Abstract
MicroRNAs (miRNAs) have recently emerged as key regulators of metabolism. For example, miR-33a and b play a crucial role in controlling cholesterol and lipid metabolism in concert with their host genes, the SREBP transcription factors. Metabolic miRNAs such as miR-103 and miR-107 regulate insulin and glucose homeostasis, while others, such as miR-34a, may be key regulators of hepatic lipid homeostasis. The discovery of circulating miRNAs has highlighted their potential as both endocrine signalling molecules and disease markers. Dysregulation of miRNAs may contribute to metabolic abnormalities, suggesting that miRNAs may potentially serve as therapeutic targets to ameliorate cardiometabolic disorders.
Introduction
Proper control of metabolic homeostasis is critical to the maintenance of human physiology and health. Accordingly, intricate and interwoven regulatory networks have evolved to monitor and respond to changes in environmental conditions and physiological states. Work over several decades has suggested that much of the orchestration of cellular and physiological responses to altered dietary and metabolic conditions occurs at the level of gene regulation in the cell nucleus. Indeed, a number of key transcription factors, including Peroxisome Proliferator-Activated Receptors (PPARs), Liver X Receptors (LXRs), Sterol Regulatory Element-Binding Proteins (SREBPs), Carbohydrate Response Element-Binding Protein ( ChREBP), CCAAT-Enhancer-Binding Protein (C/EBP), Forkhead box protein O1 (FoxO1) and others, respond directly or indirectly to nutrients and metabolic cues such as cholesterol, lipids, glucose, and insulin, to rapidly alter gene expression programs governing metabolic homeostasis1-5.
Small non-coding RNAs termed microRNAs (miRNAs) have recently been found to represent another critical regulatory layer overlaying and intersecting with transcriptional control mechanisms in guiding metabolic homeostasis. Initially discovered in the nematode Caenorhabditis elegans as regulators of developmental timing, numerous miRNAs have subsequently been found in species from plants to humans, with regulatory roles touching upon all aspects of biology. The biogenesis of microRNAs is described in Box 16, 7.
By contrast with plants, where miRNAs are often fully complementary to their mRNA targets and promote RNA cleavage and degradation, metazoan miRNAs typically exhibit only partial sequence complementarity to their mRNA targets, and initial studies suggested that they promote translational repression rather than cleavage of the mRNA8. However, it has recently become apparent that metazoan miRNAs may also affect mRNA stability by promoting mRNA deadenylation and subsequent sequestration and turnover in P-bodies9.
While functional validation is frequently lacking, target prediction databases based primarily on Watson-Crick base-pairing (e.g., TargetScan, miRanda, and Pictar10-12) have suggested that miRNAs may have hundreds of mRNA targets, thereby rivalling transcriptional mechanisms in regulatory output complexity. However, whereas transcription factors may elicit profound changes in mRNA expression levels, single miRNAs typically exert relatively modest effects on individual mRNA targets, and are thought to act primarily as “rheostats” that modulate protein expression in a nuanced fashion7. However, single miRNAs may have multiple target sites in the 3’UTRs of a particular mRNA, increasing repression efficiency, and mRNAs are predicted to be targets of many distinct miRNAs, suggesting that different miRNAs may act in a concerted manner to regulate mRNA translation and turnover13. As discussed further below, certain miRNAs have also been shown to affect multiple targets in linear pathways, or interconnected nodes in regulatory networks, thereby exerting a larger cumulative effect14. MiRNAs are also frequently found to act in feed-forward and feed-back regulation that can amplify or dampen signal output15, making timing of analysis after miRNA perturbation critical to an accurate assessment of regulatory impact. Finally, whereas miRNA functions under normal physiological conditions might be integrated into multi-layered control circuits ensuring proper development and homeostasis, dysregulation of miRNA expression or function in response to intrinsic (genetic or epigenetic) or extrinsic (environmental cues or stress) factors may contribute to aberrant gene expression patterns underlying abnormal developmental patterning or metabolic dysfunction.While it is clear that the complex mechanisms of action and impact of miRNAs on animal development, physiology, and disease need much further study, progress has been made in elucidating the individual roles of certain miRNAs in specific biological contexts.
In this review, we discuss recent advances in our understanding of the emerging roles of miRNAs in controlling cholesterol and lipid homeostasis, with particular emphasis on the well-characterized miR-122 liver-specific miRNA and the regulatory circuit comprised of the miR-33a and miR-33b miRNAs and their SREBP host genes. The role of miRNAs such as the related miR-103 and miR-107 in controlling insulin signalling and glucose homeostasis is also highlighted. The potential pathological functions of miRNAs such as miR-33 and miR-34a in conditions associated with metabolic syndrome are discussed, and we cover current efforts to therapeutically target specific metabolic miRNAs. Finally, we briefly discuss circulating miRNAs, with emphasis on new data linking miRNAs with lipoproteins and possible functions as “endocrine” signalling molecules.
MiRNA regulation of cholesterol and lipid homeostasis
While roles for miRNAs were first described in developmental regulation in metazoans, recent studies have revealed that miRNAs also play key roles in controlling metabolic homeostasis. Below, we initially discuss the roles of the liver-specific miRNA miR-122 and the miR-33-SREBP host gene circuit in cholesterol and lipid homeostasis, and highlight new and exciting data pointing to important physiological and therapeutic implications of these miRNAs in metabolic diseases.
Lipids are required as structural components of cell membranes (e.g., cholesterol and phospholipids) and serve important roles in energy storage (e.g., triglycerides), but can also act as signalling molecules (e.g., steroid hormones). Lipids such as cholesterol and fatty acids are taken up in the diet, as well as synthesized de novo, predominantly in the liver. Regulation of the biosynthesis of cholesterol, fatty acids, and phospholipids is mediated by transcription factors such as SREBPs and is under tight feedback control to maintain proper homeostasis (reviewed in16). Cholesterol and other lipids are transported in the blood by association with lipoproteins such as very high-density lipoprotein (VLDL), low-density lipoprotein (LDL) and high-density lipoproteins (HDL). LDL primarily transports cholesterol to peripheral tissues, where the LDL-receptor (LDLR) mediates LDL uptake. HDL is important for the removal of cholesterol from peripheral tissues to the liver through a system called reverse cholesterol transport (RCT). Aberrant cholesterol and lipid homeostasis represents a critical risk factor for cardiometabolic diseases prevalent in the developed world, such as metabolic syndrome (MetS), a constellation of metabolic disorders that include insulin resistance, obesity, abnormalities in circulating cholesterol and lipid profiles, and hypertension17. MetS is associated with increased risk of type 2 diabetes, and coronary artery disease (CAD). For example, the risk of atherosclerosis and CAD increases with a rise in circulating LDL levels, whereas HDL levels are inversely associated with cardiovascular disease risk17. Insulin resistance and other metabolic abnormalities can result in excess accumulation of hepatic triglycerides and fatty acids, which is associated with fatty liver diseases (hepatosteatosis) such as non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH), precursors to more severe liver diseases such as fibrosis and hepatocellular carcinoma18.
Cholesterol and lipid regulation by the liver-specific miR-122
The first miRNA linked to metabolic control, miR-122, is expressed primarily in liver and was initially shown to affect hepatic cholesterol and lipid metabolism, but has also been implicated in the maintenance of liver cell differentiation (reviewed in 19). Two early studies showed that antisense targeting of miR-122 results in significant (~25-30%) reduction in plasma cholesterol levels20, 21. First, a study employing cholesterol-conjugated antisense oligonucleotides (termed “antagomiRs”) showed that injection of antagomirs that target miR-122 into mice resulted in altered hepatic expression of a number of genes, including some involved in cholesterol biosynthesis, such as 3-hydroxy-3-methylglutaryl-coenzyme A reductase (Hmgcr), 3-hydroxy-3-methylglutaryl-coenzyme A synthase 1 (Hmgcs1), and 7-dehydrocholesterol reductase (Dhcr7)21. Accordingly, they observed significant lowering of plasma cholesterol in response to miR-122 antagonism. In a separate study, antisense-based silencing of miR-122 led to decreased hepatic cholesterol and fatty acid biosynthesis, and an increase in fatty acid β-oxidation, associated with a reduction in circulating total cholesterol and triglycerides, and decreased hepatosteatosis in mice on a high-fat diet20. In subsequent studies, miR-122 was successfully targeted by antisense inhibitors employing Locked Nucleic Acid (LNA) chemistry22 in non-human primates such as African green monkeys, resulting in lowered circulating cholesterol23. Antisense targeting of miR-122 was not associated with hepatotoxicity or adverse liver histopathology in mice or non-human primates, indicating the apparent safety of this therapeutic approach. Taken together, these pioneering studies pointed to the exciting possibility of miR-122 antisense oligonucleotides as a novel treatment strategy for lowering circulating cholesterol, and which would represent the first therapeutic targeting of a miRNA. However, the initial enthusiasms waned after it was found that miR-122 antagonism not only lowers LDL, but also causes a decline in the levels of HDL, both in mice and in non-human primates20, 23, 24. This presumably deleterious effect has called into question the therapeutic value of miR-122 as a target for the treatment of cholesterol-related disorders. Moreover, although genes involved in cholesterol and lipid metabolism are affected by miR-122 in liver, they do not appear to be direct targets of miR-12219-21, 25. This lack of a mechanistic understanding of the effects of miR-122 on cholesterol homeostasis, and the possibilities of other adverse consequences, such as hepatocellular carcinoma26, 27, has also dampened enthusiasm for the development of miR-122 antisense technologies as a therapeutic approach for the long-term management of cholesterol disorders.
Recently, miR-122 was found to be required for the propagation of the hepatitis C virus, both directly at the level of viral replication and in controlling lipid co-factors required for virus replication and assembly28-30. The therapeutic targeting of miR-122 in this context is very intriguing and represents the first miRNA-targeting drug in human clinical trials (discussed in Box 2).
The SREBP-miR-33 regulatory circuit
The SREBP family of basic-helix-loop-helix-leucine zipper transcription factors controls the expression of numerous genes involved in cholesterol and fatty acid biosynthesis and uptake, as well as in the production of phospholipids and triglycerides. The human SREBF1 gene encodes SREBP-1a and -1c, alternative splice variants that primarily regulate lipogenic genes such as fatty acid synthase (FASN), acetyl-CoA carboxylase (ACC), and stearoyl-CoA desaturase (SCD). The SREBP-2 isoform produced by the SREBF2 gene preferentially controls the expression of cholesterogenic genes, including HMGCR and LDLR3.
Intriguingly, both SREBP-encoding genes in human were recently found to be host genes to highly conserved miRNAs31-35. The hSREBF1 gene on chromosome 17 harbors miR-33b in intron 17, whereas hSREBF2 on chromosome 22 contains miR-33a in intron 16. Mature miR-33a and miR-33b only differ in 2 nucleotides and are predicted to have largely overlapping target gene sets. Of note, rodents lack miR-33b in the corresponding intron in the Srebp1 gene35; the importance of this will be discussed later. In humans, miR-33a and miR-33b appear to be extensively co-expressed with their host genes in many cells and tissues34, 36, suggesting that they derive from the same primary transcript. This also pointed to a potential coordinated function of miR-33 isoforms and their host gene products. Indeed, several studies have now revealed extensive collaboration of miR-33a and miR-33b and the SREBP gene regulators in controlling cholesterol and lipid homeostasis.
miR-33a and SREBP-2 cooperate to control cholesterol homeostasis
Several recent studies reported the discovery of the miR-33a and SREBP-2 miRNA-host gene circuit, demonstrating that miR-33a functions in concert with the SREBP-2 cholesterogenic transcription factor to boost intracellular cholesterol levels31-35. MiR-33a and b were found to play a crucial role in the post-transcriptional repression of the ATP-binding cassette transporter (subfamily A, member 1) ABCA1, which promotes the efflux of free cholesterol from within the cell to ApoA1 and is critical for the formation of high-density lipoproteins (HDL)37. Plasma HDL levels show a strong inverse relationship with cardiovascular disease risk, and intensive efforts are ongoing to find effective pharmacological approaches to increase HDL levels for the treatment of cardiovascular disease38. ABCA1 is regulated by miR-33 isoforms through its 3’UTR, confirming ABCA1 as a direct miR-33 target. In accord with regulation of ABCA1 by miR-33, modulation of miR-33a levels also results in changes in cholesterol efflux in mouse macrophages32-35, suggesting that miR-33-targeting antisense approaches might promote RCT from atherogenic macrophages and decrease atherosclerosis. Importantly, in mouse models, hepatic and macrophage ABCA1 expression and circulating HDL levels increase upon inhibition or KO ablation of miR-33a32-35. Moreover, antisense inhibition of miR-33a in LDL-receptor KO mice, a well-validated model for hypercholesterolemia and cardiovascular disease, caused reduced high-fat diet-induced atherosclerotic plaque size and lipid content and promoted RCT, verifying the therapeutic potential of miR-33 targeting for the treatment of cardiovascular disease39. However, as noted above, rodents lack miR-33b, complicating the translation and relevance of these results to humans. This difference is important because expression of SREBP-1c (and miR-33b embedded within the SREBF1 gene) is highly upregulated upon insulin stimulation36. Under elevated circulating insulin conditions (such as in insulin resistance), increased SREBP-1c and miR-33b levels in the liver could thus potentially contribute to both high VLDL levels as well as low HDL levels found in individual suffering from metabolic syndrome40. Recent studies have indeed verified that hepatic miR-33b levels are elevated in non-human primates in response to a high-carbohydrate diet, and show that antisense oligonucleotides targeting miR-33a and b are effective in increasing HDL levels and lowering VLDL-triglycerides, paving the way for future human clinical trials to evaluate the safety and efficacy of antisense-targeting miR-33a and b for the treatment of cardiometabolic disorders41 (V.R., A.M.N., et al., unpublished data). It is interesting to note that ABCA1 is also targeted by other miRNAs, such as miR-758 and miR-106b which were also shown to modulate cellular cholesterol efflux42, 43, indicating that ABCA1 and HDL regulation by miRNAs is likely complex.
Other miR-33 targets involved in cholesterol, fatty acid and lipid homeostasis
The discovery of coordinated regulation of cholesterol levels by miR-33a miRNA and the SREBP-2 host gene product has prompted investigation into whether the SREBP host genes and their intronic miRNAs might act together more broadly to control not only cholesterol, but also fatty acid and lipid homeostasis in an integrated manner. Indeed, we and others have now found that miR-33a and b also regulate intracellular fatty acid and lipid levels in concert with their SREBP host gene products14, 31, 36.
First, it was shown that miR-33a and b directly control the expression of several proteins involved in fatty acid β-oxidation, the process by which fatty acids are degraded in mitochondria and peroxisomes to produce Acetyl-CoA destined for the citric acid cycle and ATP/energy generation14, 31, 36. These include carnitine O-octanoyltransferase (CROT) which helps to break down very long-chain fatty acids (>20 carbons) in peroxisomes, as well as carnitine palmitoyltransferase 1A (CPT1A), the rate-limiting transporter of fatty acids into mitochondria, and hydroxyacyl-coenzyme A-dehydrogenase (HADHB), a subunit of the trifunctional enzyme directly involved in mitochondrial fatty acid β-oxidation14, 31, 36.
In addition to regulating fatty acid degradation, miR-33a and b have been shown to control critical upstream regulators of fatty acid and lipid homeostasis. For example, the sirtuin SIRT6, an NAD+-dependent histone deacetylase which has been shown to be a crucial regulator of glucose metabolism and stress resistance44, 45,46, 47, was found to be targeted by miR-33a and b14, 36. Interestingly, a recent study of liver-specific SIRT6 knock-out mice revealed increased hepatic glycolysis, lipogenesis and triglyceride production, resulting in hepatosteatosis48. SIRT6 was shown to directly regulate SREBP-1 target genes relevant for fatty acid production such as acetyl-CoA carboxylase 1 (Acc1), stearoyl-CoA desaturase 1 (Scd1), and Fasn. This suggests that miR-33 inhibition of SIRT6 expression may result in increased chromatin acetylation and de-repression of SREBP-dependent fatty acid biosynthesis genes, and elevated lipogenesis.
The α1 subunit of the AMP-dependent kinase (AMPKα1) has also been found to be targeted by miR-33a and b14, 36, 41. AMPK is a master regulator of cellular energy levels in response to energy stress49. It responds to low cellular energy levels (increased AMP-to-ATP ratio) and acts to reduce energy-consuming processes such as protein, fatty acid and cholesterol synthesis, as well as activating mitochondrial biogenesis, fatty acid β-oxidation and glucose uptake to promote ATP synthesis. AMPK directly phosphorylates and deactivates several key SREBP lipogenic and cholesterogenic targets such as ACC1 and HMGCR. AMPK also inhibits SREBPs themselves both indirectly, through LXR 50, and through direct phosphorylation 51. Thus, negative regulation of AMPKα1 by miR-33a and b may relieve AMPK inhibition of both SREBPs and their target genes to coordinately boost intracellular levels of cholesterol, fatty acids, and other lipids.
Insulin receptor substrate 2 (IRS-2) was also recently shown to be a miR-33 target14, 36. IRS-2 is one of two intracellular adaptor proteins for the insulin receptor that relay insulin signaling to downstream effectors (such as phosphatidylinositol 3-kinase (PI3K) and Protein Kinase B (AKT)). In mouse models, reduction of IRS-2, but not IRS-1, has been found to increase SREBP-1c expression and activity 52-54. Interestingly, in mouse NAFLD models, decreased hepatic levels of IRS-2 has been shown to result in a compensatory elevation in IRS-1 levels, which in turn activates SREBP-155. Thus, miR-33 antisense targeting could potentially result in a reversal of the hepatic changes in IRS-2 and IRS-1 expression, thereby decreasing SREBP-1c levels or activity associated with hepatosteatosis.
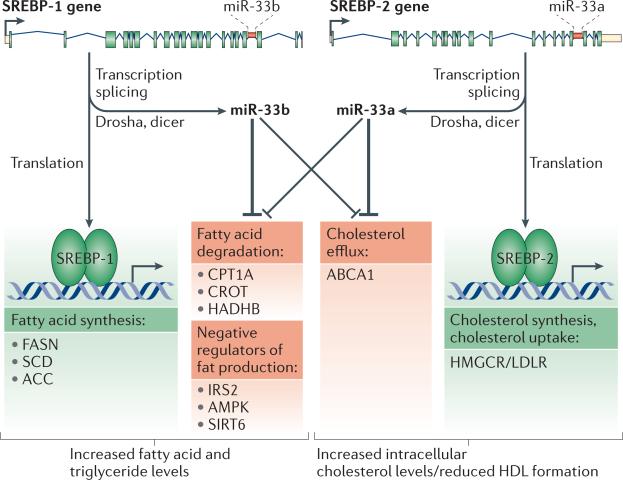
Taken together, these results reveal an extensive and integrated network of functional interactions between the SREBP transcription factors and their intronic miRNAs miR-33a and b to regulate cholesterol and lipid homeostasis (Figure 1).
Figure 1. Model of the SREBP and miR-33 circuit.
The sterol regulatory element-binding protein (SREBP) transcription factors act coordinately with their intronic miRNAs miR-33a and b to regulate fatty acid, triglyceride, and cholesterol homeostasis. Transcription of the SREBF1 and SREBF2 loci gives rise to the SREBP-1 and SREBP-2 transcription factors and the miR-33a/b miRNAs. SREBP-1 activates genes involved in fatty acid, phospholipid and triglyceride synthesis (e.g., FASN (fatty acid synthase), SCD (stearoyl-CoA desaturase) and ACC (Acetyl-CoA carboxylase)) whereas SREBP-2 activates genes involved in cholesterol synthesis/uptake, such as HMGCR (3-hydroxy-3-methylglutarylcoenzyme A reductase) and LDLR (Low-Density Lipoprotein Receptor). miR-33a and b act to repress genes functioning in fatty acid β-oxidation (e.g., CROT (carnitine O-octanoyltransferase), HADHB (hydroxyacyl– coenzyme A dehydrogenase/3-ketoacyl–coenzyme A thiolase/enoyl–coenzyme A hydratase [trifunctional protein], β subunit) and CPT1A (carnitine palmitoyltransferase 1A)), cholesterol efflux (e.g., ATP-binding cassette, subfamily A member 1 (ABCA1)), as well as negative regulators of SREBPs (e.g., IRS-2 (insulin receptor substrate 2), AMPKα1 (AMP-activated protein kinase α1 subunit) and SIRT6 (sirtuin 6)).
MiRNA regulation of insulin signalling and glucose homeostasis
A number of miRNAs have recently been implicated in controlling both insulin signalling and glucose metabolism at multiple levels56. Insulin is a hormone synthesized in and secreted by the pancreatic β-cells in response to increased nutrient levels (e.g. glucose) in the blood. It acts in concert with glucagon (a hormone produced by the pancreatic α-cells, with opposite functions to insulin) to maintain glucose homeostasis. Insulin secretion causes uptake of glucose in muscles and adipose cells (through glucose transporters such as GLUT4), blocks de novo production of glucose in the liver and increases storage of nutrients in form of fat, glycogen and protein57. Binding of insulin to the insulin receptor (INSR) promotes an intracellular signalling cascade involving the insulin receptor tyrosine kinase activity, the adaptor proteins IRS-1 and IRS-2, and a cascade of kinases including PI3K, 3-phosphoinositide-dependent protein kinase-1 (PDPK1) and AKT58. These kinases affect a complex downstream network of proteins such as glycogen synthase kinase 3β (GSK-3β), mammalian target of rapamycin complex 1 (mTORC1), and the FOXO family transcription factors59. Insulin also activates SREBF1 transcription and SREBP-1c processing and transport, stimulating lipogenesis60. Insulin resistance (a condition where tissues are no longer able to respond adequately to the action of insulin) is one of the most prevalent metabolic abnormalities in the developed world and is strongly associated with obesity, circulating cholesterol and lipid abnormalities, and NAFLD, all components of metabolic syndrome59. While the mechanisms underlying this complex metabolic disease remain to be fully elucidated, an increasing number of miRNAs have now been implicated in the control of insulin signalling and glucose homeostasis at all steps of regulation, from pancreatic islet development, β-cell differentiation, and insulin secretion, to target tissue insulin sensitivity and intracellular insulin signalling to downstream effectors (Table 1, Figure 2).
Table 1.
| miRNA | Target tissue(s) | Function | Target genes | References |
|---|---|---|---|---|
| miR-103/107 | Adipose, liver | Insulin/glucose homeostasis Adipogenesis | Cav1, Dicer | 81, 82, 85 |
| miR-122 | Liver | Hepatic lipid metabolism | CAT-1, ADAM17 | 20, 139, 140 |
| miR-124a | Pancreas | Pancreatic islet development | FoxA2, Rab27A | 66, 67 |
| miR-143 | Adipose Liver Pancreas | Adipocyte differentiation Insulin resistance | ERK5/BMK1/MAPK7 ORP8 | 77, 80, 94 |
| miR-223 | Muscle | Glucose uptake, insulin resistance | Glut4 | 75 |
| miR-27a | Adipose | Adipogenesis | [PPARγ, C/EBPα] | 141, 142 |
| miR-29 | Muscle, Adipose Liver | Glucose transport | Insig1, Cav2, Mct1, PIK3R1 | 70, 71, 73 |
| miR-33a/b | Liver Macrophage | Cholesterol/lipid/energy homeostasis | ABCA1, NPC1, CPT1A, HADHB, CROT, IRS2, SIRT6, AMPKα1 | 31-36, 39 |
| miR-335 | Pancreas Liver Adipose | Insulin production Fatty acid and TGA biosynthesis | Stxbp1 | 76, 79 |
| miR-34a | Liver, Pancreas | Lipid metabolism, B-cell exocytosis | SIRT1, VAMP2, ACSL1 | 88, 113, 143 |
| miR-375 | Pancreas | insulin secretion, pancreatic islet development | Mtpn, Usp1, Jak2, Adipor2, PDPK1 | 62-64 |
| miR-378/378* | Adipose | Adipocyte differentiation lipid synthesis | ERRγ, ? [ribosomal proteins] | 144-146 |
| miR-9 | Pancreas | Insulin secretion | Onecut2, Sirt1 | 68, 69 |
| miR-130 | Adipose | Adipogenesis | PPARγ | 147 |
| let-7 | Muscle Adipose | Insulin sensitivity | IGF-1R, INSR, IRS2 HMGA2 | 91, 148 |
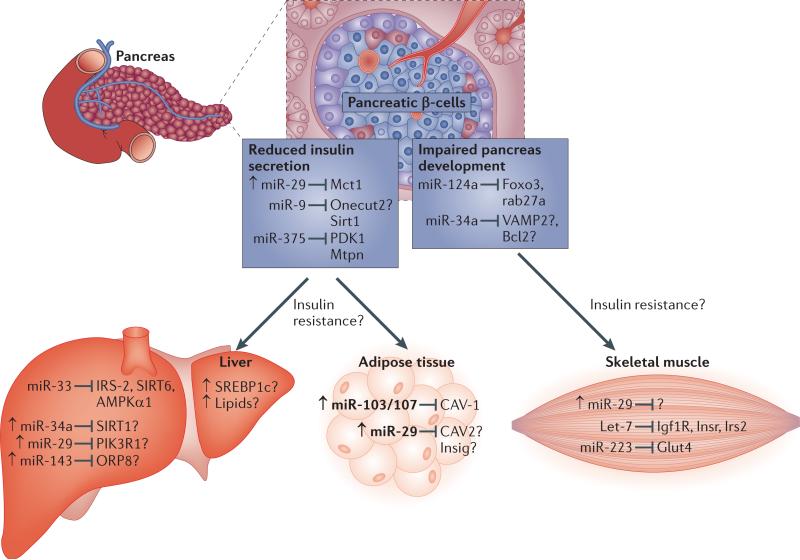
Figure 2. MiRNA regulation of insulin signalling and glucose homeostasis.
Normally, upon feeding, insulin is produced in the pancreatic β-cells and upon release will reach target tissues such as the muscle, liver and adipose to cause uptake of glucose, reduce the production of glucose and to activate fat production and storage. MiRNAs have been identified that affect diverse parts of insulin signalling in pancreas, liver, muscle and adipose tissue. miR-124a and miR-34a are involved in pancreatic development (through effects on Foxo2, Rhab27a, VAMP2, and Bcl2), whereas miR-29, miR-9 and miR-375 are involved in insulin secretion (through Mct1, Onecut2, Sirt1, PDK1, and Mtpn). miR-33, miR-34a, miR-29 and miR-143 act in the liver on targets involved in insulin signalling and its regulation (such as IRS-2, SIRT6, AMPKa1, SIRT1, PIK3R1 and ORP8). miR-103 and miR-107 and miR-29 act to modulate insulin signalling in adipose tissue (through CAV-1 and Insig1, respectively). miR-29, let-7 and miR-223 act in the muscle on insulin uptake (Glut4) and insulin signalling (Igf1R, Insr and Irs2). Known and predicted targets that lack in vivo evidence are marked with a question mark. In disease conditions such as impaired insulin secretion or insulin resistance, several miRNAs are upregulated (marked with arrow).
MiRNA regulation of pancreatic insulin production
Insulin is produced from pancreatic isletβ-cells, and the pancreatic miRNA miR-375 was shown to be required for pancreatic island development in zebrafish 61, and for the maintenance of pancreatic α and β-cell mass in mice 62. Interestingly, miR-375 may have a dual role as miR-375 was also found to decrease insulin exocytosis/secretion at least in part through the repression of myotrophin (Mtpn), a gene involved in actin depolymerization, and potentially, vesicular fusion63. It may additionally affect downstream insulin signalling through direct repression of the insulin signalling intermediate kinase PDPK164. The miR-124a miRNA is co-expressed with miR-375 and, at least in cultured cells, also targets Mtpn, suggesting coordinated control of Mtpn by several miRNAs65. MiR-124a is also involved in pancreatic islet development, potentially through regulation of the FoxA2 transcription factor involved in β-cell differentiation, as well as Rab27a, a GTPase required for insulin secretion66, 67. Other miRNA have also been implicated in controlling pancreatic insulin exocytosis. For example, miR-9 may regulate insulin secretion through its inhibition of the transcription factor one cut homeobox 2 (Onecut2) as well as through SIRT168, 69, whereas miR-29a and b, which are highly expressed in the pancreatic islets of diabetic mice, inhibit the expression of monocarboxylate transporter 1 (Mct1) and its function in insulin release70. Taken together, these findings indicate that miRNAs play key roles in controlling insulin secretion through effects on both pancreatic development and insulin exocytosis.
MiRNA modulation of insulin sensitivity, signalling and glucose metabolism in target tissues
Other miRNAs act in target tissues to regulate responses to insulin and glucose homeostasis. For example, miR-29a and b, which are upregulated in muscle, white adipose tissue, and liver of diabetic Goto-Kakizaki rats71, have been linked to insulin resistance, at least in cell culture experiments. The effects of miR-29a and b were suggested to be mediated through downregulation of proteins that promote insulin signalling such as Caveolin 2, a lipid raft-associated protein that responds to insulin levels72, as well as Insulin-induced gene 1 (Insig1), a negative regulator of SREBPs, and the insulin signalling intermediate PI3 kinase subunit p85α (PIK3R1)71, 73. Another layer of insulin regulation by miRNAs is provided by miR-126, which promotes insulin resistance through its inhibition of IRS-174. Intracellular glucose levels can also be directly regulated by miRNAs. For example, in skeletal muscle, miR-223 was found to inhibit glucose uptake through targeting of the glucose transporter GLUT475. As discussed above, miR-33a and b may also influence insulin signaling and glucose regulation by targeting IRS-2, SIRT6 and AMPKα114, 36. MiRNAs are thus acting to control insulin signalling and glucose uptake at multiple levels in target cells and tissues.
A number of miRNAs have also been implicated in metabolic disorders associated with aberrant insulin response, including obesity and NAFLD76-82. For example, the related miR-103 and miR-107 (which are located in introns in the pantothenate kinase 1-3 (PANK1-3) genes)83 were recently shown to be upregulated in livers of leptin-deficient (ob/ob) and diet-induced obese (DIO) mice81. Interestingly, antisense-mediated silencing of miR-103 and miR-107 improved insulin sensitivity and glucose homeostasis whereas overexpression (predominantly in adipose tissue) was sufficient to cause defects in glucose homeostasis in these mouse models. If confirmed in non-human primates and humans, this suggests that miR-103 and miR-107 may represent therapeutic targets to ameliorate obesity-associated insulin resistance84. These miRNAs were proposed to exert their functions on insulin and glucose regulation at least in part through the inhibition of caveolin-1 (CAV1), which influences lipid rafts and affects insulin receptor availability81. However, it is also possible that some of the effects are mediated through other miRNAs since miR-103 and miR-107 also strongly inhibit the miRNA-processing enzyme Dicer85. Indeed, partial knockdown of Dicer or pancreas-specific knockout experiments have revealed that this enzyme (and, presumably, miRNAs) is required for both the development and maintenance of the pancreas and insulin signalling86, 87.
Several additional miRNAs have been found to be elevated in obesity models and exert regulatory effects on insulin signalling and glucose homeostasis. Similar to miR-103 and miR-107, miR-143 is overexpressed in the obese leptin receptor-mutant mice (db/db) and DIO mice77. Overexpression of miR-143 reduces insulin sensitivity, presumably through its target oxysterol-binding protein-related protein 8 (ORP8), a protein involved in AKT activation. MiR-34a, a miRNA that may play important roles in cancer development through its function in a network with SIRT1 and p53, has additionally been implicated in diabetes as a target to prevent pancreatic β-cell death88-90. Finally, several members of the let-7 miRNA family have been found to be upregulated in ob/ob and DIO mice81. let-7 has well-studied tumor suppressor roles in cancer biology but was recently also shown to be involved in regulation of glucose metabolism, where its overexpression in skeletal muscle resulted in insulin resistance and impaired glucose tolerance91. This effect may be at least partially mediated by let-7 targeting of the insulin-like growth factor receptor 1 (IGF-1R), INSR, and IRS-2.
In conclusion, a number of miRNAs have been suggested to affect multiple aspects of insulin signalling and glucose homeostasis, from pancreatic insulin production to insulin sensitivity and regulation of glucose uptake in target tissues. However, much work remains to elucidate the in vivo contribution of specific miRNAs to prevalent pathological conditions such as obesity-associated insulin resistance, and to evaluate their potential for therapeutic targeting.
MiRNAs in adipogenesis, obesity, and non-alcoholic fatty liver disease
Conditions of nutritional excess and lack of physical activity can result in excessive fat storage manifesting in obesity and non-alcoholic fatty liver disease, and strongly increase the risk for cardiovascular disease and type 2 diabetes. Normal adipogenesis involves regulation by transcriptional factors such as the PPARγ and its coactivators (e.g peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α)), as well as members of the C/EBP family and Kruppel-like factors (KLFs) (reviewed in 92). MiRNAs have also recently been implicated in adipogenesis93. For instance, miR-143 has been shown to be important in adipocyte differentiation of cultured mouse 3T3-L1 pre-adipocytes, perhaps through its regulation of ERK594. Several others, including miR-204, miR-141, miR-200a-c, and miR-429, are involved in early adipocyte cell fate determination, whereas miR-17-92, miR-130, miR-27a and b, and miR-378 and 378* have been suggested to be involved in terminal differentiation and mature white adipocyte function (recently reviewed in 95). Brown adipose tissue (BAT), a highly metabolically active adipose tissue type involved in thermogenesis and energy expenditure, has been suggested to correlate inversely with obesity96. A recent study reported a key role for the miR-193b-365 miRNA cluster in brown fat differentiation, in part by repressing myogenesis97. The accumulating data thus indicate that miRNAs are acting as central modulators of normal white adipose tissue (WAT) and BAT differentiation and biology. A growing number of studies have also found obesity-associated alterations in expression levels of miRNAs in WAT82, 98-100. For example, Xie et al. showed that certain miRNAs involved in adipogenesis exhibit inverse expression patterns in obesity82. While many miRNAs have been linked to adipogenesis in vitro, or have altered expression in WAT in obese individuals and in animal obesity models, evidence for a causative role in obesity-related diseases is still lacking, and much additional detailed in vivo work is needed to clarify the regulatory roles of individual miRNAs in modulating energy balance and adipose biology, and their potential contribution to obesity.
The hepatic miR-34a and SIRT1 regulatory circuit
As mentioned above, several studies have measured miRNA expression in metabolic tissues in rodent models of obesity, type 2 diabetes and NAFLD78, 81, 82. Interestingly, a number of miRNAs that were dysregulated in these models were also changed in human patients with obesity-related NAFLD and NASH 81, 101-103 Among these, miR-34a may play a particularly relevant role in hepatic metabolic diseases. Both NAFLD and NASH patients exhibit highly elevated hepatic expression of miR-34a101, 102, and miR-34a levels were also higher in type 2 diabetic subjects as compared to healthy controls103. A plausible molecular mechanism for the function(s) of miR-34a in metabolic control has recently been suggested (reviewed in 104, summarized in Figure 3). This involves an intricate regulatory network of miR-34a, SIRT1, Farnesoid X receptor (FXR) and tumor protein 53 (p53). SIRT1 is a key sensor and regulator of metabolic states since it responds to NAD+ levels in the cell and directly deacetylates and modulates both histone and non-histone targets, including important metabolic regulators such as PGC-1α, PPARs, p53, FOXO1, LXR, FXR and SREBPs to alter the expression of transcriptional programs governing cholesterol, lipid and energy homeostasis105-112. SIRT1 was recently shown to be regulated by miR-34a, causing p53-dependent apoptosis in human colon cancer cells113. Subsequent work showed that miR-34a also inhibits SIRT1 expression in the liver114. The current data suggest that miR-34a, SIRT1, p53 and FXR constitute a regulatory loop: miR-34a inhibits SIRT1 and is itself regulated by SIRT1 through several mechanisms. First, SIRT1 directly inactivates the miR-34a promoter through histone deacetylation. Second, SIRT1 deacetylates and inactivates p53, a transcriptional activator of miR-34a115-118. Finally, SIRT1 deacetylates FXR, which then activates the liver repressor Small Heterodimer Partner (SHP). SHP sequesters p53 away from the promoter of miR-34a, again reducing miR-34a levels105, 114. Low hepatic levels of SIRT1 and high levels of miR-34a are associated with fatty liver disease. We speculate that miR-34a regulation of SIRT1 could affect other key metabolic targets of SIRT1, such as SREBPs, PPARs, PGC-1α, LXR, and FOXO1, potentially further contributing to hepatosteatosis (Figure 3). A crucial next step is to investigate the effect of antagonizing elevated miR-34a levels in models for NAFLD and obesity on SIRT1 expression, and deacetylation of SIRT1 downstream targets. SIRT1 activators, such as resveratrol and other more potent synthetic inducers, improve hepatosteatosis and insulin resistance in DIO mice111, 119-121. If verified in vivo, activation of SIRT1 through antisense inhibition of miR-34a could potentially represent an alternative therapeutic avenue for the treatment of NAFLD and other obesity-related diseases.
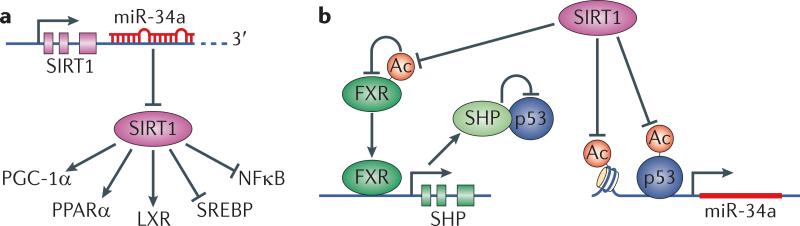
Figure 3. The regulatory loop of miR-34a, SIRT1, FXR and p53.
miR-34a is highly expressed in patients with NAFLD and NASH and type 2 diabetes. On the molecular level, miR-34a has been shown to exert its function through its effect on SIRT1. miR-34a is then in turn inhibited by SIRT1 in a regulatory loop that includes miR-34a, SIRT1, FXR and p53 to affect cholesterol, lipid and energy homeostasis as well as inflammation. A. miR-34a inhibits SIRT1 and reduces its protein level and prevents its activation of PGC-1α, PPARα and LXR (key regulators of cholesterol/lipid/energy homeostasis), and inhibition of SREBP and NF-κB (activators of lipogenesis and cholesterogenesis, and inflammation, respectively). B. SIRT1 feedback inhibits miR-34a in several ways: it deacetylates p53 and inhibits p53-dependent transcriptional activation of miR-34a. In addition, SIRT1 inhibits the miR-34a promoter through histone deacetylation. Finally, SIRT1 deacetylates and activates FXR. FXR transcriptionally activates SHP, which sequesters p53 and thus inhibits miR-34a transcription.
Circulating miRNAs as disease biomarkers and “endocrine signals”
Recent studies have found that miRNAs can be readily detected in human plasma, suggesting possibilities for novel disease biomarker discovery (reviewed in 122 and 123), as well as the intriguing notion that they may serve regulatory purposes (endocrine-miRs) in target cells. Plasma fractionation schemes initially isolated circulating miRNAs as part of microvesicles and exosomes derived from plasma membranes of “donor” cells124.
Subsequent findings indicate that miRNAs can also be found in lipid-free protein assemblies, such as in association with the miRNA-processing enzyme Ago2125-127. The discovery of circulating miRNAs represents an important advance for biomarker discovery, and altered circulating miRNA profiles have already been linked to disease states, including NAFLD, NASH and hepatic injury (miR-122)102, atherosclerosis (miR-223)128, type 2 diabetes (miR-126)129 and hypertension (let-7e)130. It is likely that additional circulating miRNAs will be identified as biomarkers for metabolic diseases, and this represents a rapidly evolving area of research.
Endocrine miRNAs?
Interestingly, circulating miRNAs have been found to traffic from donor cells to distant sites and alter the gene expression output of recipient cells131. This suggests that circulating miRNAs are not merely “vesicular passengers”, but may rather act as signaling molecules in an endocrine or paracrine fashion to deliver a regulatory “message” from donor to target cells. By contrast with the highly specific and potent action of classical endocrine hormones such as steroids132, circulating miRNAs may have only modest effects on mRNA and protein output. Similar to mRNAs taken up by cells from microvesicles133, miRNAs presumably do not require specific receptors for their action and are therefore only limited by the fact that the mRNA needs to be expressed in target cells for the miRNA to have an impact, and it is unclear whether they are regulated by intercellular feedback mechanisms. Nevertheless, this intriguing notion should be thoroughly investigated. Could specific classes of miRNAs be enriched in different types of vesicles and lipoprotein particles, with distinct cellular targeting potential depending on surface lipid and protein content, thus allowing selective messaging to specific organs or cell types? If so, might specific miRNAs highly abundant in certain vesicles and lipoprotein particles exert “mass action” effects on gene expression programs in target cells or tissues, thereby acting in a more potent manner akin to classic endocrine cues?
MiRNAs in HDL
An exciting new study revealed that miRNAs, such as miR-375 and miR-223, can also be found in HDL particles, suggesting new potential roles for HDL, and perhaps other lipoprotein assemblies, in transporting not only cholesterol and other lipids and proteins, but also RNA cargo conveying specific regulatory information128. Vickers et al. found that certain endogenous miRNAs (e.g., miR-135a*, miR-375 and miR-223) are exported from peripheral cells in HDL particles128. Export of miRNAs to HDL was shown to be controlled by neutral sphingomyelinase II (nSMase2), the rate-limiting enzyme in ceramide biosynthesis, suggesting that plasma membrane lipids such as sphingomyelin (SM) and ceramides are involved in controlling miRNA incorporation into HDL.
Once miRNAs are embedded in HDL, they are likely trafficked to the liver by the classic reverse cholesterol transport pathway for uptake in a scavenger receptor B type I (SR-BI)-dependent manner, and may exert effects on hepatic gene expression programs (Figure 4). Consistent with this notion, Vickers et al. showed that treatment of hepatic Huh-7 cells with HDL loaded with exogenous miRNAs (e.g., miR-223) could indeed impact target mRNA expression128. Interestingly, they also found that HDL miRNA profiles differ between normal subjects and atherosclerotic individuals, both in mice and humans. This warrants follow-up studies to determine whether circulating miRNAs associated with HDL in heart disease patients could participate in the disease process, or whether specific miRNAs associated with HDL might constitute part of the protective function against atherosclerosis ascribed to HDL. Finally, plasma HDL-miRNA profiling might serve as a novel biomarker tool for detecting or monitoring cardiovascular disease progression.
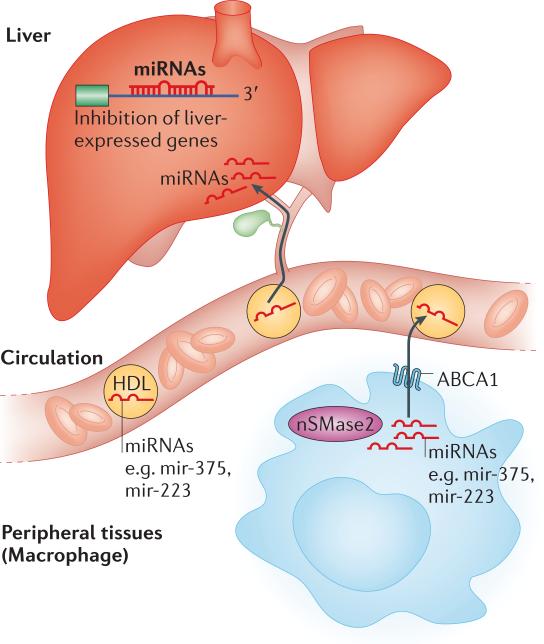
Figure 4. Model for the function of circulating miRNAs associated with HDL.
MiRNAs such as miR-375 and miR-223 are produced in peripheral tissues and incorporated in exosomes through a mechanism controlled by nSMase2, the rate limiting enzyme in ceramide biosynthesis, and transported in the blood. Some miRNAs are also bound to high-density lipoproteins (HDL), incorporation of microRNAs into HDL is controlled by the cholesterol transporter ABCA and by nSMase. Uptake of RNA and miRNAs from exosomes in target cells does not require binding to a specific receptor. MiRNAs associated with HDL are thought to be trafficked to the liver through the reverse cholesterol transport (RCT) pathway and taken up by a scavenger receptor B I-dependent mechanism. Once inside target cells, miRNAs can then exert inhibitory effects on a range of target genes.
Concluding remarks
MiRNAs have recently come to the fore as critical regulators of numerous aspects of animal development, physiology, and disease. As discussed in this review, a rapidly growing number of miRNAs have been implicated in regulation of genes and proteins involved in the control and maintenance of metabolic homeostasis. While the best-characterized roles for miRNAs in metabolic control concerns the normal maintenance of cholesterol and lipid homeostasis (e.g., miR-122 and miR-33), new data has also revealed contributions of miRNAs to insulin signaling and glucose homeostasis, as well as the potential pathological roles of aberrant miRNA expression in cardiometabolic disorders such as obesity, NAFLD, insulin resistance, type 2 diabetes, and coronary artery disease. The exciting finding that miRNAs can be found circulating in the blood not only points to their potential use as biomarkers for disease states, but also suggests that circulating miRNAs may be actively secreted from donor cells and tissues to act as signaling molecules with impact on the gene expression output in distant target organs and cells (e.g., liver in the case of HDL-associated miRNAs), akin to classical endocrine cues.
MiRNA therapeutics for the treatment of metabolic disorders
The emergence of miRNAs as important regulators of metabolism has garnered much interest not only from a scientific point of view, but also from a clinical perspective, with the potential contribution of aberrant miRNA expression and therapeutic implications for the treatment of cardiometabolic diseases raising considerable excitement. Therapeutic efforts to treat metabolic disorders have traditionally centered on inhibition of what is perceived as “druggable” targets, such as enzymes (e.g., HMG-CoA reductase targeted by cholesterol-lowering statins). However, the recent discovery by us and others that certain miRNAs may represent critical regulators of metabolism has provided important insights into metabolic regulatory networks that are already yielding novel types of therapeutic targets and strategies for the treatment of metabolic diseases. For example, the central and concerted role of miR-33a and b and their SREBP host genes in controlling metabolic homeostasis, together with promising data from mice and non-human primate models suggest that antisense targeting of miR-33a and b for the treatment of metabolic syndrome and cardiovascular disease should be explored further. It is likely that other miRNAs with regulatory roles in human metabolism (e.g., miR-103 and miR-107 in insulin resistance) may also serve as suitable candidates for therapeutic intervention.
Future directions and open questions
A number of open questions remain concerning the function of miRNAs in metabolic control (and more broadly). For example, while it is apparent that several miRNAs, including miR-33a and b, have important regulatory impact on cholesterol and lipid metabolism in vivo by themselves, it is unclear how widespread the contribution of individual miRNAs is to metabolic control. MiRNAs typically have rather modest effects on target protein levels, and, as is the case for miR-33, combinatorial action on multiple functionally related targets in linear pathways or key nodes in regulatory networks is probably required for single miRNAs to significantly influence a complex cellular and organismal process such as metabolic homeostasis. As mRNAs have multiple predicted miRNA target sites, it is likely that groups of miRNAs also act in concert to exert more potent effects on specific target mRNAs. New experimental strategies employing systems biology methodologies may be required to determine composition and regulation of such putative miRNA networks, and to elucidate how miRNA networks are coordinated with other regulatory systems (e.g., transcriptional and translational control circuits). Akin to the restrictions imposed by complex chromatin architectures on accessibility of DNA sequences to regulatory factors, could secondary and tertiary structural features of 3’UTR folding govern miRNA efficacy? Are other regulators that bind mRNAs, including proteins such as HuR and other non-coding RNAs, working together or in opposition to miRNAs in controlling mRNA folding and accessibility, stability, and translational output? Additionally, individual miRNAs typically have many targets, which gives rise to the question of whether certain miRNAs may coordinately control metabolism with other cellular and organismal processes, such as cell growth and proliferation, cellular differentiation, or organ growth? Additionally, given the likelihood that many miRNAs may regulate targets involved in multiple cellular processes, what are the possible long-term consequences of miRNA-directed therapeutics? These and many other unanswered questions concerning miRNA biology and impact on metabolic homeostasis will undoubtedly be topics for intensive and exciting research for years to come.
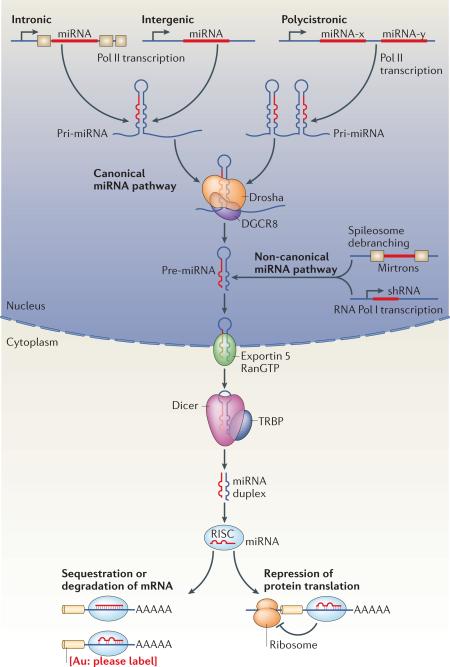
Box 1: The miRNA biogenesis pathway.
MiRNAs generated by the canonical biogenesis pathway are transcribed as precursor RNAs from intergenic, intronic, or polycistronic genomic loci by RNA polymerase II6. The primary miRNA (pri-miRNA) transcript forms a stem-loop structure that is recognized and processed by the complex of RNase III, Drosha and DGCR8 or the spliceosome apparatus in the nucleus. In the non-canonical miRNA pathway, miRNAs are transcribed directly as short endogenous hairpin RNAs (shRNAs) or derive directly through splicing from introns that can refold into hairpins (mirtrons) (reviewed in 134). The trimmed precursor (pre-miRNA) hairpins from both canonical and non-canonical miRNA pathways are then transported by an Exportin-5 and Ran-GTP-dependent process to the cytosol, where they are typically further processed by the enzyme complex of RNase III, Dicer and TRBP to form the mature double-stranded ~22 nucleotide miRNA. Argonaut proteins (e.g., Ago2) then unwind the miRNA duplex and facilitate incorporation of the miRNA-targeting strand (also known as guide strand) into the Argonaut-containing RNA-induced silencing complex (RISC). The RISC-miRNA assembly is then guided to specific target sequences in mRNAs. The initial recognition of mRNAs by the RISC-miRNA complex is driven primarily by Watson-Crick base-pairing of nucleotides 2-8 in the mature miRNA (termed the seed sequence) with specific mRNA target sequences chiefly located in the 3’UTR, with additional base-pairing affording greater affinity and targeting efficiency 7.
Box 2: miR-122 and its function is hepatitis C virus propagation.
Apart from its function in metabolic control miR-122 has also been shown to promote the propagation of hepatitis C virus (HCV) by multiple mechanisms28-30. In addition to binding the HCV 5’UTR to facilitate proper folding of the HCV RNA for translation and replication, miR-122-dependent increase in cholesterol and lipid synthesis may stimulate the production of endoplasmic reticulum-associated lipid droplets and cholesterol and lipid-rich membrane domains (termed “lipid rafts”) that are central sites of viral replication and production135. Hence, miR-122 antagonism could inhibit HCV both by preventing direct enhancement of viral RNA folding or replication as well as by lowering required cholesterol or lipid co-factors for viral packaging or extrusion, suggesting that antisense-based targeting of miR-122 could represent a powerful approach to treat chronic HCV infections which plague up to 3% of the world's population. Accordingly, weekly subcutaneous injections of miR-122-targeting LNA-based antisense oligonucleotides (miravirsen) have been found efficacious in decreasing HCV titers in chimpanzees136, and early indications from Phase II studies in humans with chronic HCV infection appear promising137. Indeed, a four-week therapy with miravirsen in treatment-naïve patients with chronic HCV genotype 1 infection provides long-lasting suppression of viremia that was maintained for more than four weeks after the last dose, did not reveal viral resistance, and was well tolerated. These studies also showed lowered total cholesterol, and further assessment of miravirsen in humans should provide important information as to whether miR-122 therapeutic targeting might have a positive impact on circulating cholesterol/lipid abnormalities linked to metabolic syndrome. HCV infection is associated with NAFLD101, and inhibition of viral propagation by miR-122-targeting antisense approaches might also ameliorate HCV-associated fatty liver disease. On the other hand, the emerging link of miR-122 to hepatocyte differentiation or cell fate determination138 may also suggest that attention should be paid to potential deleterious long-term effects, such as the development of hepatocellular carcinoma26.
“At a glance” online summary.
MiRNAs such as miR-33a/b are key regulators of cholesterol/lipid homeostasis, and represent attractive therapeutic target to raise HDL and lower triglycerides.
A role for miRNAs (e.g., miR-103 and miR-107, and let-7) in regulation of insulin signaling and control of glucose homeostasis has recently been elucidated.
An emerging link of certain miRNAs to metabolic dysregulation in adipogenesis and obesity suggests that this class of non-coding RNAs may play important roles in disorders associated with metabolic syndrome (MetS).
Aberrant hepatic miRNA expression may also contribute to other aspects of MetS, such as non-alcoholic fatty liver disease (NAFLD).
Circulating miRNAs have recently been identified in the blood, including as part of HDL. Data suggest that circulating miRNAs may exert effects on gene expression in target cells and tissues.
The link of abnormal miRNA expression to metabolic disorders has highlighted the therapeutic potential of antisense targeting of specific microRNAs (e.g., miR-122 and miR-33a/b).
Glossary terms
- Antagomir
a small synthetic cholesterol-conjugated DNA oligonucleotide that is complementary to an endogenous miRNA of interest. The cholesterol moiety allows antagomirs to enter most cell types efficiently, where they specifically bind and sequester endogenous miRNAs.
- LNA
Locked Nucleic Acid, a modified DNA oligonucleotide analogue, locked in a “N-type confirmation” capable of recognizing DNA and RNA with high affinity and is resistant to degradation.
- Metabolic syndrome (MetS)
a combination of metabolic disorders characterized by insulin resistance, obesity, abnormalities in circulating cholesterol and lipid profiles, non-alcoholic fatty liver disease, and hypertension. MetS is associated with increased risk of type 2 diabetes and cardiovascular disease (coronary artery disease and stroke).
- Lipid rafts
microdomains of the cell plasma membrane with high cholesterol and sphingolipid content that compartmentalize cellular processes by acting as platforms to co-localize proteins such as signalling molecules and receptors.
- Fatty acid β-oxidation
degradation process of fatty acids to acetyl-CoA primarily in mitochondria. Long chain fatty acids need to be transported via binding to carnitine.
- Triglycerides
esters of glycerol and fatty acids, used for the storage of energy.
- Atherogenic macrophages
cholesterol-loaded macrophages that accumulate in the arterial wall and can lead to atherosclerotic lesions.
- Exosome
small (30-90 nm) vesicle secreted from the plasma membrane of mammalian cells. They contain proteins, RNA and miRNA molecules and can be transported from cell to cell.
- Endocrine/paracrine
both are hormonal signals, secreted from a specific cell and causing a specific effect. Endocrine responses occur over large distances in distant target, paracrine signals have only local effects.
Biography
Anders M. Näär received his PhD at the University of California, San Diego, School of Medicine, Howard Hughes Medical Institute, USA, with Michael G. Rosenfeld, and performed his postdoctoral research at the University of California, Berkeley, Howard Hughes Medical Institute, USA, with Robert Tjian. He is currently an associate professor in the Department of Cell Biology at Harvard Medical School and the Massachusetts General Hospital Cancer Center, where his laboratory studies mechanisms of gene regulation.
Veerle Rottiers did her graduate studies with Adam Antebi at the Max Planck Institute for Molecular Genetics in Berlin, Germany, working on C. elegans sterol/lipid signaling pathways governing worm development, and received her PhD from Ghent University, Belgium. She is currently a postdoctoral research fellow in the laboratory of Anders M. Näär at Harvard Medical School and the Massachusetts General Hospital Cancer Center, USA, studying transcriptional and microRNA control of cholesterol/lipids.
References
- 1.Lehrke M, Lazar MA. The many faces of PPARgamma. Cell. 2005;123:993–9. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 2.Zelcer N, Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116:607–14. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–31. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakae J, Oki M, Cao Y. The FoxO transcription factors and metabolic regulation. FEBS Lett. 2008;582:54–67. doi: 10.1016/j.febslet.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 5.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–10. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–87. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 9.Fabian MR, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–80. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 13.Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 2007;8:R166. doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rottiers V, et al. MicroRNAs in Metabolism and Metabolic Diseases. Cold Spring Harbor Symposia on Quantitative Biology. 2011;76 doi: 10.1101/sqb.2011.76.011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–67. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raghow R, Yellaturu C, Deng X, Park EA, Elam MB. SREBPs: the crossroads of physiological and pathological lipid homeostasis. Trends Endocrinol Metab. 2008;19:65–73. doi: 10.1016/j.tem.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Cornier MA, et al. The metabolic syndrome. Endocr Rev. 2008;29:777–822. doi: 10.1210/er.2008-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angulo P. Obesity and nonalcoholic fatty liver disease. Nutr Rev. 2007;65:S57–63. doi: 10.1111/j.1753-4887.2007.tb00329.x. [DOI] [PubMed] [Google Scholar]
- 19.Lewis AP, Jopling CL. Regulation and biological function of the liver-specific miR-122. Biochem Soc Trans. 2010;38:1553–7. doi: 10.1042/BST0381553. [DOI] [PubMed] [Google Scholar]
- 20.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [This paper presents one of two pioneering studies utilizing antisense approaches to elucidate in vivo roles of a miRNA (miR-122).] [DOI] [PubMed] [Google Scholar]
- 21.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [This paper presents one of two pioneering studies utilizing antisense approaches to elucidate in vivo roles of a miRNA (miR-122).] [DOI] [PubMed] [Google Scholar]
- 22.Veedu RN, Wengel J. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol. 2009;6:321–3. doi: 10.4161/rna.6.3.8807. [DOI] [PubMed] [Google Scholar]
- 23.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [In this paper, locked nucleic acid (LNA)-antisense oligonucleotides were used to inhibit miR-122 in the livers of non-human primates, the first study of miRNAs in a primate model.] [DOI] [PubMed] [Google Scholar]
- 24.Elmen J, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–62. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid Res. 2010;51:1513–23. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–36. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai S, et al. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–27. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norman KL, Sarnow P. Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J Virol. 2010;84:666–70. doi: 10.1128/JVI.01156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–29. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerin I, et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J Biol Chem. 2010;285:33652–61. doi: 10.1074/jbc.M110.152090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horie T, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–6. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–32. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–9. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–3. doi: 10.1126/science.1189862. [These five papers published around the same time identify the miR-33a/b miRNAs within introns in the SREBP genes, acting in concert with their host genes as critical regulators of cholesterol/lipid homeostasis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davalos A, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–7. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang C, Oram JF. The cell cholesterol exporter ABCA1 as a protector from cardiovascular disease and diabetes. Biochim Biophys Acta. 2009;1791:563–72. doi: 10.1016/j.bbalip.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Rye KA, Bursill CA, Lambert G, Tabet F, Barter PJ. The metabolism and anti-atherogenic properties of HDL. J Lipid Res. 2009;50(Suppl):S195–200. doi: 10.1194/jlr.R800034-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rayner KJ, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011 doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown MS, Ye J, Goldstein JL. Medicine. HDL miR-ed down by SREBP introns. Science. 2010;328:1495–6. doi: 10.1126/science.1192409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rayner KJ, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–7. doi: 10.1038/nature10486. [This paper shows the therapeutic promise of antisense targeting of miR-33a/b to raise HDL and lower triglycerides in a non-human primate model.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramirez CM, et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol. 2011;31:2707–14. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J, et al. miR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp Neurol. 2011 doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–29. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 45.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–6. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao C, et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem. 2010;285:36776–84. doi: 10.1074/jbc.M110.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong L, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–93. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HS, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–36. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardie DG. AMP-activated protein kinase: a cellular energy sensor with a key role in metabolic disorders and in cancer. Biochem Soc Trans. 2011;39:1–13. doi: 10.1042/BST0390001. [DOI] [PubMed] [Google Scholar]
- 50.Yap F, Craddock L, Yang J. Mechanism of AMPK suppression of LXR-dependent Srebp-1c transcription. Int J Biol Sci. 2011;7:645–50. doi: 10.7150/ijbs.7.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–88. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsumoto M, et al. Role of the insulin receptor substrate 1 and phosphatidylinositol 3-kinase signaling pathway in insulin-induced expression of sterol regulatory element binding protein 1c and glucokinase genes in rat hepatocytes. Diabetes. 2002;51:1672–80. doi: 10.2337/diabetes.51.6.1672. [DOI] [PubMed] [Google Scholar]
- 53.Taniguchi CM, Ueki K, Kahn R. Complementary roles of IRS-1 and IRS-2 in the hepatic regulation of metabolism. J Clin Invest. 2005;115:718–27. doi: 10.1172/JCI23187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Tobe K, et al. Increased expression of the sterol regulatory element-binding protein-1 gene in insulin receptor substrate-2(−/−) mouse liver. J Biol Chem. 2001;276:38337–40. doi: 10.1074/jbc.C100160200. [DOI] [PubMed] [Google Scholar]
- 55.Kohjima M, et al. SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease. Int J Mol Med. 2008;21:507–11. [PubMed] [Google Scholar]
- 56.Guay C, Roggli E, Nesca V, Jacovetti C, Regazzi R. Diabetes mellitus, a microRNA-related disease? Transl Res. 2011;157:253–64. doi: 10.1016/j.trsl.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 57.Herman MA, Kahn BB. Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J Clin Invest. 2006;116:1767–75. doi: 10.1172/JCI29027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siddle K. Signalling by insulin and IGF receptors: supporting acts and new players. J Mol Endocrinol. 2011;47:R1–10. doi: 10.1530/JME-11-0022. [DOI] [PubMed] [Google Scholar]
- 59.Leavens KF, Birnbaum MJ. Insulin signaling to hepatic lipid metabolism in health and disease. Crit Rev Biochem Mol Biol. 2011;46:200–15. doi: 10.3109/10409238.2011.562481. [DOI] [PubMed] [Google Scholar]
- 60.Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 2009;23:2578–91. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poy MN, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813–8. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poy MN, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–30. doi: 10.1038/nature03076. [In this paper, the authors provide the first evidence for miRNA regulation of insulin secretion, suggesting that miRNAs may represent important regulators of glucose homeostasis.] [DOI] [PubMed] [Google Scholar]
- 64.El Ouaamari A, et al. miR-375 targets 3'-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–17. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baroukh NN, Van Obberghen E. Function of microRNA-375 and microRNA-124a in pancreas and brain. Febs J. 2009;276:6509–21. doi: 10.1111/j.1742-4658.2009.07353.x. [DOI] [PubMed] [Google Scholar]
- 66.Baroukh N, et al. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem. 2007;282:19575–88. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- 67.Lovis P, Gattesco S, Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 2008;389:305–12. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- 68.Ramachandran D, et al. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic beta-islets. Febs J. 2011;278:1167–74. doi: 10.1111/j.1742-4658.2011.08042.x. [DOI] [PubMed] [Google Scholar]
- 69.Plaisance V, et al. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem. 2006;281:26932–42. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 70.Pullen TJ, da Silva Xavier G, Kelsey G, Rutter GA. miR-29a and miR-29b contribute to pancreatic beta-cell-specific silencing of monocarboxylate transporter 1 (Mct1). Mol Cell Biol. 2011;31:3182–94. doi: 10.1128/MCB.01433-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol. 2007;21:2785–94. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 72.Kim S, Pak Y. Caveolin-2 regulation of the cell cycle in response to insulin in Hirc-B fibroblast cells. Biochem Biophys Res Commun. 2005;330:88–96. doi: 10.1016/j.bbrc.2005.02.130. [DOI] [PubMed] [Google Scholar]
- 73.Pandey AK, et al. miR-29a levels are elevated in the db/db mice liver and its overexpression leads to attenuation of insulin action on PEPCK gene expression in HepG2 cells. Mol Cell Endocrinol. 2011;332:125–33. doi: 10.1016/j.mce.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 74.Ryu HS, Park SY, Ma D, Zhang J, Lee W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS One. 2011;6:e17343. doi: 10.1371/journal.pone.0017343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res. 2010;86:410–20. doi: 10.1093/cvr/cvq010. [DOI] [PubMed] [Google Scholar]
- 76.Esguerra JL, Bolmeson C, Cilio CM, Eliasson L. Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One. 2011;6:e18613. doi: 10.1371/journal.pone.0018613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jordan SD, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;13:434–46. doi: 10.1038/ncb2211. [This is one of three papers linking aberrant expression of a miRNA in obesity-related disorders to impaired insulin signaling and glucose homeostasis, providing important evidence that dysregulation of miRNA expression may contribute to human metabolic diseases.] [DOI] [PubMed] [Google Scholar]
- 78.Li S, et al. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J Lipid Res. 2009;50:1756–65. doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakanishi N, et al. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun. 2009;385:492–6. doi: 10.1016/j.bbrc.2009.05.058. [DOI] [PubMed] [Google Scholar]
- 80.Takanabe R, et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun. 2008;376:728–32. doi: 10.1016/j.bbrc.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 81.Trajkovski M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–53. doi: 10.1038/nature10112. [This is one of three papers linking aberrant expression of a miRNA in obesity-related disorders to impaired insulin signaling and glucose homeostasis, providing important evidence that dysregulation of miRNA expression may contribute to human metabolic diseases.] [DOI] [PubMed] [Google Scholar]
- 82.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–7. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilfred BR, Wang WX, Nelson PT. Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab. 2007;91:209–17. doi: 10.1016/j.ymgme.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Naar AM. MiRs with a sweet tooth. Cell Metab. 2011;14:149–50. doi: 10.1016/j.cmet.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martello G, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 86.Morita S, et al. Dicer is required for maintaining adult pancreas. PLoS One. 2009;4:e4212. doi: 10.1371/journal.pone.0004212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lynn FC, et al. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–45. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 88.Lovis P, et al. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes. 2008;57:2728–36. doi: 10.2337/db07-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roggli E, et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes. 2010;59:978–86. doi: 10.2337/db09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–5. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 91.Zhu H, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [This is one of three papers linking aberrant expression of a miRNA in obesity-related disorders to impaired insulin signaling and glucose homeostasis, providing important evidence that dysregulation of miRNA expression may contribute to human metabolic diseases.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20:107–14. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 93.Alexander R, Lodish H, Sun L. MicroRNAs in adipogenesis and as therapeutic targets for obesity. Expert Opin Ther Targets. 2011;15:623–36. doi: 10.1517/14728222.2011.561317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Esau C, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–5. doi: 10.1074/jbc.C400438200. [This is the first paper linking a miRNA to the differentiation and function of a metabolically important tissue, highlighting the potential for dysregulation of miRNAs in metabolic disorders.] [DOI] [PubMed] [Google Scholar]
- 95.Romao JM, et al. MicroRNA regulation in mammalian adipogenesis. Exp Biol Med (Maywood) 2011;236:997–1004. doi: 10.1258/ebm.2011.011101. [DOI] [PubMed] [Google Scholar]
- 96.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–17. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun L, et al. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13:958–65. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kloting N, et al. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One. 2009;4:e4699. doi: 10.1371/journal.pone.0004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heneghan HM, Miller N, McAnena OJ, O'Brien T, Kerin MJ. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J Clin Endocrinol Metab. 2011;96:E846–50. doi: 10.1210/jc.2010-2701. [DOI] [PubMed] [Google Scholar]
- 100.Ortega FJ, et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS One. 2010;5:e9022. doi: 10.1371/journal.pone.0009022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheung O, et al. Nonalcoholic steatohepatitis is associated with altered hepatic MicroRNA expression. Hepatology. 2008;48:1810–20. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kong L, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48:61–9. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 104.Lee J, Kemper JK. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging (Albany NY) 2010;2:527–34. doi: 10.18632/aging.100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kemper JK, et al. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li X, et al. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 107.Motta MC, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–63. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 108.Ponugoti B, et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 2010;285:33959–70. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Purushotham A, et al. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–38. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 111.Walker AK, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–17. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yeung F, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. Embo J. 2004;23:2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–6. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee J, et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J Biol Chem. 2010;285:12604–11. doi: 10.1074/jbc.M109.094524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 118.Tarasov V, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–93. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 119.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–6. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Feige JN, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–58. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 122.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012 doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 123.Creemers EE, Tijsen AJ, Pinto YM. Circulating MicroRNAs: Novel Biomarkers and Extracellular Communicators in Cardiovascular Disease? Circ Res. 2012;110:483–95. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 124.Hunter MP, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [This paper is the first to demonstrate the presence of miRNAs in circulating microvesicles.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–59. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]; MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [This paper is the first to show miRNAs in circulating HDL, indicating that disease-associated miRNAs may traffic in lipoprotein particles and convey regulatory information to distant target tissues.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zampetaki A, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–7. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 130.Li S, et al. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124:175–84. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- 131.Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–52. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bagamasbad P, Denver RJ. Mechanisms and significance of nuclear receptor auto- and cross-regulation. Gen Comp Endocrinol. 2011;170:3–17. doi: 10.1016/j.ygcen.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [This paper identified miRNAs in circulating exosome vesicles and suggested that exosome-associated miRNAs may transmit gene regulatory cues between cells.] [DOI] [PubMed] [Google Scholar]
- 134.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Herker E, Ott M. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol Metab. 2011;22:241–8. doi: 10.1016/j.tem.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Laudadio I, et al. A Feedback Loop Between the Liver-Enriched Transcription Factor Network and Mir-122 Controls Hepatocyte Differentiation. Gastroenterology. 2011 doi: 10.1053/j.gastro.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 139.Chang J, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–13. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 140.Tsai WC, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571–82. doi: 10.1002/hep.22806. [DOI] [PubMed] [Google Scholar]
- 141.Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z. A role of miR-27 in the regulation of adipogenesis. Febs J. 2009;276:2348–58. doi: 10.1111/j.1742-4658.2009.06967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim SY, et al. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem Biophys Res Commun. 2010;392:323–8. doi: 10.1016/j.bbrc.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 143.Li WQ, et al. The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats. Febs J. 2011;278:1522–32. doi: 10.1111/j.1742-4658.2011.08075.x. [DOI] [PubMed] [Google Scholar]
- 144.Gerin I, et al. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 2010;299:E198–206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jin W, Dodson MV, Moore SS, Basarab JA, Guan LL. Characterization of microRNA expression in bovine adipose tissues: a potential regulatory mechanism of subcutaneous adipose tissue development. BMC Mol Biol. 2010;11:29. doi: 10.1186/1471-2199-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Eichner LJ, et al. miR-378( *) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab. 2010;12:352–61. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 147.Lee EK, et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2011;31:626–38. doi: 10.1128/MCB.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol. 2009;23:925–31. doi: 10.1210/me.2008-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]