Abstract
In mammals, histamine action is terminated through metabolic inactivation by histamine N-methyltransferase (HNMT) and diamine oxidase. In addition to three well-studied pharmacological functions, smooth muscle contraction, increased vascular permeability, and stimulation of gastric acid secretion, histamine plays important roles in neurotransmission, immunomodulation, and regulation of cell proliferation. The histamine receptor H1 antagonist diphenhydramine, the antimalarial drug amodiaquine, the antifolate drug metoprine, and the anticholinesterase drug tacrine (an early drug for Alzheimer’s disease) are surprisingly all potent HNMT inhibitors, having inhibition constants in the range of 10–100 nM. We have determined the structural mode of interaction of these four inhibitors with HNMT. Despite their structural diversity, they all occupy the histamine-binding site, thus blocking access to the enzyme’s active site. Near the N terminus of HNMT, several aromatic residues (Phe9, Tyr15, and Phe19) adopt different rotamer conformations or become disordered in the enzyme–inhibitor complexes, accommodating the diverse, rigid hydrophobic groups of the inhibitors. The maximized shape complementarity between the protein aromatic side-chains and aromatic ring(s) of the inhibitors are responsible for the tight binding of these varied inhibitors.
Keywords: HNMT inhibitors, diphenhydramine, amodiaquine, metoprine, tacrine
Introduction
In mammals, one-step enzymatic decarboxylation of l-histidine produces histamine. Histamine exerts its effect through the action on G-protein coupled receptors.1,2 Histamine mediates inflammatory and allergic responses via H1 and H4 receptors,3,4 gastric acid secretion via H2 receptors,5 and neurotransmitter release via H3 receptors.6 As detailed below, there is increasing evidence that histamine might become one of the important mediators (regulators) in oncology, sepsis, posttraumatic traumatic stress, embryonic development and hematopoiesis.7
In humans, decreased activity of histaminergic neurons and of histamine levels in the brain have been implicated in Alzheimer’s disease,8 attention-deficit hyperactivity disorder, and aging-related disorders.9,10 In mice, the targeted gene deletion of histidine decarboxylase (HDC), the only histamine-synthesizing enzyme, led to histamine-deficiency. These HDC knock-out mice had undetectable tissue histamine levels, and showed all of the expected abnormalities of histamine deficiency: impaired gastric acid secretion, and impaired passive and cutaneous anaphylaxis.11 Advanced studies on the HDC knock-out mice revealed roles of histamine in angiogenesis,12 sleep-wake circadian rhythm,13,14 male sex steroid production,15 elevation of bone density and protection against ovariectomy-induced bone loss,16 and bacterial eradication.17 In the latter study, when Escherichia coli was inoculated into the peritoneal cavity of wild-type and HDC-KO mice, the bacteria were much more rapidly eliminated in the knock-out mice than the wild-type. Furthermore, H1 and H2 agonists or antagonists, respectively, delayed or promoted bacterial clearance, though some other evidence suggests that the role of histamine in this case may be complex.18 These studies raise the possibility of using antihistamine drugs against bacterial infection or osteoporosis.
The physiological actions of histamine are controlled not only by the receptors but also by the inactivating enzyme histamine N-methyltransferase (HNMT) (EC 2.1.1.8). HNMT occurs ubiquitously in vertebrate species and is widely expressed in mammalian tissues with particularly high expression levels in kidney, liver, colon, prostate, ovary, and spinal cord cells.19 HNMT is a cytosolic protein that is responsible for the intracellular inactivation of histamine. In contrast to the remarkable substrate specificity of HNMT, its known potent inhibitors are diverse in terms of their chemical structure and clinical pharmacology,20 and many were originally developed to inhibit histamine receptors or other enzymes. H1-receptor antagonists (conventionally called anti-histamines) are useful for relieving the symptoms of an allergic reaction such as rhinitis, urticaria, and atopic dermatitis.21 Besides anti-histamines, several anti-malarial compounds22,23 such as quinacrine and amodiaquine,24–26 the antitumor agent metoprine,27–34 and an early drug for Alzheimer’s disease called tacrine35–37 are among the most potent HNMT inhibitors known so far. Here, we describe the structure of the H1-receptor antagonist diphenhydramine in complex with HNMT and methyl donor analog S-adenosyl-L-homocysteine (AdoHcy). Diphenhydramine is a first-generation anti-histamine that is a competitive inhibitor of histamine for H1 receptor on effector cells.38,39 We compare the mode of anti-histamine inhibition with the other three potent inhibitors (amodiaquine, metoprine, and tacrine).
Results
HNMT, a 292 residue protein, has a two-domain structure including the methyl donor S-adenosyl-l-methionine (AdoMet) binding domain and a histamine-binding domain.40 We solved four crystal structures of the human HNMT bound to four different inhibitors at resolution limits of 1.9 Å (diphenhydramine), 2.3 Å (amodiaquine), 2.48 Å (metoprine), and 2.97 Å (tacrine) (Table 1). For each structure, the bound inhibitor in the histamine-bound site is well defined and its crystallographic thermal factor is within the range of that of side-chains involved in the interactions (Table 2). Except for amodiaquine, which exhibits a mixed mode, all the other inhibitors showed a histamine-competitive mode, with inhibitor constant (Ki) values in the 10–100 nm range41–43 (see below). In comparison, both reaction products, AdoHcy and methylhistamine (1-methyl-4-[β-aminoethyl]-imidazole), are inhibitors with Ki values in the micromolar range.40
Table 1.
Data reduction and refinement statistics of X-ray diffraction, collected from X26C beamline (NSLS) at wavelength of 1.10 Å
| Inhibitor | Diphenhydramine | Amodiaquine | Metoprine | Tacrine (T105) | Tacrine (I105) |
|---|---|---|---|---|---|
| PDB code | 2AOT | 2AOU | 2AOV | 2AOX | 2AOW |
| Unit cell dimensions (Å) | a=b=131.7, c=63.8 | a=b=130.9, c=63.3 | a=b=132.9, c=65.1 | a=b=132.3, c=64.6 | a=b=132.3, c=64.6 |
| Resolution range (Å) (highest resolution shell) |
35–1.9 (1.94–1.9) | 30–2.3 (2.35–2.3) | 28–2.48 (2.52–2.48) | 25–3.12 (3.19–3.12) | 28–2.97 (3.04–2.97) |
| Measured reflections | 212,782 | 91,414 | 99,214 | 58,144 | 59,910 |
| Unique reflections | 47,507 | 26,928 | 21,368 | 11,570 | 13,697 |
| ⟨I/σ⟩ | 24.8 | 12.3 | 14.8 | 10.7 | 13.1 |
| Completeness (%) | 95.5 (95.2) | 97.4 (96.2) | 91.5 (87.7) | 99.5 (100.0) | 99.1 (98.8) |
| Rlinear=∑|I–⟨I⟩|/∑⟨I⟩ | 0.068 (0.250) | 0.076 (0.245) | 0.071 (0.191) | 0.107 (0.273) | 0.088 (0.213) |
| Rfactor=∑|Fo–Fc|/∑|Fc| | 0.221 (0.255) | 0.207 (0.234) | 0.211 (0.254) | 0.229 (0.330) | 0.219 (0.311) |
| Rfree (5% or 10% data) | 0.256 (0.282) | 0.258 (0.300) | 0.260 (0.308) | 0.285 (0.369) | 0.269 (0.356) |
| Non-hydrogen atoms (2 molecules per asymmetric unit) | |||||
| Protein | 4494 | 4479 | 4529 | 4472 | 4442 |
| Hetrogen | 90 | 104 | 56 | 30 | 30 |
| Water | 337 | 306 | 91 | 9 | 24 |
| r.m.s. deviation (Å) between the two protein molecules |
0.8 | 0.6 | 0.6 | 0.03 | 0.04 |
| r.m.s. deviation from ideality | |||||
| Bond lengths (Å) | 0.031 | 0.011 | 0.008 | 0.011 | 0.012 |
| Bond angles (°) | 1.4 | 1.4 | 1.4 | 1.3 | 1.4 |
| Dihedrals (°) | 22.7 | 22.3 | 23.0 | 22.9 | 23.2 |
| Impropers (°) | 1.1 | 0.9 | 1.6 | 0.8 | 0.8 |
| Estimated coordinate error | |||||
| From Luzzati plot (Å) | 0.24 | 0.27 | 0.31 | 0.44 | 0.38 |
| From sigma(Å) | 0.14 | 0.17 | 0.22 | 0.26 | 0.35 |
Table 2.
Average crystallographic thermal B-factor (Å2) for side-chains involved in binding of inhibitors
| Complex | Diphenhydramine | Amodiaquine | Metoprine | Tacrine (T105) | Tacrine (I105) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Molecule | A | B | A | B | A | B | A | B | A | B |
| Inhibitor | 50.9 | 50.1 | 45.9 | 31.5 | 17.0 | 16.7 | 42.3 | 39.8 | 60.2 | 55.2 |
| F9 | 60.0 | 62.6 | – | – | – | 62.6 | 57.8 | 57.3 | – | – |
| Y15 | – | 54.8 | 62.9 | 51.1 | 65.0 | 65.3 | 57.1 | 57.2 | 73.2 | 73.7 |
| F19 | 56.5 | 44.3 | 48.6 | 41.6 | 44.0 | 53.2 | 48.6 | 47.8 | 52.1 | 52.7 |
| F22 | 50.8 | 41.8 | 36.5 | 29.9 | 35.3 | 45.8 | 43.5 | 42.1 | 49.7 | 46.6 |
| E28 | 38.8 | 36.2 | 25.8 | 22.5 | 26.8 | 33.7 | 39.6 | 39.0 | 38.9 | 40.8 |
| Q143 | 17.9 | 17.9 | 16.5 | 15.3 | 17.7 | 19.7 | 31.4 | 30.7 | 17.5 | 14.8 |
| Y146 | 17.1 | 15.5 | 18.6 | 13.5 | 10.4 | 8.5 | 42.2 | 40.2 | 7.6 | 8.8 |
| Y147 | 23.5 | 20.8 | 32.5 | 28.3 | 24.6 | 28.3 | 37.9 | 35.5 | 20.6 | 19.8 |
| W179 | 24.3 | 23.3 | 23.7 | 18.1 | 13.9 | 22.2 | 40.3 | 39.6 | 25.3 | 26.5 |
| W183 | 28.8 | 27.4 | 31.6 | 23.4 | 26.9 | 24.9 | 42.9 | 42.1 | 35.2 | 34.0 |
| C196 | 37.6 | 32.9 | 43.0 | 30.9 | 46.9 | 44.3 | 54.8 | 55.0 | 57.4 | 58.6 |
| Y198 | 29.5 | 25.6 | 34.7 | 24.9 | 29.9 | 32.5 | 52.3 | 53.1 | 43.1 | 41.9 |
| F243 | 48.9 | 38.7 | 38.6 | 26.4 | 37.6 | 38.8 | 49.3 | 47.7 | 52.7 | 49.8 |
| E246 | – | 49.2 | 45.2 | 30.1 | 57.2 | – | 55.4 | 55.6 | 60.8 | 60.4 |
| N283 | 26.6 | 26.2 | 21.2 | 21.3 | 24.1 | 26.9 | 38.3 | 34.7 | 26.7 | 24.7 |
Disordered side-chain.
Antihistamine diphenhydramine
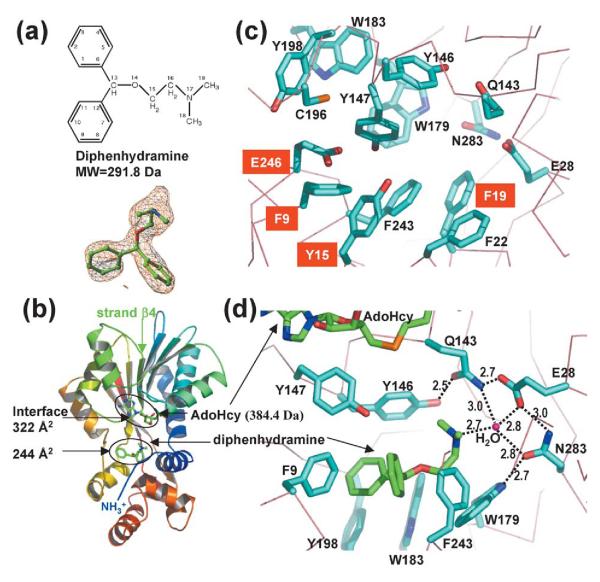
We crystallized a ternary complex containing HNMT, AdoHcy, and diphenhydramine (Figure 1(a)). AdoHcy is bound at the carboxyl ends of the parallel strands of the larger domain, as described.40 The diphenhydramine is bound in a dipolar pocket where histamine is normally bound (Figure 1(b)): a negatively charged glutamate (Glu28 or Glu246) located at each end with aromatic rings lining two sides (Figure 1(c)). The Y-shaped hydrophobic-ring structure of diphenhydramine contacts aromatic residues from different regions (Figure 1(d)): the N-terminal loop (Phe9), the loop after strand β4 of the AdoMet-binding domain (Tyr146 and Tyr147) and the histamine-binding domain (Trp179, Trp183, Tyr198, and Phe243). The phenyl ring of Phe9 points to the opening of the Y-shape (Figure 1(d)). The tail amine group of the inhibitor extends toward the polar end of Glu28, where the terminal nitrogen atom forms a hydrogen bond with an ordered water molecule. The water molecule is in the center of a network of polar interactions involving three catalytic residues, N-terminal Glu28, Gln143 of the carboxyl end of strand β4, and Asn283 of the C terminus (Figure 1(d)). The structure presented here illustrates that diphenhydramine inhibits HNMT by binding to the active site of the enzyme, thus sterically preventing substrate histamine from binding.
Figure 1.
Interactions of HNMT and diphenhydramine. (a) Chemical structure of diphenhydramine (top panel). The simulated-annealing omit electron density map is contoured at 3.5σ above the mean (bottom panel). (b) Ribbon diagram of HNMT-diphenhydramine-AdoHcy. The N terminus is in blue and the C terminus in red. (c) Active site pocket of HNMT reconstituted from the four structures. Residues adopting different rotomer conformation or disordered are labeled in white letter against red background. The side-chains of Glu246 and Tyr15 are disordered in the complex with diphenhydramine; the side-chain of Phe9 is disordered in the complexes with amodiaquine and metoprine, and adopts one rotamer in diphenhydramine and a different one in tacrine. (d) Detailed plot of HNMT–diphenhydramine interactions. The nitrogen atoms are in blue and the oxygen atoms in red. The carbon atoms are in cyan (HNMT) and in green (inhibitor). The broken lines indicate hydrogen bonds, whose distances are indicated (in Å).
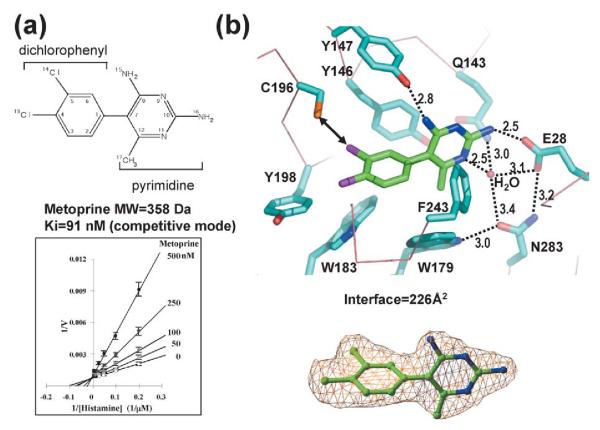
Antifolate drug metoprine
The aromatic rings of metoprine bind deep in the active site pocket, with its dichlorophenyl ring on the hydrophobic side and its pyrimidine ring near Glu28 (Figure 2). The inhibitor’s two rings are nearly perpendicular to one another, with a torsion angle of ~102°. This geometry is in close agreement with the structure of free metoprine, in which the torsion angle is 110°.44
Figure 2.
Interactions of HNMT–metoprine. (a) Chemical structure of metoprine (top panel). Lineweaver–Burke plot (bottom panel). The slope of each linear fit was plotted against the concentration of inhibitor and the intercept on the x-axis gave an estimate of Ki (91 nM). (b) Detailed plot of HNMT–diphenhydramine interactions. Replacing the chlorine atom (CL14) in the dichlorophenyl ring with sulfur might improve its interaction with Cys196 (indicated by a double-ended arrow). The simulated-annealing omit electron density map is contoured at 3.5σ above the mean (bottom panel).
The four pyrimidine nitrogen atoms, two ring and two exocyclic amino, allow direct and water-mediated hydrogen bonds. A water molecule mediates hydrogen bonds between ring nitrogen N11 and the polar side-chains of Glu28, Gln143, and Asn283. Two amide nitrogen atoms, N15 and N16, directly hydrogen bond the hydroxyl of Tyr147 and a carboxyl of Glu28, respectively. The interaction between the amide N16 and Glu28 breaks the hydrogen bond between the side-chains of Gln143 and Glu28, as also observed in the HNMT structures with diphenhydramine and amodiaquine (see below). The exocyclic methyl group (C17) of the pyrimidine makes van der Waals contacts with Val173 (not shown) and Trp179. Meanwhile, the chlorine atoms of the inhibitor’s dichlorophenyl ring make several van der Waals contacts. CL14 contacts Tyr146, Cys196, and the main chain carbonyl oxygen of Gln197 (not shown), while CL13 contacts Trp183, Tyr198, and the hydrophobic methylene group (Cγ) of Glu246 (not shown).
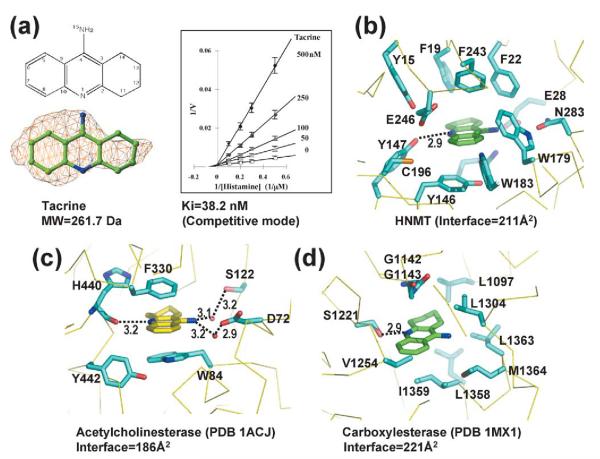
Anticholinesterase drug tacrine
Tacrine (Figure 3(a)) binds in the active site pocket closer to the polar end of Glu28 than the other inhibitors. On one side of the three-ring structure of tacrine is Tyr146, which stacks in parallel with the tacrine aromatic rings, while on the other side of the tacrine rings and perpendicular to them is Phe243 (Figure 3(b)). Along the inhibitor’s perimeter are Phe19, Phe22, Trp179, Trp183, Cys196 (the closest distance of ~5.2 Å to one of the ring carbons), and Glu246 (the closest distance of 4.2 Å to one of the ring carbon atoms). The tacrine ring nitrogen is hydrogen bonded to the hydroxyl of Tyr147, while its amino nitrogen points toward the side-chains of Glu28 and Asn283, but is not close enough to form a hydrogen bond. This is probably due to the bulky size of the three-ring structure in van der Waals contacts with the side-chains of Glu28 and Trp179, which prevent it from getting closer. Nevertheless, the amino nitrogen excludes the active site water molecule, which was seen with the other three inhibitors.
Figure 3.
Interactions of HNMT–tacrine. (a) Chemical structure of tacrine (left top panel). The simulated-annealing omit electron density map is contoured at 3.5σ above the mean (left bottom panel); Lineweaver–Burke plot (right panel). The estimate Ki is 38.2 nM. Tacrine bound in (b) HNMT, (c) acetylcholinesterase, and (d) carboxylesterase. Broken lines indicate the hydrogen bonds and distances are indicated (in Å).
There are two known structures containing tacrine, the Torpedo californica acetylcholinesterase (AcChE)45 and human carboxylesterase 1 (hCE1).35 In acetylcholinesterase (PDB 1ACJ), tacrine stacks between Trp and Phe residues; its ring nitrogen hydrogen bonded to a main-chain carbonyl oxygen, and its amino nitrogen hydrogen bonded to two water molecules (Figure 3(c)). In vivo, tacrine inhibits HNMT more potently than its purported target, acetylcholinesterase.46
While residues lining the HNMT and AcChE binding cavities are aromatic, the residues lining the carboxylesterase binding cavity are aliphatic side-chains (PDB 1MX1); the ring nitrogen of tacrine makes one hydrogen bond with this enzyme’s catalytic serine (Figure 3(d)). Comparison of the three tacrine binding proteins reveals that (1) the ring nitrogen atom forms hydrogen bonds with the target protein either via a side-chain or main-chain atom, (2) the three-ring structure is surrounded by hydrophobic residues dominated by either aromatic or hydrocarbon side-chains, and (3) developing a tacrine derivative with selectivity against just one of these enzymes would be challenging because of the high level of similarity of their active site pockets, enriched with aromatic residues. This also includes monoamine oxidase B,47 the enzyme that converts methylhistamine (the product of HNMT) to methylimidazole acetaldehyde.48 However, tacrine derivatives could provide some selectivity via interaction with the unique feature(s) of the each active site pocket (such as Cys196 and Glu246 of HNMT).
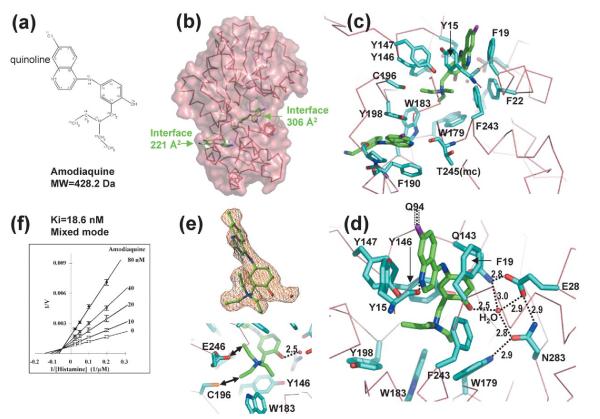
Anti-malarial drug amodiaquine
Crystals of the HNMT-amodiaquine binary complex yielded a diffraction limit of 2.3 Å. Unexpectedly, two molecules of amodiaquine (Figure 4(a)) were bound per enzyme molecule: one occupies the active site pocket and the other occupies a deep pocket on the outer surface (Figure 4(b)). The quinoline ring structures of both amodiaquine molecules show similar interactions with the enzyme: the quinoline ring in the active site pocket is sandwiched between two aromatic side-chains of Tyr15 and Phe19, while the quinoline ring in the outer surface pocket is between Tyr198 and Phe190 (Figure 4(c)). We suggest that the inhibitor bound in the active site is a competitive component, while the inhibitor bound in the outer surface pocket is an uncompetitive component. Because both competitive and uncompetitive components are present, the inhibition showed a mixed mode, and this might be the reason for its highest potency (the lowest Ki value: 7.5 nM42 or 18.6 nM (derived from Figure 4(f))) among the four inhibitors examined. An amodiaquine-Sepharose column has been used for affinity purification of HNMT,49 probably via the more-accessible outer surface-binding pocket. Interestingly, a single residue, Tyr198, acts as a “room divider”, separating the two binding pockets.
Figure 4.
Interactions of HNMT and amodiaquine. (a) Chemical structure of amodiaquine. (b) Two molecules of amodiaquine bind per HNMT; one binds in the active site pocket and the other in an outer-surface pocket. (c) Detailed plots of HNMT–amodiaquine interactions. The branch structure of amodiaquine is disordered in the outer-surface pocket. (d) A network of hydrogen bonds connects the side-chains of Tyr146, Qln143, Glu28, Asn283, Trp179, and a water molecule. The water molecule interacts with the hydroxyl group of the phenyl ring of amodiaquine. The chlorine atom of the quinoline ring of amodiaquine makes van der Waals contacts with Cγ of Gln94. (e) The two alkylene chains rotate every 120°, and make them look like three branches. The simulated-annealing omit electron density map is contoured at 3.5s above the mean (top panel). Besides the surrounding aromatic rings, one branch interacts (indicated by double-ended arrows) with the side-chain of Cys196 and another interacts with Glu246 (bottom panel). (f) Lineweaver–Burke plot. The estimated Ki is 18.6 nM.
The chlorine atom at position 11 of the amodiaquine in the active site pocket points outward and contacts the side-chain of Gln94 (Figure 4(d)), a residue normally involved in AdoMet or AdoHcy binding (see Figure 3(a) of Horton et al.40). This interaction between amodiaquine and Gln94 probably affects proper binding of AdoHcy, explaining the poor quality crystals of HNMT-amodiaquine in the presence of AdoHcy (not shown). The exocyclic hydroxyl oxygen at position 19 hydrogen bonds the active site water molecule (Figure 4(d)), which in turn interacts with three polar residues (Glu28, Gln143, and Asn283) as observed in diphenhydramine (see Figure 1(d)). The alkylamino tail has two branches at nitrogen position 21. Interestingly, we observed three identical “propeller blades” (Figure 4(e), top panel), by rotating the branch every 120°, surrounded by five aromatic residues, W179, W183, Y198, Y147, and F243. In addition, the tail methyl group of one of the branches is within hydrogen bonding distance of one of the carboxyl oxygen atoms of Glu246 (2.8 Å), while the other tail methyl group is within van der Waals contact distance of the Cys196 sulfur (3.4 Å) (Figure 4(e), bottom panel).
Discussion
A comparison between histamine-bound and inhibitor-bound HNMT structures clearly illustrates a rearrangement of the substrate-binding pocket to accommodate inhibitor molecules of various sizes. The most significant side-chain conformational changes are observed for the aromatic rings from the N terminus (Phe9, Tyr15, and Phe19), which form one wall of the active site pocket (Figure 1(c)) and can move to vary pocket size so as to accommodate either the small substrate histamine or an inhibitor as large as amodiaquine. For example, the phenyl ring of Phe9 interacts with two-ring Y-shaped structures of diphenhydramine (Figure 1(d)), while Phe9 is disordered in complexes with amodiaquine or metoprine, but in a different rotamer conformation when binding tacrine. The side-chain of Tyr15 is disordered in complexes with diphenhydramine, while its aromatic ring is stacked with the quinoline ring of amodiaquine (Figure 4(c)). While the phenyl ring of Phe19 stacks on the other side of the quinoline ring of amodiaquine, a different side-chain rotamer conformation is observed in complexes with diphenhydramine, metoprine, and tacrine. It appears that the innate flexibility of the N-terminal region of HNMT is fortuitously exploited to create a binding site for the structurally diverse inhibitor molecules.
Given this apparently promiscuous binding, how is the specificity and tight binding of HNMT inhibitors achieved? In each case, a key polar atom of the inhibitors (N1 of diphenhydramine, O19 of amodiaquine, N11 and N14 of metoprine, and N15 of tacrine) has the ability to form hydrogen bonds and occupies the HNMT active site, defined by the water-bound network of Glu28, Gln143, and Asn283. The orientation and bonding interactions clearly resemble those of the substrate complex.40 However, polar interactions between the enzyme and inhibitor are limited (three in metoprine, two in amodiaquine, and one each in diphenhydramine and tacrine).
These observations suggest that hydrophobic groups of the inhibitors and shape complementarity between the protein aromatic side-chains and aromatic ring(s) of the inhibitors are predominantly responsible for the nanomolar Ki values, despite the structural diversity of these compounds. This is possible because the protein aromatic rings in the active site pocket adopt different conformations that allow their hydrophobic groups to maximize contacts with the hydrophobic portion of the inhibitor. For example, the quinoline ring of amodiaquine is sandwiched face-to-face between Tyr15 and Phe19 (Figure 4(c)); the pyrimidine ring of metoprine is sandwiched between Tyr146 (face-to-face) and Phe243 (face-to-edge) (Figure 2); the three-ring structure of tacrine is sandwiched between Tyr146 (face-to-face) and Phe243 (face-to-edge) (Figure 3(b)).
A comparison of the four HNMT–inhibitor structures illustrates differences in their modes of binding, and suggests possible means of improving specificity and affinity. First, consider interactions with Glu246, the negatively charged residue at the opposite end of the pocket from Glu28 (Figure 1(c)). In binding diphenhydramine, the side-chain of Glu246 is disordered. In binding amodiaquine, one of the carboxyl oxygen atoms is in hydrogen bonding distance from the tail methyl group (Figure 4(e)); if the tail methyl group were replaced with an amino group, a hydrogen bond could be formed. In binding quinacrine, one of the chlorine atoms of the dichlorophenyl ring is in van der Waals contact with the Cβ atom of Glu246 (not shown); if the chlorine atom were eliminated and the ring carbon atom replaced with a ring nitrogen atom, a hydrogen bond could be formed.
Next consider nearby Cys196 (which is probably responsible for the inhibitory effect of SH-group reagents on HNMT50). In binding amodiaquine, the Cys196 sulfur is in van der Waals contact with one of the tail methyl groups of the inhibitor (Figure 4(e)). If the methyl group were replaced with a sulfhydryl group, a disulfide bond could be formed. Similarly, for the covalent disulfide bond attachment of a metoprine-like compound, replacing one the chlorine atoms in the dichlorophenyl ring with sulfur (Figure 2(b)) might improve its potency.
The structures of HNMT in complex with drugs, originally developed for diseases unrelated to histamine, should aid in the design of new drugs with selectivity for their intended targets over HNMT, or vice versa. It is also possible that visualization of diphenhydramine bound to HNMT will assist in the development of new generation H1-receptor antagonists with “dual” actions. A recent study demonstrated that inducible histamine reduced the excessive innate immune response through the stimulation of H2-receptors,51 suggesting anti-inflammatory effects of histamine. Similarly, a new class of potent and selective histamine H3 receptor ligands with combined inhibitory activity at HNMT has been designed.52 The resulting dual effect of an antagonist (via H3 receptor) and an enzyme inhibitor (via HNMT) could increase the levels of histamine in the synaptic space in a synergistic manner, and thus greatly enhance the histaminergic neurotransmission in psychiatric and neurodegenerative diseases.53
Summary
Our studies reveal how four different drugs bind to and inhibit human HNMT, an enzyme that is not their normal target. The bulky, hydrophobic portions of these drugs occupy the histamine-binding pocket, with the polar portions, atom(s) with potential to form hydrogen bonds, occupying the center of the active site. Thus, access of the substrate histamine to HNMT is blocked when any of these drugs is bound. Their tight binding is probably due to the large number of van der Waals interactions with HNMT. The structurally diverse, rigid hydrophobic groups of the drugs are accommodated by different rotamers of the HNMT aromatic rings. These interactions suggest avenues for improving the pharmacologically important HNMT inhibitors.
Materials and Methods
HNMT was expressed, purified and concentrated as described.40 HNMT inhibitors were obtained from Sigma-Aldrich as hydrochloride salts (except for metoprine) and dissolved in 15 mM Tris–HCl (pH 7.4), 1 mM DTT, and 1% (w/v) polyethylene glycol (PEG) 400. We obtained metoprine as a gift from Dr Nicol of Wellcome Research Laboratories, Research Triangle Park, NC. It was first wetted with concentrated HCl, and kept at room temperature for 15 min before water was added. The pH was adjusted with NaOH to approximately pH 4–5, and a 5 mM stock solution was made.
HNMT was pre-incubated with inhibitors (at two–threefold molar excess to protein) and AdoHcy (at an approximately 2:1 ratio of AdoHcy to enzyme) for at least 2 h before crystallization. Crystals were grown using the hanging-drop method with conditions being 19–26% PEG 6000, 100 mM Mes pH (5.5–6.2) and 5% ethylene glycol. All crystals had space group P6, with two molecules in the asymmetric unit. The structures of HNMT–inhibitor complexes were solved by molecular replacement of REPLACE,54 utilizing coordinates of the protein component from the ternary structure with histamine and AdoHcy (PDB 1JQD).40 Inhibitor models were built manually into densities of difference maps using the program O.55 Refinement proceeded with the program CNS.56 Non-crystallographic restraints were imposed during the refinement and were released at the later cycles for higher resolution structures (diphenhydramine, amodiaquine, and metoprine). The r.m.s. deviations for residues 5 to 292 between the two protein molecules of the asymmetric unit were 0.6–0.8 (diphenhydramine, amodiaquine, and metoprine) or 0.03 (tacrine) (Table 1); thus, only one monomer structure is described in the text. The protein–inhibitor interface area was calculated with the CCP4 program areaimol.57
A radiometric procedure was utilized to measure HNMT activity as described.40 Inhibitor (for each indicated concentration) was pre-incubated with HNMT (15 nM) and AdoMet (40 μM). The reactions were started by the addition of histamine (5, 10, 20, and 40 μM). The slope of each linear fit was plotted against the concentration of inhibitor and the intercept on the x-axis gave an estimate of the Ki value.
Figures were drawn using the program Pymol, a user-sponsored molecular modeling system with an OPENSOURCE foundation.†
Acknowledgements
We thank Dr Robert M. Blumenthal (Medical University of Ohio) and Stanley Hattman (University of Rochester) for comments on the manuscript, and Dr Xing Zhang for discussion throughout the project. The study was partly supported by the US National Science Foundation (INT-0003815), Emory University Research Committee, Georgia Research Alliance Eminent Scholar Challenge Grand Awards (No. RGA.CG04.300.D and No. GRA.CG05.F). The Cheng laboratory is currently supported by the National Institutes of Health grant GM068680. X-ray data for this study were measured at beamlines X26C of the National Synchrotron Light Source. Financial support for the beamlines comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health.
Abbreviations used
- HNMT
histamine N-methyltransferase
- AdoMet
S-adenosyl-l-methionine
- AdoHcy
S-adenosyl-l-homocysteine
Footnotes
Protein Data Bank accession codes Atomic coordinates have been deposited in the RCSB Protein Data Bank with accession numbers 2AOT (diphenhydramine), 2AOU (amodiaquine), 2AOV (metoprine), 2AOX and 2AOW (tacrine).
References
- 1.Hill SJ, Ganellin CR, Timmerman H, Schwartz JC, Shankley NP, Young JM, et al. International union of pharmacology XIII. Classification of histamine receptors. Pharmacol. Rev. 1997;49:253–278. [PubMed] [Google Scholar]
- 2.Hough LB. Genomics meets histamine receptors: new subtypes, new receptors. Mol. Pharmacol. 2001;59:415–419. [PubMed] [Google Scholar]
- 3.Hofstra CL, Desai PJ, Thurmond RL, Fung-Leung WP. Histamine H4 receptor mediates chemotaxis and calcium mobilization of mast cells. J. Pharmacol. Expt. Ther. 2003;305:1212–1221. doi: 10.1124/jpet.102.046581. [DOI] [PubMed] [Google Scholar]
- 4.Thurmond RL, Desai PJ, Dunford PJ, Fung-Leung WP, Hofstra CL, Jiang W, et al. A potent and selective histamine H4 receptor antagonist with anti-inflammatory properties. J. Pharmacol. Expt. Ther. 2004;309:404–413. doi: 10.1124/jpet.103.061754. [DOI] [PubMed] [Google Scholar]
- 5.Black JW, Duncan WA, Durant CJ, Ganellin CR, Parsons EM. Definition and antagonism of histamine H 2-receptors. Nature. 1972;236:385–390. doi: 10.1038/236385a0. [DOI] [PubMed] [Google Scholar]
- 6.Arrang JM, Garbarg M, Schwartz JC. Auto-inhibition of brain histamine release mediated by a novel class (H3) of histamine receptor. Nature. 1983;302:832–837. doi: 10.1038/302832a0. [DOI] [PubMed] [Google Scholar]
- 7.Falus A, Grosman N, Darvas Z. Histamine: Biology and Medical Aspects. SpringMed Publishing Ltd./S. Karger AG; Budapest/Basel: 2004. [Google Scholar]
- 8.Panula P, Rinne J, Kuokkanen K, Eriksson KS, Sallmen T, Kalimo H, et al. Neuronal histamine deficit in Alzheimer’s disease. Neuroscience. 1998;82:993–997. doi: 10.1016/s0306-4522(97)00353-9. [DOI] [PubMed] [Google Scholar]
- 9.Ligneau X, Lin J, Vanni-Mercier G, Jouvet M, Muir JL, Ganellin CR, et al. Neurochemical and behavioral effects of ciproxifan, a potent hista-mine H3-receptor antagonist. J. Pharmacol. Expt. Ther. 1998;287:658–666. [PubMed] [Google Scholar]
- 10.Morisset S, Rouleau A, Ligneau X, Gbahou F, Tardivel-Lacombe J, Stark H, et al. High constitutive activity of native H3 receptors regulates histamine neurons in brain. Nature. 2000;408:860–864. doi: 10.1038/35048583. [DOI] [PubMed] [Google Scholar]
- 11.Ohtsu H, Tanaka S, Terui T, Hori Y, Makabe-Kobayashi Y, Pejler G, et al. Mice lacking histidine decarboxylase exhibit abnormal mast cells. FEBS Letters. 2001;502:53–56. doi: 10.1016/s0014-5793(01)02663-1. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh AK, Hirasawa N, Ohtsu H, Watanabe T, Ohuchi K. Defective angiogenesis in the inflammatory granulation tissue in histidine decarboxylase-deficient mice but not in mast cell-deficient mice. J. Expt. Med. 2002;195:973–982. doi: 10.1084/jem.20011782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubota Y, Ito C, Sakurai E, Watanabe T, Ohtsu H. Increased methamphetamine-induced locomotor activity and behavioral sensitization in histamine-deficient mice. J. Neurochem. 2002;83:837–845. doi: 10.1046/j.1471-4159.2002.01189.x. [DOI] [PubMed] [Google Scholar]
- 14.Parmentier R, Ohtsu H, Djebbara-Hannas Z, Valatx JL, Watanabe T, Lin JS. Anatomical, physiological, and pharmacological characteristics of histidine decarboxylase knock-out mice: evidence for the role of brain histamine in behavioral and sleep-wake control. J. Neurosci. 2002;22:7695–7711. doi: 10.1523/JNEUROSCI.22-17-07695.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pap E, Racz K, Kovacs JK, Varga I, Buzas E, Madarasz B, et al. Histidine decarboxylase deficiency in gene knockout mice elevates male sex steroid production. J. Endocrinol. 2002;175:193–199. doi: 10.1677/joe.0.1750193. [DOI] [PubMed] [Google Scholar]
- 16.Fitzpatrick LA, Buzas E, Gagne TJ, Nagy A, Horvath C, Ferencz V, et al. Targeted deletion of histidine decarboxylase gene in mice increases bone formation and protects against ovariectomy-induced bone loss. Proc. Natl Acad. Sci. USA. 2003;100:6027–6032. doi: 10.1073/pnas.0934373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hori Y, Nihei Y, Kurokawa Y, Kuramasu A, Makabe-Kobayashi Y, Terui T, et al. Accelerated clearance of Escherichia coli in experimental peritonitis of histamine-deficient mice. J. Immunol. 2002;169:1978–1983. doi: 10.4049/jimmunol.169.4.1978. [DOI] [PubMed] [Google Scholar]
- 18.Malaviya R, Ikeda T, Abraham SN. Contribution of mast cells to bacterial clearance and their proliferation during experimental cystitis induced by type 1 fimbriated E. coli. Immunol. Letters. 2004;91:103–111. doi: 10.1016/j.imlet.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Schwelberger HG. Histamine N-methyltransferase (HNMT) enzyme and gene. In: Falus A, Grosman N, Darvas Z. s., editors. Histamine N-methyltransferase. Enzymology and functional aspects. SpringMed Publishing Ltd/S. Karger AG; Budapest/Basel: 2004. pp. 53–59. [Google Scholar]
- 20.Verburg KM, Henry DP. Histamine N-methyltransferase. Enzymology and functional aspects. In: Boulton AA, Baker GB, Yu PH, editors. Histamine receptors: specific ligands, receptor biochemistry, and signal transduction. Humana Press; Clifton, NJ: 1986. pp. 147–204. [Google Scholar]
- 21.Bakker RA, Timmerman H, Leurs R. Histamine receptors: specific ligands, receptor biochemistry, and signal transduction. Clin. Allergy Immunol. 2002;17:27–64. [PubMed] [Google Scholar]
- 22.O’Neill PM, Mukhtar A, Stocks PA, Randle LE, Hindley S, Ward SA, et al. Isoquine and related amodiaquine analogues: a new generation of improved 4-aminoquinoline antimalarials. J. Med. Chem. 2003;46:4933–4945. doi: 10.1021/jm030796n. [DOI] [PubMed] [Google Scholar]
- 23.Taylor WR, White NJ. Antimalarial drug toxicity: a review. Drug Saf. 2004;27:25–61. doi: 10.2165/00002018-200427010-00003. [DOI] [PubMed] [Google Scholar]
- 24.Barth H, Lorenz W, Troidl H. Effect of amodiaquine on gastric histamine methyltransferase and on histamine-stimulated gastric secretion. Br. J. Pharmacol. 1975;55:321–327. doi: 10.1111/j.1476-5381.1975.tb06934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nowak JZ, Zandarowska E. Effect of amodiaquine on histamine level and histaminemethyltransferase activity in the rat brain. Arch. Immunol. Ther. Expt. (Warsz) 1980;28:927–930. [PubMed] [Google Scholar]
- 26.Harle DG, Baldo BA. Structural features of potent inhibitors of rat kidney histamine N-methyltransferase. Biochem. Pharmacol. 1988;37:385–388. doi: 10.1016/0006-2952(88)90203-1. [DOI] [PubMed] [Google Scholar]
- 27.Zawilska J, Nowak JZ. Changes in the rat brain histamine content following metoprine and other histamine-methyltransferase (HMT) inhibitors. Pol. J. Pharmacol. Pharm. 1985;37:821–830. [PubMed] [Google Scholar]
- 28.Hough LB, Khandelwal JK, Green JP. Inhibition of brain histamine metabolism by metoprine. Biochem. Pharmacol. 1986;35:307–310. doi: 10.1016/0006-2952(86)90530-7. [DOI] [PubMed] [Google Scholar]
- 29.Sakai N, Onodera K, Maeyama K, Yanai K, Watanabe T. Effects of (S)-alpha -fluoromethylhistidine and metoprine on locomotor activity and brain histamine content in mice. Life Sci. 1992;51:397–405. doi: 10.1016/0024-3205(92)90406-f. [DOI] [PubMed] [Google Scholar]
- 30.Lecklin A, Jarvikyla M, Tuomisto L. The effect of metoprine on glucoprivic feeding induced by 2-deoxy-D-glucose. Pharmacol. Biochem. Behav. 1994;49:853–857. doi: 10.1016/0091-3057(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 31.Lecklin A, Tuomisto L, MacDonald E. Metoprine, an inhibitor of histamine N-methyltransferase but not catechol-O-methyltransferase, suppresses feeding in sated and in food deprived rats. Methods Find, Expt, Clin. Pharmacol. 1995;17:47–52. [PubMed] [Google Scholar]
- 32.Irman-Florjanc T. Effects of tele-methylhistamine and metoprine on plasma levels of histamine and on its elimination from cat plasma. Inflamm. Res. 1997;46(Suppl 1):S89–S90. [PubMed] [Google Scholar]
- 33.Lecklin A, Eriksson L, Leppaluoto J, Tarhanen J, Tuomisto L. Metoprine-induced thirst and diuresis in Wistar rats. Acta Physiol. Scand. 1999;165:325–333. doi: 10.1046/j.1365-201X.1999.00513.x. [DOI] [PubMed] [Google Scholar]
- 34.Malmberg-Aiello P, Ipponi A, Bartolini A, Schunack W. Antiamnesic effect of metoprine and of selective histamine H(1) receptor agonists in a modified mouse passive avoidance test. Neurosci. Letters. 2000;288:1–4. doi: 10.1016/s0304-3940(00)01176-9. [DOI] [PubMed] [Google Scholar]
- 35.Bencharit S, Morton CL, Hyatt JL, Kuhn P, Danks MK, Potter PM, Redinbo MR. Crystal structure of human carboxylesterase 1 complexed with the Alzheimer’s drug tacrine. From binding promiscuity to selective inhibition. Chem. Biol. 2003;10:341–349. doi: 10.1016/s1074-5521(03)00071-1. [DOI] [PubMed] [Google Scholar]
- 36.Cumming P, Vincent SR. Inhibition of histamine-N-methyltransferase (HNMT) by fragments of 9- amino-1,2,3,4-tetrahydroacridine (tacrine) and by beta-carbolines. Biochem. Pharmacol. 1992;44:989–992. doi: 10.1016/0006-2952(92)90133-4. [DOI] [PubMed] [Google Scholar]
- 37.Freeman SE, Dawson RM. Tacrine: a pharmacological review. Prog. Neurobiol. 1991;36:257–277. doi: 10.1016/0301-0082(91)90002-i. [DOI] [PubMed] [Google Scholar]
- 38.Taylor KM, Snyder SH. Histamine methyltransferase: inhibition and potentiation by antihistamines. Mol. Pharmacol. 1972;8:300–310. [PubMed] [Google Scholar]
- 39.Nowak JZ, Zawilska J. Histamine in the rat brain: effect of acute and chronic treatment with tricyclic antidepressants and H1-antihistaminics. Pol. J. Pharmacol. Pharm. 1985;37:147–162. [PubMed] [Google Scholar]
- 40.Horton JR, Sawada K, Nishibori M, Zhang X, Cheng X. Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons. Structure (Camb.) 2001;9:837–849. doi: 10.1016/s0969-2126(01)00643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taraschenko OD, Barnes WG, Herrick-Davis K, Yokoyama Y, Boyd DL, Hough LB. Actions of tacrine and galanthamine on histamine-N-methyltransferase. Methods Find. Expt. Clin. Pharmacol. 2005;27:161–165. doi: 10.1358/mf.2005.27.3.890872. [DOI] [PubMed] [Google Scholar]
- 42.Nishibori M, Oishi R, Itoh Y, Saeki K. 9-Amino-1,2,3,4-tetrahydroacridine is a potent inhibitor of histamine N-methyltransferase. Jpn. J. Pharmacol. 1991;55:539–546. doi: 10.1254/jjp.55.539. [DOI] [PubMed] [Google Scholar]
- 43.Cumming P, Reiner PB, Vincent SR. Inhibition of rat brain histamine-N-methyltransferase by 9-amino-1,2,3,4-tetrahydroacridine (THA) Biochem. Pharmacol. 1990;40:1345–1350. doi: 10.1016/0006-2952(90)90402-7. [DOI] [PubMed] [Google Scholar]
- 44.Cody V. Crystallographic studies of the antineoplastic antifolate 2,4-diamino-5-(3′,4′-dichlorophenyl)-6-methylpyrimidine (DDMP) ethanesulfonate salt. Cancer Biochem. Biophys. 1983;6:173–177. [PubMed] [Google Scholar]
- 45.Harel M, Schalk I, Ehret-Sabatier L, Bouet F, Goeldner M, Hirth C, et al. Quaternary ligand binding to aromatic residues in the active-site gorge of acetylcholinesterase. Proc. Natl Acad. Sci. USA. 1993;90:9031–9035. doi: 10.1073/pnas.90.19.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morisset S, Traiffort E, Schwartz JC. Inhibition of histamine versus acetylcholine metabolism as a mechanism of tacrine activity. Eur. J. Pharmacol. 1996;315:R1–R2. doi: 10.1016/s0014-2999(96)00738-8. [DOI] [PubMed] [Google Scholar]
- 47.Edmondson DE, Mattevi A, Binda C, Li M, Hubalek F. Structure and mechanism of monoamine oxidase. Curr. Med. Chem. 2004;11:1983–1993. doi: 10.2174/0929867043364784. [DOI] [PubMed] [Google Scholar]
- 48.Lin JS, Fort P, Kitahama K, Panula P, Denney RM, Jouvet M. Immunohistochemical evidence for the presence of type B monoamine oxidase in histamine-containing neurons in the posterior hypothalamus of cats. Neurosci. Letters. 1991;128:61–65. doi: 10.1016/0304-3940(91)90760-q. [DOI] [PubMed] [Google Scholar]
- 49.Tahara A, Nishibori M, Ohtsuka A, Sawada K, Sakiyama J, Saeki K. Immunohistochemical localization of histamine N-methyltransferase in guinea pig tissues. J. Histochem. Cytochem. 2000;48:943–954. doi: 10.1177/002215540004800707. [DOI] [PubMed] [Google Scholar]
- 50.Harvima RJ, Kajander EO, Harvima IT, Fraki JE. Purification and partial characterization of rat kidney histamine-N- methyltransferase. Biochim. Biophys. Acta. 1985;841:42–49. doi: 10.1016/0304-4165(85)90272-7. [DOI] [PubMed] [Google Scholar]
- 51.Yokoyama M, Yokoyama A, Mori S, Takahashi HK, Yoshino T, Watanabe T, et al. Inducible histamine protects mice from P. acnes-primed and LPS-induced hepatitis through H2-receptor stimulation. Gastroenterology. 2004;127:892–902. doi: 10.1053/j.gastro.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 52.Apelt J, Ligneau X, Pertz HH, Arrang JM, Ganellin CR, Schwartz JC, et al. Development of a new class of nonimidazole histamine H(3) receptor ligands with combined inhibitory histamine N-methyltransferase activity. J. Med. Chem. 2002;45:1128–1141. doi: 10.1021/jm0110845. [DOI] [PubMed] [Google Scholar]
- 53.Stark H, Arrang JM, Ligneau X, Garbarg M, Ganellin CR, Schwartz JC, Schunack W. The histamine H3 receptor and its ligands. Prog. Med. Chem. 2001;38:279–308. doi: 10.1016/s0079-6468(08)70096-1. [DOI] [PubMed] [Google Scholar]
- 54.Tong L, Rossmann MG. Rotation function calculations with GLRF program. Methods Enzymol. 1997;276:594–611. [PubMed] [Google Scholar]
- 55.Jones TA. Interactive electron-density map interpretation: from INTER to O. Acta Crystallog. sect. D. 2004;60:2115–2125. doi: 10.1107/S0907444904023509. [DOI] [PubMed] [Google Scholar]
- 56.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallog. sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 57.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]