Abstract
Fertilization triggers a complex cellular programme that transforms two highly specialized meiotic germ cells, the oocyte and the sperm, into a totipotent mitotic embryo: linkages between sister chromatids are remodelled to support the switch from reductional meiotic to equational mitotic divisions; the centrosome, which is absent from the egg, needs to be reintroduced; the axis of cell division is shifted from extremely asymmetric to symmetric; genomic imprinting is selectively erased and re-established; and protein expression shifts from translational control back to transcriptional control. Recent work has started to reveal how this remarkable transition from meiosis to mitosis is achieved.
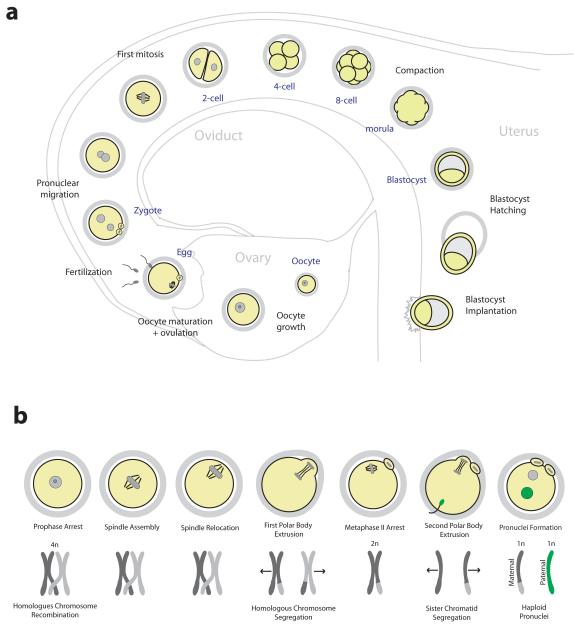
During fertilization of an egg with a sperm, the haploid genomes of each parent are unified to form the diploid genome of a new and unique individual. Yet the road to reproduction begins a long time before the fusion of male and female gametes. In females, egg precursor cells, termed oocytes, are stored in the ovary from before birth. It is generally thought that oocytes are not replenished after birth, but this dogma of developmental biology has recently been challenged1,2. The oocytes have already undergone meiotic DNA replication and recombination that ensures the genetic diversity of potential offspring (FIG. 1b). The stored oocytes are arrested in meiotic prophase and surrounded by somatic cells in a functional unit termed primordial follicle. Periodically some primordial follicles initiate a prolonged growth phase. The somatic cells that surround the oocyte divide and feed the oocyte with precursors of macromolecules through gap junctions3. The oocyte increases in size and accumulates all the storage material necessary to support the development of the early embryo. Once every menstrual cycle a surge of gonadotropins induces the meiotic maturation of these fully grown oocytes into fertilizable eggs. Still in the follicle, the oocyte nucleus breaks down and a microtubule spindle assembles around the chromosomes. The spindle then migrates to the oocyte surface and segregates half the homologous chromosomes into a small cell termed polar body. The remaining chromosomes are captured by a second meiotic spindle, and the egg remains arrested at this stage awaiting fertilization by a sperm (FIG. 1b). While the oocyte matures, which takes 12-14 hours in mice and more than 24 hours in humans, a mucified matrix develops between the somatic cells of the follicle. The matrix expands and ruptures the surface of the follicle so that the egg can be released into the oviduct.
Figure 1. From oocyte to embryo.
a | An overview of preimplantation development. Following ovulation, eggs are fertilized in the oviduct to form the zygote. After several mitotic divisions, the embryo undergoes compaction to form the morula. A fluid-filled cavity develops inside the embryo forming the blastocyst, which hatches from the zona pellucida to implant into the uterine wall. b | Stages of oocyte maturation and corresponding chromosome configuration. Following nuclear envelope breakdown the spindle assembles, relocates to the oocyte surface and segregates half the homologous chromosomes into a polar body. A spindle assembles around the remaining chromosomes and the egg arrests in metaphase II awaiting fertilization. Upon sperm binding, the egg segregates half of the sister chromatids into a second polar body. The resulting zygote contains haploid pronuclei from the mother and the father.
In the oviduct, the sperm binds to the zona pellucida, a glycoprotein matrix that surrounds the egg, and the gametes fuse to form the zygote. The egg resumes meiosis and segregates half of the remaining sister chromatids into a second polar body (FIG. 1b). The male and female haploid pronuclei form and migrate towards each other before the first mitotic spindle assembles around the now diploid zygotic genome. A series of mitotic cell divisions then produce smaller embryonic cells, termed blastomeres. The blastomeres start to adhere to each other in the 8 cell stage and undergo compaction to form a solid ball of cells known as morula. The two subsequent cell divisions generate two different populations of cells; those that occupy the inside of the embryo, which contribute to the embryo proper, and those that occupy the outside, which will give rise to the extraembryonic tissue that is required to support embryo development in the uterus. At the 32 cell stage, a fluid-filled cavity begins to form inside the embryo. This cavity continues to grow as the embryo matures into a blastocyst. By this time, the embryo has migrated into the uterus. The blastocyst hatches from the zona pellucida and implants into the uterine wall, where the embryo continues to develop (FIG. 1a).
The transition from egg to embryo is perhaps one of the most dramatic and complex cell transformations in human biology: two highly differentiated gametes fuse, and the resulting cell, the zygote, is capable of dividing to generate all the cells of the human body. In this review we discuss the remarkable changes to the cellular machinery that govern this transition. We begin by reviewing our current knowledge of the fertilization process, with emphasis on sperm-egg binding and exit from meiotic arrest. We then discuss the recent advances in our understanding of the egg to embryo transition, with particular focus on the shift from meiosis to mitosis. Fertilization-triggered changes to chromosomes, the microtubule spindle, cell division symmetry and gene expression regulation will all be discussed. We mainly focus on mammals, but refer to findings from non-mammalian organisms when relevant for understanding the human condition.
Fertilization
Sperm-egg binding
Oscar Hertwig first recognised that fertilization involves the fusion of sperm and egg cells (BOX 1). Sperm initially bind to the egg’s zona pellucida4 (FIG. 2a), which is made up of just a few glycoproteins, ZP1 to ZP3 in mice and ZP1 to ZP4 in humans, yet the precise molecules that mediate mammalian sperm-egg binding have remained elusive to researchers. Early studies identified ZP3 as a potential primary sperm receptor5,6, however, it seems unlikely that ZP3 alone is sufficient for sperm binding, because mouse eggs in which mouse ZP3 is replaced with the human ZP3 are unable to bind human sperm7.
Box 1. History of the discovery of fertilization.
How is an entirely new human being generated following sexual intercourse? This question has evoked human thinking since ancient times. Hippocrates (460-370 BC) argued that man and woman each contributed semen that mixed in the uterus to form the embryo, whereas Aristotle (384-322 BC) favoured a more male-centred view that the woman merely provided fertile ground for the male seed to grow. These ideas dominated thinking until the 17th century, when the combined work of William Harvey, Jan van Horne, Jan Swammerdam, Neils Stensen, Regner de Graaf and Francesco Redi led to the theory that all female organisms, including humans, produced eggs139. Indeed, Harvey went so far as declaring “Ex Ova Omnia”, or everything from the egg140. Soon after, in 1677, Antonii van Leeuwenhoek built a microscope to study human semen and made the remarkable discovery of spermatozoa141. Therefore by the late 17th century, both key components of fertilization – the egg and sperm – had been realised, yet the relative contributions that each made to the embryo remained unclear for almost 200 years. In the early 19th century, Karl Ernst von Baer first confirmed the presence of the mammalian egg under the microscope142, and Schleiden and Schwann postulated that egg and sperm are somewhat equivalent in that they are both cells. At around the same time it was becoming apparent that the widely observed phenomenon of heredity must somehow involve factors that are contained within egg and sperm respectively143. Finally, it was in 1876 that Oscar Hertwig made the seminal discovery of fertilization in sea urchins144. Hertwig observed that the nuclei of sperm and egg fused during fertilization144, thereby providing a conceptual basis for genetic inheritance and settling the long standing debate over the role of the egg and sperm in the generation of new life.
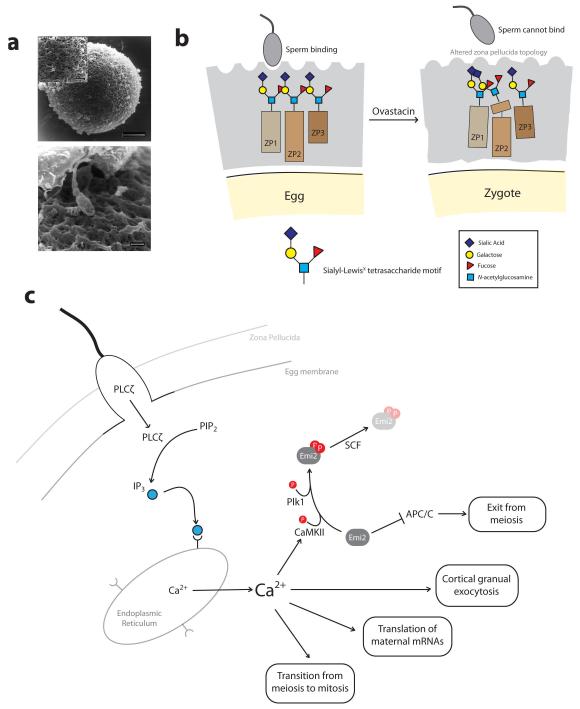
Figure 2. Fertilization: Sperm-egg binding and exit from meiosis.
a | Scanning electron microscopy of a human egg surrounded by the zona pellucida (top). A human sperm can be seen approaching the surface of the zona pellucida (bottom). b | Schematic of sperm-egg binding. The zona pellucida (grey) consists of glycoproteins that are modified with oligosaccharide chains. Sperm can bind to the zona pellucida of unfertilized eggs. Following fertilization, cleavage of ZP2 by ovastacin renders the zona pellucida non-permissive for sperm binding. c | Following sperm-egg membrane fusion, sperm-derived PLCζ promotes the production of IP3, which binds to receptors on the endoplasmic reticulum causing release of Ca2+. Ca2+ activates CaMKII which, together with Plk1, phosphorylates Emi2 promoting its targeting for destruction by the SCF ubiquitin ligase. This releases Emi2-mediated inhibition of the APC/C, allowing the APC/C to promote exit from meiosis. The rise in intracellular Ca2+ also triggers cortical granule exocytosis, translation of maternal mRNAs and ultimately the transition from meiosis to mitosis. Images in part a are reproduced, with permission, from REF. 145 © (1998) Oxford Journals.
An alternative possibility is that the zona pellucida proteins together adopt a three-dimensional structure that presents a binding site for sperm8. Such a binding site would be predicted to be lost upon the cleavage of ZP2 that occurs after fertilization9, providing a possible mechanism for preventing polyspermy. Indeed, the protease responsible for ZP2 cleavage, Ovastacin, was recently identified. Ovastacin is a component of cortical granules, which fuse with the egg plasma membrane and release their contents after fertilization10. Crucially, sperm can bind to 2-cell embryos isolated from mice expressing non-cleavable ZP2, but not to wild type embryos whose ZP2 is presumably cleaved. These findings are consistent with a model in which sperm-egg binding depends on the cleavage status of ZP211, 12 (FIG. 2b). In the future it would be interesting to utilize the tobacco etch virus (TEV) protease technology13 to test if ZP2 cleavage is sufficient to prevent sperm binding.
The zona pellucida proteins are glycoproteins modified with oligosaccharide sequences at asparagine (N-linked) and/or threonine (O-linked) residues4. A recent study of unfertilized human eggs found that the majority of oligosaccharides terminated with the Sialyl-LewisX tetrasaccharide motif14. Consistent with a function of the Sialyl-LewisX motif in sperm-egg binding, Sialyl-LewisX oligosaccharides or anti Sialyl-LewisX antibodies interfere with sperm-egg binding in vitro, and the removal of sialic acid from solubilized zona pellucida reduces its affinity for sperm14. An important challenge for the future will be to identify the sperm-bound receptor responsible for binding to the Sialyl-LewisX motif.
Although a complete understanding of mammalian sperm-egg interaction is still lacking, one model that encompasses much of the available data is that the three-dimensional architecture of the zona pellucida, which depends on the cleavage state of ZP2, serves to present oligosaccharide chains in a way that permits sperm binding15 (FIG. 2b.). Further work will be necessary to test this model, and how variations on a theme ensure species-specific sperm-egg-binding. Once bound, the sperm releases a specialised secretory vesicle, the acrosome, which contains a mix of hydrolytic and proteolytic enzymes that once released pave a way through the zona pellucida promoting plasma membrane fusion16.
Ca2+ triggers exit from meiosis
The fusion of the sperm with the egg’s plasma membrane triggers the completion of the second meiotic division and the transition to mitosis. How does the sperm trigger this transition? The sperm induces a rise in free Ca2+ in the egg, which was first observed more than 30 years ago in medaka and sea urchin eggs and subsequently confirmed in all species studied to date17, 18. In mammals, a series of Ca2+ oscillations triggers a temporally ordered sequence of events, including the release of cortical granules, the completion of the second meiotic division, translation of maternal mRNAs and ultimately the transition from meiosis to mitosis19, 20. The increase in Ca2+ in mammalian eggs is likely to be initiated by the phospholipase C-zeta (PLCζ)21, which is introduced into the egg by the sperm. PLCζ promotes the production of inositol 1,4,5-trisphosphate (IP3), which then binds to receptors on the endoplasmic reticulum, causing the release of Ca2+22, 23 (FIG. 2c). Recent studies have shed light on how the increase in Ca2+ triggers the exit from meiosis. All vertebrate eggs, including human eggs, arrest in metaphase of the second meiotic division while they await fertilization. This arrest is mediated by an activity termed cytostatic factor (CSF) that was first found by Masui and Markert in 197124. An essential mediator of CSF activity is Emi2. Emi2 maintains metaphase II arrest by inhibiting the anaphase-promoting complex / cyclosome (APC/C), which targets cyclin B and the separase inhibitor securin for degradation25-29. A recent study revealed that Emi2 also regulates APC/C activity during the early mitotic divisions of Xenopus embryos30, and it will be interesting to investigate why the function of Emi2 as an APC/C regulator continues beyond meiosis into the mitotic divisions. The increase in intracellular Ca2+ upon fertilization activates Ca2+/calmodulin-dependent protein kinase II (CaMKII), which phosphorylates Emi226. Emi2 is further phosphorylated by Plk126, 27, 31, and subsequently targeted for degradation by the SCF ubiquitin ligase31 (FIG. 2c). This leads to the activation of the APC/C, destruction of cyclin B and securin, elimination of half of the sister chromatids into the second polar body and the formation of the female pronucleus.
From meiotic to mitotic chromosomes
Upon fertilization, the parental genomes must be reprogrammed for totipotency, and the cell cycle machinery must switch from meiotic (reductional) to mitotic (equational) chromosome segregation. In this section we will discuss recent work that has begun to provide a molecular understanding of fertilization-triggered changes to chromosomes.
Chromatin remodelling
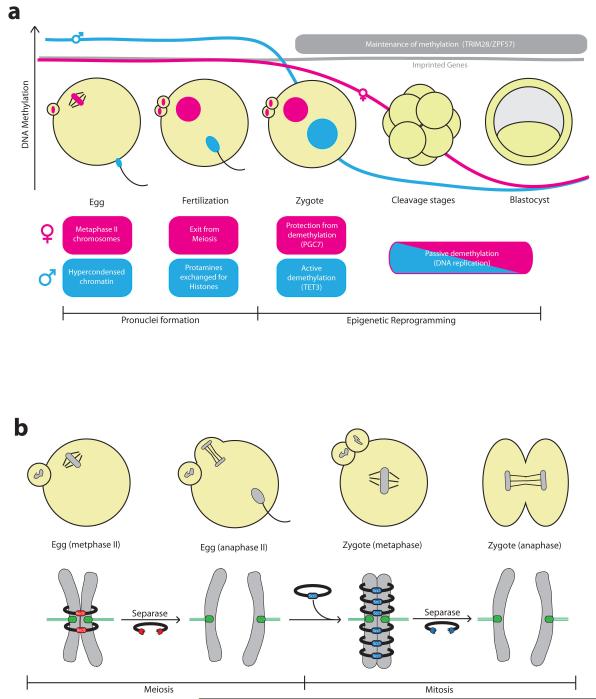
At fertilization, the maternal genome is contained within individualised chromosomes arrested in metaphase II. The paternal genome is tightly compacted for packaging into the sperm head. This is achieved by replacing most nucleosomes with protamines that are rich in positively charged amino acids which form a nucleoprotamine complex with the negatively charged DNA32. Therefore in the zygote, there initially exists a striking difference in the chromatin state of the parental genomes that must be resolved to ensure accurate segregation of chromosomes during the first mitotic division. Almost immediately after fertilization this disparity begins to be resolved: the increase in intracellular Ca2+ triggers exit from meiosis and the formation of a female pronucleus, whilst the sperm genome undergoes decompaction. Protamines are rapidly removed from the sperm pronucleus and the DNA is re-wrapped around nucleosomes that contain the histone H3 variant H3.3, which is replaced with canonical histone H3 during DNA replication33, 34 (FIG. 3a). Although now equivalently structured in nucleosomes, the two pronuclei retain some parent-specific histone methylation patterns, particularly at pericentromeric heterochromatin regions, which are equilibrated gradually during the first embryonic divisions35.
Figure 3. Transition from meiotic to mitotic chromosomes.
a | Schematic of chromatin remodelling during fertilization and early embryo development. Following fertilization, the egg exits from meiosis and assembles a haploid pronucleus. At the same time the sperm genome, which enters the egg in a highly compact state, undergoes decompaction. Sperm protamines are replaced by histones and the male haploid pronucleus is formed. Maternal and paternal genomes are methylated during gametogenesis, including parental-specific methylation at imprinted genes. The sperm pronucleus undergoes rapid active demethylation in the zygote mediated by Tet3. The maternal pronucleus is largely protected from this active demethylation by the action of PGC7. Following pronuclear fusion, the zygotic genome undergoes passive demethylation during the blastomeres cleavage stages, reaching a minimum in the blastocyst. The maintenance of methylation at imprinted regions depends on the ZPF57/TRIM28 DNA-binding complex. b | Sister chromatids in the egg are held together by Rec8-containing cohesin complex. Rec8 cleavage by separase triggers second polar body extrusion following fertilization. Scc1-containing cohesin complexes are loaded onto chromosomes immediately in the zygote, and Scc1 cleavage by separase triggers sister chromatid segregation during the first mitotic division.
For the zygote to acquire totipotency, the parental genomes must also undergo extensive epigenetic reprogramming, which involves global DNA demethylation. The sperm genome is highly methylated compared to that of the egg36, perhaps reflecting terminal differentiation and dense chromatin packaging within the sperm. Within hours of fertilization, however, the sperm pronucleus undergoes rapid active demethylation prior to DNA replication37, 38. Recently, this was found to be mediated by the oxidation of 5-methyl cytosine by the DNA dioxygenase Tet339-41, yet the precise mechanisms involved remain poorly understood. Significantly, Tet3-null mice show developmental failure, suggesting that active paternal demethylation is crucial for proper embryonic development41. Maternal chromatin is largely protected from this initial active demethylation by the action of the DNA binding protein PGC742. PGC7 binds to H3K9 methylation that is present mainly on maternal chromatin43, 44. During the cleavage stages of the early embryo, both parental genomes undergo passive, replication-dependent demethylation45. Methylation finally reaches a minimum at the blastocyst stage, prior to cell lineage specification46 (FIG. 3a).
DNA demethylation in the embryo, however, must not go to completion. A small proportion of mammalian genes are imprinted, meaning that they are expressed from only one of the two parental chromosomes. Imprinted genes are established by differential methylation marks laid down in the sperm and egg during gametogenesis47, which must be maintained during epigenetic reprogramming in the embryo. Imprinted loci marked by H3K9 methylation recruit PGC7, which protects against active demethylation immediately after fertilization44. A chromatin-modifier complex that assembles via the interaction of ZPF57 and TRIM28 also ensures that imprinted loci are spared from passive demethylation during blastomere cleavage stages48-50.
Chromosome Segregation
Not only the chromatin itself is remodelled upon fertilization, but also the physical linkages between chromosomes have to be reorganized to support the reductional segregation of homologous chromosomes during the first meiotic division, the segregation of sister chromatids during the second meiotic division and finally the equational segregation of replicated sister chromatids during the first mitotic division of the embryo (FIG. 1b). An elegant set of experiments has found that the transition from meiosis to mitosis involves a dramatic change in the composition of the chromosomal cohesin complex13. The multiprotein cohesin complex forms a ring that entraps sister chromatids following DNA replication. Chromosome segregation in anaphase is triggered by cleavage of cohesin’s kleisin subunit by the protease separase. This destroys the cohesin ring and therefore sister chromatid cohesion51. Mitotic cohesin complexes contain the Scc1 kleisin subunit. In meiotic cohesin complexes, Scc1 is largely replaced by the meiosis-specific Rec8 protein, which is essential for reductional chromosome segregation52. By engineering mice to express versions of Scc1 and Rec8 that can be artificially cleaved by the TEV protease, it was possible to test the relative contributions of Scc1- and Rec8-containing cohesin complexes in holding sister chromatids together before and after fertilization. Artificial Rec8 cleavage could trigger meiosis II chromosome segregation in the egg but not mitotic chromosome segregation in the zygote, whereas Scc1 cleavage had the opposite effect13. Therefore, the switch from meiotic to mitotic cohesin complex occurs immediately after fertilization in the zygote (FIG. 3b).
How might such a rapid switch be facilitated? The transition from egg to embryo occurs in the absence of transcription (see below). Thus, it seems likely that some proteins required for the mitotic divisions are already present before fertilization. In fact, Scc1 protein is abundant in oocytes53, 54 and so may be readily available for loading onto chromosomes during DNA replication in the zygote. A recent genome-wide polysome profiling study revealed a 10-fold increase in translation of the mitotic cohesin proteins STAG1 and STAG2 in eggs compared to prophase oocytes55. This suggests that translational upregulation of the mitotic machinery just before fertilization may also contribute to the transition from meiosis to mitosis. Because all Rec8 protein is likely cleaved by separase in meiosis II, and subsequently degraded by the N-end rule pathway56, meiotic cohesin complexes may be rapidly inactivated after fertilization, further facilitating a rapid switch to mitosis in the zygote.
Intriguingly, however, it seems that not all aspects of mitotic chromosome segregation are adopted immediately in the zygote. Mitotic cells lose chromosome cohesion in a step-wise manner: cohesion between chromosome arms is eliminated during prophase, whereas centromere cohesion is protected, giving rise to the characteristic X-shape (or V-shape in telocentric species) of metaphase chromosomes57, 58. Metaphase chromosomes of zygotes, however, retain chromosome arm cohesion13. Furthermore, mitotic chromosome spreads during early embryonic divisions suggest that arm cohesion may not be lost until the 8-cell stage of the embryo59. This raises the interesting question as to how cohesin regulatory mechanisms are re-tuned during the egg to embryo transition.
From meiotic to mitotic spindle
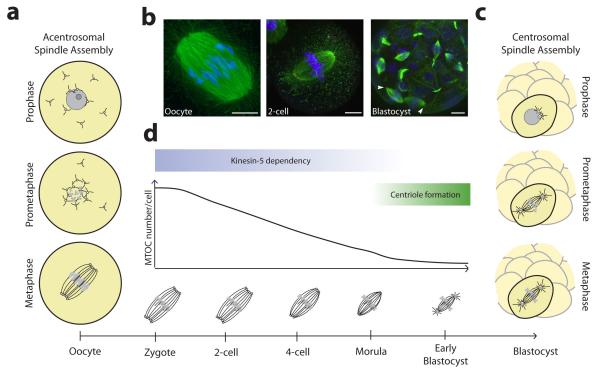
Chromosome segregation is mediated by microtubules, which assemble a bipolar spindle structure that captures and aligns chromosomes before distributing them equally between daughter cells during cell division. In somatic cells, the major microtubule organising centre (MTOC) is the centrosome, an organelle consisting of a pair of centrioles surrounded by a cloud of pericentriolar material60. Centrosomes are instrumental in spindle assembly during mitosis, with each centrosome forming one of the poles of the bipolar spindle (FIG. 4c). Female meiotic cells, however, are unique in that they do not contain canonical centriole-containing centrosomes61. In this section we discuss the mechanism of acentrosomal spindle assembly in mammalian oocytes, and how the transition from acentrosomal to centrosomal spindle assembly is achieved after fertilization.
Figure 4. Transition from acentrosomal to centrosomal spindle assembly.
a | Acentrosomal spindle assembly in mouse oocytes is driven by multiple acentriolar microtubules organising centres (aMTOCs) that self-assemble a bipolar spindle. b | An acentrosomal spindle with unfocused poles in the mouse oocyte (left) and 2-cell embryo (middle). Cells in the blastocyst assemble from centrosomes and have focused spindle poles as indicated by arrow heads (right). Scale bars: 10 μm. c | Centrosomal spindle assembly involves the separation of two centrosomes that act as MTOCs and form the spindle poles. d | The transition from acentrosomal to centrosomal spindle assembly during mouse development is a gradual process that is completed by the blastocyst stage. Image of oocyte in part b is courtesy of the author D.Clift. Images of 2-cell embryo and blastocyst in part b are reproduced, with permission, from REF. 73 © (2012) Rockefeller University Press.
Acentrosomal spindle assembly in oocytes
In almost all species studied to date, including humans, centrosomes are absent from oocyte meiotic spindles61. Electron microscopy of mouse oocytes showed that centrioles are present in fetal oocytes at pachytene stage, but not thereafter, indicative of active centriole elimination during oogenesis62. However, the underlying mechanisms of centriole elimination still remain poorly understood. A recent study in C. elegans identified a role for the mRNA stabilizing factor CGH-1 in this process63, although factors directly affecting centriole elimination have yet to be identified.
So how do mammalian oocytes assemble a meiotic spindle without centrosomes? At the onset of meiotic maturation, mouse oocytes contain numerous acentriolar MTOCs (aMTOCs) that exhibit similar microtubule nucleation properties as centrosomes64. These aMTOCs appear to consist of an electron dense material62, and contain gamma-tubulin and pericentrin, suggesting that they may be similar to the pericentriolar material of centrosomes, although their exact composition and structure is unclear. Upon nuclear envelope breakdown, GTP-bound Ran promotes a massive increase in microtubule nucleation from aMTOCs64, 65. The microtubules then self-assemble into a bipolar spindle through the action of the plus-end directed motor protein Kinesin-5, with aMTOCs distributed to spindle poles64, 66 (FIG 4a). An important challenge for the future will be to address whether human oocytes employ a similar mechanism of acentrosomal spindle assembly.
Transition to centrosomal spindle assembly
When and how does the transition from acentrosomal to centrosomal spindle assembly occur following fertilization? During mouse development, this transition is a gradual process. In rodents, sperm centrioles degenerate during spermatogenesis, and the sperm brings little or no centrosome material into the egg at fertilization67, 68. The first three embryonic divisions in the mouse are meiosis-like, with the spindle assembling from multiple aMTOCs similar to the oocyte69-73. During the subsequent three divisions (morula-blastocyst), the number of aMTOCs from which the spindles are assembled gradually decreases, and some cells begin to show centrin-positive staining73, consistent with the first appearance of centrioles72, 74. Also the spindle morphology gradually adopts the typical mitotic appearance: the length of the spindle decreases and the spindle poles become more focussed as the embryo develops (FIG. 4b,d). Finally, in the blastocyst, all cells assemble a spindle from two centrin-positive MTOCs, indicative of a typical centrosome-driven mitosis73. At about the same time, maintenance of spindle bipolarity shifts from kinesin-5-dependent to – independent75, consistent with the observation that centrosomal spindles do not rely on kinesin-5 to maintain bipolarity76. Therefore, the transition from the meiotic to mitotic spindle machinery occurs gradually in mice, and involves the de novo production of centrioles in the blastocyst (FIG. 4d).
Whether a similar gradual transition occurs during human development, however, is unclear. In humans, sperm centrioles are not completely lost, and so the human sperm delivers one intact and one partially degenerate centriole to the egg at fertilization77. Indeed, centrioles can be detected at spindle poles in the human zygote78, suggestive of a centrosomal mechanism of spindle assembly and therefore a switch from meiotic to mitotic spindle function immediately after fertilization. However, whether the human zygotic spindle is assembled exclusively by a centrosomal mechanism, or if residual aMTOCs from the egg continue to contribute to spindle assembly in the embryo has yet to be tested. In this regard, it is intriguing that sperm centrioles are not required for the initial embryonic divisions, because human parthenotes develop to the blastocyst stage79, 80.
From asymmetric to symmetric divisions
One of the most dramatic transitions that is triggered by fertilization is the switch from the extremely asymmetric meiotic divisions of the oocyte to the symmetric division of the one cell embryo. In this section, we will review our current understanding of the molecular and cellular machineries that support these two contrasting types of divisions at the transition from meiosis to mitosis.
Asymmetric spindle positioning in oocytes
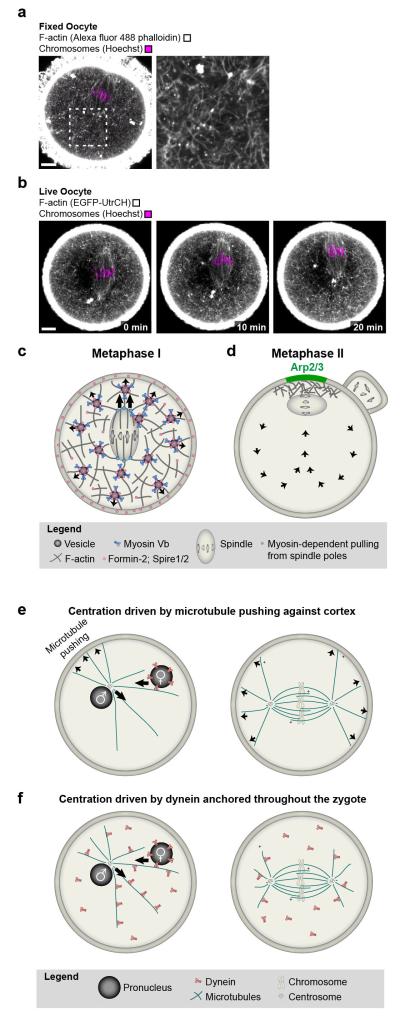
The meiotic divisions of the oocyte are the most asymmetric divisions that are known in mammals. They are essential to form a large egg, which contains sufficient storage material for embryo development. To divide asymmetrically, mammalian oocytes have to move the spindle from their centre to the cortex using an actin-dependent mechanism81 (reviewed in82). Several recent studies in mouse oocytes showed that asymmetric spindle positioning requires a dynamic actin network that fills the cytoplasm and is nucleated by cooperation between the actin nucleators Formin-2, Spire1 and Spire283-87 (FIG. 5a,c). The actin network includes Rab11a-positive vesicles as building blocks which recruit the network’s nucleators and assemble actin filaments from the vesicles’ surface88, 89. The vesicles also recruit the motor protein myosin-Vb, which continuously transports the vesicles along the network’s actin filaments towards the plasma membrane88 and drives the dynamics of the actin network89 (FIG. 5d).
Figure 5. Transition from asymmetric to symmetric spindle positioning.
a | A cytoplasmic F-actin network mediates asymmetric spindle positioning during meiosis I in mouse oocytes. F-actin (grey; Alexa fluor 488 phalloidin) and chromosomes (magenta; Hoechst) in fixed mouse oocyte. Scale bar: 10 μm. b | Autofluorescence (green) around spindle region and chromosomes (magenta; Hoechst) in fixed mouse oocyte. Scale bar: 10 μm. c | F-actin (grey; EGFP-UtrCH) and chromosomes (magenta; Hoechst) in live oocyte during asymmetric spindle positioning. Scale bar: 10 μm. d | Model for vesicle-actin network mediated asymmetric spindle positioning during the first meiotic division e | Model for how the Arp2/3-complex helps to maintain the metaphase II spindle in cortical proximity while the egg awaits fertilization. f | Model for centration of pronuclei and first mitotic spindle by pushing of astral microtubules against the cortex. g | Model for centration of pronuclei and first mitotic spindle driven by dynein that is anchored throughout the cytoplasm.
Recent work has shed light on how the vesicle-actin network might mediate asymmetric spindle positioning. The spindle poles locally contract the network’s actin filaments in a myosin light chain kinase-dependent manner and thereby pull on the network85. This pulling could couple the spindle to the outward-directed movement of the Rab11a-positive vesicles and their associated actin filaments (FIG. 5d). Consistent with this model, Rab11a-positive vesicles89, myosin Vb89, Myosin light chain kinase activity85 and the dynamics of the vesicle-actin network89 are required for asymmetric spindle positioning. In addition, Mos90, 91, Cdc4292 and the Arp2/3 complex93-95 have been implicated in asymmetric spindle positioning, but their precise function remains to be investigated.
A previous study suggested that the spindle might be pushed to the cortex by an actin cloud at the back of the spindle96. However, the actin reporter that was used in this study was later found to be unsuitable for labeling intracellular actin structures in oocytes97. The actin cloud signal might have represented autofluoresence from FAD2+ in the oocyte’s mitochondria instead98, which are clustered around the spindle99 (FIG. 5b). Consistent with autofluorescence, the oocyte’s prominent actin cortex was not labeled in the figure and movie that were shown as evidence for the actin cloud (Figure 3c-g and Movie 596). Other reporters for actin in live oocytes as well as staining of fixed oocytes with fluorescent phalloidin do not detect an actin cloud pushing the spindle, but reveal the extensive actin network that fills the oocyte (FIG. 5a,c)83, 85-87.
Also the metaphase II spindle needs to be positioned asymmetrically before polar body extrusion. The metaphase II spindle forms close to the cortex because it assembles from the spindle half that remains in the egg upon polar body extrusion. The maintenance of the metaphase II spindle at the cortex is actin-dependent and requires Rac1100 and the Arp2/3 complex97 (FIG. 5e). The Arp2/3 complex becomes locally activated in the cortical region overlying the spindle where it nucleates a thick cortical actin layer termed ‘actin cap’101, 102. The Arp2/3-dependent actin cap is essential for cytoplasmic flows that have been suggested to push the metaphase II spindle against the cortex97. Analogously, Arp2/3-dependent cytoplasmic flows have also been suggested to promote the late stages of asymmetric spindle positioning during meiosis I103.
Spindle centration in the embryo
In stark contrast to the asymmetric meiotic divisions, the first mitotic division after fertilization needs to be symmetrical to equally distribute storage material between the two blastomeres of the 2-cell embryo. How is this rapid switch to spindle centration achieved? A key step towards centering the first mitotic spindle is the migration of the male and female pronuclei towards the zygote’s centre (FIG. 5f,g). The movement of both the male and the female pronucleus is microtubule-dependent. In most species, including humans, the male pronucleus is associated with the centrosome that is contributed by the sperm during fertilization. The centrosome nucleates a large microtubule aster, termed sperm aster104. The female pronucleus is likely to associate with the sperm aster and to move towards the male pronucleus in a dynein-dependent manner101,106,107. But how do both pronuclei move towards the zygote’s centre? Early studies suggested that pushing of sperm aster microtubules against the zygote’s cortex could move the male pronucleus towards the centre (reviewed in104). However, cortical interactions are dispensable for pronuclear movement in sanddollar eggs105, and microtubules emanating from the sperm aster are too short to interact with the cortex in large zygotes such as those of Xenopus and zebrafish106, 107. Several recent studies support a model in which pulling by dynein that is anchored throughout the zygote mediates the movement of the male pronucleus towards the zygote’s centre106-109. In C. elegans embryos, dynein has been suggested to be anchored on intracellular organelles109, but also the endoplasmic reticulum or other cytoskeletal structures could serves as sites for dynein anchoring107. How is the pronucleus moving inwards if dynein is homogenously distributed throughout the cytoplasm? A length-dependent microtubule pulling mechanism105 could explain this point: the sperm aster’s microtubules are longer towards the centre of zygote so that a higher number of dynein motors should pull the aster inward rather than outward106, 110 (FIG. 5g).
Upon nuclear envelope breakdown, the first mitotic spindle assembles. In C. elegans embryos, astral microtubules interact with the polarized cortex to position the mitotic spindle. In Xenopus and zebrafish zygotes, the spindle’s astral microtubules are too short to reach the cortex, but cytoplasmic dynein has been suggested to help position the spindle106. Whether dynein centers the spindle in mammalian one cell embryos, remains to be investigated. It is also unclear if the actin-dependent mechanisms that drive asymmetric spindle positioning in mammalian oocytes are completely switched off in the one cell embryo. Interestingly, data from mouse111 and pig112 zygotes suggest that actin is also required for pronuclear movement and spindle centration113, but the mechanism by which actin promotes spindle centration in mammalian embryos is still unclear.
As the distinct cell lineages of the early embryo develop, asymmetry needs to re-established (reviewed in114). It will be interesting to analyze whether the spindle positioning mechanisms that act in the oocyte and one cell embryo are adapted to promote symmetric and asymmetric spindle positioning during later stages of early embryo development as well.
Control of gene expression
Transcription comes to a halt towards the end of the growth phase of the oocyte prior to maturation, when the chromatin is highly condensed and surrounding the nucleolus115. It is not until after fertilization, at the 2-cell stage in mice116 or 4-cell stage in humans117, that transcription fully resumes. Therefore, oocyte maturation, fertilization and the multitude of cellular modifications that drive the transition from egg to embryo all occur in the absence of transcriptional regulation. In this section, we discuss how posttranscriptional control of gene expression in mammalian oocytes ensures the storage and timely activation of maternal factors necessary for the egg to embryo transition. We further discuss the mechanisms by which the regulation of gene expression is transferred from maternal to embryonic control after fertilization.
Translational control
During oocyte growth, the genome is actively transcribed. Some transcripts are readily translated into proteins118. However, many transcripts must be stockpiled and their translation prevented until oocyte maturation or until after fertilization. Indeed, there is a dramatic difference in polyribosome-associated mRNAs between prophase and MII oocytes55, and between MII oocytes and zygotes119, suggestive of stage-specific translation initiation of a multitude of transcripts.
Many oocyte mRNAs contain cytoplasmic polyadenylation elements (CPEs) in their 3′ untranslated region120. These CPEs recruit a translation-repressing complex of CPE-binding protein (CPEB) and its binding partner Maskin121. Such transcripts appear to be stored in oocyte-specific subcortical aggregates that may be functionally related to P bodies122. These dormant mRNAs can then be activated for translation by phosphorylation of CPEB, which occurs at the onset of oocyte maturation. This displaces Maskin and promotes the assembly of the translation initiation complex121. The importance of CPEB-mediated translation regulation is underscored by CPEB1-depleted mouse oocytes, which exhibit severe defects in oocyte development123.
A key question is how are different mRNAs activated for translation at different times during oocyte maturation? Recently it was shown that CPEB1 activates the translation of the RNA-binding protein Dazl55. Dazl in turn directs its own translation as well as that of an additional subset of mRNAs that are important for meiotic spindle assembly and early embryonic development55. The sequential activation of two RNA binding proteins, CPEB1 and Dazl, may therefore provide one mechanism by which mammalian oocytes temporally regulate the translation of different subsets of transcripts55. An important challenge for the future will be to determine how fertilization triggers additional changes to the translational landscape in the zygote119.
Eliminating the maternal stockpile
The handover from maternal to embryonic control of gene expression requires that the stockpile of maternal transcripts is significantly reduced. However, oocyte mRNAs are inherently stable, remaining in the oocyte for up to a few weeks prior to ovulation124. There must therefore be a dramatic change in the stability of the maternal transcript pool at some point during the fertilization process. It turns out that the degradation of excess maternal mRNA begins at the onset of oocyte maturation125, and continues beyond fertilization, with mRNA levels reaching a minimum at the 2-cell stage in mice126 (FIG. 6). In fact, maternal transcripts are degraded in a timely and selective manner. During oocyte maturation, transcripts involved in processes related to earlier events such as oocyte growth and prophase arrest are rapidly degraded, whereas those transcripts important for maintaining the metaphase II arrest remain intact127. Similarly, transcripts involved in meiotic processes, but not embryonic development, are rapidly degraded upon fertilization128.
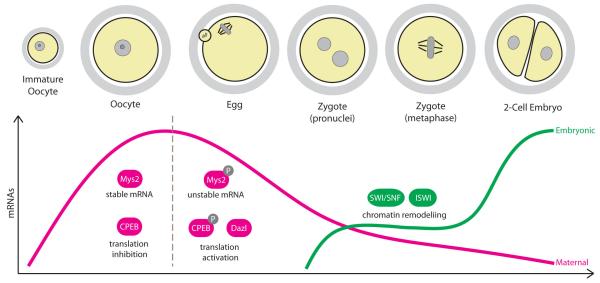
Figure 6. Regulation of gene expression.
During the growing phase, oocytes accumulate large amounts of mRNA necessary to drive the transcriptionally silent phase of oocyte maturation and fertilization. Global mRNA stability in the oocyte is ensured by the RNA-binding protein Mys2 and many mRNAs are held in a translationally-repressed state by binding of CPEB to their 3′UTRs. Maternal mRNAs begin to be translated and/or degraded during oocyte maturation, which involves the phosphorylation of Mys2 and CPEB, and mRNA levels reach a minimum at the 2-cell stage. Transcription from the embryonic genome is initiated firstly in the zygote followed by a robust activation of transcription in the 2-cell embryo, and involves extensive chromatin remodelling.
How are maternal transcripts degraded in mammals? Work from Schultz and colleagues have identified a critical function for the RNA-binding protein Mys2 in this process. Mouse oocytes lacking Mys2 contain less total mRNA and the stability of reporter mRNAs injected into these oocytes is substantially reduced, suggesting that Mys2 promotes global mRNA stability in oocytes129. A clue that regulation of Mys2 may play a role in mRNA degradation came from the finding that Mys2 is phosphorylated in a Cdk1-dependent manner during meiotic maturation130.
Importantly, expression of a non-phosphorylatable Mys2 prevents maturation-induced mRNA degradation, whereas expression of a phosphomimetic mutant can induce mRNA degradation even in prophase-arrested oocytes130. Therefore, phosphorylation of Mys2 during meiotic maturation may inactivate Mys2 and trigger a transition from mRNA stability to instability129. Further work is required to elucidate the mechanism by which Mys2 binding confers mRNA stability and how phosphorylation inactivates Mys2 during oocyte maturation. What regulates the selectivity of transcript degradation during the oocyte to embryo transition also remains completely unknown127, 128.
Reactivating the genome
The final step in the transition from maternal to embryonic control of gene expression involves the activation of transcription from the newly formed embryonic genome, a process termed zygotic genome activation. The vital importance of zygotic genome activation for embryo development was first realised in the 1970s, when it was found that inhibition of transcription using the RNA polymerase inhibitor α-amantin caused mouse embryos to arrest at the 2-cell stage131, 132. Subsequent studies have shown that zygotic genome activation begins in the zygote, where there is a low level of transcription primarily from the paternal pronucleus133, followed by a major wave of transcription in the 2-cell embryo116 (FIG. 6).
In general, transcription is associated with an ‘open’ chromatin state, which allows transcription factors to easily access the DNA134. Therefore, it has long been proposed that chromatin remodelling plays a key role in the initiation of zygotic genome activation135. Consistently, mouse embryos lacking the maternal copy of Brg1, the catalytic subunit of the SWI/SNF chromatin remodelling complex, arrest at the 2-cell stage with reduced transcription of 30% of expressed genes136. Brg1-depleted embryos also show decreased H3K4 dimethylation, which is a marker for transcriptionally active chromatin136. The localization of both Brg1 and the catalytic subunit of the ISWI chromatin remodelling complex, SNF2H, is perturbed in embryos where TIF1α (transcription intermediary factor 1α) is inhibited, and such embryos exhibit misregulated transcription of a subset of genes in the zygote137. Therefore, chromatin remodelling by maternal complexes appears to be an important step for zygotic genome activation in mice. The identification of additional maternal factors essential for zygotic genome activation will be necessary to further our understanding of how the genome is reactivated after fertilization.
Conclusions and perspectives
Recent work has begun to shed light on the complexity of the cellular programmes that transform two highly specialized cells, the sperm and the egg, into a totipotent mitotic embryo. Reprogramming takes place on all levels: the highly condensed chromatin of the sperm head is re-wrapped around nucleosomes; genomic imprinting is selectively erased and re-established; protein expression shifts from translational control back to transcriptional control; linkages between sister chromatids are remodelled to support the switch from reductional meiotic to equational mitotic divisions; the centrosome, which is absent from the egg, needs to be reintroduced; and the axis of cell division is shifted from extremely asymmetric to symmetric. And all this is triggered by the fusion of the sperm with egg, causing an increase in intracellular Ca2+ levels.
How abrupt or gradual these transitions are achieved, varies for each aspect. Several features of mitosis are adopted immediately in the zygote, for example mitotic cohesin complexes immediately replace their meiotic counterparts and cell division rapidly switches from asymmetric to symmetric. Other mitotic features develop more gradually. For example, the switch from translational to transcriptional control of protein expression is gradually achieved over the first mitotic divisions; and the switch to canonical mitotic centrosomal spindle assembly is not observed until the blastocyst stage during mouse development.
Although parts of the complex transition puzzle are beginning to emerge, we are still far from having a clear picture. What are the molecules and mechanisms involved in re-packaging the paternal genome? How is methylation at imprinted regions preserved during global reprogramming? What are the mechanisms responsible for degradation of maternal transcripts? How is transcription re-initiated in the embryo? These are just a few of the many unanswered questions that will need to be addressed in the future.
Exploring the mechanisms that drive the reprogramming of the highly specialized meiotic germ cells towards a totipotent embryo is not only of great interest from a fundamental science point of view, but also directly relevant to understanding the development of embryonic stem cells and their potential for therapeutic use in the future138. Independently of where you come from, the transition from meiosis to mitosis certainly represents an exciting and underexplored field for future research.
Glossary
- Zona pellucida
a specialized extracellular matrix that forms a thick coat surrounding the egg.
- Polyspermy
an egg is fertilized by more than one sperm.
- TEV protease technology
technique of introducing the consensus sequence for the tobacco etch virus (TEV) protease into proteins so that they may be artificially cleaved in vivo by ectopic expression of TEV protease.
- Anaphase Promoting Complex/Cyclosome
an E3 ubiquitin ligase that triggers anaphase by ubiquitinating cyclin B and securin, targeting them for proteolysis by the proteasome.
- Separase
a cysteine protease that triggers chromosome segregation by cleaving chromosomal cohesin complexes
- Pronucleus
the haploid nucleus of either sperm or egg within the zygote
- Reductional chromosome segregation
During meiosis I, pairs of homologous chromosomes are segregated so that each daughter cell contains half the number of chromosomes as the parent.
- Equational chromosome segregation
During meiosis II and mitosis, sister chromatids are segregated so that each daughter cell contains the same number of chromosomes as the parent.
- Totipotency
the ability of a cell to divide and produce all the differentiated cells of a complete organism.
- Polysome
a cluster of ribosomes bound to an mRNA molecule, indicative of active translation.
- N-end rule pathway
a ubiquitin-proteosome proteolysis pathway that regulates the half-life of cellular proteins based on their N-terminal amino acid residue.
- Telocentric
centromeres located at the ends of chromosomes close to telomeres
Footnotes
Competing interests statement: The authors declare no competing interests.
References
- 1.Johnson J, Canning J, Kaneko T, Pru JK, Tilly JL. Germline stem cells and follicular renewal in the postnatal mammalian ovary. Nature. 2004;428:145–150. doi: 10.1038/nature02316. [DOI] [PubMed] [Google Scholar]
- 2.White YAR, et al. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat Med. 2012;18:413–421. doi: 10.1038/nm.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li R, Albertini DF. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat Rev Mol Cell Biol. 2013;14:141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- 4.Wassarman PM, Litscher ES. Mammalian fertilization: the egg’s multifunctional zona pellucida. Int J Dev Biol. 2008;52:665–76. doi: 10.1387/ijdb.072524pw. [DOI] [PubMed] [Google Scholar]
- 5.Bleil JD, Wassarman PM. Mammalian sperm-egg interaction: identification of a glycoprotein in mouse egg zonae pellucidae possessing receptor activity for sperm. Cell. (1980;20:873–82. doi: 10.1016/0092-8674(80)90334-7. [DOI] [PubMed] [Google Scholar]
- 6.Vazquez MH, Phillips DM, Wassarman PM. Interaction of mouse sperm with purified sperm receptors covalently linked to silica beads. J Cell Sci. 1989;92(Pt 4):713–22. doi: 10.1242/jcs.92.4.713. [DOI] [PubMed] [Google Scholar]
- 7.Rankin TL, et al. Human ZP3 restores fertility in Zp3 null mice without affecting order-specific sperm binding. Development. (1998;125:2415–24. doi: 10.1242/dev.125.13.2415. [DOI] [PubMed] [Google Scholar]
- 8.Dean J. Reassessing the molecular biology of sperm–egg recognition with mouse genetics. BioEssays. 2004;26:29–38. doi: 10.1002/bies.10412. [DOI] [PubMed] [Google Scholar]
- 9.Bleil JD, Beall CF, Wassarman PM. Mammalian sperm-egg interaction: Fertilization of mouse eggs triggers modification of the major zona pellucida glycoprotein, ZP2. Developmental Biology. 1981;86:189–197. doi: 10.1016/0012-1606(81)90329-8. [DOI] [PubMed] [Google Scholar]
- 10.Burkart AD, Xiong B, Baibakov B, Jimenez-Movilla M, Dean J. Ovastacin, a cortical granule protease, cleaves ZP2 in the zona pellucida to prevent polyspermy. J Cell Biol. 2012;197:37–44. doi: 10.1083/jcb.201112094. [This study identifies Ovastacin as the protease responsible for ZP2 cleavage, and shows that it is a component of cortical granules] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gahlay G, Gauthier L, Baibakov B, Epifano O, Dean J. Gamete recognition in mice depends on the cleavage status of an egg’s zona pellucida protein. Science. 2010;329:216–9. doi: 10.1126/science.1188178. [Using mouse genetics, this study demonstrates that sperm-egg binding depends on the cleavage status of the zona pellucida protein ZP2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baibakov B, Boggs NA, Yauger B, Baibakov G, Dean J. Human sperm bind to the N-terminal domain of ZP2 in humanized zonae pellucidae in transgenic mice. The Journal of Cell Biology. 2012;197:897–905. doi: 10.1083/jcb.201203062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tachibana-Konwalski K, et al. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev. 2010;24:2505–16. doi: 10.1101/gad.605910. [This study shows that there is a rapid switch from meiotic to mitotic cohesin complexes upon fertilization in mice] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pang P-C, et al. Human Sperm Binding Is Mediated by the Sialyl-Lewisx Oligosaccharide on the Zona Pellucida. Science. 2011;333:1761–1764. doi: 10.1126/science.1207438. [DOI] [PubMed] [Google Scholar]
- 15.Clark GF. The role of carbohydrate recognition during human sperm–egg binding. Human Reproduction. 2013;28:566–577. doi: 10.1093/humrep/des447. [DOI] [PubMed] [Google Scholar]
- 16.Wassarman PM, Jovine L, Litscher ES. A profile of fertilization in mammals. Nat Cell Biol. 2001;3:E59–64. doi: 10.1038/35055178. [DOI] [PubMed] [Google Scholar]
- 17.Ridgway EB, Gilkey JC, Jaffe LF. Free calcium increases explosively in activating medaka eggs. Proc Natl Acad Sci U S A. 1977;74:623–7. doi: 10.1073/pnas.74.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinhardt R, Zucker R, Schatten G. Intracellular calcium release at fertilization in the sea urchin egg. Dev Biol. 1977;58:185–96. doi: 10.1016/0012-1606(77)90084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ducibella T, et al. Egg-to-embryo transition is driven by differential responses to Ca(2+) oscillation number. Dev Biol. 2002;250:280–91. [PubMed] [Google Scholar]
- 20.Horner VL, Wolfner MF. Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev Dyn. 2008;237:527–44. doi: 10.1002/dvdy.21454. [DOI] [PubMed] [Google Scholar]
- 21.Saunders CM, et al. PLC zeta: a sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development. 2002;129:3533–44. doi: 10.1242/dev.129.15.3533. [DOI] [PubMed] [Google Scholar]
- 22.Miyazaki S, et al. Block of Ca2+ wave and Ca2+ oscillation by antibody to the inositol 1,4,5-trisphosphate receptor in fertilized hamster eggs. Science. 1992;257:251–5. doi: 10.1126/science.1321497. [DOI] [PubMed] [Google Scholar]
- 23.Miyazaki S, Shirakawa H, Nakada K, Honda Y. Essential role of the inositol 1,4,5-trisphosphate receptor/Ca2+ release channel in Ca2+ waves and Ca2+ oscillations at fertilization of mammalian eggs. Dev Biol. 1993;158:62–78. doi: 10.1006/dbio.1993.1168. [DOI] [PubMed] [Google Scholar]
- 24.Masui Y, Markert CL. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J Exp Zool. 1971;177:129–145. doi: 10.1002/jez.1401770202. [DOI] [PubMed] [Google Scholar]
- 25.Rauh NR, Schmidt A, Bormann J, Nigg EA, Mayer TU. Calcium triggers exit from meiosis II by targeting the APC/C inhibitor XErp1 for degradation. Nature. 2005;437:1048–52. doi: 10.1038/nature04093. [A must read paper that elucidates the molecular mechanism by which a rise in intracellular calcium triggers exit from meiosis/metaphase II arrest] [DOI] [PubMed] [Google Scholar]
- 26.Hansen DV, Tung JJ, Jackson PK. CaMKII and polo-like kinase 1 sequentially phosphorylate the cytostatic factor Emi2/XErp1 to trigger its destruction and meiotic exit. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:608–13. doi: 10.1073/pnas.0509549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Maller JL. Calcium elevation at fertilization coordinates phosphorylation of XErp1/Emi2 by Plx1 and CaMK II to release metaphase arrest by cytostatic factor. Current biology: CB. 2005;15:1458–68. doi: 10.1016/j.cub.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 28.Shoji S, et al. Mammalian Emi2 mediates cytostatic arrest and transduces the signal for meiotic exit via Cdc20. The EMBO journal. 2006;25:834–45. doi: 10.1038/sj.emboj.7600953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madgwick S, Hansen DV, Levasseur M, Jackson PK, Jones KT. Mouse Emi2 is required to enter meiosis II by reestablishing cyclin B1 during interkinesis. The Journal of cell biology. 2006;174:791–801. doi: 10.1083/jcb.200604140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tischer T, Hormanseder E, Mayer TU. The APC/C inhibitor XErp1/Emi2 is essential for Xenopus early embryonic divisions. Science. 2012;338:520–4. doi: 10.1126/science.1228394. [This study demonstrates that Emi2 regulates APC/C activity during early embryonic divisions of Xenopus laevis] [DOI] [PubMed] [Google Scholar]
- 31.Schmidt A, Rauh NR, Nigg EA, Mayer TU. Cytostatic factor: an activity that puts the cell cycle on hold. Journal of cell science. 2006;119:1213–8. doi: 10.1242/jcs.02919. [DOI] [PubMed] [Google Scholar]
- 32.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–8. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 33.van der Heijden GW, et al. Asymmetry in Histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mechanisms of Development. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol. 2006;50:455–61. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- 35.Puschendorf M, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411–20. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi H, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 38.Oswald J, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–8. doi: 10.1016/s0960-9822(00)00448-6. [References 37 and 38 were the first to show that the paternal genome is actively demethylated independently of DNA replication following fertilization] [DOI] [PubMed] [Google Scholar]
- 39.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–7. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wossidlo M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 41.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–10. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura T, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 43.Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280:225–36. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura T, et al. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–9. doi: 10.1038/nature11093. [This paper reveals the mechanism by which PGC7 protects maternal chromatin from active demethylation in the zygote] [DOI] [PubMed] [Google Scholar]
- 45.Rougier N, et al. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–13. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–44. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 48.Li X, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–57. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quenneville S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–72. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Messerschmidt DM, et al. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- 51.Nasmyth K, Haering CH. Cohesin: Its Roles and Mechanisms. Annual Review of Genetics. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 53.Parra MT, et al. Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J Cell Sci. 2004;117:1221–34. doi: 10.1242/jcs.00947. [DOI] [PubMed] [Google Scholar]
- 54.Xu H, et al. A new role for the mitotic RAD21/SCC1 cohesin in meiotic chromosome cohesion and segregation in the mouse. EMBO Rep. 2004;5:378–84. doi: 10.1038/sj.embor.7400121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen J, et al. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25:755–66. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao H, Uhlmann F, Nasmyth K, Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–9. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- 57.Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- 58.Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–62. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–78. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 61.Manandhar G, Schatten H, Sutovsky P. Centrosome reduction during gametogenesis and its significance. Biol Reprod. 2005;72:2–13. doi: 10.1095/biolreprod.104.031245. [DOI] [PubMed] [Google Scholar]
- 62.Calarco PG, Donahue RP, Szollosi D. Germinal vesicle breakdown in the mouse oocyte. J Cell Sci. 1972;10:369–85. doi: 10.1242/jcs.10.2.369. [DOI] [PubMed] [Google Scholar]
- 63.Mikeladze-Dvali T, et al. Analysis of centriole elimination during C. elegans oogenesis. Development. 2012;139:1670–9. doi: 10.1242/dev.075440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–98. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 65.Dumont J, et al. A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J Cell Biol. 2007;176:295–305. doi: 10.1083/jcb.200605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Breuer M, et al. HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J Cell Biol. 2010;191:1251–60. doi: 10.1083/jcb.201005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woolley DM, Fawcett DW. The degeneration and disappearance of the centrioles during the development of the rat spermatozoon. Anat Rec. 1973;177:289–301. doi: 10.1002/ar.1091770209. [DOI] [PubMed] [Google Scholar]
- 68.Manandhar G, Sutovsky P, Joshi HC, Stearns T, Schatten G. Centrosome reduction during mouse spermiogenesis. Dev Biol. 1998;203:424–34. doi: 10.1006/dbio.1998.8947. [DOI] [PubMed] [Google Scholar]
- 69.Calarco-Gillam PD, Siebert MC, Hubble R, Mitchison T, Kirschner M. Centrosome development in early mouse embryos as defined by an autoantibody against pericentriolar material. Cell. 1983;35:621–9. doi: 10.1016/0092-8674(83)90094-6. [DOI] [PubMed] [Google Scholar]
- 70.Maro B, Howlett SK, Webb M. Non-spindle microtubule organizing centers in metaphase II-arrested mouse oocytes. The Journal of Cell Biology. 1985;101:1665–1672. doi: 10.1083/jcb.101.5.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hiraoka L, Golden W, Magnuson T. Spindle-pole organization during early mouse development. Developmental Biology. 1989;133:24–36. doi: 10.1016/0012-1606(89)90293-5. [DOI] [PubMed] [Google Scholar]
- 72.Gueth-Hallonet C, et al. gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J Cell Sci. 1993;105(Pt 1):157–66. doi: 10.1242/jcs.105.1.157. [DOI] [PubMed] [Google Scholar]
- 73.Courtois A, Schuh M, Ellenberg J, Hiiragi T. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol. 2012;198:357–70. doi: 10.1083/jcb.201202135. [Shows that the transition from meiotic to mitotic spindle assembly occurs gradually in early mouse embryos, where the centrosome is not introduced by the sperm] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Howe K, FitzHarris G. A non-canonical mode of microtubule organization operates throughout pre-implantation development in mouse. Cell Cycle. 2013;12:1616–1624. doi: 10.4161/cc.24755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fitzharris G. A shift from kinesin 5-dependent metaphase spindle function during preimplantation development in mouse. Development. 2009;136:2111–9. doi: 10.1242/dev.035089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. Probing Spindle Assembly Mechanisms with Monastrol, a Small Molecule Inhibitor of the Mitotic Kinesin, Eg5. The Journal of Cell Biology. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manandhar G, Simerly C, Schatten G. Highly degenerated distal centrioles in rhesus and human spermatozoa. Human Reproduction. 2000;15:256–263. doi: 10.1093/humrep/15.2.256. [DOI] [PubMed] [Google Scholar]
- 78.Sathananthan AH, et al. Centrioles in the beginning of human development. Proc Natl Acad Sci U S A. 1991;88:4806–10. doi: 10.1073/pnas.88.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paffoni A, et al. In vitro development of human oocytes after parthenogenetic activation or intracytoplasmic sperm injection. Fertil Steril. 2007;87:77–82. doi: 10.1016/j.fertnstert.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 80.de Fried EP, et al. Human parthenogenetic blastocysts derived from noninseminated cryopreserved human oocytes. Fertil Steril. 2008;89:943–7. doi: 10.1016/j.fertnstert.2007.04.045. [DOI] [PubMed] [Google Scholar]
- 81.Longo FJ, Chen DY. Development of cortical polarity in mouse eggs: involvement of the meiotic apparatus. Developmental biology. 1985;107:382–94. doi: 10.1016/0012-1606(85)90320-3. [DOI] [PubMed] [Google Scholar]
- 82.Chaigne A, Verlhac MH, Terret ME. Spindle positioning in mammalian oocytes. Experimental cell research. 2012;318:1442–7. doi: 10.1016/j.yexcr.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 83.Azoury J, et al. Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr Biol. 2008;18:1514–9. doi: 10.1016/j.cub.2008.08.044. [DOI] [PubMed] [Google Scholar]
- 84.Leader B, et al. Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat Cell Biol. 2002;4:921–8. doi: 10.1038/ncb880. [DOI] [PubMed] [Google Scholar]
- 85.Schuh M, Ellenberg J. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol. 2008;18:1986–92. doi: 10.1016/j.cub.2008.11.022. [References 83 and 85 were the first studies demonstrating that asymmetric spindle positioning relies on a Fmn2-dependent cytoplasmic actin network in mouse oocytes] [DOI] [PubMed] [Google Scholar]
- 86.Azoury J, Lee KW, Georget V, Hikal P, Verlhac MH. Symmetry breaking in mouse oocytes requires transient F-actin meshwork destabilization. Development. 2011;138:2903–8. doi: 10.1242/dev.060269. [DOI] [PubMed] [Google Scholar]
- 87.Pfender S, Kuznetsov V, Pleiser S, Kerkhoff E, Schuh M. Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Current biology: CB. 2011;21:955–60. doi: 10.1016/j.cub.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schuh M. An actin-dependent mechanism for long-range vesicle transport. Nature cell biology. 2011;13:1431–6. doi: 10.1038/ncb2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Holubcová Z, Schuh M. A new function for vesicles: modulators of an actin network for asymmetric spindle positioning. Nat Cell Biol. 2013;15:937–947. doi: 10.1038/ncb2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Verlhac MH, Lefebvre C, Guillaud P, Rassinier P, Maro B. Asymmetric division in mouse oocytes: with or without Mos. Curr Biol. 2000;10:1303–6. doi: 10.1016/s0960-9822(00)00753-3. [DOI] [PubMed] [Google Scholar]
- 91.Choi T, et al. The Mos/mitogen-activated protein kinase (MAPK) pathway regulates the size and degradation of the first polar body in maturing mouse oocytes. Proc Natl Acad Sci U S A. 1996;93:7032–5. doi: 10.1073/pnas.93.14.7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Na J, Zernicka-Goetz M. Asymmetric positioning and organization of the meiotic spindle of mouse oocytes requires CDC42 function. Curr Biol. 2006;16:1249–54. doi: 10.1016/j.cub.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 93.Sun SC, et al. Arp2/3 complex regulates asymmetric division and cytokinesis in mouse oocytes. PLoS One. 2011;6:e18392. doi: 10.1371/journal.pone.0018392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun SC, et al. WAVE2 regulates meiotic spindle stability, peripheral positioning and polar body emission in mouse oocytes. Cell cycle. 2011;10:1853–60. doi: 10.4161/cc.10.11.15796. [DOI] [PubMed] [Google Scholar]
- 95.Sun SC, Sun QY, Kim NH. JMY is required for asymmetric division and cytokinesis in mouse oocytes. Molecular human reproduction. 2011;17:296–304. doi: 10.1093/molehr/gar006. [DOI] [PubMed] [Google Scholar]
- 96.Li H, Guo F, Rubinstein B, Li R. Actin-driven chromosomal motility leads to symmetry breaking in mammalian meiotic oocytes. Nature cell biology. 2008;10:1301–8. doi: 10.1038/ncb1788. [DOI] [PubMed] [Google Scholar]
- 97.Yi K, et al. Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nature cell biology. 2011;13:1252–8. doi: 10.1038/ncb2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dumollard R, Ward Z, Carroll J, Duchen MR. Regulation of redox metabolism in the mouse oocyte and embryo. Development. 2007;134:455–465. doi: 10.1242/dev.02744. [DOI] [PubMed] [Google Scholar]
- 99.Dalton CM, Carroll J. Biased inheritance of mitochondria during asymmetric cell division in the mouse oocyte. J Cell Sci. 2013 doi: 10.1242/jcs.128744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Halet G, Carroll J. Rac activity is polarized and regulates meiotic spindle stability and anchoring in mammalian oocytes. Developmental cell. 2007;12:309–17. doi: 10.1016/j.devcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 101.Deng M, Suraneni P, Schultz RM, Li R. The Ran GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Developmental cell. 2007;12:301–8. doi: 10.1016/j.devcel.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 102.Dehapiot B, Halet G. Ran promotes oocyte polarization by regulating ERM (Ezrin/Radixin/Moesin) activation. Cell Cycle. 2013;12 doi: 10.4161/cc.24901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yi K, et al. Sequential actin-based pushing forces drive meiosis I chromosome migration and symmetry breaking in oocytes. The Journal of cell biology. 2013;200:567–76. doi: 10.1083/jcb.201211068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reinsch S, Gonczy P. Mechanisms of nuclear positioning. Journal of cell science. 1998;111(Pt 16):2283–95. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 105.Hamaguchi MS, Hiramoto Y. Analysis of the Role of Astral Rays in Pronuclear Migration in Sand Dollar Eggs by the Colcemid-UV Method. Development, Growth & Differentiation. 1986;28:143–156. doi: 10.1111/j.1440-169X.1986.00143.x. [Using elegant colcemid-UV experiments in sand dollar eggs, Hamaguchi et al. suggest a microtubule length-dependent pulling mechanism for pronuclear centration] [DOI] [PubMed] [Google Scholar]
- 106.Wuhr M, Tan ES, Parker SK, Detrich HW, 3rd, Mitchison TJ. A model for cleavage plane determination in early amphibian and fish embryos. Current biology: CB. 2010;20:2040–5. doi: 10.1016/j.cub.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wuhr M, Dumont S, Groen AC, Needleman DJ, Mitchison TJ. How does a millimeter-sized cell find its center? Cell cycle. 2009;8:1115–21. doi: 10.4161/cc.8.8.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kimura A, Onami S. Computer simulations and image processing reveal length-dependent pulling force as the primary mechanism for C. elegans male pronuclear migration. Developmental cell. 2005;8:765–75. doi: 10.1016/j.devcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 109.Kimura K, Kimura A. Intracellular organelles mediate cytoplasmic pulling force for centrosome centration in the Caenorhabditis elegans early embryo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:137–42. doi: 10.1073/pnas.1013275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shinar T, Mana M, Piano F, Shelley MJ. A model of cytoplasmically driven microtubule-based motion in the single-celled Caenorhabditis elegans embryo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10508–13. doi: 10.1073/pnas.1017369108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schatten G, et al. Latrunculin inhibits the microfilament-mediated processes during fertilization, cleavage and early development in sea urchins and mice. Exp Cell Res. 1986;166:191–208. doi: 10.1016/0014-4827(86)90519-7. [DOI] [PubMed] [Google Scholar]
- 112.Kim NH, Simerly C, Funahashi H, Schatten G, Day BN. Microtubule organization in porcine oocytes during fertilization and parthenogenesis. Biology of reproduction. 1996;54:1397–404. doi: 10.1095/biolreprod54.6.1397. [DOI] [PubMed] [Google Scholar]
- 113.Chew TG, Lorthongpanich C, Ang WX, Knowles BB, Solter D. Symmetric cell division of the mouse zygote requires an actin network. Cytoskeleton. 2012;69:1040–6. doi: 10.1002/cm.21062. [DOI] [PubMed] [Google Scholar]
- 114.Wennekamp S, Mesecke S, Nedelec F, Hiiragi T. A self-organization framework for symmetry breaking in the mammalian embryo. Nat Rev Mol Cell Biol. 2013;14:452–459. doi: 10.1038/nrm3602. [DOI] [PubMed] [Google Scholar]
- 115.Bouniol-Baly C, et al. Differential transcriptional activity associated with chromatin configuration in fully grown mouse germinal vesicle oocytes. Biol Reprod. 1999;60:580–7. doi: 10.1095/biolreprod60.3.580. [DOI] [PubMed] [Google Scholar]
- 116.Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–31. doi: 10.1016/s1534-5807(03)00373-3. [A comprehensive analysis of gene expression during mouse development that identifies waves of maternal transcript degradation and zygotic genome activation] [DOI] [PubMed] [Google Scholar]
- 117.Braude P, Bolton V, Moore S. Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature. 1988;332:459–61. doi: 10.1038/332459a0. [DOI] [PubMed] [Google Scholar]
- 118.Wang S, et al. Proteome of mouse oocytes at different developmental stages. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:17639–17644. doi: 10.1073/pnas.1013185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Potireddy S, Vassena R, Patel BG, Latham KE. Analysis of polysomal mRNA populations of mouse oocytes and zygotes: dynamic changes in maternal mRNA utilization and function. Dev Biol. 2006;298:155–66. doi: 10.1016/j.ydbio.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 120.Oh B, Hwang S, McLaughlin J, Solter D, Knowles BB. Timely translation during the mouse oocyte-to-embryo transition. Development. 2000;127:3795–3803. doi: 10.1242/dev.127.17.3795. [First study to show that transcript stability and translation during oocyte maturation in mammals is regulated by CPEs in the 3′ UTR] [DOI] [PubMed] [Google Scholar]
- 121.Vasudevan S, Seli E, Steitz JA. Metazoan oocyte and early embryo development program: a progression through translation regulatory cascades. Genes & Development. 2006;20:138–146. doi: 10.1101/gad.1398906. [DOI] [PubMed] [Google Scholar]
- 122.Flemr M, Ma J, Schultz RM, Svoboda P. P-Body Loss Is Concomitant with Formation of a Messenger RNA Storage Domain in Mouse Oocytes. Biology of Reproduction. 2010;82:1008–1017. doi: 10.1095/biolreprod.109.082057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Racki WJ, Richter JD. CPEB controls oocyte growth and follicle development in the mouse. Development. 2006;133:4527–37. doi: 10.1242/dev.02651. [DOI] [PubMed] [Google Scholar]
- 124.Jahn CL, Baran MM, Bachvarova R. Stability of RNA synthesized by the mouse oocyte during its major growth phase. Journal of Experimental Zoology. 1976;197:161–171. doi: 10.1002/jez.1401970202. [DOI] [PubMed] [Google Scholar]
- 125.Paynton BV, Rempel R, Bachvarova R. Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Developmental Biology. 1988;129:304–314. doi: 10.1016/0012-1606(88)90377-6. [DOI] [PubMed] [Google Scholar]
- 126.Piko L, Clegg KB. Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Developmental Biology. 1982;89:362–378. doi: 10.1016/0012-1606(82)90325-6. [DOI] [PubMed] [Google Scholar]
- 127.Su Y-Q, et al. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Developmental Biology. 2007;302:104–117. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Alizadeh Z, Kageyama S, Aoki F. Degradation of maternal mRNA in mouse embryos: selective degradation of specific mRNAs after fertilization. Mol Reprod Dev. 2005;72:281–90. doi: 10.1002/mrd.20340. [DOI] [PubMed] [Google Scholar]
- 129.Medvedev S, Pan H, Schultz RM. Absence of MSY2 in Mouse Oocytes Perturbs Oocyte Growth and Maturation, RNA Stability, and the Transcriptome. Biology of Reproduction. 2011;85:575–583. doi: 10.1095/biolreprod.111.091710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Medvedev S, Yang J, Hecht NB, Schultz RM. CDC2A (CDK1)-mediated phosphorylation of MSY2 triggers maternal mRNA degradation during mouse oocyte maturation. Developmental Biology. 2008;321:205–215. doi: 10.1016/j.ydbio.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Golbus MS, Calarco PG, Epstein CJ. The effects of inhibitors of RNA synthesis (alpha-amanitin and actinomycin D) on preimplantation mouse embryogenesis. J Exp Zool. 1973;186:207–16. doi: 10.1002/jez.1401860211. [DOI] [PubMed] [Google Scholar]
- 132.Warner CM, Versteegh LR. In vivo and in vitro effect of alpha-amanitin on preimplantation mouse embryo RNA polymerase. Nature. 1974;248:678–80. doi: 10.1038/248678a0. [DOI] [PubMed] [Google Scholar]
- 133.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 134.Workman JL. Nucleosome displacement in transcription. Genes & Development. 2006;20:2009–2017. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 135.Schultz RM, Worrad DM. Role of chromatin structure in zygotic gene activation in the mammalian embryo. Seminars in Cell Biology. 1995;6:201–208. doi: 10.1006/scel.1995.0028. [DOI] [PubMed] [Google Scholar]
- 136.Bultman SJ, et al. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes & Development. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Torres-Padilla ME, Zernicka-Goetz M. Role of TIF1α as a modulator of embryonic transcription in the mouse zygote. The Journal of Cell Biology. 2006;174:329–338. doi: 10.1083/jcb.200603146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tachibana M, et al. Human Embryonic Stem Cells Derived by Somatic Cell Nuclear Transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lopata A. History of the Egg in Embryology. Journal of Mammalian Ova Research. 2009;26:2–9. [Google Scholar]
- 140.Harvey W, Whitteridge G. Disputations touching the generation of animals. Blackwell Scientific; Oxford; Boston; St. Louis, Missouri: 1981. [Google Scholar]
- 141.Leeuwenhoek A, Observationes D. Anthonii Lewenhoeck, de natis e’ semini genitali animalcules. Phil Trans. 1677;12:1040–1046. [Google Scholar]
- 142.Karl Ernst von B, O’Malley CD. On the Genesis of the Ovum of Mammals and of Man. Isis. 1956;47:117–153. [Google Scholar]
- 143.Cobb M. Heredity before genetics: a history. Nat Rev Genet. 2006;7:953–8. doi: 10.1038/nrg1948. [DOI] [PubMed] [Google Scholar]
- 144.Hertwig O. Beiträge zur Kenntniss der Bildung, Befruchtung und Theilung des thierischen Eies. Morphol Jahrb. 1876;1:347–434. [Google Scholar]
- 145.Magerkurth C, Töpfer-Petersen E, Schwartz P, Michelmann HW. Scanning electron microscopy analysis of the human zona pellucida: influence of maturity and fertilization on morphology and sperm binding pattern. Human Reproduction. 1999;14:1057–1066. doi: 10.1093/humrep/14.4.1057. [DOI] [PubMed] [Google Scholar]