ABSTRACT
Downregulation of the mammalian target of rapamycin (mTOR) pathway by its inhibitor rapamycin is emerging as a potential pharmacological intervention that mimics the beneficial effects of dietary restriction. Modulation of mTOR has diverse effects on mitochondrial metabolism and biogenesis, but the role of the mitochondrial genotype in mediating these effects remains unknown. Here, we use novel mitochondrial genome replacement strains in Drosophila to test the hypothesis that genes encoded in mitochondrial DNA (mtDNA) influence the mTOR pathway. We show that rapamycin increases mitochondrial respiration and succinate dehydrogenase activity, decreases H2O2 production and generates distinct shifts in the metabolite profiles of isolated mitochondria versus whole Drosophila. These effects are disabled when divergent mitochondrial genomes from D. simulans are placed into a common nuclear background, demonstrating that the benefits of rapamycin to mitochondrial metabolism depend on genes encoded in the mtDNA. Rapamycin is able to enhance mitochondrial respiration when succinate dehydrogenase activity is blocked, suggesting that the beneficial effects of rapamycin on these two processes are independent. Overall, this study provides the first evidence for a link between mitochondrial genotype and the effects of rapamycin on mitochondrial metabolic pathways.
KEY WORDS: Rapamycin, Metabolism, Mitochondrial genotype
INTRODUCTION
Mitochondria are specialized organelles that convert metabolic substrates into ATP, the energy currency of cells, through the process of oxidative phosphorylation (OXPHOS). During OXPHOS, NADH and FADH2, which are derived from the mitochondrial tricarboxylic acid (TCA) cycle and fatty-acid oxidation, pass electrons to complex I (NADH dehydrogenase) and complex II (succinate dehydrogenase) of the electron transport chain (ETC), respectively. Electrons are then transferred through complex III (cytochrome bc1) to complex IV (cytochrome c oxidase), where they are passed to oxygen and produce water (Scheffler, 2007). This process generates a proton gradient and establishes a membrane potential (Δψm) across the inner-mitochondrial membrane. Ultimately, the proton gradient is dissipated either at complex V, producing ATP, or through uncoupling proteins producing heat (Scheffler, 2007). Reactive oxygen species (ROS) are generated during electron transport as a by-product of OXPHOS. Levels of ROS are tightly regulated because ROS serve as a secondary messenger to mediate signal transduction and metabolism (Cheng and Ristow, 2013), and excess ROS can damage DNA, lipids and proteins (Balaban et al., 2005; Murphy et al., 2011). In addition to ATP production, mitochondria play crucial roles in amino acid metabolism, carbohydrates metabolism and fatty acid oxidation.
Although mitochondria contain their own genome and function as distinct organelles separated by lipid membranes, mitochondrial processes are crucially dependent on nuclear-encoded gene products and environmental signals in order to accommodate metabolic cellular requirements (Finley and Haigis, 2009; Liu and Butow, 2006; Woodson and Chory, 2008). These interactions require continuous communication between mitochondria and the cytosol (Woodson and Chory, 2008). A crucial link between cytosolic and mitochondrial metabolism is the enzyme succinate dehydrogenase (complex II of the ETC). Succinate dehydrogenase is the only enzyme shared by the TCA cycle and ETC, and it lies at the intersection of pathways that connect cell metabolism with mitochondrial respiration (Scheffler, 2007). In the ETC, succinate dehydrogenase acts as an entry point for electrons from FADH2, produced through fatty-acid oxidation; in the TCA cycle it oxidizes succinate to fumarate. Notably, it is the only complex of the ETC with no mitochondrial DNA (mtDNA)-encoded subunits.
A pathway that has been identified as a potential mediator of cross-talk between mitochondria and the cytosol is the mammalian target of rapamycin (mTOR) (Finley and Haigis, 2009; Schieke and Finkel, 2006). The mTOR pathway is highly conserved and regulates diverse functions related to nutritional cues and cellular stress (Baltzer et al., 2010; Finley and Haigis, 2009). In yeast, reduced TOR signaling enhances mitochondrial respiration and modulates ROS production to extend chronological life span (Bonawitz et al., 2007; Pan et al., 2012). In mammals, the role of the mTOR pathway in mitochondrial function has tissue-specific effects (Bentzinger et al., 2008; Ramanathan and Schreiber, 2009; Cunningham et al., 2007; Paglin et al., 2005; Polak et al., 2008; Schieke et al., 2006; Düvel et al., 2010). The best characterized function of mTOR is the regulation of translation initiation by mTOR complex 1 (mTORC1). This involves mTORC1-mediated phosphorylation of the eukaryotic initiation factor 4E binding protein 1 (eIF4E-BP1) and ribosomal protein S6 kinase 1 (S6K1) in order to upregulate 5′-cap-dependent protein translation (Richter and Sonenberg, 2005). In addition to modulation of protein translation, the mTOR pathway has also been implicated in cell growth, autophagy, longevity and metabolism (Finley and Haigis, 2009; Laplante and Sabatini, 2012; Soliman, 2011; Mathew and White, 2011), all key biological processes in health and homeostasis. Indeed, dysregulation of mTOR signaling is a common condition in human diseases (Dazert and Hall, 2011).
There is increasing interest in identifying the genetic and cellular mechanisms underlying the connection between mTOR signaling and mitochondrial function. The majority of genes that function in mitochondria are encoded by the nuclear genome, but a small and crucial fraction of genes have been retained in the mitochondrial genome since the endosymbiotic origin of the eukaryotic cell roughly two billion years ago (Lane, 2005). The majority of studies seeking to understand the role of the mTOR pathway in mitochondrial function have focused on these nuclear-encoded proteins that are translated on cytosolic ribosomes and imported in to the mitochondrion. The genes encoded in the mitochondrial genome have received little attention in experimental work related to the mechanisms of action in the mTOR pathway.
Here, we test the hypothesis that the relationship between the mTOR pathway and mitochondrial physiology is modified by genes encoded in alternative mitochondrial genomes. To accomplish this we used strains of Drosophila in which different mtDNAs from both D. melanogaster and D. simulans were placed on a single D. melanogaster nuclear background (Montooth et al., 2010). We posit that this genetic manipulation will compromise the co-evolved nature of mitochondria–nuclear cross talk and provide a novel hypomorphic context for analyses of how rapamycin influences mTOR functions. We show that treatment with rapamycin, a specific mTORC1 inhibitor, increases mitochondrial oxidative capacity and respiration rate, as well as decreasing the production of mitochondrial H2O2, one of the ROS produced in the mitochondria. This effect was pronounced in D. melanogaster genotypes carrying the native D. melanogaster mtDNAs, but these beneficial effects of rapamycin were significantly reduced in the strains carrying the ‘foreign’ D. simulans mitochondrial genotypes in the D. melanogaster nuclear background. Using comparative metabolomics analyses, we show that whole fly homogenates have a strong signature of response to rapamycin with only subtle effects of mtDNA genotypes. In contrast, metabolite profiles of isolated mitochondria show strong mtDNA-genotype-dependent responses to rapamycin. The coevolved genotypes carrying the D. melanogaster mtDNAs show clear reprogramming of their carbohydrate and amino acid profiles under rapamycin treatment, but the foreign D. simulans mtDNAs shift these metabolite levels such that rapamycin has little additional effect. We further show that rapamycin treatment specifically affects the ETC at the level of the activity of complex II and does so only for the D. melanogaster mtDNA genotypes. Overall, our experiments confirm that rapamycin has multiple beneficial effects on mitochondrial function and show for the first time that the full effects of these benefits are dependent on the metabolic plasticity mediated by genes encoded in the mtDNA.
RESULTS
Rapamycin increases mitochondrial respiration and decreases H2O2 production
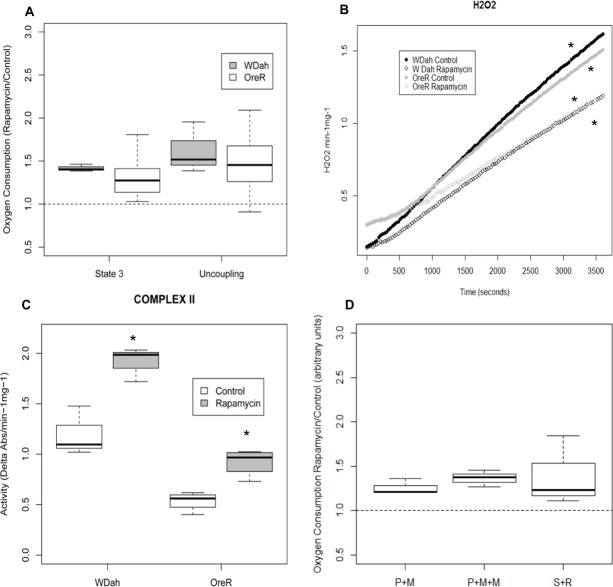
To determine the effect of rapamycin on mitochondrial function, we measured the OXPHOS capacity of mitochondria isolated from two standard laboratory D. melanogaster strains: (1) white Dahomey (wDah), and (2) Oregon R (OreR). Flies were fed food containing 200 µM rapamycin, a concentration shown to inhibit the phosphorylation of TOR target S6K at T398 and to affect protein translation (supplementary material Fig. S1; Bjedov et al., 2010). We observed that after ADP stimulation (state 3 respiration), mitochondria from flies treated with rapamycin had a higher rate of oxygen consumption than from vehicle-treated flies (Fig. 1A), suggesting that rapamycin increases mitochondrial oxidative capacity. Uncoupled mitochondrial respiration rates were measured after the addition of the chemical uncoupler FCCP. Flies treated with rapamycin displayed increased uncoupling, indicative of a higher maximum oxidative capacity (Fig. 1A). We observed that the absolute values for oxygen consumption could vary significantly from one day to the next, but the relative values of rapamycin-treated flies to that of control flies showed consistent differences in oxygen consumption rates, hence we report ratios to show the rapamycin effect.
Fig. 1.
Effects of rapamycin on mitochondrial functions. (A) Oxygen consumption during state 3 (ADP added) and after addition of FCCP uncoupler (uncoupling) in two D. melanogaster laboratory strains (wDah and OreR) fed with rapamycin for 10 days. Results are shown as the ratio of rapamycin (Rapa) to vehicle control treatment. Ratios were assessed for significance by a Wilcoxon signed rank test. State-3, V = 0, P = 0.00; uncoupling, V = 1, P = 0.00. V, sum of ranks in which the ratio rapamycin:control is below 1. (B) H2O2 production of mitochondria isolated from wDah and OreR flies treated with rapamycin or vehicle control for 10 days. *P<0.05 versus control as determined by ANCOVA: wDah, treatment: F value, 835.9, P<0.00; OreR, treatment: F value, 161.3, P<0.00. (C) Enzymatic activity of complex II in isolated mitochondria from wDah and OreR flies treated with rapamycin or vehicle control for 10 days. Enzymatic activity was normalized to sample protein content. *P<0.05 versus control as determined by Student's t-test. wDah, P = 0.004; OreR, P = 0.003. (D) Effects of rapamycin on complex-I- and complex-II-mediated respiration. Oxygen consumption during state 3 (ADP added) of complex I (pyruvate and malate, or pyruvate, malate and malonate), complex II (succinate and rotenone) of mitochondria from flies fed with rapamycin for 10 days. Results are shown as the ratio of rapamycin to vehicle control. Ratios were assessed for significance by a Wilcoxon signed rank test (V = 0, P = 0.004). The whiskers represent the 10th–90th percentile; the box represents the 25th–75th percentile; the middle line is the median. Abs, absorbance.
As mitochondrial respiration is the main source of ROS in cells, we measured H2O2 in rapamycin-treated flies. In both strains, rapamycin decreased the production of H2O2 (Fig. 1B).
Effects of rapamycin on ETC complexes and mitochondrial membrane potential
The protein complexes I, III, IV and V of the ETC are jointly encoded by nuclear and mitochondrial genes and work together in the process of OXPHOS. In order to test whether the effect of rapamycin on mitochondrial oxygen consumption is due to an effect in one or more of the ETC complexes, we studied the in vitro activity of the respiratory chain enzyme complexes (complex I, complex II, complex III and complex IV) (Fig. 1C; supplementary material Fig. S2A–C) and the enzymatic activity of citrate synthase, as a marker of mitochondrial content and integrity (supplementary material Fig. S2D). Rapamycin-fed flies show increased complex II enzymatic activity (Fig. 1C). No effect was observed in any of the other enzyme complexes (supplementary material Fig. S2A–C). Interestingly, complex II is the only complex of the ETC that is solely encoded by nuclear genes, and is an integrator of nutrient catabolism between glycolytic and respiratory pathways that contribute different reducing equivalents (NADH versus FADH) (Scheffler, 2007). As an indicator of hydrogen ion pumping across the inner membrane during the process of electron transport and oxidative phosphorylation, we measured changes in membrane potential (Δψm). Rapamycin had no significant effect on mitochondrial (Δψm) (supplementary material Fig. S2E).
Rapamycin effects on respiration are independent of increased complex II activity
We then tested the hypothesis that the effect of rapamycin on mitochondrial respiration is mediated by the increased activity of complex II. We measured mitochondrial complex-I- and complex-II-dependent oxygen consumption in flies treated with rapamycin. Oxygen consumption was quantified with substrates and inhibitors that permit complex I respiration but block complex II: with pyruvate and malate alone versus pyruvate and malate plus malonate (a competitive inhibitor of complex II activity). The reciprocal experiment was also performed in which oxygen consumption was studied with the complex II substrate succinate, plus rotenone as an inhibitor of complex I. Rapamycin increased oxygen consumption of both complex-I- and complex-II-mediated respiration in the presence of inhibitors of either complex (Fig. 1D).
Divergent mitochondrial genotypes on a common nuclear background respond differently to rapamycin treatment
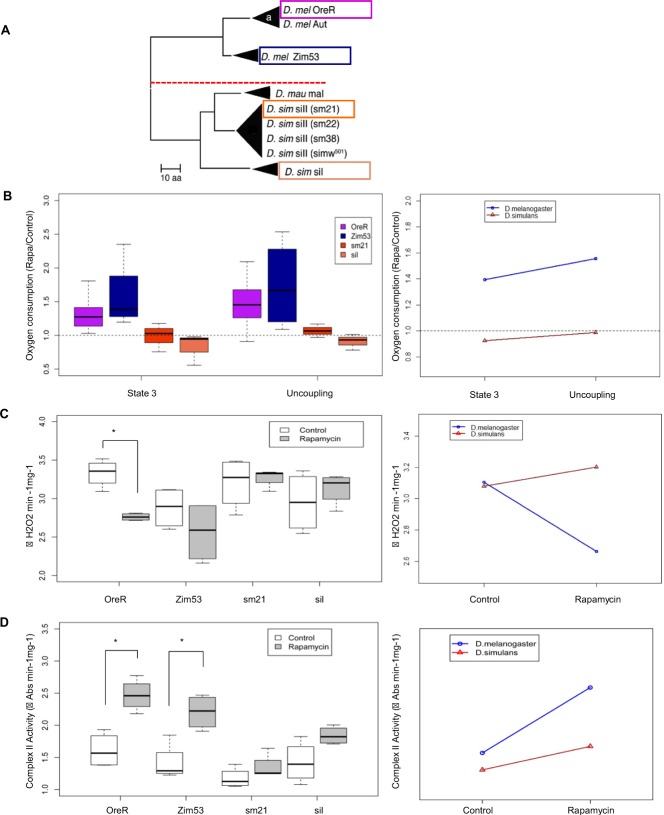
To test the hypothesis that the effects of rapamycin are dependent on genes encoded in the mtDNA, four distinct mtDNAs from different strains of D. melanogaster (OreR and Zim53) and D. simulans (sm21 and si1) were introgressed onto the single D. melanogaster nuclear background OreR (Fig. 2A).
Fig. 2.
Effects of rapamycin on Drosophila from divergent mitochondrial and nuclear lineages. (A) mtDNA phylogeny between the four mitochondrial haplotype groups used (in boxes). The tree is based on amino acid sequences. Species names are followed by the mitochondrial haplotype. In parentheses are the lines from which the mtDNA was isolated (modified from Montooth et al., 2010). Oxygen consumption during state 3 and after adding FCCP uncoupler (B), H2O2 production (C), and enzymatic activity of complex II (D) of mitochondria isolated from the divergent mitochondrial and nuclear lineages after 10 days treatment with rapamycin. Enzymatic activity was normalized to sample protein content. *P<0.0125 versus control as determined by Student's t-test: P<0.00 (C); OreR, P = 0.004, Zim53, P = 0.007 (D). The whiskers represent the 10th–90th percentile; the box represents the 25th–75th percentile; the middle line is the median. The graphs presented to the right of B–D represent the effect of rapamycin in mitochondrial respiration (B), ROS production (C) and complex II activity (D) from D. melanogaster and D. simulans mitochondria. ANOVA: state 3, F value, 10.258, P = 0.004; uncoupling, F value, 9.9567, P = 0.005; H2O2 production, Df:1, F value, 9.9567, Pr(F), 0.005; complex II, F value, 6.918, P = 0.01. Abs, absorbance.
Offspring from crosses between D. simulans and D. melanogaster are sterile. However, a rescue of this sterility can be obtained when females from the D. simulans C167.4 strain are crossed to D. melanogaster males (Davis et al., 1996). To transfer mtDNA from D. simulans to D. melanogaster, sm21 and si1 females were backcrossed for several generations to D. simulans C167.4 males. Subsequently, females from these backcrosses were crossed to D. melanogaster males to obtain the mtDNA replacement strains (Montooth et al., 2010). A series of backcrosses using balancer chromosome stocks and the OreR strain of D. melanogaster replaced all D. simulans nuclear alleles with OreR nuclear alleles in each of the desired cytoplasmic (mtDNA) backgrounds (Montooth et al., 2010).
Given that all mtDNAs were placed on the same OreR nuclear background, differences among the introgressed genotypes could only result from main effects of the distinct mtDNAs or specific interactions of the OreR nuclear background with the different mtDNAs. We observed that rapamycin significantly affected mitochondrial respiration (Fig. 2B), H2O2 production (Fig. 2C) and complex II activity (Fig. 2D) in flies harboring mtDNA from D. melanogaster (OreR and Zim53) but this rapamycin effect was significantly reduced in flies carrying D. simulans mtDNA (sm21 and si1).
Rapamycin and mitochondrial genotype alter metabolite profiles of whole flies and isolated mitochondria
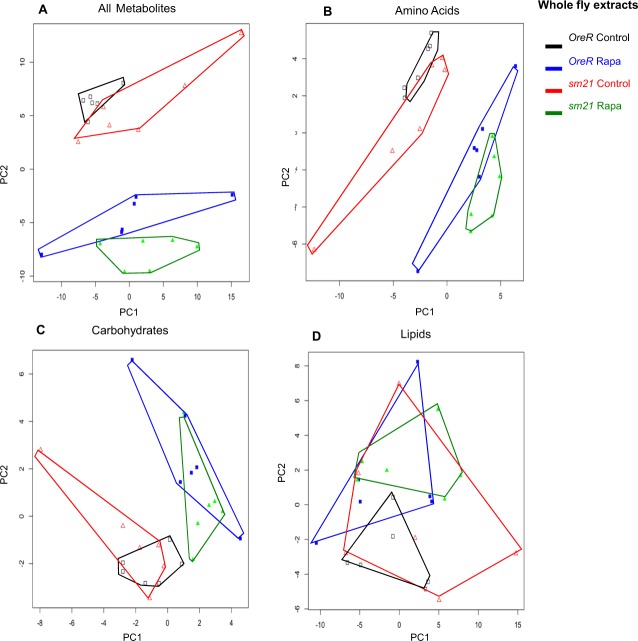
We performed comparative metabolomics analyses of rapamycin on a subset of our strains, the ‘responding’ (OreR) and ‘non-responding’ (sm21) mitochondrial genotypes both on the common OreR nuclear background after treatment with rapamycin. Metabolites from whole-fly extracts and from mitochondrial extracts were obtained by using gas chromatography mass spectrometry (GC/MS) and liquid chromatography-tandem mass spectrometry (LC-MS/MS). We detected 210 metabolites in the whole-fly extracts and 230 metabolites in the mitochondrial extracts. These metabolites were separated into four categories, those involved in amino acid metabolism, carbohydrate metabolism, lipid metabolism and those involved in energy homeostasis (cofactors, vitamins and TCA cycle intermediates) (Figs. 3–5; supplementary material Tables S1, S2). The data were subjected to a principle components analysis (PCA) to visualize the clustering of treatments in metabolite space. For the whole-fly extracts, the mtDNA genotype had subtle effects on the metabolite profile as revealed by the proximity of the OreR control-treated and the sm21 control-treated samples in PCA space (Fig. 3A; Fig. 5A, open symbols). However, rapamycin treatment has a significant impact of the metabolite profiles, as was evident from the displacement of the rapamycin-treated samples to lower values of PC2 (Fig. 3A; Fig. 5A, solid symbols). Bivariate normal ellipses defining a 95% confidence area for the rapamycin-treated samples for each genotype did not overlap, but the ellipses from the two mtDNA genotype samples in either the control or the rapamycin treatment were broadly overlapping (supplementary material Fig. S3A). The predominance of the rapamycin effect, and the weak genotype effect, was evident in the similar relative positions of the sample envelopes for genotypes in the amino acid and carbohydrate PCA plots, which mirrored the patterns seen for all metabolites (Fig. 3B,C; supplementary material Fig. S3C). The lipid analysis showed little differentiation of the samples (Fig. 3D), and the 95% ellipses were broadly overlapping for lipid metabolites indicating no significant differentiation (data not shown).
Fig. 3.
Metabolic reprogramming of whole-fly extracts by mtDNA genotype and rapamycin treatment. Principle components analysis was performed on a sample of 210 metabolites detected in the whole-fly extracts from flies carrying OreR and sm21 mtDNA genotypes treated with vehicle control or 200 µm rapamycin. (A) PCA of 210 metabolites identified. (B). PCA of 50 amino acids or amino acid derivatives. (C). PCA of 26 carbohydrates. (D). PCA of 80 lipids. Complete lists of these metabolites are provides in supplementary material Table S1. Black open squares, OreR mtDNA on control diet; blue solid squares, OreR mtDNA on rapamycin; red open triangles, sm21 mtDNA on control diet; green solid triangles, sm21 mtDNA on rapamycin. Polygons surrounding points are intended to aid the visualization of the six replicate samples for each treatment. See text and supplementary material Fig. S2 for statistical analyses.
Fig. 4.
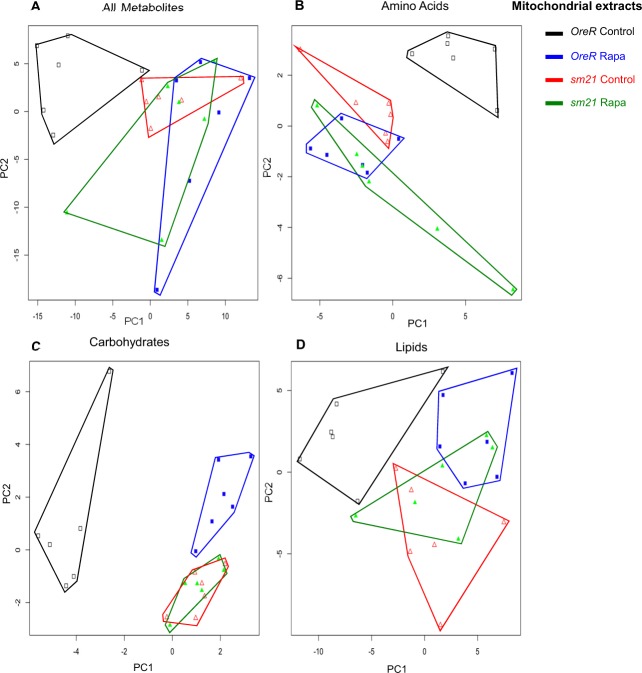
Metabolic reprogramming of mitochondrial extracts by mtDNA genotype and rapamycin treatment. PCA was performed on a sample of 230 metabolites detected on the mitochondrial extracts from flies carrying OreR and sm21 mtDNA genotypes treated with vehicle control or 200 µm rapamycin. (A) PCA of all 230 metabolites. (B) PCA of 39 amino acids or amino acid derivatives. (C) PCA of 20 carbohydrates. (D) PCA of 83 lipids. Complete lists of these metabolites are provides in supplementary material Table S1. Black open squares, OreR mtDNA on control diet; blue solid squares, OreR mtDNA on rapamycin; red open triangles, sm21 mtDNA on control diet; green solid triangles, sm21 mtDNA on rapamycin. Polygons surrounding points are intended to aid the visualization of the six replicate samples for each treatment. See text and supplementary material Fig. S2 for statistical analyses.
Fig. 5.
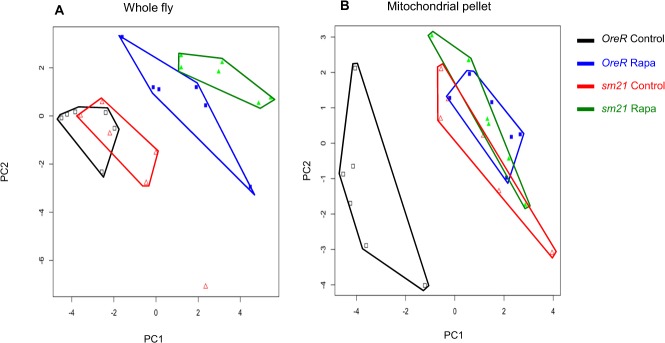
PCA analysis of metabolites involved in energy homeostasis (cofactors, vitamins and TCA cycle intermediates). (A) Results from whole-fly extracts and (B) mitochondrial extracts. Complete lists of these metabolites are provides as supplementary material Table S1. Black open squares, OreR mtDNA on control diet; blue solid squares, OreR mtDNA on rapamycin; red open triangles, sm21 mtDNA on control diet; green solid triangles, sm21 mtDNA on rapamycin. Polygons surrounding points are intended to aid the visualization of the six replicate samples for each treatment.
The metabolite profiles from mitochondrial isolates analyses were distinct from those of whole-fly extracts and identify a genotype-by-rapamycin interaction effect (Fig. 4; Fig. 5B). Rapamycin had a strong effect on the metabolite profiles of the responsive mtDNA genotypes (OreR). The OreR rapamycin-treated samples were displaced along the PC1 axis (Fig. 4A; Fig. 5B). Bivariate normal ellipses defining a 95% confidence area surrounding the OreR control treated samples did not overlap with other treatment classes (supplementary material Fig. S3B). For the ‘non-responding’ sm21 mtDNA genotype, rapamycin had noticeably less of an effect on the shift of the metabolite landscapes (Fig. 4; Fig. 5B). The 95% confidence ellipses for control and rapamycin-treated samples were broadly overlapping for this genotype (supplementary material Fig. S3B). This pattern was most evident in the PCA plots based on amino acids (Fig. 4B), carbohydrates (Fig. 4C; supplementary material Fig. S3D) and metabolites involved in energy homeostasis (cofactors, vitamins and TCA cycle intermediates) (Fig. 5B), where rapamycin induced a substantial shift in OreR but had virtually no impact on the sm21 genotypes. This pattern was similar but not as pronounced in the lipid analysis (Fig. 4D).
An analysis of the individual metabolites that were differentially affected in OreR and sm21 flies after rapamycin treatment reflected an imbalance of several mitochondrial pathways. The TCA metabolite fumarate is decreased, whereas malate is increased in OreR flies after rapamycin treatment (Table 1). Rapamycin-treated OreR mitochondria showed significantly lower levels of fatty acids, reduced hydroxybutyrate (BHBA) and other ketone bodies, and increased carnitine levels. Because the oxidation of exogenous fatty acids is enhanced by rapamycin treatment (Brown et al., 2007), we hypothesized that this imbalance in lipid metabolism might partially result from a decline in β-oxidation as a consequence of the depletion of available endogenous lipids by this time point under rapamycin treatment. Notably, basal levels of BHBA and other ketone bodies significantly differed between OreR and sm21 flies, suggesting differences in basal lipid oxidation between haplotypes (data not shown).
Table 1. Percentage mitochondrial metabolite change between haplotypes.
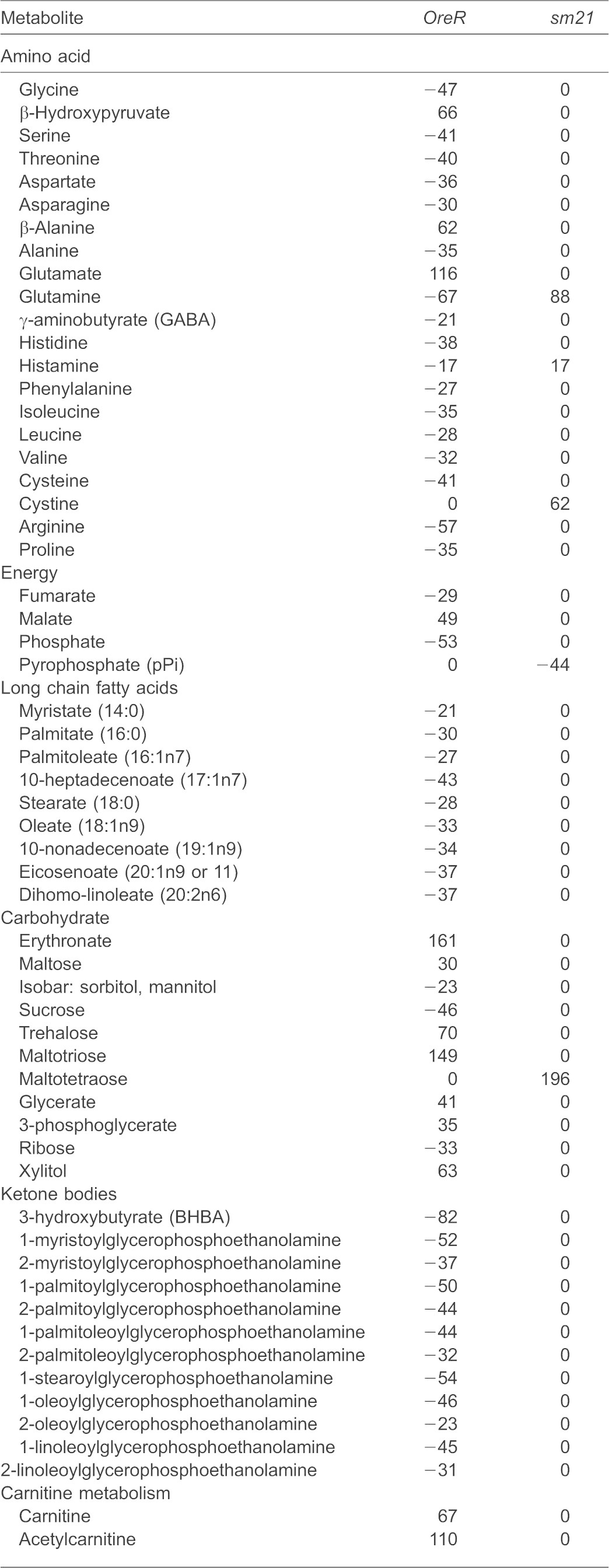
A list of metabolites within the mitochondrial extract that differs between OreR and sm21 haploypes after rapamycin treatment. Results are shown as the percentage of change after rapamycin treatment. Positive and negative numbers indicate increased and decreased abundance respectively. 0 represents no change. P<0.05 for all metabolites represented.
Within the amino acids, β-hydroxypyruvate, β-alanine and glutamate levels increased after rapamycin treatment in OreR but not sm21 flies. The rest of amino acids that changed, including glutamine, which has been recently shown to be regulated by mTOR (Csibi et al., 2013), were downregulated in OreR mitochondria. In sm21 mitochondria, rapamycin increased the levels of glutamine, histamine and cystine (Table 1).
DISCUSSION
Mitochondria–nuclear interactions are the result of a two billion year old symbiosis between two genomes with a history of gene transfer from the ancestral mitochondrial genome into the emerging nuclear genome, and a system to import proteins back into the mitochondrial organelle (Rand et al., 2004; Ballard and Rand, 2005; Pesole et al., 2012). As an outcome of this co-adaptation the mitochondria and cytosol have established circuits of signaling that ensure homeostasis through cellular plasticity in response to altered environmental conditions. Cytosolic pathways signal to modulate mitochondrial activity (anterograde signaling) and mitochondria signal to the cytosol to alert of changes in mitochondrial metabolism (retrograde signaling) (Liu and Butow, 2006; Woodson and Chory, 2008). Therefore, it is not surprising, that mitochondrial dysfunction is associated with the onset of many diseases (Ristow, 2006; Wiederkehr and Wollheim, 2006; Fukui and Moraes, 2008; Tatsuta and Langer, 2008) and might have an important role in the aging process (Masoro et al., 1982; Tatsuta and Langer, 2008; Finley and Haigis, 2009; Raffaello and Rizzuto, 2011; Vendelbo and Nair, 2011). However, there is controversy over the mechanisms by which mitochondrial activity modulates longevity. Traditionally ROS production in the mitochondria has been postulated as a cause of aging (Harman, 1956), and manipulations that either increase ROS scavenging or decrease ROS production have extended longevity. However, recent studies have challenged the role of ROS as only detrimental in aging, given evidence that ROS is also an important factor promoting longevity through signaling and homeostasis (Ristow and Schmeisser, 2011).
In this study, we identify complex II as the only member of the ETC that displays altered activity upon rapamycin treatment. The significant effect on complex II and lack of effects on other complexes points to the interaction of mitochondrial and metabolic energy transduction as an important aspect of mTOR signaling. Participating in both the TCA and ETC, complex II is uniquely situated to coordinate both pathways. Furthermore, complex II is stimulated by FADH2, which is mainly derived from fatty-acid oxidation. mTOR metabolism has been previously described to be connected to lipid metabolism regulation (Laplante and Sabatini, 2012; Soliman, 2011). Moreover, levels of ketone bodies, carnitine metabolism and free fatty acids significantly differed between OreR and sm21 rapamycin-treated flies, suggesting that lipid oxidation is an important metabolic pathway that mediates the beneficial effects of rapamycin on mitochondrial physiology. Dysfunction of complex II, as well as upregulation of the mTOR pathway, is associated with precocious aging and cancer (Brière et al., 2005; King et al., 2006; Ishii et al., 2007; Zoncu et al., 2011). Thus, our data suggest that some of the anti-carcinogenic and anti-aging effects of rapamycin treatment (Sharp and Richardson, 2011) can be explained by its effect on succinate dehydrogenase activity.
The relationship between complex II stimulation and mitochondrial respiration warrants further study. Our results show that rapamycin treatment increases the oxygen consumption respiration mediated by complex I and complex II, even though in vitro complex I activity is not affected by treatment with rapamycin (supplementary material Fig. S2A). This implies that the effects of rapamycin are mediated through the actions of other mitochondrially encoded proteins, which agrees with the data obtained from the metabolomic profiling. Considering this, we hypothesize that metabolic shifts that lead to changes in the pool of reducing equivalents NADH and FADH2 might underlie the effect of rapamycin on the in vitro activity of complex II. To test this hypothesis, we measured the NAD+:NADH ratio on mitochondrial isolates from the D. melanogaster (OreR) and D. simulans (sm21) mtDNA haplotypes. Rapamycin decreased the NAD+:NADH ratio only in OreR mitochondria (supplementary material Fig. S2F). Given that NADH is a free cofactor to complex I and is added at non-limiting concentrations to the in vitro reaction, rapamycin would not affect the in vitro activity of complex I. Unlike NADH, FADH2 is attached to complex II through a covalent flavin linkage (Cecchini, 2003), and changes in FADH2 abundance will affect both in vivo and in vitro complex II activity.
The beneficial effect of rapamycin is diminished in flies carrying D. simulans mitochondrial genomes, which have ∼100 amino acid substitutions among the 13 protein coding subunits of OXPHOS complexes. The PCA analysis of metabolites extracted from mitochondrial isolates of sm21 (D. simulans) mitochondrial genotypes showed that ‘non-responsive’ mitochondria shifted the metabolite profile in a manner that did not permit additional shifts from rapamycin treatment. However, the analysis of metabolites extracted from whole flies showed a substantial effect of rapamycin treatment and a parallel metabolic shift of both the ‘responsive’ and ‘non-responsive’ genotypes. Overall our data show that mitochondrially encoded genes are important for rapamycin benefits. These genes are mediating the ability of the drug to reprogram mitochondrial metabolism and increase mitochondrial efficiency. In this study, the enzyme activity and respiration analyses were performed on mitochondria isolated from whole flies. It is possible that different tissues have distinct mitochondrial responses to rapamycin, but it is unlikely that the effects we report here are due to artifacts of tissue-specific activities. For example, if different tissues have opposing effects of rapamycin, this would reduce our ability to uncover the effects we see using whole-fly mitochondrial isolation. If some tissues are responsive and others are unresponsive, again, the signal from whole-fly extractions would be reduced compared to mitochondrial assays from a single tissue. At a minimum, the mtDNA-dependent abrogation of the rapamycin effects we have described must be operating in the majority of cells in the fly because our assays capture the activity of mitochondria in proportion to the biomass of cell types in a whole fly. Future studies addressing the specific effects of rapamycin on different tissues will allow us to better understand the benefits of this drug and identify key tissues that mediate different haplotype responses.
The results presented here have uncovered an effect of rapamycin treatment on the homeostatic nature of metabolic networks with a systems-level response. We described new roles for mtDNA-encoded genes in mediating the effects of rapamycin, and offer a new set of genetic reagents to examine the complex interactions governing TOR and mitochondrial signaling. We have uncovered a novel association between complex II stimulation, mitochondrial respiration, and rapamycin treatment that is modified by genes encoded in the mitochondrial genome. Our experiments established that the stimulation of respiration by rapamycin is not functionally dependent on the enhanced complex II activity, implying that rapamycin may act on these two processes independently or indirectly through shifts in metabolic state. These alternatives remain important questions for future studies in this system. It would be fruitful to explore the variety of epistatic interactions between pairs of nuclear and mtDNA genotypes that encode the proteins mediating the mTOR and OXPHOS pathways.
MATERIALS AND METHODS
Fly stocks and husbandry
The white Dahomey (wDah) strain is as described previously (Bjedov et al., 2010). Mitochondria–nuclear substitution strains are as described previously (Montooth et al., 2010). All stocks were maintained and conducted under standard conditions [25°C, 12 h light and 12 h dark on normal medium (11% sugar, 2% autolyzed yeast, 5.2% cornmeal, agar 0.79% (w/v in water), and 0.2% tegosept-methyl 4-hydroxybenzoate, from Sigma-Aldrich, St Louis, MO, USA]. All stocks were density controlled in replicate vials using 48-hour egg lays by five pairs of parents for two generations prior to collection of flies for experimental assays.
Rapamycin treatment
Rapamycin was purchased from LC Laboratories (Woburn, MA, USA.). Rapamycin was dissolved in ethanol and added to food at the final concentration of 200 µM, as described in Bjedov et al., 2010. Newly eclosed adult flies were collected within 48 h and maintained on rapamycin-treated or ethanol vehicle control food for 10 days.
Western immunoblotting
20 female flies per treatment were homogenized in homogenization buffer (1% Triton X-100, 10 mM Tris-HCl base pH 7.6, 5 mM EDTA, 50 mM NaCl, 30 mM Na pyrophosphate, 50 mM NaF, 100 µM orthovanadate, complete protease inhibitor cocktail (Roche, Basel, Switzerland). Following homogenization, samples were incubated on ice for 30 min and centrifuged at 16,000 g for 20 min. The supernatant was quantified for protein abundance using the BCA Protein Assay Kit from Thermo Scientific (Rockford, IL, USA). 30 µg of total protein was loaded per lane. Proteins were separated on SDS–polyacrylamide gels, transferred to polyvinylidene difluoride membranes, and subjected to immunoblotting. Antibody to Drosophila S6K1 were a gift from Thomas Neufeld (College of Biological Sciences, University of Minnesota, MN, USA), and to phospho-S6K1 was purchased from Cell Signaling Technology (Danvers, MA, USA).
Mitochondrial and cytosolic isolation, and DNA quantification
30 female flies were gently homogenized in 1 ml chilled isolation buffer (225 mM mannitol, 75 mM sucrose, 10 mM MOPS, 1 mM EGTA and 0.5% fatty acid free BSA, pH 7.2) using a glass-teflon dounce homogenizer. The extracts were centrifuged at 300 g for 5 min at 4°C. The obtained supernatant was then centrifuged at 6000 g for 10 min at 4°C to enrich for mitochondria. The pellet was resuspended in 100 µl of respiration buffer (225 mM mannitol, 75 mM sucrose, 10 mM KCl, 10 mM Tris-HCl, 5 mM KH2PO4, pH 7.2). The supernatant was taken as the cytosolic fraction. Freshly prepared mitochondrial isolates were used for respiration and ROS assays, or were aliquoted and frozen at −80°C for later enzymatic activity assays. Quantification of mitochondrial protein obtained from the isolation was determined by use of Bradford reagent (Sigma-Aldrich, St. Louis, MO, USA) following the manufacturer's protocol. For DNA quantification, RNA from the mitochondrial and cytosolic fractions was removed by RNase A treatment (Qiagen, Valencia CA, USA), and the amount of DNA was determined spectrophotometrically (NanoDrop ND-1000).
35S-methionine protein incorporation
Protein synthesis was measured as the incorporation of 35S-methionine into protein. 20 female flies were treated with rapamycin for 10 days and maintained in 100 µCi/ml 35S-methionine plus rapamycin or vehicle control food. After 48 h of incubation, flies were sorted to separate mitochondrial and cytosolic fraction as described above. To quantify proteins in mitochondria and cytosol isolates using the same method (BCA in this case) a different isolation buffer that lacks BSA (210 mM mannitol, 70 mM sucrose, 5 mM Hepes, 1 mM EDTA, pH 7.35) was used to homogenize flies. Proteins were precipitated in 10% trichloracetic acid (TCA) and the quantity of radiolabeled methionine incorporated into protein was measured in both fractions using a Beckam LS 6500 multi porpoise scintillation counter.
Mitochondrial respiration and H2O2 measurement
Respiration rates were determined by oxygen consumption using a Clark-type electrode and metabolic chamber (Hansatech Instruments, Norfolk, UK). 5 µM of pyruvate plus 5 µM malate, or 10 µM succinate plus 0.5 µM rotenone was added to an isolated mitochondrial suspension in 1 ml of respiration buffer held in the respiration chamber at 30°C. 125 nmol of ADP was added to generate state 3 respiration rates. Uncoupled respiration rates, indicative of maximal rate of oxygen consumption, were generated by the addition of 0.5 nM carbonyl cyanide 4-(trifluoro-methoxy) phenylhydrazone (FCCP), a chemical uncoupler.
H2O2 production was measured using an Amplex Red and horseradish peroxidase assay (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. 20 µM of glycerol 3-phosphate or 5 µM of pyruvate plus 5 µM malate were used as substrate. Qualitatively identical results were obtained with both substrates.
Enzymatic assay and measurement of the membrane potential and NAD+:NADH ratio
Enzymatic assays were modified from Barrientos et al. (Barrientos, 2002) and are as previously described (Meiklejohn et al., 2013). The specific activity of complex I was determined as the rotenone-sensitive rate following the oxidation of NADH at 340 nm with the coenzyme Q analog decylubiquinone as the electron acceptor (reaction mixture containing 35 mM NaH2PO4, 5 mM MgCl2, 2.5 mg/ml BSA, 2 mM KCN, 2 µg/ml antimycin A, 100 µM NADH and 100 µM decylubiquinone (2 mM rotenone to inhibit reaction)]. The catalytic activity of complex II was monitored by monitoring the reduction of 2,6-dichlorophenolindophenol (DCPIP) at 600 nm [reaction mixture containing 30 mM NaH2PO4, 100 µM EDTA, 2 mM KCN, 2 µg/ml antimycin A, 2 µg/ml rotenone, 750 µM BSA, 10 mM succinate, 100 µM DCPIP, 100 µM decylubiquinone (400 mM malonate to inhibit reaction)]. Complex III activity was measured by monitoring the reduction of cytochrome c at 550 nm [reaction mixture containing 35 mM NaH2PO4, 2.5 mg/ml BSA, 5 mM MgCl2, 2 mM KCN, 2 µg/ml rotenone, 50 µM cytochrome c, 25 µM decylubiquinol (5 µg/ml antimycin A to inhibit reaction)]. Potassium borohydride was used to reduce decylubiquinone. Complex IV activity was measured by determining the rate of oxidation of reduced cytochrome c at 550 nm [reaction mixture containing 5 mM MgCl2, 2 µg/ml rotenone, 2 µg/ml antimycin A, 1 mM DDM, 45 µM cytochrome c (4 mM KCN to inhibit reaction)]. Sodium dithionite was used to reduce cytochrome c. To measure citrate synthase activity, the rate-limiting reaction of citrate synthase was coupled to a chemical reaction in which DTNB reacts with CoA-SH and the absorbance of the product is measured at 412 nm (reaction mixture containing 100 µM DTNB, 300 µM acetyl-CoA, 100 mM Tris-HCl, 300 µM oxaloacetic acid).
JC-1 indicator dye from AnaSpec (Fremont, CA, USA) was used to measure membrane potential in isolated mitochondria. 30 female flies were gently homogenized and the mitochondria were extracted as described above. The mitochondrial pellet obtained was resuspended in 300 µl of respiration buffer. 3 µl of a 1 µg/µl solution of JC-1 dissolved in DMSO was added to the suspension. Mitochondrial samples were incubated for 15 min at 37 °C protected from light. Samples were centrifuged for an additional 2 min at 6000 g and resuspended in 600 µl of fresh respiration buffer. Fluorescence was measured for red (excitation 550 nm, emission 600 nm) and green (excitation 485 nm, emission 535 nm) corresponding to the monomeric and aggregate forms of JC-1.
NAD+:NADH ratios and NADH levels were measured with the Amplite TM Flourimetric NAD/NADH Ratio assay kit (catalog number 15263) from AAT Bioquest (Sunnyvale, CA, USA). Mitochondria from 20 females were extracted as described above. The reaction was performed following the manufacturer's protocol.
Metabolomic profiling
Metabolomic profiles of rapamycin-treated flies were assessed by using a Metabolon (Durham, NC) with Metabolon's standard solvent extraction methods (proprietary information). The extracted samples were split into equal parts for analysis on the GC/MS and LC-MS/MS platforms. A total of six replicates per sample were performed. Technical replicate samples were created from a homogeneous pool containing a small amount of all study samples. Values for each sample were corrected by Bradford protein quantification. Each compound was then normalized to the median value for each run-day block (block normalization). Missing values were imputed with the observed minimum for that particular compound.
Statistical analysis
Comparisons between two treatments were performed using the unpaired Student's t-test and linear regression as noted in the figure legends. Analysis of variance was used in cases of multiple comparisons. Differences were considered significant at P<0.05, P<0.025 and P<0.0125 as noted in the figure legends. Statistical analyses were performed using the R statistical package unless otherwise specified.
Supplementary Material
Acknowledgments
We thank Thomas Neufeld for the total Drosophila S6K antibody, and Jim Mossman and other members of the Rand laboratory for advice and discussion.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
E.V.C. conceived of and designed experiments, performed the experiments, analyzed the data and wrote the paper. M.A.H. performed the experiments. D.M.R. conceived of experiments, analyzed the data and wrote the paper.
Funding
This work is supported by National Institutes of Aging [grant number 5R01AG027849 to D.M.R.]; and the National Institute of General Medical Sciences [grant number R01GM067862 to D.M.R.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.142026/-/DC1
References
- Balaban R. S., Nemoto S., Finkel T. (2005). Mitochondria, oxidants, and aging. Cell 120, 483–495 10.1016/j.cell.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Ballard J. W. O., Rand D. M. (2005). The population biology of mitochondrial dna and its phylogenetic implications. Annu. Rev. Ecol. Evol. Syst. 36, 621–642 10.1146/annurev.ecolsys.36.091704.175513 [DOI] [Google Scholar]
- Baltzer C., Tiefenböck S. K., Frei C. (2010). Mitochondria in response to nutrients and nutrient-sensitive pathways. Mitochondrion 10, 589–597 10.1016/j.mito.2010.07.009 [DOI] [PubMed] [Google Scholar]
- Barrientos A. (2002). In vivo and in organello assessment of OXPHOS activities. Methods 26, 307–316 10.1016/S1046--2023(02)00036--1 [DOI] [PubMed] [Google Scholar]
- Bentzinger C. F., Romanino K., Cloëtta D., Lin S., Mascarenhas J. B., Oliveri F., Xia J., Casanova E., Costa C. F., Brink M. et al. (2008). Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 8, 411–424 10.1016/j.cmet.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Bjedov I., Toivonen J. M., Kerr F., Slack C., Jacobson J., Foley A., Partridge L. (2010). Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46 10.1016/j.cmet.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N. D., Chatenay-Lapointe M., Pan Y., Shadel G. S. (2007). Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 5, 265–277 10.1016/j.cmet.2007.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brière J-J., Favier J., El Ghouzzi V., Djouadi F., Bénit P., Gimenez A. P., Rustin P. (2005). Succinate dehydrogenase deficiency in human. Cell. Mol. Life Sci. 62, 2317–2324 10.1007/s00018--005--5237--6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N. F., Stefanovic-Racic M., Sipula I. J., Perdomo G. (2007). The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism 56, 1500–1507 10.1016/j.metabol.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Cecchini G. (2003). Function and structure of complex II of the respiratory chain. Annu. Rev. Biochem. 72, 77–109 10.1146/annurev.biochem.72.121801.161700 [DOI] [PubMed] [Google Scholar]
- Cheng Z., Ristow M. (2013). Mitochondria and metabolic homeostasis. Antioxid. Redox Signal. 19, 240–242 10.1089/ars.2013.5255 [DOI] [PubMed] [Google Scholar]
- Csibi A., Fendt S-M., Li C., Poulogiannis G., Choo A. Y., Chapski D. J., Jeong S. M., Dempsey J. M., Parkhitko A., Morrison T. et al. (2013). The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 153, 840–854 10.1016/j.cell.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham J. T., Rodgers J. T., Arlow D. H., Vazquez F., Mootha V. K., Puigserver P. (2007). mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736–740 10.1038/nature06322 [DOI] [PubMed] [Google Scholar]
- Dazert E., Hall M. N. (2011). mTOR signaling in disease. Curr. Opin. Cell Biol. 23, 744–755 10.1016/j.ceb.2011.09.003 [DOI] [PubMed] [Google Scholar]
- Düvel K., Yecies J. L., Menon S., Raman P., Lipovsky A. I., Souza A. L., Triantafellow E., Ma Q., Gorski R., Cleaver S. et al. (2010). Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley L. W. S., Haigis M. C. (2009). The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res. Rev. 8, 173–188 10.1016/j.arr.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H., Moraes C. T. (2008). The mitochondrial impairment, oxidative stress and neurodegeneration connection: reality or just an attractive hypothesis? Trends Neurosci. 31, 251–256 10.1016/j.tins.2008.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. (1956). Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 11, 298–300 10.1093/geronj/11.3.298 [DOI] [PubMed] [Google Scholar]
- Ishii N., Ishii T., Hartman P. S. (2007). The role of the electron transport SDHC gene on lifespan and cancer. Mitochondrion 7, 24–28 10.1016/j.mito.2006.11.012 [DOI] [PubMed] [Google Scholar]
- King A., Selak M. A., Gottlieb E. (2006). Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene 25, 4675–4682 [DOI] [PubMed] [Google Scholar]
- Lane N. (2005). Power, Sex, Suicide: Mitochondria and the Meaning of Life New York, NY: Oxford University Press [Google Scholar]
- Laplante M., Sabatini D. M. (2012). mTOR signaling in growth control and disease. Cell 149, 274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Butow R. A. (2006). Mitochondrial retrograde signaling. Annu. Rev. Genet. 40, 159–185 10.1146/annurev.genet.40.110405.090613 [DOI] [PubMed] [Google Scholar]
- Masoro E. J., Yu B. P., Bertrand H. A. (1982). Action of food restriction in delaying the aging process. Proc. Natl. Acad. Sci. USA 79, 4239–4241 10.1073/pnas.79.13.4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., White E. (2011). Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr. Opin. Genet. Dev. 21, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn C. D., Holmbeck M. A., Siddiq M. A., Abt D. N., Rand D. M., Montooth K. L. (2013). An incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet. 9, e1003238 10.1371/journal.pgen.1003238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth K. L., Meiklejohn C. D., Abt D. N., Rand D. M. (2010). Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila. Evolution 64, 3364–3379 10.1111/j.1558--5646.2010.01077.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. P., Holmgren A., Larsson N-G., Halliwell B., Chang C. J., Kalyanaraman B., Rhee S. G., Thornalley P. J., Partridge L., Gems D. et al. (2011). Unraveling the biological roles of reactive oxygen species. Cell Metab. 13, 361–366 10.1016/j.cmet.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paglin S., Lee N-Y., Nakar C., Fitzgerald M., Plotkin J., Deuel B., Hackett N., McMahill M., Sphicas E., Lampen N. et al. (2005). Rapamycin-sensitive pathway regulates mitochondrial membrane potential, autophagy, and survival in irradiated MCF-7 cells. Cancer Res. 65, 11061–11070 10.1158/0008--5472.CAN--05--1083 [DOI] [PubMed] [Google Scholar]
- Pan Y., Nishida Y., Wang M., Verdin E. (2012). Metabolic regulation, mitochondria and the life-prolonging effect of rapamycin: a mini-review. Gerontology 58, 524–530 10.1159/000342204 [DOI] [PubMed] [Google Scholar]
- Pesole G., Allen J. F., Lane N., Martin W., Rand D. M., Schatz G., Saccone C. (2012). The neglected genome. EMBO Rep. 13, 473–474 10.1038/embor.2012.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P., Cybulski N., Feige J. N., Auwerx J., Rüegg M. A., Hall M. N. (2008). Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 8, 399–410 10.1016/j.cmet.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Raffaello A., Rizzuto R. (2011). Mitochondrial longevity pathways. Biochim. Biophys. Acta 1813, 260–268 10.1016/j.bbamcr.2010.10.007 [DOI] [PubMed] [Google Scholar]
- Ramanathan A., Schreiber S. L. (2009). Direct control of mitochondrial function by mTOR. Proc. Natl. Acad. Sci. USA 106, 22229–22232 10.1073/pnas.0912074106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand D. M., Haney R. A., Fry A. J. (2004). Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19, 645–653 10.1016/j.tree.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Richter J. D., Sonenberg N. (2005). Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477–480 10.1038/nature03205 [DOI] [PubMed] [Google Scholar]
- Ristow M. (2006). Oxidative metabolism in cancer growth. Curr. Opin. Clin. Nutr. Metab. Care 9, 339–345 10.1097/01.mco.0000232892.43921.98 [DOI] [PubMed] [Google Scholar]
- Ristow M., Schmeisser S. (2011). Extending life span by increasing oxidative stress. Free Radic. Biol. Med. 51, 327–336 10.1016/j.freeradbiomed.2011.05.010 [DOI] [PubMed] [Google Scholar]
- Scheffler I. E. (2007). Mitochondria (Scheffler, Mitochondria) Wiley-Liss [Google Scholar]
- Schieke S. M., Finkel T. (2006). Mitochondrial signaling, TOR, and life span. Biol. Chem. 387, 1357–1361 10.1515/BC.2006.170 [DOI] [PubMed] [Google Scholar]
- Schieke S. M., Phillips D., McCoy J. P., Jr, Aponte A. M., Shen R-F., Balaban R. S., Finkel T. (2006). The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 281, 27643–27652 10.1074/jbc.M603536200 [DOI] [PubMed] [Google Scholar]
- Sharp Z. D., Richardson A. (2011). Aging and cancer: can mTOR inhibitors kill two birds with one drug? Target. Oncol. 6, 41–51 10.1007/s11523--011--0168--7 [DOI] [PubMed] [Google Scholar]
- Soliman G. A. (2011). The integral role of mTOR in lipid metabolism. Cell Cycle 10, 861–862 10.4161/cc.10.6.14930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T., Langer T. (2008). Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 27, 306–314 10.1038/sj.emboj.7601972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendelbo M. H., Nair K. S. (2011). Mitochondrial longevity pathways. Biochim. Biophys. Acta 1813, 634–644 10.1016/j.bbamcr.2011.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A., Wollheim C. B. (2006). Minireview: implication of mitochondria in insulin secretion and action. Endocrinology 147, 2643–2649 10.1210/en.2006--0057 [DOI] [PubMed] [Google Scholar]
- Woodson J. D., Chory J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395 10.1038/nrg2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., Sabatini D. M. (2011). mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.