Abstract
The four Toll/IL-1R (TIR) domain-containing adaptor proteins MyD88, MAL, TRIF and TRAM are well established as essential mediators of TLR signaling and gene induction following microbial detection. In contrast, the function of the fifth, most evolutionarily conserved TIR adaptor sterile alpha and HEAT/Armadillo motif-containing protein (SARM) has remained more elusive. Recent studies of Sarm−/− mice have highlighted a role for SARM in stress-induced neuronal cell death and immune responses in the CNS. However, whether SARM has a role in immune responses in peripheral myeloid immune cells is less clear. Thus, we characterized TLR-induced cytokine responses in SARM-deficient murine macrophages, and discovered a requirement for SARM in CCL5 production, while gene induction of TNF, IL1β, CCL2 and CXCL10 were SARM-independent. SARM was not required for TLR-induced activation of MAPKs or of transcription factors implicated in CCL5 induction, namely NF-κB and IFN regulatory factors, nor for Ccl5 mRNA stability or splicing. However, SARM was critical for the recruitment of transcription factors and of RNA polymerase II to the Ccl5 promoter. Strikingly, the requirement of SARM for CCL5 induction was not restricted to TLR pathways, as it was also apparent in cytosolic RNA and DNA responses. Thus, this study identifies a new role for SARM in CCL5 expression in macrophages.
Introduction
Innate immune cells such as macrophages are key players in initiating an immune response following the detection of invading pathogens via pattern-recognition receptors (PRRs). Engagement of a PRR with its microbial ligand initiates signaling cascades leading to activation of MAPKs and transcription factors, such as NF-κB and IFN regulatory factors (IRFs) (1). These transcription factors subsequently translocate to the nucleus and bind to specific promoter elements of proinflammatory cytokines (e.g. TNF, IL1β), chemokines (e.g. CCL5, CCL2, CXCL10) and type I IFNs (IFN-α and IFN-β) to upregulate gene expression.
One major family of PRRs are the membrane-bound TLRs, which signal through homotypic interactions via distinct Toll/IL-1R (TIR) domain-containing adaptor proteins, namely MyD88, MyD88-adaptor like protein (MAL), TIR domain-containing adaptor inducing IFN-β (TRIF) and TRIF-related adaptor molecule (TRAM) (2). TLR4 is located on the plasma membrane, where it detects LPS. TLR4 utilizes all four adaptor proteins for signaling, activating a MAL/MyD88-dependent pathway from the plasma membrane, followed by TLR4 internalization and TRAM/TRIF-signaling from endolysosomes. In contrast, the microbial nucleic acid-sensing TLRs, namely TLR3, TLR7 and TLR9, are expressed on endosomes, where they sense viral dsRNA, ssRNA or CpG DNA, respectively (1). A single TIR adaptor is then recruited to mediate downstream signaling, that is TRIF for TLR3 and MyD88 for TLR7 and TLR9. Apart from the TLRs, ubiquitously expressed RNA and DNA sensors are found in the cytosol of many different cell types to detect intracellular viruses (3). Such cytosolic PRRs include the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) RIG-I and melanoma differentiation-associated gene 5 (MDA5) which sense viral RNA and signal via the adaptor mitochondrial antiviral signaling (MAVS), and cytosolic DNA sensors such as IFN-inducible protein 16 (IFI16) and cyclic-GMP-AMP synthase (cGAS) which signal via the endoplasmic reticulum-resident protein stimulator of IFN genes (STING) (3).
Sterile α and HEAT/Armadillo motif-containing protein (SARM) has a C-terminal TIR domain, and was therefore expected to function in the TLR pathway (4). Notably, SARM is highly conserved between mammals, Drosophila and worm, and is in fact the most evolutionary conserved member of the cytosolic TIR-containing proteins (5). Unlike the other four TIR adaptor proteins, early studies showed that overexpressed SARM did not lead to NF-κB and IRF3 activation (6, 7). Instead, SARM was found to negatively regulate TRIF-dependent TLR3 and TLR4 signaling in human cells through direct interaction with TRIF (7). However, studies of Sarm−/− mice have mainly revealed neuronal functions for SARM. In an initial study by Kim et al., SARM was discovered to trigger stress-induced neuronal cell death in the CNS after oxygen and glucose deprivation (8), while SARM was recently also implicated in mediating neuronal cell death during infection with neurotropic La Crosse virus (9). In both cases, SARM was found to trigger neuronal cell death via apoptosis, while SARM was also discovered to be required for Wallerian degeneration, whereby injured neurons die via a non-apoptotic cell death pathway (10, 11). Furthermore, SARM was found to regulate neuronal morphology through controlling dendritic aborisation and outgrowth (12). From an evolutionary perspective, these newly discovered functions of mammalian SARM are consistent with the role of the SARM C. elegans ortholog TIR-1 in neuronal development and anoxic death (13, 14). TIR-1 functions upstream of a MAPK cascade in the worm, and has also been shown to have a role in worm immunity, by controlling the induction of antimicrobial peptides (6, 15). SARM was also demonstrated to have a role in mammalian immunity in the CNS, for example Szretter et al. reported that Sarm−/− mice infected with neurotropic West Nile Virus (WNV) displayed a defect in viral clearance and reduced TNF production in the brainstem (16). However, whether SARM has any role in peripheral innate immunity in the mouse has remained unclear.
Given the importance of macrophages in peripheral innate immune defense, we investigated a potential role for mouse SARM in cytokine responses in bone marrow-derived macrophages (BMDMs). We found that SARM was required for optimal CCL5 production in response to TLR4 and TLR7 stimulation, while the induction of TNF and of some other proinflammatory cytokines and chemokines was SARM-independent. Surprisingly therefore, the key transcription factors implicated in CCL5 induction, namely NF-κB, IRF3 and IRF1, were activated normally in the absence of SARM, as were the MAPKs, p38, JNK and ERK. Rather, chromatin immunoprecipitation (ChIP) analysis of Sarm−/− BMDMs revealed a requirement of SARM for the recruitment of transcription factors and RNA polymerase II (Pol II) to the Ccl5 promoter, a crucial step in the transcriptional induction of Ccl5 (17). Accordingly, non-TLR pathways to CCL5 induction were also dependent on SARM. Thus SARM has an essential role in CCL5 chemokine induction in mouse BMDMs.
Materials and Methods
Mice and cell culture
Generation of the Sarm−/− mice on the C57BL/6 background has been previously described (8). Femurs and tibia of wild type (WT) and Sarm−/− mice were obtained from mice bred and maintained at the University of Massachusetts Medical School, USA, in accordance with the guidelines set forth by the University of Massachusetts Medical School Department of Animal Medicine and the Institutional Animal Care and Use Committee. Primary BMDMs were differentiated from bone marrow cells for 7d in complete DMEM (DMEM + GlutaMAX, 10% (v/v) FCS, 10μg/ml ciprofloxacin) supplemented with 15-20% (v/v) L929 supernatant as a source of M-CSF. On day 7, cells were trypsinized and seeded for experiments in complete DMEM. Immortalized WT and Sarm−/− BMDMs were generated with J2 recombinant retrovirus carrying ν-myc and ν-raf/mil oncogenes as previously described (18, 19). Thioglycollate elicited peritoneal macrophages were isolated from mice 4 days after i.p. injection of sterile 3% thioglycollate (Remel, Lenexa, KS). NIH3T3 cells and HEK293T cells were purchased from Sigma and ECACC, respectively, and maintained in complete DMEM.
Receptor agonists and cell transfection
Ultrapure LPS from E. coli, Serotype EH100 (>99.9% pure in respect to contaminating protein and DNA with agonistic TLR activity) was from Alexis Biochemicals, CLO75 was from InvivoGen, mouse rIFN-α was from Miltenyi Biotech, polyinosinic-polycytidylic acid (Poly(I:C)) was from Sigma. The vaccinia virus (VACV) 70bp dsDNA oligonucleotide (dsVACV 70mer) was synthesized by MWG Biotech as previously described (20), and complementary strands were annealed by heating the mixture to 99°C and slow cooling to room temperature. Cells were transfected with Poly(I:C) or dsVACV 70mer using Lipofectamine 2000 (Invitrogen).
Antibodies
Primary Abs used for immunoblotting were anti-β-actin (AC-74) and anti-Flag (M2) from Sigma, anti-IκBα (a gift from Prof. R. Hay, Dundee University, UK), anti-phospho-JNK (Thr183/Tyr185, 44-682G) from Biosource, anti-p38 (#9212), anti-phospho-p38 (Thr180/Tyr182, #9211), anti-JNK (#9252), anti-phospho-ERK1/2 (Thr202/Tyr204, #4377) and anti-phospho-STAT1 (Tyr701, #9171) from Cell Signaling Technology. The secondary Abs for immunoblotting were IRDye 680LT anti-mouse, IRDye 800CW anti-rabbit and IRDye 680LT anti-goat (LI-COR Biosciences). Primary Abs for confocal microscopy were anti-p65 (F-6, sc-8008) from Santa Cruz and anti-IRF3 (#51-3200) from Invitrogen, while secondary Abs were Alexa Fluor 647 anti-mouse or Alexa Fluor 488 anti-rabbit (Invitrogen). Abs used for ChIP were anti-Pol II (N-20, sc-899X), anti-p65 (C-20, sc-372X), anti-IRF1 (M-20, sc-640X) and anti-IRF8 (C-19, sc-6058X) from Santa Cruz, anti-IRF3 (#51-3200) from Invitrogen and isotype-control rabbit IgG (Sigma). The neutralizing IFNAR1 Ab (MAR1-5A3, #16-5945) was purchased from eBioscience.
Quantitative RT-PCR
For mRNA expression analysis, cells were seeded at 4 × 105 cells/ml in 24-well plates and stimulated as indicated the next day. Total RNA was extracted using the High Pure RNA Isolation Kit (Roche), and reversed transcribed with random hexamers (MWG Biotech) using M-MLV Reverse Transcriptase (Promega) according to the manufacturer's instructions. cDNA was analyzed by quantitative RT-PCR using the SYBR Green qPCR Master Mix GoTaq (Promega) or KAPA SYBR FAST Universal (Kapa Biosystems), and gene-specific primer pairs (Table 1). Relative mRNA expression was calculated using the comparative CT method, normalizing the gene of interest to the housekeeping gene β-actin, and comparing it to an untreated sample as calibrator.
Table 1.
PCR primer sequences
| Target gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| β-actin mRNA | TCCAGCCTTCCTTCTTGGGT | GCACTGTGTTGGCATAGAGGTC |
| Ccl5 mRNA | CTCACCATATGGCTCGGACA | ACAAACACGACTGCAAGATTGG |
| Tnf mRNA | TCCCCAAAGGGATGAGAAGTT | GTTTGCTACGACGTGGGCTAC |
| Il1b mRNA | GTGAAATGCCACCTTTTGACAGTGATGAG | CTGCTGCGAGATTTGAAGCTGGATG |
| Ccl2 mRNA | AACTGCATCTGCCCT | ACGGGTCAACTTCAC |
| Cxcl10 mRNA | TCTGAGTGGGACTCAAGGGAT | TCGTGGCAATGATCTCAACACG |
| Ifit2 mRNA | GACTTAGAGGTGCTGCACAG | CTCGTTGTACTCATGACTGCTG |
| Irf7 mRNA | TTGGATCTACTGTGGGCCCA | CTTGCCAGAAATGATCCTGGG |
| Ccl5 pre-mRNA1 | GTGTTGACCTTCCTCTCTCC | CCTCTATCCTAGCTCATCTCC |
| Ccl5 mRNA1 | CTTGCAGTCGTGTTTGTCACTC | CCTCTATCCTAGCTCATCTCC |
| Ccl5 promoter (ChIP) | GCAGTTAGAGGCAGAGTCATAC | CCAGGGTAGCAGAGGAAGTG |
| Tnf promoter (ChIP) | GATTCCTTGATGCCTGGGTG | GCTCTCATTCAACCCTCGGA |
| total Sarm mRNA | CGCTGCCCTGTACTGGAGG | CTTCAGGAGGCTGGCCAGCT |
| Sarm_724/_764 mRNA | CCTTCGCCAGCTACGCTACTTG | CTTCAGGAGGCTGGCCAGCT |
| wt Sarm locus (Genotyping) | ACGCCTGGTTTCTTACTCTACGA | GCTGGGGCCTCCTTACCTCTT |
| mutated Sarm locus (Genotyping) | CAGGTAGCCGGATCAAGCGTATGC | CCTGTCCGGTGCCCTGAATGAACT |
To distinguish unspliced transcripts (pre-mRNA) and spliced transcripts (mRNA), the forward primer was either designed to bind within an intron (unspliced) or intron spanning (spliced), while the reverse primer targeted an exon.
ELISA
Cell culture supernatants were assayed for CCL5 or TNF protein using ELISA kits (R&D Systems) according to the manufacturer's instructions.
Immunoblotting
Cells were seeded at 4 × 105 cells/ml in 6-well plates and stimulated as indicated the next day. Cells were washed with ice-cold PBS, freeze-thawed once at -80°C, and then scraped into 80μl Lysis buffer [50mM Tris pH7.4, 150mM NaCl, 30mM NaF, 5mM EDTA, 10% (v/v) glycerol, 40mM β-Glycerophosphate, 1% (v/v) Triton X-100; containing the inhibitors 1mM Na3VO4, 1mM PMSF and 1% (v/v) Aprotinin] and left on ice for 45min. Cleared lysates were mixed with 3X Sample buffer [187.5mM Tris pH6.8, 6% (w/v) SDS, 30% (v/v) glycerol, 0.3% (w/v) bromophenol blue, 150mM DTT] and boiled for 5min at 99°C. 20μl lysates were resolved on 10% SDS-PAGE, transferred to PVDF membrane (Millipore), blocked for 1h in 3% (w/v) BSA in PBS and probed overnight with primary Ab (1/1000 dilution in blocking solution). The next day, membranes were incubated with secondary Abs (1/8000 dilution in blocking solution) and blots were visualized using the Odyssey Imaging system (LI-COR Biosciences).
Confocal microscopy
Cells were seeded at 3 × 105 cells/ml on glass coverslips in 24-well plates and stimulated the next day as indicated. Cells were fixed for 12min in 4% (w/v) paraformaldehyde and permeabilized for 15min with 0.5% (v/v) Triton X-100 in PBS. Coverslips were blocked for 1h in 5% (w/v) BSA/ 0.05% (v/v) Tween-20 in PBS and stained overnight with primary Abs (1/200 dilution in blocking solution). The following day, coverslips were incubated for 3h with secondary Abs (1/500 dilution in blocking solution) and mounted in Mowiol 4-88 (Calbiochem) containing 1μg/ml DAPI. Images were obtained with an Olympus FV1000 confocal microscope using a 60x oil-immersion objective.
Reporter gene assay
The 724 aa murine SARM (SARM_724) was subcloned from pGW1-SARM1 (a gift from YP. Hsueh, Academia Sinica, Taiwan), and the 764 aa murine SARM (SARM_764) was subcloned from pUNO-mSARM1B (InvivoGen). Both isoforms were cloned with a C-terminal Flag-tag attached into the mammalian expression vector pEF-BOS. The -190 to +57 Ccl5 gene promoter region (transcriptional start site designated +1) was cloned from genomic DNA of primary WT BMDMs into the pGL3-Control Vector (Promega), replacing the SV40 promoter, and generating a pGL3-Ccl5 promoter firefly luciferase reporter gene construct. The murine (-1260) pGL3-Tnf promoter luciferase reporter gene construct was a gift from DV. Kuprash, Russian Academy of Sciences, Russia. For reporter gene assays, NIH3T3 cells were seeded at 0.8 × 105 cells/ml in 96-well plates, and transfected 16h later with 150ng SARM_724, SARM_764 or pEF-BOS empty vector control, 60ng pGL3-Ccl5 or pGL3-Tnf promoter reporter, and 20ng pGL3-renilla luciferase using GeneJuice transfection reagent (Novagen). 48h after transfection, cells were lysed in Passive Lysis Buffer (Promega) and analyzed for luciferase activity. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency.
Retroviral transduction
SARM_724 was subcloned from pGW1-SARM1, and SARM_764 was subcloned from pUNO-mSARM1B, with a C-terminal Flag-tag attached into the retroviral expression vector pMSCV-neo. Retroviral particles were produced in HEK293T cells: Cells were seeded at 2 × 105 cells/ml in 10cm-dishes, and transfected the next day with 3μg pMSCVSARM_724, pMSCV-SARM_764 or pMSCV empty vector control, together with 1μg VSV-G and 1μg Gag-Pol plasmids using 15μl GeneJuice according to the manufacturer's instructions. Medium was then replaced 8h later. Retroviral supernatants were harvested 48h and 72h after transfection, centrifuged and filtered through a 0.45μm filter. NIH3T3 cells were seeded at 1 × 105 cells/ml in 10cm-dishes, and transduced the next day with viral supernatant mixed 1:1 with fresh medium and 6μg/ml polybrene (Sigma). The transduction was repeated 24h later, and after a further 48h cells were selected for 10d in medium containing 1mg/ml G418 (Biosciences). For transduction of immortalized BMDMs, cells were seeded at 3 × 105 cells/ml in 6-well plates, and spinoculated the next day with viral supernatant containing 8μg/ml polybrene at 2000g for 1h at 20°C. Spinoculation was repeated 24h later, and cells were cultured as described for NIH3T3 cells. Expression of exogenous protein was confirmed by immunoblotting.
ChIP
The protocol was adapted from Nelson et al. (21). 1.5 × 107 cells were seeded into 15cm-dishes, and stimulated as indicated the next day prior to harvesting. Formaldehyde was added to the medium to a final concentration of 1% (v/v) to fix the cells for 10min at room temperature, and then the reaction was quenched for 5min with 125mM glycine (from 1M stock solution). Cell monolayers were washed with ice-cold PBS, and scraped into 10ml PBS. Cell pellets were washed again with 1ml PBS, before lysis in 1ml ChIP buffer [50mM Tris-HCl pH7.5, 150mM NaCl, 5mM EDTA, 0.5% (v/v) Nonidet P-40, 1% (v/v) Triton X-100; containing the inhibitors 1mM Na3VO4, 1mM PMSF and 1% (v/v) Aprotinin]. Lysates were immediately centrifuged, and nuclear pellets were washed with 1ml ChIP buffer. The pellet was resuspended in 1ml ChIP buffer and sonicated using a Branson Sonifier 250 with a microtip attached. For BMDMs, 8 cycles of 18x 1s pulses (power output 3, 90% duty cycle), with 1min rest on ice between each cycle, was found to generate optimal DNA fragment sizes ranging from 200-1000bp. Sheared chromatin was cleared by centrifugation. For each IP, sheared chromatin equivalent to 2 × 106 cells was made up to 300μl with ChIP buffer and incubated overnight at 4°C with 2μg specific Ab or an isotype control (IgG), while rotating. The following day, Protein A- or Protein A/G-sepharose beads (Sigma) were blocked for 45min with 100μg salmon sperm DNA (Invitrogen) and 0.5mg BSA per 1ml beads (50% slurry in ChIP buffer), then washed once in ChIP buffer. 40μl blocked beads (50% slurry) were incubated with cleared chromatinimmunocomplexes for 1h at 4°C, while rotating. Then beads were washed 5 times with ChIP buffer (without inhibitors), before 100μl Chelex (10% (w/v) slurry in H20; Bio-Rad) was added to the beads. Samples were vortexed and boiled for 10min at 99°C. After cooling, 1μl Proteinase K (20μg/ml; Qiagen) was added and incubated for 40min at 55°C, while shaking. Samples were boiled again, centrifuged, and 70μl supernatant (containing DNA) was collected. In a different protocol, 250μl 1% (w/v) SDS/ 0.1M NaHCO3 were added to the washed bead pellets instead of Chelex, and complexes were eluted for 2h at 65°C, while shaking. Eluates were collected and incubated overnight at 65°C to reverse cross-links. The next day, an equal volume of TE buffer and 1μl RNase (100mg/ml; Qiagen) were added and incubated for 1h at 37°C. Then, samples were incubated with 3μl Proteinase K for 2h at 55°C, followed by boiling. DNA fragments were purified with Wizard SV Gel and PCR Clean-Up System (Promega) and eluted in 40μl H2O. 2μl DNA was applied to quantitative RT-PCR using SYBR Green KAPA Universal, and primer pairs specific to the proximal region of the gene promoter (Table 1).
Statistical analysis
All data were analyzed with Prism (Graphpad Software, Inc.) and statistical significance was determined using the two-tailed unpaired Student's t-test.
Results
SARM is required for optimal TLR-induced CCL5 expression in macrophages
To investigate a potential function of SARM in murine macrophages, we generated BMDMs from WT and Sarm−/− mice. The genotype of the cells was confirmed by PCR analysis of the genomic Sarm locus (Supplemental Fig. S1A). As previously reported, SARM protein expression was difficult to assess in peripheral cells with currently available antibodies (data not shown, (8, 16)). However, analysis of Sarm mRNA revealed clear expression of SARM in WT BMDMs (Supplemental Fig. S1B). Two mouse SARM isoforms have been reported, of 724aa and 764aa in length (here termed SARM_724 and SARM_764, respectively), the latter being a splice variant with an extended region between the second sterile α motif (SAM) and the TIR domain. Further PCR analysis determined that BMDMs predominantly expressed SARM_724, while SARM_764 was barely detectable (Supplemental Fig. S1C), which was in agreement with the expression profile of SARM in murine T cells (22).
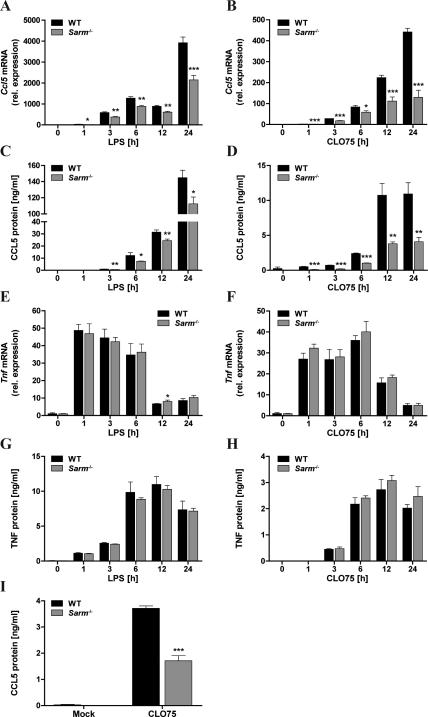
In order to determine whether SARM had a role in TLR-mediated cytokine induction in macrophages, TLR4- and TLR7-dependent responses were examined, since TLR4 signaling is partly TRIF-dependent while TLR7 is solely MyD88-dependent. BMDMs were treated with the TLR4 agonist LPS or the TLR7 agonist CL075 and cytokine mRNA induction was measured over 24h. Strikingly, Ccl5 mRNA induction was significantly impaired in SARM-deficient BMDMs compared to WT cells, both for LPS and CL075 (Fig. 1A, B), as was CCL5 protein production (Fig. 1C, D). At the same time, SARM deficiency did not affect the induction of TNF mRNA or protein in response to TLR stimulation (Fig. 1E-H), nor did it affect TLR-stimulated induction of Il1b, Ccl2 or Cxcl10 mRNA (Supplemental Fig. S2). Importantly, the requirement for SARM was not restricted to bone marrow-derived macrophages, since CL075-induced CCL5 production was also significantly impaired in SARM-deficient peritoneal macrophages compared to WT cells (Fig. 1I).
Figure 1. SARM is required for optimal TLR-induced CCL5 expression, but dispensable for TNF.
Primary WT and Sarm−/− BMDMs were stimulated with 100ng/ml LPS (A,C,E,G) or 5μg/ml CLO75 (B,D,F,H) for the indicated times, or medium as control. Ccl5 (A,B) or Tnf (E,F) mRNA were assayed by quantitative RT-PCR, normalized to the housekeeping gene β-actin and are presented relative to the untreated WT control. Supernatants were assayed for CCL5 (C,D) or TNF (G,H) protein by ELISA. I, Peritoneal macrophages were generated from WT and Sarm−/− mice and stimulated with 10μg/ml CL075 for 24h. Supernatants were assayed for CCL5 by ELISA. The data are mean ± SD of triplicate samples and are representative of at least three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with WT (Student's t-test).
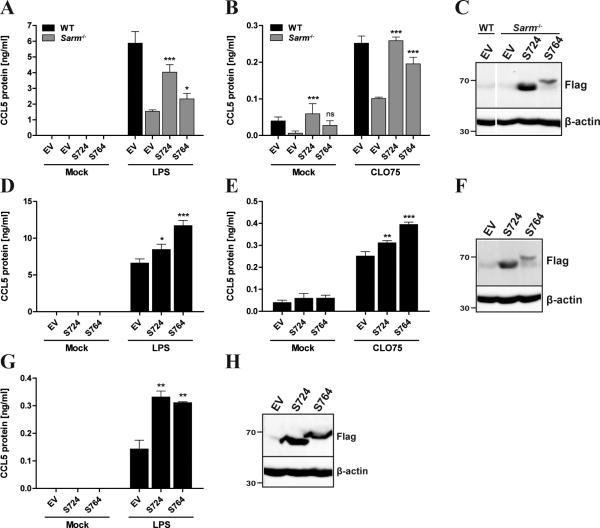
To confirm the role of SARM in CCL5 induction, we reconstituted Sarm−/− BMDMs with either SARM_724 or SARM_764 using retroviral transduction. Sarm−/− BMDMs reconstituted with SARM_724 completely restored TLR7-induced CCL5 production, and also augmented CCL5 to almost WT levels in response to TLR4 (Fig. 2A, B). SARM_764-expressing cells also regained some ability to produce CCL5 in response to TLR stimulation (Fig. 2A, B), but to a lesser extent than SARM_724-reconstituted cells, which correlated with the lower expression of the SARM_764 protein compared to SARM_724 (Fig. 2C). Next, we overexpressed both isoforms of SARM in WT BMDMs, which significantly elevated TLR4- and TLR7-induced CCL5 level, compared to the empty vector control (Fig. 2D-F). In a similar manner, we examined TLR4-induced CCL5 protein in NIH3T3 cells stably overexpressing SARM. Consistent with the results in BMDMs, this different murine cell type also showed significantly enhanced TLR4-induced CCL5 expression when either SARM isoform was ectopically expressed (Fig. 2G, H). Together these data clearly show a role for SARM in TLR-induced CCL5 expression in peripheral mouse cells.
Figure 2. Retroviral expression of SARM rescues CCL5 production in Sarmf−/− BMDMs and augments CCL5 expression in WT cells.
Immortalized WT and Sarm−/− BMDMs (A-C), immortalized WT BMDMs (D-F) or NIH3T3 cells (G,H) were stably transfected with SARM_724-Flag (S724), SARM_764-Flag (S764) or pMSCV empty vector (EV) control using a retroviral system. Cells were stimulated for 24h with 1ng/ml LPS (A), 100ng/ml LPS (D,G), 5μg/ml CLO75 (B,E) or medium as control (mock), and supernatants were assayed for CCL5 protein by ELISA. The data are mean ± SD of triplicate samples and are representative of at least three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with Sarm−/− EV (A,B) or WT EV (D,E,G) (Student's t-test). C,F,H, Cell lysates were subjected to SDS-PAGE and immunoblotted for SARM using a specific antibody to Flag, or β-actin as a loading control. The molecular weight markers are indicated to the left of the gel (in kDa).
SARM is dispensable for the activation of transcription factors implicated in CCL5 expression
To gain insights into the mechanism of SARM-mediated CCL5 regulation, we first defined the signaling pathways that upregulated CCL5 expression in BMDMs. TLR stimulation leads to activation of MAPKs and several transcription factors, including NF-κB and IRF3, which collectively induce the expression of primary response genes (PRGs). PRGs themselves mediate further responses, and some are involved in the induction of secondary response genes (SRGs). A particular example of this is TLR-induced type I IFNs, which subsequently signal via the IFN-α/β receptor (IFNAR) to upregulate numerous IFN-stimulated genes (ISGs) (23). To determine whether TLR-induced CCL5 was a PRG and/or a SRG in BMDMs, we pretreated WT BMDMs with the protein synthesis inhibitor cycloheximide (CHX) before eliciting CCL5 production through TLR4 stimulation. As expected, the ISG Ifit2, which is known to be upregulated through type I IFNs secreted in response to TLR4 stimulation (24), showed reduced mRNA expression in CHX-treated cells (Supplemental Fig. S3B). In contrast, the induction of Ccl5 mRNA was not impaired in the presence of CHX (Supplemental Fig. S3A), suggesting the direct transcriptional induction of Ccl5 as a PRG after TLR4 stimulation. Furthermore, blocking type I IFN-mediated responses with an IFNAR blocking antibody did not affect CCL5 induction by TLR4 (Supplemental Fig. S3C), whereas this did inhibit induction of the ISGs Ifit2 and Irf7 (25), as it inhibited type I IFN signaling (Supplemental Fig. S3D-F). Thus, concordant with published data (26), Ccl5 is a PRG in TLR-stimulated BMDMs.
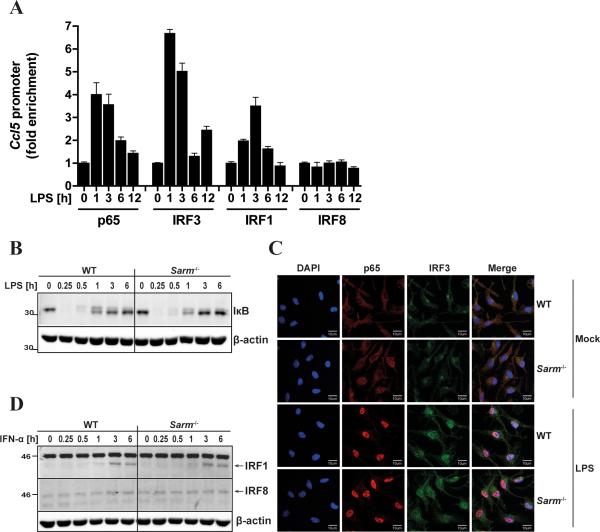
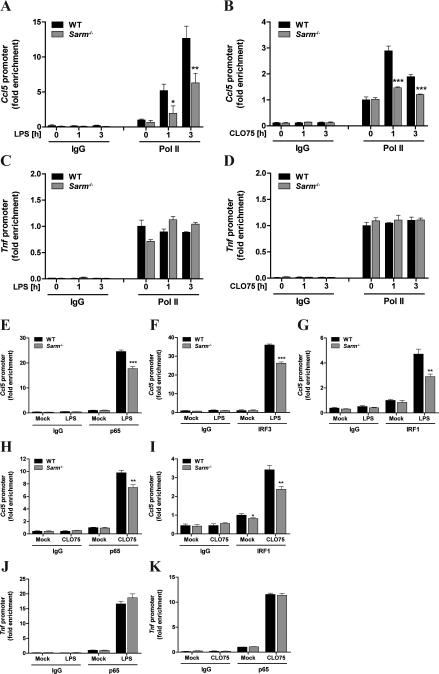
Consequently, SARM was predicted to directly regulate the TLR signaling pathway to promote CCL5 expression, and we therefore tested whether SARM was required for the activation of transcription factors required for CCL5 induction. To define the transcription factors relevant for TLR4-mediated CCL5 induction in BMDMs, we assessed their binding to the Ccl5 promoter using ChIP analysis. Among the transcription factors previously implicated in CCL5 induction in different cell types by different stimuli (27-30), we observed a transient recruitment of the NF-κB p65 subunit, IRF3 and IRF1, but not IRF8, to the Ccl5 promoter in BMDMs in response to LPS (Fig. 3A). Recruitment of p65 and IRF3 to the promoter peaked after 1h LPS stimulation, while IRF1 promoter occupancy was maximal at 3h post-LPS (Fig. 3A). Because IRF1 is induced as an SRG in response to TLR stimulation, via type I IFN induction (31), it was unlikely to be essential for TLR4- induced CCL5 since Ccl5 mRNA induction was type I IFN-independent (Supplemental Fig. S3C). Thus we examined whether the activation of NF-κB or IRF3 was impaired in the absence of SARM. The activation of NF-κB was first assessed indirectly by monitoring the LPS-induced degradation of the cytosolic inhibitor of NF-κB, IκB, which was found to be unaffected by the absence of SARM (Fig. 3B). Furthermore confocal microscopy showed that LPS-induced translocation of p65 to the nucleus was also normal in cells lacking SARM (Fig. 3C). This was also the case for IRF3 translocation to the nucleus (Fig. 3C). We also demonstrated no role for SARM in the upregulation of IRF1 protein in response to type I IFN (Fig. 3D), while IRF8, which was not recruited to the Ccl5 promoter in response to LPS stimulation, was not induced by TLR4 or IFNAR signaling (Fig. 3D, and data not shown).
Figure 3. SARM is dispensable for the activation of transcription factors that are recruited to the Ccl5 promoter.
A, Immortalized WT BMDMs were stimulated with 100ng/ml LPS for the indicated times or medium as control. Sheared chromatin lysates were subjected to ChIP with antibodies specific to NF-κB p65, IRF3, IRF1 and IRF8, or an isotype-control antibody (IgG; not shown). Precipitated DNA and Input DNA were analyzed by quantitative RT-PCR using primer pairs specific for the Ccl5 promoter. Results are normalized to Input and presented as fold enrichment of each transcription factor at the Ccl5 promoter relative to the untreated control. The data are mean ± SD of PCR technical triplicates and are representative of at least two independent experiments. B,D, Primary WT and Sarm−/− BMDMs were stimulated with 100ng/ml LPS (B) or 1000U/ml IFN-α (D) for the indicated times, or medium as control. Cell lysates were subjected to SDS-PAGE and immunoblotted with specific antibodies to IκB (B) or IRF1 and IRF8 (D; arrow indicates the specific band). β-actin served as loading control. The molecular weight markers are shown to the left of the gel (in kDa). C, Primary WT and Sarm−/− BMDMs grown on glass coverslips were stimulated for 1h with 100ng/ml LPS or medium as control (mock), then fixed and stained with antibodies specific to NF-κB p65 (red) or IRF3 (green). Nuclei were visualized using the DNA-intercalating dye DAPI (blue). The scale bars represent 10μm. The data B-D are representative of at least three independent experiments.
Given that neither NF-κB nor IRF3 activation by TLR4 were found to be SARM-dependent, and in light of the role of worm SARM (TIR-1) upstream of a p38 MAPK pathway in C. elegans (6), we additionally assessed LPS-induced activation of the three main MAPKs, p38, JNK and ERK, which might contribute to Ccl5 promoter activity (28). However, we observed a similar degree of MAPK phosphorylation in Sarm−/− BMDMs compared to the corresponding WT (Supplemental Fig. S4), suggesting that mammalian SARM does not regulate MAPK pathways in BMDMs. Together, the data demonstrated that SARM was not required for the activation of the main transcription factors implicated in TLR4-stimulated CCL5 expression, nor was it involved in activating the MAPKs p38, JNK and ERK.
SARM regulates the recruitment of transcription factors and Pol II to the Ccl5 promoter
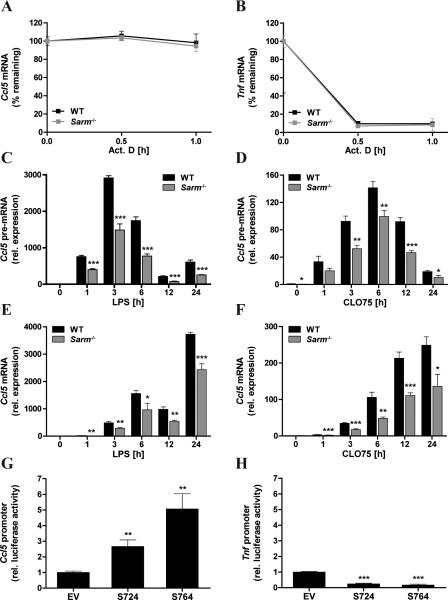
The finding that SARM was not involved in the activation of NF-κB and IRF3 in BMDMs was consistent with the lack of requirement for SARM for the induction of other NF-κB- and IRF3-dependent TLR-induced genes (Fig. 1E-H and Supplemental Fig. S2) and suggested a more direct gene-specific role for SARM in CCL5 regulation. We therefore next examined a potential role for SARM in post-transcriptional regulation of CCL5 expression. In contrast to the induction of Tnf mRNA, which is rapid and transient, Ccl5 mRNA induction is more delayed and gradually increases over a longer time course ((32) and Fig.1A, B, E, F). The different mRNA induction kinetics are mainly due to Tnf mRNA being unstable due to AU-rich elements in the 3’ untranslated region, whereas Ccl5 mRNA lacked those elements and was found to be particularly stable (32). In order to test whether SARM had any role in maintaining Ccl5 mRNA stability after immune stimulation, the stability of LPS-induced Ccl5 mRNA was monitored in WT and Sarm−/− BMDMs after transcription was blocked with Actinomycin D (Act. D). As expected Tnf mRNA showed rapid decay after Act. D treatment, while no decay was observed for Ccl5 mRNA over 60min (Fig. 4A, B). Importantly, the absence of SARM did not provoke any instability of Ccl5 mRNA, indicating that SARM did not exert its CCL5-regulatory function at the level of mRNA stability.
Figure 4. SARM regulates CCL5 expression at the promoter and not post-transcriptionally.
A,B, Primary WT and Sarm−/− BMDMs were prestimulated for 3h with 100ng/ml LPS, and then treated with 5μg/ml Actinomycin D (Act. D) for the indicated times or DMSO as control. Ccl5 (A) or Tnf (B) mRNA were assayed by quantitative RTPCR, normalized to the housekeeping gene β-actin and are presented as percentage remaining compared with each WT or Sarm−/− control. C-F, Primary WT and Sarm−/−BMDMs were stimulated with 100ng/ml LPS (C,E) or 5μg/ml CLO75 (D,F) for the indicated times, or medium as control. Ccl5 pre-mRNA (C,D) or mature mRNA (E,F) were assayed by quantitative RT-PCR, normalized to the housekeeping gene β-actin and are presented relative to the untreated WT control. G,H, NIH3T3 cells were transfected for 48h with 150ng SARM_724-Flag (S724), SARM_764-Flag (S764) or pEF-BOS empty vector (EV) control, along with the pGL3-Ccl5 promoter luciferase reporter gene (G) or pGL3-Tnf promoter luciferase reporter gene (H), and the control pGL3-renilla luciferase gene. Then luciferase activity was measured, normalized to Renilla and is presented relative to EV control. All data are mean ± SD of triplicate samples and are representative of at least two independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with WT (C-F) or EV (G,H) (Student's t-test).
A recent study demonstrated that the delayed expression of genes, such as Ccl5, resulted from a prolonged splicing time rather than a lag in transcriptional initiation (33). Thus, we asked whether there would be a defect in regulation of Ccl5 RNA splicing in the absence of SARM. If so, Ccl5 precursor (pre)-mRNA would be expected to accumulate in Sarm−/− BMDMs in comparison to WT cells. Thus, we designed specific primer pairs to distinguish unspliced Ccl5 RNA (intron-containing) from spliced transcripts, and performed a time course to monitor the expression kinetics of both forms following TLR4 and TLR7 activation. We observed peak expression of Ccl5 pre-mRNA 3h after LPS and 6h after CLO75 stimulation, which gradually decreased thereafter (Fig. 4C, D). The decrease in unspliced transcripts coincided with a steady increase of spliced Ccl5 mRNA that continued up to 24h post stimulation (Fig. 4E, F). However Ccl5 pre-mRNA did not accumulate in SARM-deficient cells, but rather similar to mature mRNA, it was significantly reduced in the absence of SARM compared to WT cells (Fig. 4C, D). This excluded a role for SARM in RNA splicing, and suggested SARM regulated CCL5 at or prior to transcriptional induction of the Ccl5 promoter.
Thus we next examined the potential of SARM ectopic expression to activate the Ccl5 promoter directly, using a luciferase reporter gene under the control of the Ccl5 promoter in mouse NIH3T3 cells. The expression of either isoform of mouse SARM significantly triggered Ccl5 promoter induction, while having no positive effect on Tnf promoter induction (Fig. 4G, H), supporting a role for SARM in stimulating the transcriptional initiation of the Ccl5 promoter. To further explore a role for SARM in inducing the endogenous Ccl5 promoter, we performed ChIP analysis in WT and Sarm−/− BMDMs for the recruitment of Pol II. This was a key experiment, as, in the case of CCL5, stimulus-induced Pol II recruitment to the promoter is a hallmark of transcriptional induction (17). The analysis revealed that SARM-deficient cells were significantly impaired in their ability to recruit Pol II to the Ccl5 promoter in response to TLR4 and TLR7 signaling (Fig. 5A, B). To determine the specificity of this observation, we also examined Pol II occupancy at the Tnf promoter. In contrast to CCL5, for TNF Pol II is known to be already present at the Tnf promoter in the absence of cell stimulation (17), and this was also evident in our ChIP study, where Pol II was readily detectable at the Tnf promoter in unstimulated cells, and did not significantly increase with TLR4 or TLR7 stimulation over 3h (Fig. 5C, D). In contrast to the Ccl5 promoter, Pol II abundance at the Tnf promoter was comparable between WT and SARM-deficient BMDMs (Fig. 5C, D). This was consistent with the fact that TNF induction is independent of SARM (Fig. 1E, F).
Figure 5. SARM is required for optimal Pol II recruitment and the assembly of transcription factors at the Ccl5 promoter.
A-D, Primary WT and Sarm−/− BMDMs were stimulated with 100ng/ml LPS (A,C) or 5μg/ml CLO75 (B,D) for the indicated times, or medium as control. Sheared chromatin lysates were subjected to ChIP with an antibody specific to RNA Polymerase II (Pol II) or an isotype-control antibody (IgG). Precipitated DNA and Input DNA were analyzed by quantitative RT-PCR using primer pairs specific for the Ccl5 or Tnf promoter. Results are normalized to Input and are presented as fold enrichment of Pol II at the Ccl5 promoter (A,B) or Tnf promoter (C,D) relative to the untreated WT control. E-K, Primary WT and Sarm−/− BMDMs were stimulated for 1h with 100ng/ml LPS (E-G,J), 5μg/ml CLO75 (H,I,K) or medium as control (mock). Sheared chromatin lysates were subjected to ChIP with antibodies specific to NF-кB p65 (E,H,J,K), IRF3 (F), IRF1 (G,I) or IgG control. Samples were analyzed as in A-D and are presented as fold enrichment of each specific transcription factor at the Ccl5 promoter (E-I) or Tnf promoter (J,K) relative to mock treated WT cells. All data are mean ± SD of PCR technical triplicates and are representative of at least three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with WT (Student's t-test).
Recruitment of Pol II to the Ccl5 promoter is known to be tightly controlled, and highly dependent on transcription factor binding and chromatin remodeling (34, 35). Thus although transcription factor activation was found to be unaffected by SARM ablation (Fig. 3), we performed ChIP analysis to investigate whether SARM would be required for transcription factor binding to the Ccl5 promoter. Intriguingly, both TLR4- and TLR7-mediated recruitment of transcription factors to the Ccl5 promoter was significantly impaired in Sarm−/− BMDMs compared to the corresponding WT cells (Fig. 5E-I). Hence optimal recruitment of p65, IRF3 and IRF1 to the Ccl5 promoter in response to TLR4 stimulation, and of p65 and IRF1 to the promoter in response to TLR7 all depended on SARM (Fig. 5E-I). IRF3 recruitment was not examined for TLR7, as IRF3 is generally not activated by this pathway (1). In contrast to the case for the Ccl5 promoter, p65 recruitment to the Tnf promoter after LPS or CL075 stimulation was normal in the absence of SARM (Fig. 5J, K). Overall, the ChIP data showed that SARM regulated Ccl5 transcription by mediating the proper assembly of transcription factors at the Ccl5 promoter and subsequently recruitment of Pol II, and thus the formation of the transcriptional machinery on the Ccl5 promoter.
SARM is also required for optimal CCL5 induction in response to non-TLR pathways
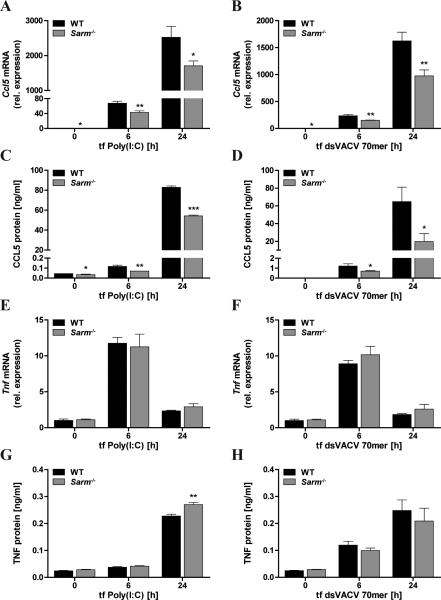
In light of the promoter-regulatory role of SARM for CCL5 induction we wondered whether SARM had an intrinsic stimulus-independent role in CCL5 induction and therefore examined CCL5 expression elicited by non-TLR cytosolic PRRs. Transfection of the synthetic dsRNA Poly(I:C) was used to stimulate the cytosolic RNA sensor MDA5 (3), while the VACV 70bp dsDNA oligonucleotide (dsVACV 70mer) was transfected into cells to stimulate STING-dependent DNA sensing pathways involving sensors such as the IFI16 homolog p204 and cGAS (20, 36). CCL5 and TNF induction in response to the transfected nucleic acids was then compared in Sarm−/− and WT BMDMs. Interestingly, this revealed that SARM was required for optimal CCL5 mRNA and protein expression downstream of cytosolic receptor stimulation (Fig. 6A-D), while it was dispensable for TNF induction (Fig. 6E-H), concordant with the data obtained for TLR-induced CCL5 and TNF.
Figure 6. SARM is also required for optimal CCL5 expression of non-TLR pathways.
Primary WT and Sarm−/− BMDMs were transfected with 2μg/ml Poly(I:C) (A,C,E,G) or 1μg/ml dsVACV 70mer (B,D,F,H) for the indicated times, or medium as control. Ccl5 (A,B) or Tnf (E,F) mRNA were assayed by quantitative RT-PCR, normalized to the housekeeping gene β-actin and are presented relative to the untreated WT control. Supernatants were assayed for CCL5 (C,D) or TNF (G,H) protein by ELISA. The data are mean ± SD of triplicate samples and are representative of at least three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001 compared with WT (Student's t-test).
Cumulatively, the data reveals a new role for SARM in induction of CCL5 expression in macrophages by both TLR and non-TLR pathways, and that SARM is required for the proper assembly of transcription factors and for subsequent RNA Pol II recruitment at the Ccl5 promoter.
Discussion
It has become increasingly apparent recently that SARM might execute quite different roles compared to the other TIR adaptors, MyD88, MAL, TRIF and TRAM, which initiate downstream signaling from TLRs. Several groups have demonstrated that mouse SARM was predominantly expressed in neurons of the brain, and that it had a role in regulating neuronal morphology, stress-induced neuronal cell death and axon degeneration (8-12). The role of SARM in the nervous system was concordant with its evolutionary counterpart TIR-1 in C. elegans (13, 14). However, there is much less data on SARM functions in the periphery.
Here, we have identified a crucial role for SARM in macrophages in the regulation of CCL5 expression by PRRs. This regulatory function of SARM was found to be quite selective for CCL5, as other cytokines and chemokines tested, namely TNF, IL1b, CCL2 and CXCL10, were not affected by SARM ablation. A previous study by Kim et al. showed that TLR-induced cytokine responses were normal in Sarm−/− BMDMs, however that study only examined TNF and CCL2 and not CCL5 (8), and so is consistent with our data here. SARM function had been suggested to be likely CNS-restricted, due to its abundant expression in the brain compared to other tissues and immune cells (8), but apart from our study here, another recent paper showed a role for SARM in peripheral cells. That report demonstrated that SARM mediated T cell death in immune-activated and expanded T cells to restore homeostasis after infection (22).
Roles for SARM in the regulation of cytokine and chemokine induction in the CNS have also emerged. Thus infecting Sarm−/− mice with neurotropic WNV indicated a requirement of SARM for virus-induced TNF production in the brainstem (16). Furthermore Hou et al. demonstrated that SARM-deficient mice showed a significant defect in upregulating cytokines in the CNS in response to vesicular stomatitis virus (VSV) infection (37). However, the range of cytokines dependent on SARM seems to be much broader in the CNS compared to our data for peripheral macrophages here, and includes IL-6, TNF, CCL2, CCL5 and type I IFNs. A broader requirement for SARM for cytokine gene induction in the CNS might result from the higher SARM expression in CNS-resident cells.
Of note, therefore, SARM has been found to regulate CCL5 both in the CNS and in peripheral macrophages, which could have particular implications in disease pathogenesis. For example, CCL5 and its receptor CCR5 were shown to be particularly important for leukocyte recruitment to the CNS, and subsequent control of WNV infection in mice (38, 39), while humans with a homozygous CCR5 mutation (CCR5Δ32) have an increased risk for symptomatic WNV infection (40). Strikingly, Sarm−/− mice were shown to be more susceptible to WNV (16). Clearly SARM is required for immune cell infiltration into the brain, since after either WNV (16) or VSV infection (37) there were reduced macrophage numbers in the brain of mice lacking SARM compared to normal mice, while macrophage/monocyte infiltration of lesioned nerves was suppressed in Sarm−/− mice (10). Whether SARM-dependent CCL5 production contributes to virus- and insult-stimulated cellular infiltration would need to be studied in detail.
Here we focused on understanding mechanistically how SARM regulated CCL5 expression in BMDMs. A key observation was that SARM function was not restricted to TLR pathways, as would have been expected for a cytosolic TIR domain-containing protein, and given the role of human SARM in regulating TRIF signaling (7). Instead, SARM also regulated CCL5 induction downstream of cytosolic RLRs and DNA sensors, which signal via the non-TIR domain-containing adaptor proteins MAVS and STING respectively (1). However the TLR-independent function of SARM here is actually consistent with the fact that its ortholog TIR-1 in C. elegans triggered immune responses independent of the sole C. elegans TLR ortholog, TOL-1 (15). Of interest, TLR-independent roles for other mammalian TIR adaptors have recently emerged in that MyD88 and TRIF apparently function as signaling adaptors downstream of DExD/H-box helicase proteins implicated in cytosolic RNA and DNA sensing (41, 42).
To define how SARM was regulating CCL5 induction here, we first confirmed the mechanism by which PRRs induced CCL5 in BMDMs. This showed that PRR-induced CCL5 was a PRG upregulated directly in response to TLR stimulation independently of any secondary-induced factor or of type I IFN signaling. Consequently, SARM was predicted to modulate the PRR pathway directly, but the main transcription factors implicated in CCL5 induction were activated normally in the absence of SARM, namely NF-κB, IRF3 and IRF1. Also the activation of the MAPKs, p38, JNK and ERK, was normal in Sarm−/− BMDMs. Apart from transcriptional regulation, Ccl5 mRNA expression was recently demonstrated to be controlled by regulated RNA splicing (33). But we could clearly exclude a role for SARM in the post-transcriptional events of Ccl5 RNA splicing, as well as mRNA stability. Instead, SARM was found to enhance Ccl5 promoter activity, indicating its regulatory function operated at the transcriptional level. This was confirmed for the endogenous promoter, where SARM was required for optimal Pol II recruitment to the Ccl5 promoter, while dispensable for Pol II binding to the Tnf promoter. Notably, the accumulation of Pol II at the Ccl5 promoter is regarded as the rate-limiting step in Ccl5 transcription, and depends on prior transcription factor binding and chromatin remodeling to open the otherwise tightly closed promoter (17, 34, 35). A detailed ChIP analysis revealed that recruitment of NF-κB p65, IRF3 and IRF1 to the Ccl5 promoter was significantly impaired in the absence of SARM. Reduced transcription factor recruitment to the promoter likely explains the impaired Pol II recruitment to Ccl5 in the absence of SARM, since IRF3 in particular is implicated in stimulating chromatin remodeling at the Ccl5 promoter (35).
Thus, our study in macrophages provides compelling evidence that SARM promotes CCL5 expression via controlling the proper recruitment of transcription factors, and subsequently Pol II, to the Ccl5 promoter. Future experiments will aim to elucidate the exact mechanism used by SARM to achieve this. Given the TLR-independent nature of this SARM function, other protein motifs in SARM apart from the TIR domain, might be involved in mediating Ccl5 promoter induction. For example, SAM domains, which are known to mediate diverse protein-protein interactions (43), may recruit a transcriptional regulator to the Ccl5 promoter. Also, SARM may recruit a DExD/H-box protein to the Ccl5 promoter, since such helicases can interact with TIR adaptors (41, 42), and are also know to regulate gene promoters (44). Further, it will be interesting to define whether SARM acts in close proximity to the Ccl5 promoter, or whether it functions in a more upstream role. In support of the latter model is the fact that SARM is reported to be expressed in the cytosol and mostly located at the mitochondria (8, 45). However, we and others (46) have also detected SARM in the nucleus, although we have no evidence to date of PRR-stimulated re-localisation of SARM (data not shown). Apart from elucidating the exact mechanism of Ccl5 transcriptional regulation by SARM in macrophages, it will also be of interest to determine whether SARM-dependent cytokine induction in the CNS (37) is also due to regulation of recruitment of transcription factors and Pol II to selected gene promoters.
Overall this study has identified a novel function of mouse SARM in selectively promoting PRR-induced CCL5 expression in macrophages. Thereby, the data contribute to establishing SARM as a positive regulator of cytokine responses, concordant with previous murine in vivo studies. Not least, the report highlights the TLR-independent role of another mammalian TIR adaptor protein.
Supplementary Material
Acknowledgements
We thank Dmitry Kuprash for the gift of the Tnf promoter luciferase reporter gene.
Grant support: This work was supported by Science Foundation Ireland grant 11/PI/1056 (to A.G.B.) and National Institutes of Health grant AI067497 (to K.A.F.).
Abbreviations used
- BMDM
bone marrow-derived macrophage
- ChIP
chromatin immunoprecipitation
- CHX
cycloheximide
- IRF
IFN regulatory factor
- ISG
interferon stimulated gene
- PRG
primary response gene
- PRR
pattern-recognition receptor
- Pol II
RNA polymerase II
- SARM
sterile alpha and HEAT/Armadillo motif-containing protein
- SAM
sterile α motif
- SRG
secondary response gene
- TRIF
TIR domain-containing adaptor inducing IFN-β
- TIR
Toll/IL-1R
- WNV
West Nile Virus
- WT
wild type
References
- 1.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 3.Gurtler C, Bowie AG. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013 doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Neill LA, Fitzgerald KA, Bowie AG. The Toll-IL-1 receptor adaptor family grows to five members. Trends Immunol. 2003;24:286–290. doi: 10.1016/s1471-4906(03)00115-7. [DOI] [PubMed] [Google Scholar]
- 5.Mink M, Fogelgren B, Olszewski K, Maroy P, Csiszar K. A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo/beta-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics. 2001;74:234–244. doi: 10.1006/geno.2001.6548. [DOI] [PubMed] [Google Scholar]
- 6.Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci U S A. 2004;101:6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carty M, Goodbody R, Schroder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y, Zhou P, Qian L, Chuang JZ, Lee J, Li C, Iadecola C, Nathan C, Ding A. MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J Exp Med. 2007;204:2063–2074. doi: 10.1084/jem.20070868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee P, Woods TA, Moore RA, Peterson KE. Activation of the innate signaling molecule MAVS by bunyavirus infection upregulates the adaptor protein SARM1, leading to neuronal death. Immunity. 2013;38:705–716. doi: 10.1016/j.immuni.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, MacDonald JM, Ziegenfuss JS, Milde S, Hou YJ, Nathan C, Ding A, Brown RH, Jr., Conforti L, Coleman M, Tessier-Lavigne M, Zuchner S, Freeman MR. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdts J, Summers DW, Sasaki Y, DiAntonio A, Milbrandt J. Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J Neurosci. 2013;33:13569–13580. doi: 10.1523/JNEUROSCI.1197-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CY, Lin CW, Chang CY, Jiang ST, Hsueh YP. Sarm1, a negative regulator of innate immunity, interacts with syndecan-2 and regulates neuronal morphology. J Cell Biol. 2011;193:769–784. doi: 10.1083/jcb.201008050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuang CF, Bargmann CI. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev. 2005;19:270–281. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayakawa T, Kato K, Hayakawa R, Hisamoto N, Matsumoto K, Takeda K, Ichijo H. Regulation of anoxic death in Caenorhabditis elegans by mammalian apoptosis signal-regulating kinase (ASK) family proteins. Genetics. 2011;187:785–792. doi: 10.1534/genetics.110.124883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- 16.Szretter KJ, Samuel MA, Gilfillan S, Fuchs A, Colonna M, Diamond MS. The immune adaptor molecule SARM modulates tumor necrosis factor alpha production and microglia activation in the brainstem and restricts West Nile Virus pathogenesis. J Virol. 2009;83:9329–9338. doi: 10.1128/JVI.00836-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci U S A. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberson SM, Walker WS. Immortalization of cloned mouse splenic macrophages with a retrovirus containing the v-raf/mil and v-myc oncogenes. Cell Immunol. 1988;116:341–351. doi: 10.1016/0008-8749(88)90236-5. [DOI] [PubMed] [Google Scholar]
- 19.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 22.Panneerselvam P, Singh LP, Selvarajan V, Chng WJ, Ng SB, Tan NS, Ho B, Chen J, Ding JL. T-cell death following immune activation is mediated by mitochondria-localized SARM. Cell Death Differ. 2013;20:478–489. doi: 10.1038/cdd.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noppert SJ, Fitzgerald KA, Hertzog PJ. The role of type I interferons in TLR responses. Immunol Cell Biol. 2007;85:446–457. doi: 10.1038/sj.icb.7100099. [DOI] [PubMed] [Google Scholar]
- 24.Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. Contribution of interferon-beta to the murine macrophage response to the toll-like receptor 4 agonist, lipopolysaccharide. J Biol Chem. 2006;281:31119–31130. doi: 10.1074/jbc.M604958200. [DOI] [PubMed] [Google Scholar]
- 25.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 26.Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 27.Genin P, Algarte M, Roof P, Lin R, Hiscott J. Regulation of RANTES chemokine gene expression requires cooperativity between NF-kappa B and IFN-regulatory factor transcription factors. J Immunol. 2000;164:5352–5361. doi: 10.4049/jimmunol.164.10.5352. [DOI] [PubMed] [Google Scholar]
- 28.Casola A, Garofalo RP, Haeberle H, Elliott TF, Lin R, Jamaluddin M, Brasier AR. Multiple cis regulatory elements control RANTES promoter activity in alveolar epithelial cells infected with respiratory syncytial virus. J Virol. 2001;75:6428–6439. doi: 10.1128/JVI.75.14.6428-6439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Guan X, Ma X. Interferon regulatory factor 1 is an essential and direct transcriptional activator for interferon {gamma}-induced RANTES/CCl5 expression in macrophages. J Biol Chem. 2005;280:24347–24355. doi: 10.1074/jbc.M500973200. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Ma X. Interferon regulatory factor 8 regulates RANTES gene transcription in cooperation with interferon regulatory factor-1, NF-kappaB, and PU.1. J Biol Chem. 2006;281:19188–19195. doi: 10.1074/jbc.M602059200. [DOI] [PubMed] [Google Scholar]
- 31.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 32.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao S, Baltimore D. RNA splicing regulates the temporal order of TNF-induced gene expression. Proc Natl Acad Sci U S A. 2013;110:11934–11939. doi: 10.1073/pnas.1309990110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao TS, Fitzgerald KA. The cGAS-STING pathway for DNA sensing. Mol Cell. 2013;51:135–139. doi: 10.1016/j.molcel.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou YJ, Banerjee R, Thomas B, Nathan C, Garcia-Sastre A, Ding A, Uccellini MB. SARM is required for neuronal injury and cytokine production in response to central nervous system viral infection. J Immunol. 2013;191:875–883. doi: 10.4049/jimmunol.1300374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosking MP, Lane TE. The role of chemokines during viral infection of the CNS. PLoS Pathog. 2010;6:e1000937. doi: 10.1371/journal.ppat.1000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim JK, Murphy PM. Chemokine control of West Nile virus infection. Exp Cell Res. 2011;317:569–574. doi: 10.1016/j.yexcr.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim JK, Louie CY, Glaser C, Jean C, Johnson B, Johnson H, McDermott DH, Murphy PM. Genetic deficiency of chemokine receptor CCR5 is a strong risk factor for symptomatic West Nile virus infection: a meta-analysis of 4 cohorts in the US epidemic. J Infect Dis. 2008;197:262–265. doi: 10.1086/524691. [DOI] [PubMed] [Google Scholar]
- 41.Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, Liu YJ. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim CA, Bowie JU. SAM domains: uniform structure, diversity of function. Trends Biochem Sci. 2003;28:625–628. doi: 10.1016/j.tibs.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Panneerselvam P, Singh LP, Ho B, Chen J, Ding JL. Targeting of pro-apoptotic TLR adaptor SARM to mitochondria: definition of the critical region and residues in the signal sequence. Biochem J. 2012;442:263–271. doi: 10.1042/BJ20111653. [DOI] [PubMed] [Google Scholar]
- 46.Sethman CR, Hawiger J. The innate immunity adaptor SARM translocates to the nucleus to stabilize lamins and prevent DNA fragmentation in response to pro-apoptotic signaling. PLoS One. 2013;8:e70994. doi: 10.1371/journal.pone.0070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.