Abstract
Background:
Current histopathological staging procedures in colon carcinomas depend on midline division of the lymph nodes with one section of haematoxylin & eosin (H&E) staining only. By this method, tumour deposits outside this transection line may be missed and could lead to understaging of a high-risk group of stage UICC II cases, which recurs in ∼20% of cases. A new diagnostic semiautomated system, one-step nucleic acid amplification (OSNA), detects cytokeratin (CK) 19 mRNA in lymph node metastases and enables the investigation of the whole lymph node. The objective of this study was to assess whether histopathological pN0 patients can be upstaged to stage UICC III by OSNA.
Methods:
Lymph nodes from patients who were classified as lymph node negative after standard histopathology (single (H&E) slice) were subjected to OSNA. A result revealing a CK19 mRNA copy number >250, which makes sure to detect mainly macrometastases and not isolated tumour cells (ITC) or micrometastases only, was regarded as positive for lymph node metastases based on previous threshold investigations.
Results:
In total, 1594 pN0 lymph nodes from 103 colon carcinomas (median number of lymph nodes per patient: 14, range: 1–46) were analysed with OSNA. Out of 103 pN0 patients, 26 had OSNA-positive lymph nodes, resulting in an upstaging rate of 25.2%. Among these were 6/37 (16.2%) stage UICC I and 20/66 (30.3%) stage UICC II patients. Overall, 38 lymph nodes were OSNA positive: 19 patients had one, 3 had two, 3 had three, and 1 patient had four OSNA-positive lymph nodes.
Conclusions:
OSNA resulted in an upstaging of over 25% of initially histopathologically lymph node-negative patients. OSNA is a standardised, observer-independent technique, allowing the analysis of the whole lymph node. Therefore, sampling bias due to missing investigation of certain lymph node tissue can be avoided, which may lead to a more accurate staging.
Keywords: colon cancer, OSNA, CK19, staging, lymph node, minimal residual disease
Lymph node metastases are among the most important prognostic indicators in colon carcinoma patients and are associated with increased loco-regional recurrence rates as well as worse 5-year survival (Merkel et al, 2001a; Croner et al, 2009; Kennedy et al, 2011). Adjuvant chemotherapy represents the standard treatment in lymph node-positive (stage UICC III) colon carcinoma patients. Adjuvant treatment improves the 8-year overall survival by ∼10% in these cases (Sargent et al, 2009). Interestingly, some subgroups of lymph node-negative tumours (stage UICC II) have a worse prognosis compared with stage UICC III carcinomas, for example, patients suffering from pT4 N0 tumours have a lower 5-year survival compared with those suffering from pT1-2 N1 tumours. Furthermore, the recurrence rate of stage UICC II colon carcinoma patients after complete (R0) resection amounts to 20% (Croner et al, 2009; Kennedy et al, 2011; Salazar et al, 2011). Nevertheless, adjuvant treatment is not generally recommended for stage UICC II colon carcinomas (Merkel et al, 2001b). There are ongoing discussions on whether lymph node-negative patients with high risk exist and how these patients can be identified for proper postoperative treatment. Several clinical and molecular classifiers within the primary tumour have been identified to select these cases (Rosenberg et al, 2002; Meyer et al, 2009; Sargent et al, 2009; Gray et al, 2011; Salazar et al, 2011). But none of these molecular classifiers have made it to clinical routine so far.
A different approach to identify stage I and II patients who may benefit from adjuvant chemotherapy is a thorough assessment of lymph nodes, which were determined as metastases-free by routine haematoxylin & eosin (H&E) analysis. However, step sectioning and immunohistochemistry are not commonly carried out for lymph node investigations and most pathologists use only a single slice of H&E staining. Therefore, small tumour deposits outside the midline transaction line of the lymph node may be missed during routine histopathological work-up and result in understaging of patients (Iddings et al, 2006; Davies et al, 2008).
One-step nucleic acid amplification (OSNA) is a novel technique to amplify mRNA directly from tissue lysates in an isothermal manner and has been developed to provide quantitative measurement of cytokeratin 19 (CK19) mRNA expression. In contrast to current routine histopathological procedures, it enables the workup of the whole lymph node in sufficient time under clinical conditions. Lymph node metastases in breast and colorectal carcinomas can be correctly identified by this procedure (Croner et al, 2010; Cserni, 2012; Guller et al, 2012; Yamamoto et al, 2013).
The objective of the present prospective, European multicentre study was to assess whether histopathological pN0 staged patients can be upstaged to stage UICC III by OSNA analyses. Patients recruited for this study were not included in recent single-institutional investigations demonstrating the accuracy of OSNA for lymph node staging in colorectal carcinomas (Croner et al, 2010; Guller et al, 2012).
Patients and Methods
Patients
The study was carried out after ethical approval was obtained from the included European study centres from Germany (Erlangen, Munich), Switzerland (Baden, Basel, Berne, St Gallen) and Spain (Lleida). Patients with histopathology-proven colon adenocarcinoma ⩾18 years between 2010 and 2012 were recruited after obtaining informed consent. Patient characteristics are listed in Table 1. Patients with synchronous second colon carcinomas, distant metastases (M+) or neoadjuvant treatment prior to surgery were excluded from this study. Tumours with a distance of at least 16 cm proximal to the anal verge measured with a rigid endoscope were classified as colon carcinomas. Colon resection was performed either by open complete mesocolic excision or by laparoscopic techniques, with a standardised regional lymphadenectomy (West et al, 2010; Lee et al, 2012). Immediately after surgery, the entire tumour-bearing specimen was delivered to an experienced pathologist who harvested the lymph nodes from the fresh mesocolon, following a standardised study protocol.
Table 1. Patient demographics, tumour characteristics and histopathological criteria,
| |
Patients |
OSNA negative |
OSNA positive |
|
|||
|---|---|---|---|---|---|---|---|
| Characteristic | n | % | n | % | n | % | P |
| Total |
103 |
100 |
77 |
74.8 |
26 |
25.2 |
|
|
Gender | |||||||
| Female | 49 | 47.6 | 35 | 71.4 | 14 | 28.6 | |
| Male |
54 |
52.4 |
42 |
77.8 |
12 |
22.2 |
0.50 |
|
Age | |||||||
| Median (range) |
71 (23–92) |
71 (23–90) |
71 (50–92) |
0.8 |
|||
|
Surgery | |||||||
| Open | 81 | 78.6 | 58 | 71.6 | 23 | 28.4 | |
| Laparoscopic |
22 |
21.4 |
19 |
86.4 |
3 |
13.6 |
0.27 |
|
Length of resected colon (cm) | |||||||
| Median (range) |
29.5 (8–73) |
27 (8–73) |
33.5 (15–65) |
0.27 |
|||
|
Tumour localisation | |||||||
| Coecum | 20 | 19.4 | 12 | 60 | 8 | 40 | |
| Ascending colon | 34 | 33 | 27 | 79.4 | 7 | 20.6 | |
| Right flexure | 2 | 1.9 | 2 | 100 | 0 | 0 | |
| Transverse colon | 7 | 6.8 | 5 | 71.4 | 2 | 28.6 | |
| Left flexure | 1 | 1 | 1 | 100 | 0 | 0 | |
| Descending colon | 9 | 8.7 | 5 | 55.6 | 4 | 44.4 | |
| Sigmoid colon |
30 |
29.1 |
25 |
83.3 |
5 |
16.7 |
0.34 |
|
Tumour size (cm) | |||||||
| <5 cm | 65 | 63.1 | 50 | 76.9 | 15 | 23.1 | 0.65 |
| ⩾5 cm |
38 |
36.9 |
27 |
71.1 |
11 |
28.9 |
|
| Median, cm (range) |
4 (0.5–16) |
4 (0.7–12) |
4.5 (0.5–16) |
|
|||
|
T-category | |||||||
| T1 | 9 | 8.7 | 7 | 77.8 | 2 | 22.2 | |
| T2 | 28 | 27.2 | 24 | 85.7 | 4 | 14.3 | |
| T3 | 58 | 56.3 | 41 | 70.7 | 17 | 29.3 | |
| T4 |
8 |
7.8 |
5 |
62.5 |
3 |
37.5 |
0.18 |
|
Histological grade | |||||||
| Well (G1) | 5 | 4.9 | 4 | 80 | 1 | 20 | |
| Moderate (G2) | 80 | 77.7 | 57 | 71.2 | 23 | 28.8 | |
| Poor (G3) |
18 |
17.5 |
16 |
88.9 |
2 |
11.1 |
0.32 |
|
Lymphatic invasion | |||||||
| Yes | 9 | 8.7 | 6 | 66.7 | 3 | 33.3 | |
| No |
94 |
91.3 |
71 |
75.5 |
23 |
24.5 |
0.69 |
|
Vascular invasion | |||||||
| Yes | 7 | 6.8 | 5 | 71.4 | 2 | 28.6 | |
| No |
96 |
93.2 |
72 |
75 |
24 |
25 |
1.0 |
|
Perineural invasion (Pn) | |||||||
| Yes | 11 | 14.1 | 7 | 63.6 | 4 | 36.4 | |
| No | 67 | 85.9 | 53 | 79.1 | 14 | 20.9 | 0.27 |
| Not determined |
25 |
|
|
|
|
|
|
|
R (resection status) | |||||||
| R0 | 103 | 100 | 77 | 74.8 | 26 | 25.2 | NS |
| R1 |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
LNs from fresh tissue for OSNA | |||||||
| Median, n (range) |
14 (1–46) |
12 (1–39) |
18 (4–46) |
|
|||
| <12 LN | 43 | 41.7 | 37 | 86.0 | 6 | 14 | |
| ⩾12 LN |
60 |
58.3 |
40 |
66.7 |
20 |
33.3 |
0.04 |
|
LNs from remaining formalin-fixed tissue | |||||||
| Median, n (range) |
5 (0–28) |
5 (0–28) |
4 (0–15) |
|
|||
| <7 | 60 | 58.3 | 43 | 71.7 | 17 | 28.3 | |
| ⩾7 | 43 | 41.7 | 34 | 79.1 | 9 | 20.9 | 0.49 |
Abbreviations: LNs=lymph nodes; OSNA=one-step nucleic acid amplification.
Study design
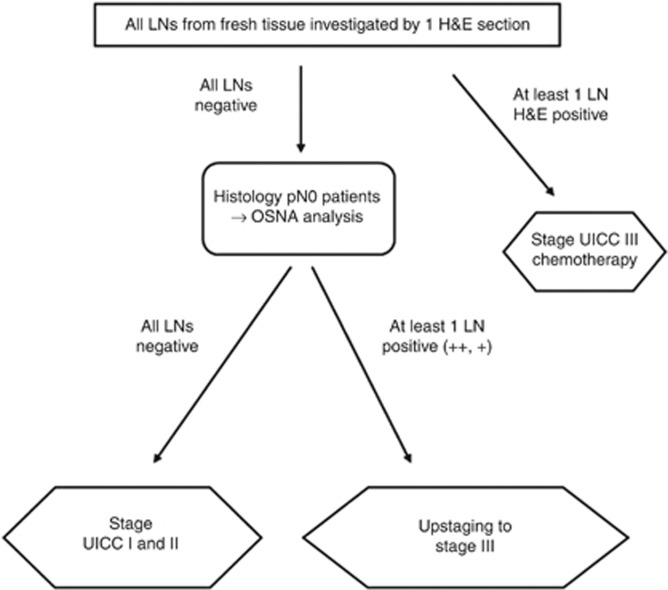
This study was designed to assess the potential upstaging rate of colon carcinoma patients by OSNA from stage UICC I/II after standard histopathology (one level of H&E staining) to stage UICC III (Figure 1). In this prospective study, the decision whether adjuvant treatment was administered was based on the histopathology results only, not on the OSNA results.
Figure 1.
Study design for upstaging of histopathology lymph node-negative colon carcinomas into lymph node-positive tumours by one-step nucleic acid amplification (OSNA) technology (++) has a CK19 mRNA copy number ⩾5000, a (+) between 250–4999. Abbreviations: LN=lymph node; H&E=haematoxylin & eosin.
Lymph node workup
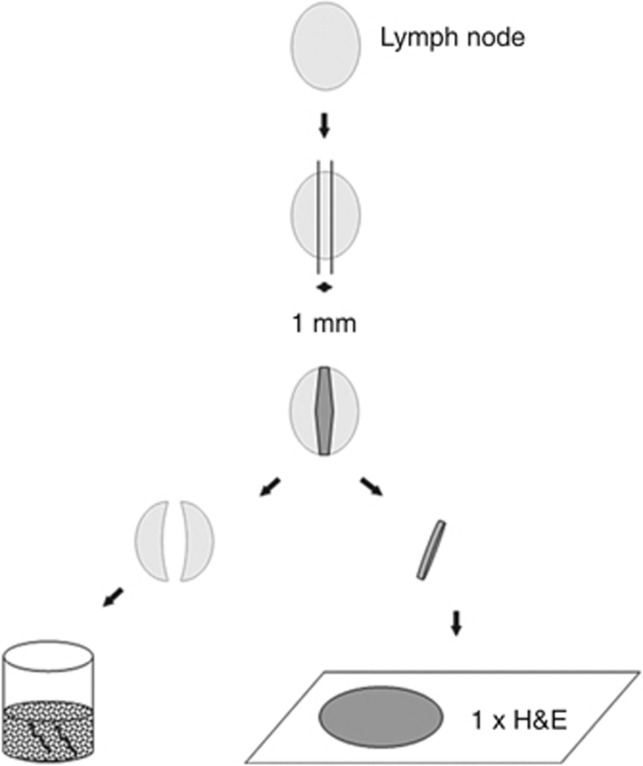
Lymph nodes were harvested as fresh specimens immediately after surgery. Involved pathologists underwent a special training for this procedure. Upon preparation of a lymph node, a 1 mm middle slice was immediately cut, and the two other lymph node pieces designated for OSNA were placed in a sterile tube and put into liquid nitrogen or on ice (Figure 2). Initially, the 1 mm middle slices of all lymph nodes were investigated by standard histology (one section of H&E staining) and the remaining parts of the lymph nodes were stored at −80 °C until OSNA analysis was performed. Owing to limited amount of tissue, lymph nodes <3 mm in diameter were excluded from OSNA analysis, but were investigated by histopathology only. If all lymph nodes were negative by histopathology (pN0), the remaining frozen parts of the lymph node were investigated by OSNA at the study centres in Erlangen and Lleida. Olten samples were analysed in Basel, and Munich samples were shipped to Erlangen, Basel, and Luebeck for OSNA analysis. If at least one lymph node was positive by histopathology (macro-metastasis or micro-metastasis), no OSNA analysis was performed. If at least one lymph node was positive by OSNA in a patient who was pN0 after standard H&E analysis, the patient was regarded as upstaged. If all remaining lymph nodes were negative by OSNA, the patient was not regarded as upstaged (Figure 1).
Figure 2.
Lymph node preparation for histopathology and OSNA work-up. The middle part of the lymph node was analysed by histopathology (haematoxylin & eosin (H&E) staining of one slice) for lymph nodes >6 mm in greatest diameter. For lymph nodes with 4–6 mm the lymph node was bisected and one half underwent OSNA analysis. If the lymph node was ⩽3 mm, it was excluded from OSNA analysing. In case of negative H&E staining, the lateral parts or the other half of the lymph nodes were transferred to OSNA analysing.
Histopathology and CK19 immunohistochemistry
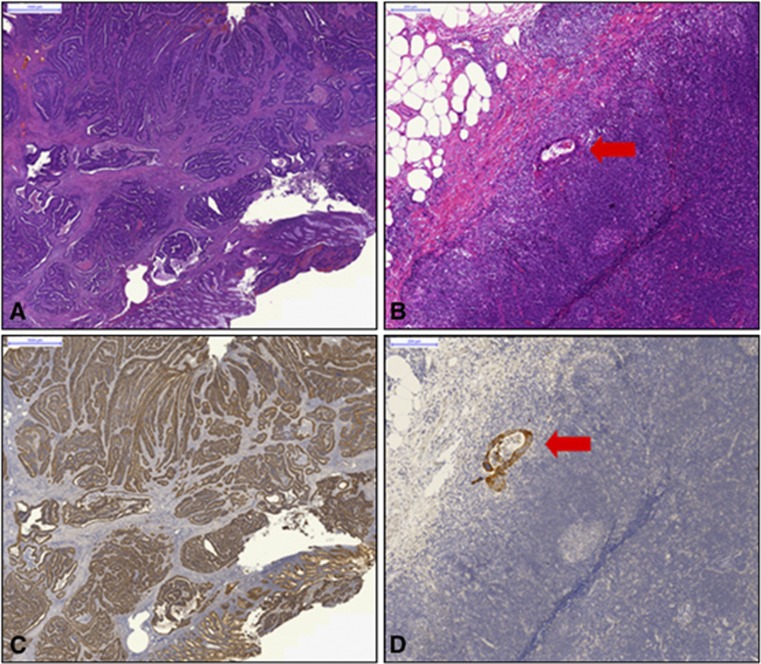
For all primary tumours, immunohistochemistry was performed for CK19 receptor (CK19 mouse monoclonal primary antibody) by the participating centres (Baloch et al, 1999; Uen et al, 2007). Sections of 1 μm size were cut from paraffin blocks mounted on superfrost glass slides, put in an incubator, and stained automatically. Tissue sections were then counterstained in the automated stainer with haematoxylin and treated with a bluing reagent. Evaluation of each immunohistochemical staining was performed by light microscopy at a magnification of 10–40-fold. A positive membranous expression with or without cytoplasmic staining in ⩾10% of neoplastic cells qualified the case as ‘positive (+)' for CK19. Every single colon carcinoma was stained and scored for CK19 as well as a fraction of randomly selected lymph nodes that were histologically detected positive (Figure 3).
Figure 3.
Haematoxylin & Eosin (H&E) staining (A, B) and CK19 immunohistochemistry (C, D) of the primary tumour (A, C) and lymph node metastasis (red arrow: B, D) of colon carcinoma.
One-step nucleic acid amplification
The lymph node tissue designated for OSNA (maximum weight of 600 mg per sample run) was shock frozen and stored at −80 °C until further use. OSNA analysis was performed no later than 6 months after sampling. If the tissue from one lymph node exceeded 600 mg, it was divided into portions weighing <600 mg. Each portion was analysed as a separate OSNA sample. Rapid RNA detection was achieved by homogenising slices of the dissected lymph node and directly amplifying CK19 mRNA without prior extraction or purification of nucleic acids (DNA and/or RNA). A designated reagent system was used for this method, according to the instructions for use of the manufacturer (Sysmex, Kobe, Japan). On the basis of previous investigations, an OSNA result designated as (++) has a CK19 mRNA copy number ⩾5000, and that designated as (+), between 250–4999. Less than 250 copies were considered as a negative result. This cutoff was defined in previous studies to make sure that mainly macrometastases (>0.2 mm) and not only micrometastases (⩽0.2 mm) or isolated tumour cells were scored as metastastic disease (Tsujimoto et al, 2007; Croner et al, 2010; Guller et al, 2012).
Statistical analysis
All statistics have been calculated using SAS 9.2 software (SAS Institute Inc., 2010b). The Cochran-Armitage test for trend was appropriate for a two-way table where one variable has two levels and the other variable is ordinal (P, 1955). The two-level variable represents the response, and the other variable represents an explanatory variable with ordered levels. The trend test is based on the regression coefficient for the weighted linear regression of the binomial proportions on the scores of the explanatory variable levels. The exact option was used to calculate P-values (SAS Institute Inc., 2010a). The Exact Fisher Test was used for calculating P-values for all frequency tables. Arithmetic means of metric variables were compared by t-test statistics using the pooled variance for calculating the error term. In all cases, two-sided P-values were reported. After adjustment for multiple testing according to Holm–Bonferroni procedure, the critical value for significance that warrants a global alpha of 0.05 becomes 0.003.
Results
Patient characteristics and tumour histopathology
A total of 103 patients with histopathological pN0 tumours (stage UICC I, II) were included in this prospective, European, multicentre study. Median age was 71 (range 23–92) years. Eighty-one patients (78.6%) underwent open surgery and 22 (21.4%) underwent laparoscopic procedures. All patients underwent a complete removal of the tumour-bearing tissue (R0 resection). The median length of the resected colon was 29.5 cm (range 8–73). Most tumours were localised in the coecum (19.4%), ascending colon (33%), or sigmoid colon (20.4%). Median tumour size was 4 cm (range: 0.5–16). Most patients had T3 (56.3%) and T2 (27.2%) tumours. Most carcinomas (80%) were moderately or well differentiated (G2) and showed neither lymphatic invasion (91.3%) nor vascular (93.2%) or perineural invasion (85.9%) (Table 1).
As a quality control of CK19 positivity, all primary tumours were analysed by immunohistochemistry. All included samples were positive for CK19 protein expression.
Lymph node harvesting
A median of 14 lymph nodes (range 1–46) was harvested from fresh specimens. In patients undergoing laparoscopic surgery (mean: 11), less number of lymph nodes were sampled from fresh specimen compared with open procedures (mean: 17; P=0.007). After formalin fixation of the remaining tissue, an additional median of 5 lymph nodes (range 0–28) were identified and embedded for histopathological workup only. The median total number of harvested lymph nodes (fresh and formalin fixed) for open and laparoscopic surgery was thus 22 and 16, respectively. In total, 1594 lymph nodes from 103 colon carcinoma specimens (median number per patient: 14, range: 5–46) were analysed with OSNA. In all but two patients (10 and 11 lymph nodes) ⩾12 lymph nodes were harvested in fresh tissue and after formalin fixation of the specimens.
OSNA positivity in histopathological pN0 tumours
Of the 103 histologically pN0 patients, 26 were positive with OSNA, resulting in an upstaging rate of 25.2%. Of these, 6/37 (16.2%) stage I UICC colon carcinoma patients and 20/66 (30.3%) stage UICC II patients were upstaged. Nineteen patients had one OSNA-positive lymph node, three had two, three had three, and one had four OSNA-positive lymph nodes (Table 2). Patients with T1, T2, and T4 tumours had only one positive lymph node, whereas all multiple OSNA-positive lymph nodes were found in patients with T3 tumour. Furthermore, 9/10 patients with a (++) OSNA result were found in the group of T3 tumour patients (Table 2).
Table 2. T-status of colon carcinomas and OSNA-positive lymph node (LN) results.
| T-category | Patients; n | Patients upstaged by OSNA; n | OSNA-positive LNs; n total (n per patient) | OSNA+ or ++ |
|---|---|---|---|---|
| T1 | 9 | 2 | 2 (1) | 1+ |
| |
|
|
|
1++ |
| T2 |
28 |
4 |
4 (1) |
4+ |
| T3 | 58 | 17 | 10 (1) | 8+ |
| 3 (2) | 9++ | |||
| 3 (3) | ||||
| |
|
|
1 (4) |
|
| T4 | 8 | 3 | 3 (1) | 3+ |
Abbreviations: LNs=lymph nodes; OSNA=one-step nucleic acid amplification.
OSNA result designated as (++) has a CK19 mRNA copy number ⩾5000, (+) between 250–4999.
Of the 38 OSNA-positive lymph nodes, 28 had CK19 mRNA copy number between 250–4999/μl (+), indicating a small tumour load, and 10 lymph nodes had copy numbers ⩾5000, indicating a higher tumour load (Croner et al, 2010). Correlation between OSNA upstaged patients and clinicopathological criteria was identified for gender, age, length of resected specimen, grade, tumour stage, tumour size and vascular/lymphatic or perineural invasion with OSNA positivity (Table 1). In patients with <12 lymph nodes, which were mainly found in the group undergoing laparoscopic procedures, the percentage of OSNA-positive cases was considerably lower than in patients with ⩾12 lymph nodes investigated (P=0.04). Of the 38 OSNA-positive lymph nodes, 22 were located ⩽5 cm and 7 were located >5 cm from the primary tumour. No information regarding the localisation is available in 9 OSNA-positive lymph nodes.
Discussion
In the present prospective, European, multicentre study – the largest so far published trial – OSNA resulted in an upstaging of over 25% of initially nodal-negative patients after conventional H&E analysis. OSNA provides a standardised technique allowing the analysis of the whole lymph node, thereby avoiding sampling bias due to missing investigation of certain lymph nodes and leading to more accurate staging. During the current clinical routine, only one slice in the midline area of the lymph node is investigated by H&E staining. Tumour deposits outside this area may be missed.
Tumour recurrence in stage UICC II colon carcinomas amounts to 20% after surgery with complete removal of the carcinoma-bearing specimen. Based on this unsettling fact, there is an ongoing debate if stage UICC II patients should receive chemotherapy. Several studies identified prognostic genes that may select high-risk patients for adjuvant treatment (Croner et al, 2008; Gray et al, 2011; Kennedy et al, 2011; Salazar et al, 2011; Nitsche et al, 2012; Maak et al, 2013). But none of these marker panels have made it into clinical practice so far. During routine histopathology, following H&E sectioning of lymph nodes, metastatic spread beneath the sectioned area may be missed and therefore such patients are understaged as pN0. Hence, methods are urgently needed that enable the investigation of the whole lymph node in a timely and feasible manner. OSNA is a novel molecular technology for CK19 mRNA-based identification of lymph node metastases in breast cancer and colorectal carcinomas (Tsujimoto et al, 2007; Croner et al, 2010; Yamamoto et al, 2011, 2013; Guller et al, 2012). The enormous advantage of this method is that the complete lymph node can be homogenised and transferred to OSNA without prior preparation, thus reducing the risk of potential contamination. The procedure is standardised and has a flat learning curve, which makes the procedure sufficient for clinical use. Furthermore, the results are ready within 20 min for one lymph node and within 30–40 min for 3–4 lymph nodes. Serial sections of the whole node for histology or immunohistochemistry are time consuming and can therefore not be performed routinely. Hence OSNA is more appropriate for clinical routine procedures. We found that the upstaging rate correlates with the number of lymph nodes analysed and with tumour stage. Notably, the OSNA upstaging rate in the present investigation for stage UICC I and II patients was 16.2% and 30.3%, respectively. The overall upstaging was 25.2%, which is within the reported recurrence rates for stage I and II colon carcinoma patients. Therefore, it can be hypothesised that stage UICC I and II patients who suffer from recurrent disease were understaged by conventional H&E analysis and were in reality in stage UICC III. To make sure that we identified not only micro-metastases or ITCs, which are still under discussion regarding their prognostic value, we defined a copy number >250 CK19 mRNA copies as a positive value (Guller et al, 2012). This threshold was identified in previous feasibility studies where ITCs were negative for OSNA in over 95% of cases respecting this cutoff. In histopathological workup, lymph nodes are completely embedded, but often only a single H&E-stained slide is assessed for each lymph node. Hence, nonsectioned tumour deposits to the left and right of this area, deeper in the paraffin block, may well be missed and staging biases may occur. To prove that OSNA upstaging of node-negative patients is associated with a higher recurrence rate, all patients in the present study will be followed for at least 5 years, with survival analysis after 3 and 5 years. However, these results are not yet mature. The T3 patient group was not only the largest group (56.3%) among the pN0 patients in the present study, but it also seems to have the most important upstaging potential. This is intuitive, as the T1/T2 group with a more favourable disease is expected to have a lower rate of lymph node metastases. Furthermore, it is evident that metastatic tumour cell spread does not follow a linear lymphatic drain of the carcinoma, but is more frequent in lymph nodes closely related to the primary tumour. Whether these lymph nodes are conditioned by tumour-related cytokines or chemokines and are therefore more recipient for the seeding of malignant cells remains a matter of debate (Langheinrich et al, 2012). A statistically significant correlation of histopathological parameters with OSNA was not reached. One explanation might be the relatively small study cohort. For instance, 46 out of 77 patients (59%) in the OSNA-negative cohort were classified as T3 or T4, whereas this percentage was higher in the OSNA-positive category (20/26 patients, 77%). Similarly, 7/60 OSNA-negative (12%) vs 4/18 OSNA-positive patients (22%) had perineural invasion. These findings indicate a relationship of OSNA with advanced tumours.
We would like to acknowledge the limitations of the present study. A disadvantage of OSNA is that in homogenised lymph node tissue-specific lymphadenopathic changes such as an infiltration by any other tumour or other important changes, for example, sarcomatoid-like lesions, cannot be detected. However, these are rare phenomena. Second, fresh or frozen material is necessary, which renders the lymph node harvesting challenging and time consuming, because the lymph node preperation in the mesenterial fat prior to formalin fixation requires experience (Guller et al, 2012). However, the potential benefit of precise staging by OSNA outweighs these shortcomings.
In summary, the present prospective, European, multicentre study – the largest one in the literature – OSNA demonstrated an upstaging in about one-fourth of colon carcinoma patients previously diagnosed as pN0 by routine histopathology. These findings confirm the technical feasibility of lymph node staging of colon carcinoma patients as recently described (Croner et al, 2010; Guller et al, 2012; Yamamoto et al, 2013). OSNA appears to be a more accurate method of staging colon carcinoma patients, and identifies pN0 individuals who may benefit from adjuvant treatment. As a next step, a randomised controlled trial is planned, allocating patients to either OSNA (entire node without H&E) vs standard H&E pathological analyses to evaluate OSNA as a method of routine staging in clinical practice.
Acknowledgments
This study was sponsored by Sysmex, the German Research Foundation (DFG 136/3-2), the German Federal Ministry of Education and Research (BMBF, Polyprobe-Study), and the ELAN-Fond of the University Erlangen-Nuremberg. We thank Dr E Breit, PhD, for technical support and assistance in writing the manuscript and Karin Erdtman and Anja Engelbrecht for study assistance during patient recruitment. Tissue samples were obtained in agreement with the institutional policies of each centre (Lleida, RD09/0076/0059, Munich 3006/10, and Erlangen 4214).
Author Contributions
Conception and design: RS Croner, C-I Geppert, M Zuber. Provision of study materials or patients: RS Croner, C-I Geppert, FG Bader, U Nitsche, C Späth, R Rosenberg, A Zettl, X Matias-Guiu, J Tarragona, M Zuber Collection and assembly of data: RS Croner, C-I Geppert, FG Bader, U Nitsche, C Späth, R Rosenberg, A Zettl, X Matias-Guiu, J Tarragona, U Güller, M Stürzl, M Zuber. Data analysis and interpretation: RS Croner, C-I Geppert, FG Bader, U Nitsche, C Späth, R Rosenberg, A Zettl, X Matias-Guiu, J Tarragona, U Güller, M Stürzl, M Zuber Manuscript writing: R S Croner, C-I Geppert, U Güller, M Stürzl, M Zuber. Final approval of manuscript: All authors.
The authors declare no conflict of interest.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Armitage P. Trends for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- Baloch ZW, Abraham S, Roberts S, LiVolsi VA. Differential expression of cytokeratins in follicular variant of papillary carcinoma: an immunohistochemical study and its diagnostic utility. Hum Pathol. 1999;30:1166–1171. doi: 10.1016/s0046-8177(99)90033-3. [DOI] [PubMed] [Google Scholar]
- Croner RS, Fortsch T, Bruckl WM, Rodel F, Rodel C, Papadopoulos T, Brabletz T, Kirchner T, Sachs M, Behrens J, Klein-Hitpass L, Sturzl M, Hohenberger W, Lausen B. Molecular signature for lymphatic metastasis in colorectal carcinomas. Ann Surg. 2008;247:803–810. doi: 10.1097/SLA.0b013e31816bcd49. [DOI] [PubMed] [Google Scholar]
- Croner RS, Merkel S, Papadopoulos T, Schellerer V, Hohenberger W, Goehl J. Multivisceral resection for colon carcinoma. Dis Colon Rectum. 2009;52:1381–1386. doi: 10.1007/DCR.0b013e3181ab580b. [DOI] [PubMed] [Google Scholar]
- Croner RS, Schellerer V, Demund H, Schildberg C, Papadopulos T, Naschberger E, Sturzl M, Matzel KE, Hohenberger W, Schlabrakowski A. One step nucleic acid amplification (OSNA) - a new method for lymph node staging in colorectal carcinomas. J Transl Med. 2010;8:83. doi: 10.1186/1479-5876-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserni G. Intraoperative analysis of sentinel lymph nodes in breast cancer by one-step nucleic acid amplification. J Clin Pathol. 2012;65:193–199. doi: 10.1136/jclinpath-2011-200301. [DOI] [PubMed] [Google Scholar]
- Davies M, Arumugam PJ, Shah VI, Watkins A, Roger Morgan A, Carr ND, Beynon J. The clinical significance of lymph node micrometastasis in stage I and stage II colorectal cancer. Clin Transl Oncol. 2008;10:175–179. doi: 10.1007/s12094-008-0176-y. [DOI] [PubMed] [Google Scholar]
- Gray RG, Quirke P, Handley K, Lopatin M, Magill L, Baehner FL, Beaumont C, Clark-Langone KM, Yoshizawa CN, Lee M, Watson D, Shak S, Kerr DJ. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29:4611–4619. doi: 10.1200/JCO.2010.32.8732. [DOI] [PubMed] [Google Scholar]
- Guller U, Zettl A, Worni M, Langer I, Cabalzar-Wondberg D, Viehl CT, Demartines N, Zuber M. Molecular investigation of lymph nodes in colon cancer patients using one-step nucleic acid amplification (OSNA): a new road to better staging. Cancer. 2012;118 (24:6039–6045. doi: 10.1002/cncr.27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iddings D, Ahmad A, Elashoff D, Bilchik A. The prognostic effect of micrometastases in previously staged lymph node negative (N0) colorectal carcinoma: a meta-analysis. Ann Surg Oncol. 2006;13:1386–1392. doi: 10.1245/s10434-006-9120-y. [DOI] [PubMed] [Google Scholar]
- Kennedy RD, Bylesjo M, Kerr P, Davison T, Black JM, Kay EW, Holt RJ, Proutski V, Ahdesmaki M, Farztdinov V, Goffard N, Hey P, McDyer F, Mulligan K, Mussen J, O'Brien E, Oliver G, Walker SM, Mulligan JM, Wilson C, Winter A, O'Donoghue D, Mulcahy H, O'Sullivan J, Sheahan K, Hyland J, Dhir R, Bathe OF, Winqvist O, Manne U, Shanmugam C, Ramaswamy S, Leon EJ, Smith WI, Jr., McDermott U, Wilson RH, Longley D, Marshall J, Cummins R, Sargent DJ, Johnston PG, Harkin DP. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J Clin Oncol. 2011;29:4620–4626. doi: 10.1200/JCO.2011.35.4498. [DOI] [PubMed] [Google Scholar]
- Langheinrich MC, Schellerer V, Perrakis A, Lohmuller C, Schildberg C, Naschberger E, Sturzl M, Hohenberger W, Croner RS. Molecular mechanisms of lymphatic metastasis in solid tumors of the gastrointestinal tract. Int J Clin Exp Pathol. 2012;5:614–623. [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Delaney CP, Lipman JM. Current state of the art in laparoscopic colorectal surgery for cancer: update on the multi-centric international trials. Ann Surg Innov Res. 2012;6:5. doi: 10.1186/1750-1164-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maak M, Simon I, Nitsche U, Roepman P, Snel M, Glas AM, Schuster T, Keller G, Zeestraten E, Goossens I, Janssen KP, Friess H, Rosenberg R. Independent validation of a prognostic genomic signature (ColoPrint) for patients with Stage II colon cancer. Ann Surg. 2013;257:1053–1058. doi: 10.1097/SLA.0b013e31827c1180. [DOI] [PubMed] [Google Scholar]
- Merkel S, Mansmann U, Papadopoulos T, Wittekind C, Hohenberger W, Hermanek P. The prognostic inhomogeneity of colorectal carcinomas Stage III: a proposal for subdivision of Stage III. Cancer. 2001;92:2754–2759. [PubMed] [Google Scholar]
- Merkel S, Mansmann U, Siassi M, Papadopoulos T, Hohenberger W, Hermanek P. The prognostic inhomogeneity in pT3 rectal carcinomas. Int J Colorectal Dis. 2001;16:298–304. doi: 10.1007/s003840100309. [DOI] [PubMed] [Google Scholar]
- Meyer A, Merkel S, Bruckl W, Schellerer V, Schildberg C, Campean V, Hohenberger W, Croner RS. Cdc2 as prognostic marker in stage UICC II colon carcinomas. Eur J Cancer. 2009;45:1466–1473. doi: 10.1016/j.ejca.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Nitsche U, Rosenberg R, Balmert A, Schuster T, Slotta-Huspenina J, Herrmann P, Bader FG, Friess H, Schlag PM, Stein U, Janssen KP. Integrative marker analysis allows risk assessment for metastasis in stage II colon cancer. Ann Surg. 2012;256:763–771. doi: 10.1097/SLA.0b013e318272de87. [DOI] [PubMed] [Google Scholar]
- Rosenberg R, Hoos A, Mueller J, Baier P, Stricker D, Werner M, Nekarda H, Siewert JR. Prognostic significance of cytokeratin-20 reverse transcriptase polymerase chain reaction in lymph nodes of node-negative colorectal cancer patients. J Clin Oncol. 2002;20:1049–1055. doi: 10.1200/JCO.2002.20.4.1049. [DOI] [PubMed] [Google Scholar]
- Salazar R, Roepman P, Capella G, Moreno V, Simon I, Dreezen C, Lopez-Doriga A, Santos C, Marijnen C, Westerga J, Bruin S, Kerr D, Kuppen P, van de Velde C, Morreau H, Van Velthuysen L, Glas AM, Van't Veer LJ, Tollenaar R. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29:17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- Sargent D, Sobrero A, Grothey A, O'Connell MJ, Buyse M, Andre T, Zheng Y, Green E, Labianca R, O'Callaghan C, Seitz JF, Francini G, Haller D, Yothers G, Goldberg R, de Gramont A. Evidence for cure by adjuvant therapy in colon cancer: observations based on individual patient data from 20,898 patients on 18 randomized trials. J Clin Oncol. 2009;27:872–877. doi: 10.1200/JCO.2008.19.5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc. 2010SAS/STAT 922 User's Guide. SAS Proprietary Software 9.2. Cary, NC, USA.
- SAS Institute Inc. 2010SAS/STAT 9.2 User's Guide: Introduction to Statistical Modeling with SAS/STAT Software. SAS Proprietary Software 9.2. Cary, NC, USA.
- Tsujimoto M, Nakabayashi K, Yoshidome K, Kaneko T, Iwase T, Akiyama F, Kato Y, Tsuda H, Ueda S, Sato K, Tamaki Y, Noguchi S, Kataoka TR, Nakajima H, Komoike Y, Inaji H, Tsugawa K, Suzuki K, Nakamura S, Daitoh M, Otomo Y, Matsuura N. One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients. Clin Cancer Res. 2007;13:4807–4816. doi: 10.1158/1078-0432.CCR-06-2512. [DOI] [PubMed] [Google Scholar]
- Uen YH, Lin SR, Wu DC, Su YC, Wu JY, Cheng TL, Chi CW, Wang JY. Prognostic significance of multiple molecular markers for patients with stage II colorectal cancer undergoing curative resection. Ann Surg. 2007;246:1040–1046. doi: 10.1097/SLA.0b013e318142d918. [DOI] [PubMed] [Google Scholar]
- West NP, Hohenberger W, Weber K, Perrakis A, Finan PJ, Quirke P. Complete mesocolic excision with central vascular ligation produces an oncologically superior specimen compared with standard surgery for carcinoma of the colon. J Clin Oncol. 2010;28:272–278. doi: 10.1200/JCO.2009.24.1448. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Sekimoto M, Oya M, Yamamoto N, Konishi F, Sasaki J, Yamada S, Taniyama K, Tominaga H, Tsujimoto M, Akamatsu H, Yanagisawa A, Sakakura C, Kato Y, Matsuura N. OSNA-based novel molecular testing for lymph node metastases in colorectal cancer patients: results from a multicenter clinical performance study in Japan. Ann Surg Oncol. 2011;18:1891–1898. doi: 10.1245/s10434-010-1539-5. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Daito M, Hiyama K, Ding J, Nakabayashi K, Otomo Y, Tsujimoto M, Matsuura N, Kato Y. An optimal mRNA marker for OSNA (One-step Nucleic Acid Amplification) based lymph node metastasis detection in colorectal cancer patients. Jpn J Clin Oncol. 2013;43:264–270. doi: 10.1093/jjco/hys227. [DOI] [PubMed] [Google Scholar]