualified reads from the wild horse and Mongolian horse were mapped to the X chromosome of Equus caballus, and all repetitive and low complexity regions were excluded. At each nucleotide position, the predominant call was assumed to be true, and all others were considered to be errors.
Synteny analysis
We used the MAUVE program44 to construct the synteny map for chromosome 5 of domestic horses (Thoroughbred and Mongolian horse) and chromosomes 23 and 24 of the wild horse. We masked out all repetitive sequences, and unique sequences were preserved. Then, we used the Mauve Contig Mover (MCM) to order the draft genomes of the wild horse and Mongolian horse relative to the Thoroughbred horse genome (Equcab 2.0). The synteny analysis used progressiveMAUVE.
We used MUMmer45 to perform the synteny analysis for the whole genomes of the wild horse and Mongolian horse, in addition to the reference genome. Four types of rearrangements (BRK, DUP, INV, JMP) were identified using the “nucmer” module. The parameter was Options “-c 800 -g300 –l 100”.
Repetitive sequence analysis
We screened DNA sequences for interspersed repeats and low complexity DNA sequences using RepeatMasker (http://www.repeatmasker.org/) in the collinearity and rearrangement regions. Collinearity regions, which were larger than 100 kb, were used for following analysis, and rearrangement loci plus 2 kb extended flanking regions were treated as rearrangement regions. The “T-test” was performed using R software.
SNP calling and heterozygosity rate estimation
We utilized the BWA program46 to map the usable reads from the pair-end libraries (500 bp) of the wild horse and Mongolian horse to the genome sequences of Thoroughbred horse (Equcab 2.0). The parameters chosen for mapping were as follows: seed length of 32, and the maximum occurrences for extending a long deletion of 10. Duplicated reads were removed by SAMtools47. SNPs and InDels were called using the Genome Analysis Toolkit48 according to the guidelines as described.
The heterozygosity rate was estimated as the density of heterozygous SNPs for the whole genome. For the estimation of local heterozygosity rate, sliding windows of 50 kb with 80% overlap between adjacent windows were used to scan the genome.
Author Contributions
M.D., H.M., Ya.Z., Z.S. designed and managed the project. J.H., W.Y., Ya.Z., H.M. designed and performed the genome assembly and analyses. J.H., H.M., Ya.Z., M.D. wrote the paper. D.D., D.B., J.H., Yi.Z., H.H., H.B., Y.B. collected samples and prepared the nucleic acid samples. Yi.Z., W.S., B.L., D.B., L.Y., B.J., Q.Z., Y.G., J.W., W.B., A.L., Yu.Z., L.Z., Z.Z., W.Z. performed the genomes sequencing.
Additional information
Data Access The Whole Genome Shotgun project has been deposited in DDBJ/EMBL/GenBank as project accession PRJNA200657 and PRJNA200654 of wild horse and Mongolian horse, respectively. The genome assembly of wild horse has been deposited at DDBJ/EMBL/GenBank under the accession ATBW00000000 and this version described in this paper is version ATBW01000000. The genome assembly of Mongolian horse has been deposited at DDBJ/EMBL/GenBank under the accession ATDM00000000 and the version described in this paper is version ATDM01000000. Transcript sequencing data have been deposited under Short Read Archive (SRA) accession SRR1014663.
Supplementary Material
Supplementary material
Supplementary table S8
Supplementary table S9
Acknowledgments
This work was supported by Ministry of Science and Technology of the People's Republic of China specific scientific and technological cooperation with Russia (2011DFR30860), Inner Mongolia major scientific and technological projects (NK-20120395), National Natural Science Foundation of China (31360538), Inner Mongolia key laboratory project (20130902), National Natural Science Foundation of China (31160446), Inner Mongolia Agricultural University Equine Science and Industrialization Innovation Team Support Program (NDTD2010-12), and Ministry of Agriculture of the People's Republic of China public welfare specific scientific and technological projects (201003075). We thank Dr. Liqing Zhang (Department of Computer Science, Virginia Tech, USA) for valuable comments to this manuscript.
References
- Outram A. K. et al. The earliest horse harnessing and milking. Science 323, 1332–1335 (2009). [DOI] [PubMed] [Google Scholar]
- Lindsay E. H., Opdyke N. D. & Johnson N. M. Pliocene dispersal of the horse Equus and late Cenozoic mammalian dispersal events. Nature 287, 135–138 (1980). [Google Scholar]
- Bush G. L., Case S. M., Wilson A. C. & Patton J. L. Rapid speciation and chromosomal evolution in mammals. Proc Natl Acad Sci U S A 74, 3942–3946 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobigny G., Aniskin V. & Volobouev V. Explosive chromosome evolution and speciation in the gerbil genus Taterillus (Rodentia, Gerbillinae): a case of two new cryptic species. Cytogenet Genome Res 96, 117–124 (2002). [DOI] [PubMed] [Google Scholar]
- Koehler U., Bigoni F., Wienberg J. & Stanyon R. Genomic reorganization in the concolor gibbon (Hylobates concolor) revealed by chromosome painting. Genomics 30, 287–292 (1995). [DOI] [PubMed] [Google Scholar]
- Murphy W. J. et al. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309, 613–617 (2005). [DOI] [PubMed] [Google Scholar]
- Muller S., Hollatz M. & Wienberg J. Chromosomal phylogeny and evolution of gibbons (Hylobatidae). Hum Genet 113, 493–501 (2003). [DOI] [PubMed] [Google Scholar]
- Trifonov V. A. et al. Multidirectional cross-species painting illuminates the history of karyotypic evolution in Perissodactyla. Chromosome Res 16, 89–107 (2008). [DOI] [PubMed] [Google Scholar]
- Piras F. M. et al. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet 6, e1000845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benirschke K., Malouf N., Low R. J. & Heck H. Chromosome Complement: Differences between Equus caballus and Equus przewalskii, poliakoff. Science 148, 382–383 (1965). [DOI] [PubMed] [Google Scholar]
- Koulischer L. & Frechkop S. Chromosome Complement: A Fertile Hybrid between Equus priewalskii and Equus caballus. Science 151, 93–95 (1966). [DOI] [PubMed] [Google Scholar]
- Myka J. L., Lear T. L., Houck M. L., Ryder O. A. & Bailey E. FISH analysis comparing genome organization in the domestic horse (Equus caballus) to that of the Mongolian wild horse (E. przewalskii). Cytogenet Genome Res 102, 222–225 (2003). [DOI] [PubMed] [Google Scholar]
- Ahrens E. & Stranzinger G. Comparative chromosomal studies of E. caballus (ECA) and E. przewalskii (EPR) in a female F1 hybrid. J Anim Breed Genet 122 Suppl 1, 97–102 (2005). [DOI] [PubMed] [Google Scholar]
- Orlando L. et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78 (2013). [DOI] [PubMed] [Google Scholar]
- Goto H. et al. A massively parallel sequencing approach uncovers ancient origins and high genetic variability of endangered Przewalski's horses. Genome Biol Evol 3, 1096–1106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl G. B. & Pucek Z. Genetic depletion in the European bison (Bison bonasus) and the significance of electrophoretic heterozygosity for conservation. Conservation biology 8, 167–174 (1994). [Google Scholar]
- Hoelzel A. R. et al. Elephant seal genetic variation and the use of simulation models to investigate historical population bottlenecks. J Hered 84, 443–449 (1993). [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M. & O'Brien S. J. Dating the genetic bottleneck of the African cheetah. Proc Natl Acad Sci U S A 90, 3172–3176 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornstad G., Nilsen N. O. & Roed K. H. Genetic relationship between Mongolian and Norwegian horses? Anim Genet 34, 55–58 (2003). [DOI] [PubMed] [Google Scholar]
- Li L. F., Guan W. J., Hua Y., Bai X. J. & Ma Y. H. Establishment and characterization of a fibroblast cell line from the Mongolian horse. In Vitro Cell Dev Biol Anim 45, 311–316 (2009). [DOI] [PubMed] [Google Scholar]
- Parra G., Bradnam K. & Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007). [DOI] [PubMed] [Google Scholar]
- Jirimutu et al. Genome sequences of wild and domestic bactrian camels. Nat Commun 3, 1202 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q. H. et al. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res 23, 1091–1105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet 45, 701–706 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. S. et al. The tiger genome and comparative analysis with lion and snow leopard genomes. Nat Commun 4, 2433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y. et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat Biotechnol 31, 135–141 (2013). [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N. & Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudsepp T. et al. A detailed physical map of the horse Y chromosome. Proc Natl Acad Sci U S A 101, 9321–9326 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade C. M. et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326, 865–867 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- Mikkelsen T. S. et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447, 167–177 (2007). [DOI] [PubMed] [Google Scholar]
- Webber C. & Ponting C. P. Hotspots of mutation and breakage in dog and human chromosomes. Genome Res 15, 1787–1797 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L. et al. Two replication fork maintenance pathways fuse inverted repeats to rearrange chromosomes. Nature 501, 569–572 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R. H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002). [DOI] [PubMed] [Google Scholar]
- Kirkness E. F. et al. The dog genome: survey sequencing and comparative analysis. Science 301, 1898–1903 (2003). [DOI] [PubMed] [Google Scholar]
- Elsik C. G. et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 324, 522–528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M. A. et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491, 393–398 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet 6, 137–143 (1980). [DOI] [PubMed] [Google Scholar]
- Engel E. Uniparental disomies in unselected populations. Am J Hum Genet 63, 962–966 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzot D. Complex and segmental uniparental disomy (UPD): review and lessons from rare chromosomal complements. J Med Genet 38, 497–507 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20, 265–272 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L. et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18, 188–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. I. et al. A highly annotated whole-genome sequence of a Korean individual. Nature 460, 1011–1015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B. & Perna N. T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS one 5, e11147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A. L. et al. Alignment of whole genomes. Nucleic Acids Res 27, 2369–2376 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary table S8
Supplementary table S9