Abstract
The chemical synthesis of ribonucleic acids (RNA) with novel chemical modifications is largely driven by the motivation to identify eligible functional probes for the various applications in life sciences. To this end, we have a strong focus on the development of novel fluorinated RNA derivatives that are powerful in NMR spectroscopic analysis of RNA folding and RNA ligand interactions. Here, we report on the synthesis of 2′-SCF3 pyrimidine nucleoside containing oligoribonucleotides and the comprehensive investigation of their structure and base pairing properties. While this modification has a modest impact on thermodynamic stability when it resides in single-stranded regions, it was found to be destabilizing to a surprisingly high extent when located in double helical regions. Our NMR spectroscopic investigations on short single-stranded RNA revealed a strong preference for C2′-endo conformation of the 2′-SCF3 ribose unit. Together with a recent computational study (L. Li, J. W. Szostak, J. Am. Chem. Soc. 2014, 136, 2858–2865) that estimated the extent of destabilization caused by a single C2′-endo nucleotide within a native RNA duplex to amount to 6 kcal mol−1 because of disruption of the planar base pair structure, these findings support the notion that the intrinsic preference for C2′-endo conformation of 2′-SCF3 nucleosides is most likely responsible for the pronounced destabilization of double helices. Importantly, we were able to crystallize 2′-SCF3 modified RNAs and solved their X-ray structures at atomic resolution. Interestingly, the 2′-SCF3 containing nucleosides that were engaged in distinct mismatch arrangements, but also in a standard Watson–Crick base pair, adopted the same C3′-endo ribose conformations as observed in the structure of the unmodified RNA. Likely, strong crystal packing interactions account for this observation. In all structures, the fluorine atoms made surprisingly close contacts to the oxygen atoms of the corresponding pyrimidine nucleobase (O2), and the 2′-SCF3 moieties participated in defined water-bridged hydrogen-bonding networks in the minor groove. All these features allow a rationalization of the structural determinants of the 2′-SCF3 nucleoside modification and correlate them to base pairing properties.
Introduction
Fluorine is hardly encountered in biomolecules and because of this pronounced bioorthogonality, it becomes a highly attractive reporter group. In particular for magnetic resonance spectroscopy, fluorine represents an excellent probe. Many applications, from structure and dynamics investigations to cellular imaging, have been reported over the past decade.1−15 Concerning ribonucleic acids (RNA), the potential of fluorine has been explored, mainly relying on labeling patterns with fluorine atoms that were attached at the 5-positions of pyrimidine nucleobases,8−12 or alternatively, at the ribose 2′-positions along the backbone.13−15 Although being powerful, these reporter units rely on a single fluorine atom, and thus limitations with respect to sensitivity could potentially be encountered. To find a solution for the sensitivity problem, trifluoromethylation of appropriate nucleoside positions seemed a logical consequence; however, efficient CF3 labeling approaches for RNA have not been available until recently.16
We have originally reported on 2′-trifluoromethylthio-2′-deoxy(2′-SCF3) uridine as a potential candidate to achieve RNA trifluoromethylation patterns in a straightforward manner.16 A first set of NMR spectroscopic applications using this label was indeed significant and diverse.16 The very preliminary observation that the novel modification, however, decreased the stability of a double helix to a very significant extent, brings up the questions on the generality of this behavior which is indeed surprising when compared to related derivatives. Many other small-size C2′ nucleoside modifications (e.g., 2′-OCH3,17 2′-O(CH2)2OCH3,17 2′-OCF3,18 2′-F,17) increase pairing stability, and the remaining leave it largely unaltered (e.g., 2′-N3),19,20 or only cause a minor decrease (e.g., 2′-CH3,21 2′-NH2,22 2′-SeCH3,23). Clearly, more comprehensive studies are warranted to explore the properties of 2′-SCF3 modified RNA and shed light on their molecular basis. To address some of the open questions, we made a combined effort involving chemical synthesis, UV-spectroscopy, isothermal titration calorimetry (ITC), NMR spectroscopy and X-ray crystallography. We present the synthesis of the novel 2′-trifluoromethylthio-2′-deoxy(2′-SCF3) cytidine phosphoramidite (C7) for RNA solid-phase synthesis and thereby further expand the site-specific introduction of the 2′-SCF3 modification into RNA. We provide a detailed thermodynamic analysis of duplex and hairpin stabilities and discuss the pairing properties in the light of sequence context and modified ribose conformations, analyzed by solution NMR spectroscopic means. Importantly, we have solved the X-ray structures of RNA with 2′-SCF3 modified nucleosides in three distinct base pair situations, at atomic resolution, to disclose crucial structural features such as ribose puckers, hydrogen-bonding networks, and hydration patterns of the 2′-SCF3 RNA modification, and to correlate them to base pairing properties.
Results and Discussion
Synthesis of 2′-SCF3 Cytidine
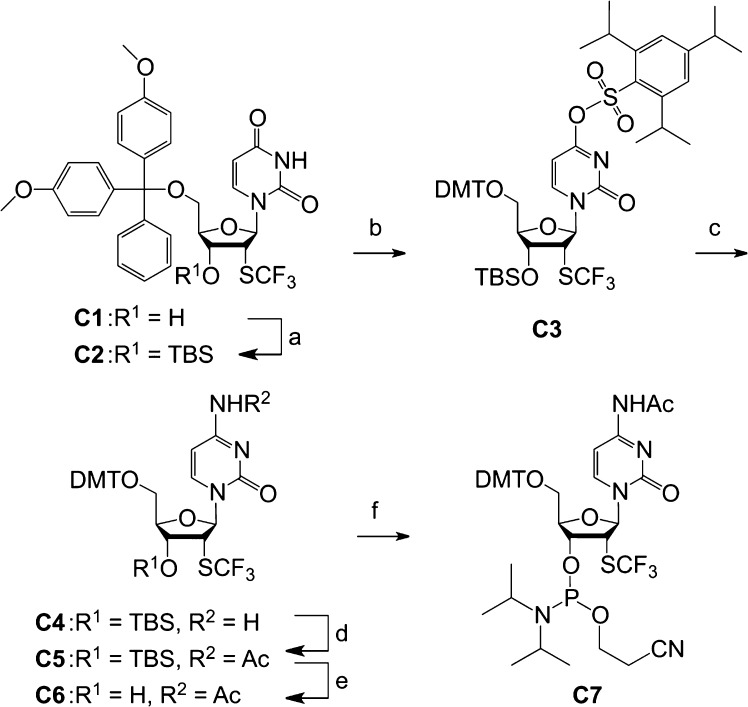
For building block C7 (Scheme 1), we started the synthesis from the 2′-trifluoromethylthio-2′-deoxyuridine derivative C1, which was readily obtained from 2′-deoxy-2′-mercaptouridine.16 The 3′-OH of compound C1 was protected using tert-butyldimethylsilyl (TBS) chloride and imidazole in dimethylformamide (DMF) to furnish derivative C2. Then, the reaction of C2 with 2,4,6-triisopropylbenzenesulfonyl chloride in the presence of triethylamine and 4-dimethylaminopyridine (DMAP) in dichloromethane resulted in regioselective O4-trisylation. After work-up, the trisylated derivative C3 can be used without further purification and directly converted into C4 upon treatment with aqueous ammonium hydroxide in tetrahydrofuran (THF) in 88% yield over the two steps. Acetylation of the amino function was then achieved with acetic anhydride in pyridine to provide C5, followed by cleavage of the 3′-O-TBS group with 1 M tetrabutylammonium fluoride (TBAF) and 0.5 M acetic acid in THF to give C6. Finally, conversion into the corresponding phosphoramidite C7 was achieved in good yields by reaction with 2-cyanoethyl N,N-diisopropylchlorophosphoramidite. Starting with compound C1, our route provides C7 in a 33% overall yield in six steps with four chromatographic purifications; in total, 1.2 g of 7 was obtained in the course of this study.
Scheme 1. Synthesis of 2′-SCF3 Cytidine Phosphoramidite C7.
Reaction conditions: (a) 5.0 equiv TBSCl, 6.0 equiv imidazole, 1.0 equiv AgNO3, in DMF, room temp, 16 h, 92%; (b) 1.5 equiv 2,4,6-triisopropylbenzenesulfonyl chloride, 10.0 equiv NEt3, 0.12 equiv DMAP, in CH2Cl2, room temp, 1.5 h, 66%; (c) 32% aqueous NH3, in THF, room temp, 16 h, 88%; (d) 2.5 equiv acetic anhydride, in pyridine, 0 °C to room temp, 90 min, 90%; (e) 1 M TBAF/0.5 M acetic acid, in THF, room temp, 2.5 h, 86%; (f) 1.5 equiv 2-cyanoethyl N,N-diisopropylchlorophosphoramidite, 10.0 equiv EtN(iPr)2, CH2Cl2, room temp, 3 h, 80%.
Synthesis of the 2′-SCF3-Modified RNA
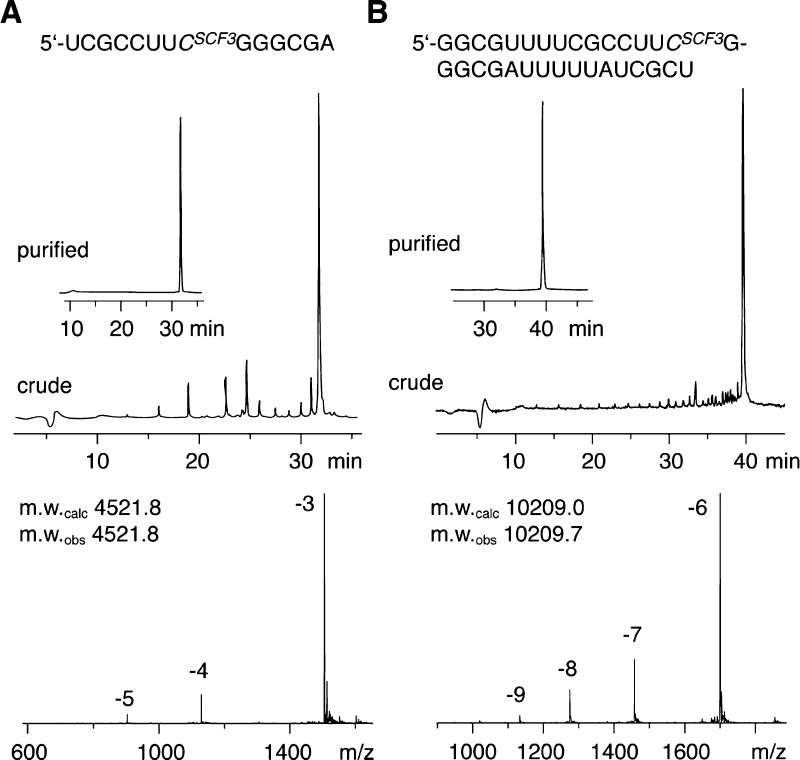
The solid-phase synthesis of RNA with site-specific 2′-SCF3 modifications was performed following the 2′-O-[(Triisopropylsilyl)oxy]methyl (TOM) approach.24,25 Coupling yields of the novel building block were higher than 98% according to the trityl assay. Cleavage of the oligonucleotides from the solid support and their deprotection were performed using CH3NH2 in ethanol/H2O, followed by treatment with TBAF in THF. Salts were removed by size-exclusion chromatography on a Sephadex G25 column, and RNA sequences were purified by anion-exchange chromatography under strong denaturating conditions (6 M urea, 80 °C; Figure 1). The molecular weights of the purified RNA molecules were confirmed by liquid-chromatography (LC) electrospray-ionization (ESI) mass spectrometry (MS). Synthesized RNA sequences containing 2′-SCF3 labels are listed in Supporting Information, Table S1. Noteworthy, the 2′-SCF3 label was completely stable under repetitive oxidative conditions (20 mM aqueous iodine solution) required during RNA solid-phase synthesis for transformation of P(III) to P(V) (Figure 1).
Figure 1.
Analysis of 2′-SCF3 modified RNA: anion-exchange HPLC traces (top) of 14 nt RNA (A) and 32 nt RNA (B), and respective LC-ESI mass spectra (bottom). HPLC conditions: Dionex DNAPac column (4 × 250 mm), 80 °C, 1 mL min–1, 0 to 60% buffer B in 45 min. Buffer A: Tris-HCl (25 mM), urea (6 M), pH 8.0. Buffer B: Tris-HCl (25 mM), urea (6 M), NaClO4 (0.5 M), pH 8.0. For LC-ESI MS conditions, see the Supporting Information.
Thermodynamic Stability of 2′-SCF3 Modified RNA
A single 2′-SCF3 modified nucleoside can exhibit an extraordinary attenuation of RNA duplex stability if the modification is located in the Watson–Crick base pairing region. UV melting profile analysis of the palindromic RNA 5′-GG(2′-SCF3-C)UAGCC (Figure 2A) revealed an average decrease of 28 °C in Tm-values for RNA concentrations in the micromolar range (ΔG°, −9.5 kcal mol–1; ΔH°, −75.2 kcal mol–1; ΔS°, −220 cal mol–1 K–1), compared to the unmodified counterpart (ΔG°, −18.4 kcal mol–1; ΔH°, −92.9 kcal mol–1; ΔS°, −254 cal mol–1 K–1) (Table 1). As a second example, the hairpin-forming RNA 5′-GAAGG-GCAA-C(2′-SCF3–C)UUCG (Figure 2B) also showed a pronounced decrease (19 °C) of Tm-values determined at micromolar RNA concentrations (ΔG°, −4.4 kcal mol–1; ΔH°, −50.4 kcal mol–1; ΔS°, −154 cal mol–1 K–1), compared to the unmodified counterpart (ΔG°, −7.1 kcal mol–1; ΔH°, −52.1 kcal mol–1; ΔS°, −151 cal mol–1 K–1) (Table 1). We hypothesized that the destabilization may stem—at least in part—from an inherent preference of the modified nucleoside to adopt the C2′-endo conformation. To provide evidence for such a hypothesis, we synthesized short, single-stranded RNAs, 5′-GU(2′-SCF3–U)CG, and 5′-UG(2′-SCF3–C)UCG, and determined 3J (H1′–H2′) coupling constants by 2D 1H,1H exclusive correlation spectroscopy (ECOSY) (Figure 3) and 1H,1H DQF COSY NMR experiments (Supporting Information, Figure S1). For both 2′-SCF3 uridine and -cytidine, values around 10.4 Hz were determined, accounting for a population of 100% of C2′-endo ribose conformation in single stranded RNA. As a consequence, this observation is a strong hint that forcing a 2′-SCF3 nucleoside into a C3′-endo ribose pucker, as mandatory for an A-form RNA double helix to avoid steric interference of the 2′ substituent, would introduce an energetic penalty. At this point we mention that 1H NMR spectra of RNAs with the 2′-SCF3 modification in double helical regions showed significantly broadened imino proton signals in that region, indicating accelerated exchange rates of the NH imino protons with bulk water (Supporting Information, Figure S2), and hence increased structural dynamics.
Figure 2.
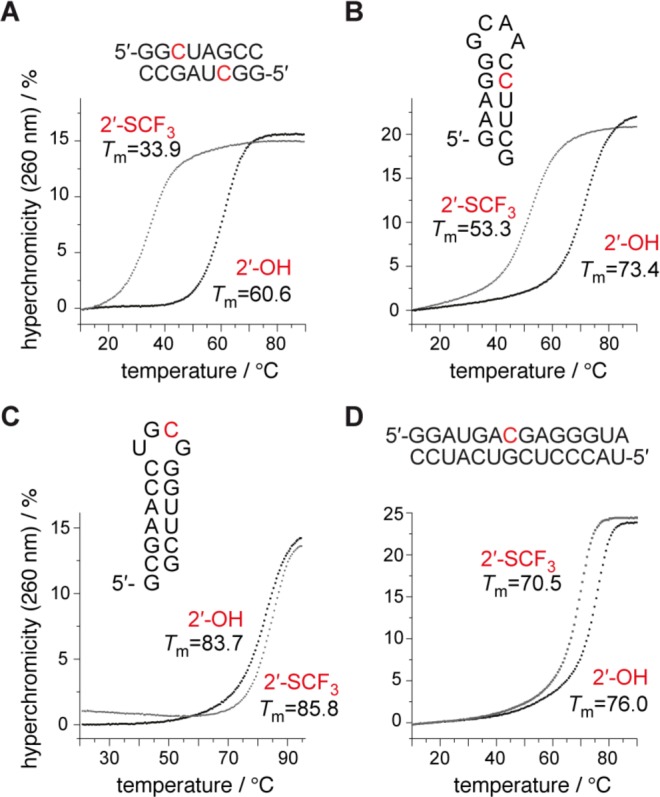
Thermal stabilities of unmodified and 2′-SCF3 modified oligoribonucleotides. UV-melting profiles of (A) self-complementary 8 nt RNA, (B) 15 nt RNA hairpin, (C) 17 nt RNA hairpin, and (D) 14 nt RNA duplex. Conditions: cRNA = 8 μM for profiles A and B, 4 μM for profiles C and D; 10 mM Na2HPO4, 150 mM NaCl, pH 7.0. Nucleotide abbreviations in red indcate the 2′-SCF3 modified position.
Table 1. Thermodynamic Parameters of 2′-SCF3-Modified RNA Obtained by UV Melting Profile Analysisa.
| sequence (5′→3′) | nt | ΔG298° [kcal mol–1] | ΔH° [kcal mol–1] | ΔS° [cal mol–1 K–1] |
|---|---|---|---|---|
| GGCUAGCC | 8 | –18.4 | –92.9 | –254 |
| GGCUAGCC | 8 | –9.5 | –75.2 | –220 |
| GGUCGACC | 8 | –15.4 | –84.8 | –233 |
| GGUCGACC | 8 | –9.2 | –58.3 | –165 |
| GAAGG-GCAA-CCUUCG | 15 | –7.1 | –52.1 | –151 |
| GAAGG-GCAA-CCUUCG | 15 | –4.4 | –50.4 | –154 |
| GCGAACC-UGCG-GGUUCG | 17 | –8.3 | –52.4 | –148 |
| GCGAACC-UGCG-GGUUCG | 17 | –8.9 | –53.2 | –149 |
| GGAUGACGAGGGUA/UACCCUCGUCAUCC | 14, 14 | –30.0 | –154.1 | –417 |
| GGAUGACGAGGGUA/UACCCUCGUCAUCC | 14, 14 | –22.4 | –114.4 | –309 |
U, 2′-SCF3 uridine; C, 2′-SCF3 cytidine. Buffer: 10 mM Na2HPO4, 150 mM NaCl, pH 7.0. ΔH and ΔS values were obtained by van’t Hoff analysis according to refs (27) and (28). Errors for ΔH and ΔS, arising from noninfinite cooperativity of two-state transitions and from the assumption of a temperature-independent enthalpy, are typically 10–15%. Additional error is introduced when free energies are extrapolated far from melting transitions; errors for ΔG are typically 3–5%.
Figure 3.
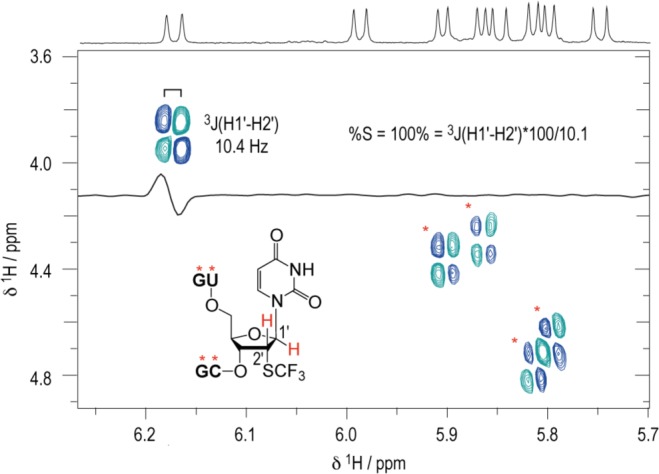
ECOSY NMR spectrum of the single-stranded RNA 5′-GU(2′-SCF3-U)CG. For the 2′-SCF3 uridine moiety, the 3-bond scalar coupling constant of H1′ and H2′ (3JH1′-H2′) was extracted from the corresponding crosspeak and amounted to 10.4 Hz. Assuming a pure C2′/C3′-endo equilibrium, this value is correlated to a C2′-endo (South) population of 100%.29,30 For the other single-stranded RNA nucleotides, coupling constants of 8.5 to 9.0 Hz were measured corresponding to C2′-endo populations between 84 to 89%. Conditions: cRNA = 0.3 mM; 25 mM sodium cacodylate, pH 7.0, 298 K.
Further support for the, C2′-endo hypothesis stems from a very recent computational study by Li and Szostak who developed a new free energy calculation method for molecular dynamics simulations.26 The calculated free energy landscape revealed that the C2′-endo conformation of a single nucleoside within a native A-form RNA duplex is significantly less stable by 6 kcal mol−1 compared to the C3′-endo conformer.26 This large value can be rationalized by the observation that the adoption of the C2′-endo pucker mode destabilizes the A-form because it disrupts the planar base pair structure, therefore weakening stacking and hydrogen-bonding interactions.26
On the basis of these results, we speculated that the 2′-SCF3 modification may also carry the potential to increase the thermodynamic stability of an RNA fold, namely if the C2′-endo ribose conformation were already present in the unmodified RNA and became further stabilized by the replacement of the 2′-OH group with 2′-SCF3. We therefore synthesized the hairpin 5′-GCGAACG-UGCG-GGUUCG (Figure 2C) which contains a UNCG tetranucleotide loop motif; the cytidine in such loops is known to adopt a C2′-endo conformation which was confirmed for the particular sequence used here by solution NMR spectroscopy.31 The concentration-independent Tm value of this hairpin was 83.7 °C (ΔG°, −8.3 kcal mol–1; ΔH°, −52.4 kcal mol–1; ΔS°, −148 cal mol–1 K–1). Indeed, the modified variant 5′-GCGAACG-UG(2′-SCF3-C)G-GGUUCG revealed a Tm value of 85.8 °C (ΔG°, −8.9 kcal mol–1; ΔH°, −53.2 kcal mol–1; ΔS°, −149 cal mol–1 K–1) (Table 1), clearly higher than the unmodified counterpart. To verify this observation, we analyzed the shorter sequence analogue 5′-AAGC-UGCG-GGUUC, and additionally, 5′-ACG-UUCG-GCU, both RNAs possessing lower Tm values (69.7 and 43.4 °C) allowing for a more reliable determination of thermodynamic parameters (Supporting Information, Figure S3). The corresponding modified counterparts comprising a -UG(2′-SCF3-C)G- and -UU(2′-SCF3-C)G-loop, respectively, were indeed thermodynamically more stable (increase in Tm values by three and four degrees: 73.5 and 47.3 °C) (Supporting Information, Figure S3).
The slight stabilizing effect through a C2′-endo adopting 2′-SCF3-cytosine in UNCG-loop motifs of hairpins (∼0.6 kcal mol−1, see Table 1), was furthermore evaluated independently for a bistable RNA. A bistable RNA consists of two defined secondary structures in dynamic equilibrium, and in the simplest case, involves competing hairpins.32 Here, we included the 17 nt stem-loop sequence discussed above (Figure 2C) into a bistable 34 nt RNA construct (Figure 4). Indeed, we observed the expected shift of the secondary structure equilibrium position from 6:4 for the unmodified RNA (Figure 4D; see also references (32) and (31)) to 9:1 (Figure 4B) for the modified counterpart toward fold A that comprises the 2′-SCF3 group within the UNCG loop. We suggest that the (single-stranded) loop becomes favorably preformed because of the 2′-SCF3 cytosine in C2′-endo conformation, and that this loop preorganization results in an improved orientation of the strand portions for double helix nucleation.
Figure 4.
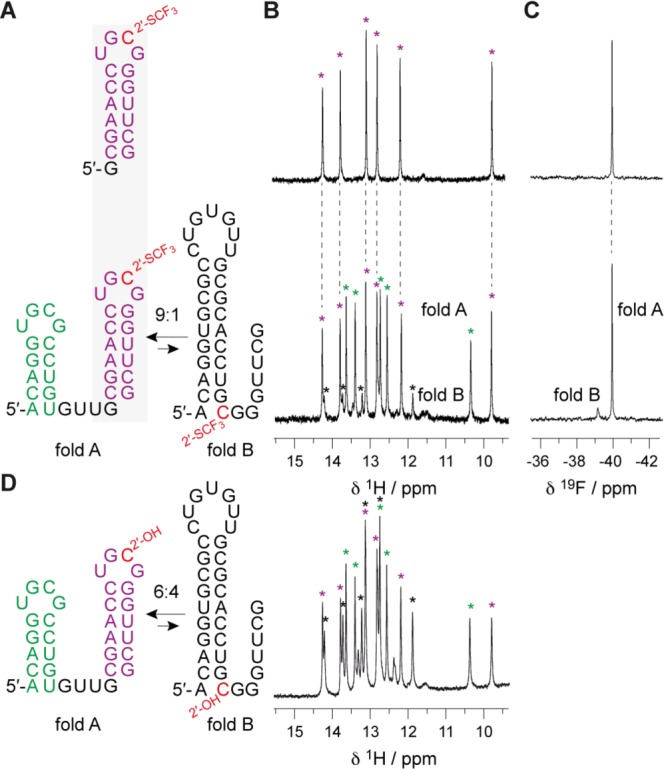
NMR spectroscopic analysis of 2′-SCF3 modified RNAs. (A) 17 nt RNA hairpin (fold A reference) (top) and 34 nt bistable RNA (bottom) and corresponding 1H imino proton (B) and 19F (C) NMR spectra. (D) 1H imino proton spectrum of the unmodified 34 nt RNA reference. Conditions: cRNA = 0.3 mM; 25 mM Na2HAsO4, pH 7.0, 25 °C. Nucleotide abbreviations in red indicate the 2′-SCF3 position.
We note that a stabilizing effect of the 2′-SCF3 group has been observed so far only in the specific context of a UNCG loop. Other bistable RNA with the 2′-SCF3 group in single-stranded regions of both mutually exclusive folds provide the same equilibrium position as observed for the unmodified counterpart (for an example see ref (16)). Consistently, when the 2′-SCF3 modification is placed in a manner that it resides in the single-stranded region of one fold but in a double helical region of the alternative fold, the latter becomes dramatically lower populated, or not observable at all (Supporting Information, Figure S4).
In addition, we investigated the influence of a single 2′-SCF3 group at a base pair in the center of an extended duplex, providing a 6 bp stretch upstream and a 7 bp stretch downstream of the modification (Figure 2D). In this case, the degree of duplex destabilization caused by the modification was smaller and amounted to 6 °C (Table 1). A possible explanation is that the minimal number of canonical base pairs required for double helix nucleation, that is three to four,33,34 is provided by both Watson–Crick base pair stretches that neighbor the modification while this criterion was not fulfilled for the oligoribonucleotides described above. For the RNAs that experienced very pronounced destabilization, the number of base pairs next to the 2′-SCF3 modification was typically one to three.
Finally, we investigated the impact of the 2′-SCF3 modification on thermodynamic duplex stability by isothermal titration calorimetry (ITC).35 The destabilizing effect of the 2′-SCF3 group in the asymmetric 14 bp RNA duplex was well reflected in the obtained thermodynamic parameters, ΔHITC and ΔSITC (Figure 5). A direct comparison of ITC with thermally derived enthalpy values, however, has to be taken with caution.36 Although the same buffer/salt conditions (as for the UV spectroscopic experiments) were used, ΔHITC values were smaller compared to the corresponding ΔHUV values (Figure 5, Table 1), for the unmodified RNA duplex even significantly smaller. This phenomenon has been observed also by others37 and may account for the difference in single strand folding and unfolding contributions for the distinct experimental setups. In the UV melting experiment, single strands are significantly unfolded at the Tm, so further temperature-dependent unfolding of those strands will be modest, though not absent. In contrast, perturbation of single-stranded structure (e.g., nucleobase stacking, but also mismatched hairpin formation) across the lower temperature ranges typically sampled in ITC experiments can be significant,37 as observed here.
Figure 5.
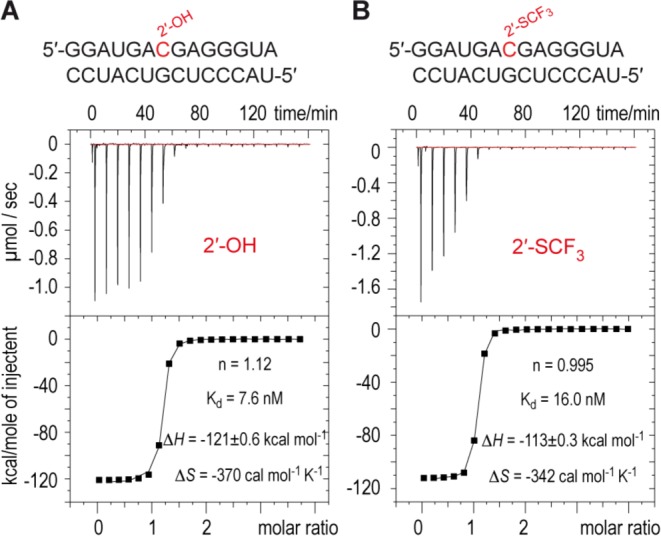
Exemplary isothermal titration calorimetry (ITC) experiments for unmodified (A) and 2′-SCF3 modified (B) 14 bp RNA duplexes. Conditions: A 58 (A) and 165 (B) μM solution of lower strand was titrated into 0.3 mL of 4.1 (A) and 5.5 (B) μM upper strand equilibrated at 25 °C. Both RNAs were in 10 mM Na2HPO4, 150 mM NaCl, pH 7.0. The experiments depicted yielded fitting parameters as indicated. Unmodified RNA: ΔH = −116 ± 9 kcal mol−1, ΔS = −356 ± 30 cal mol–1 K–1. 2′-SCF3 RNA: ΔH = −106 ± 7 kcal mol−1, ΔS = −323 ± 25 cal mol–1 K–1 (from at least two independent measurements).
X-ray Analysis of the 2′-SCF3-Modified RNA
We set out for the X-ray analysis of a 2′-SCF3 modified RNA and focused on the 27 nt fragment of the E. coli 23 S rRNA sarcin–ricin loop (SRL) (Figure 6A).38 The SRL RNA is known to be a robust and well behaved crystallization scaffold that can accommodate small modifications.19,38 For the modification of interest we first considered nucleotide U2656 which forms a Hoogsteen base pair with A2665 and is involved in a base triplet together with G2655. As a second target for 2′-SCF3 labeling, we selected C2667 which forms a water-mediated base pair with U2653. Both nucleosides adopt C3′-endo conformations and should be well available for modifications at the ribose 2′ position, according to our previous analysis of the unmodified SRL structure (Protein Data Bank [PDB] identification no. 3DVZ) that showed that the 2′-OH groups of U2656 and C2653 are not involved in crystal contacts.38 Also, UV melting experiments were encouraging as exemplified by the melting profile of the 2′-SCF3 U2667 modified SRL RNA which revealed a high Tm value albeit destabilization compared to the unmodified counterpart (Supporting Information, Figure S5A). Crystallization trials (at 293 K) for both 2′-SCF3-modified RNAs were successful, providing crystals diffracting to atomic resolution (Table 2).
Figure 6.
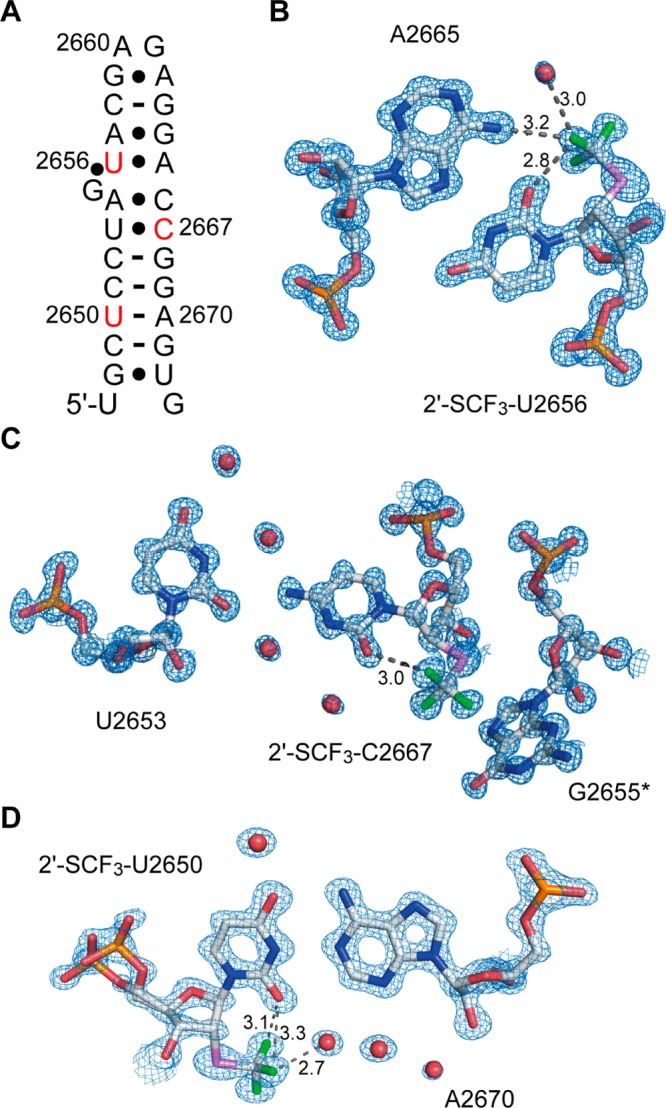
X-ray structures of RNAs with a single 2′-SCF3 modification at atomic resolution. (A) E. coli sarcin–ricin stem-loop (SRL) RNA used for crystallization; secondary structure; nucleosides that were modified are indicated in red. 2Fobs – Fcalc electron density maps showing (B) the A2665/2′-SCF3-U2656, (C) the U2653/2′-SCF3-C2667, and (D) the 2′-SCF3-U2650/A2670 nucleobase interactions. Water molecules are shown as red spheres. The CF3 group in (C) stacks on G2655 of the neighboring hairpin in the crystal (indicated by asterisk). Distances are in Å.
Table 2. X-ray Data Collection and Refinement Statistics.
| SRL RNA derivative | 2′-SCF3-U2656 | 2′-SCF3-C2667 | 2′-SCF3-U2650 |
|---|---|---|---|
| PDB ID | 4NMG | 4NLF | 4NXH |
| space group | P43 | P21 | P43 |
| a (Å) | 29.57 | 29.17 | 29.56 |
| b (Å) | 29.57 | 39.57 | 29.56 |
| c (Å) | 76.52 | 29.92 | 76.73 |
| β | 90° | 90.92° | 90° |
| beamline | PX III-X06DA | PX III-X06DA | PX III-X06DA |
| resolution range (Å) | 30–1.01 | 30–1.00 | 30–1.16 |
| no. frames | 1800 | 7200 | 3600 |
| oscillation angle | 0.2° | 0.1° | 0.2° |
| wavelength | 0.8 | 0.8 | 1.0 |
| average redundancy | 6.5 | 5.7 | 12.1 |
| completeness1 | 99.6% (97.6%) | 95.4% (91.2%) | 99.3% (93.3%) |
| Rmerged1 | 4.7% (108.6%) | 3.1% (9.3%) | 7.1% (96.2%) |
| CC1/21 | 100% (65%) | 100% (99.5%) | 100% (69.8%) |
| average I/σ1 | 18.8 (1.5) | 38.8 (15.8) | 17.8 (2.1) |
| ISa | 27 | 29 | 14 |
| R/Rfree | 12.0/14.1 | 9.9/11.6 | 12.0/14.8 |
| coordinate error (Å) | 0.09 | 0.03 | 0.10 |
| Wilson B | 9.3 | 5.5 | 11.1 |
Values for last resolution shell are shown in parentheses.
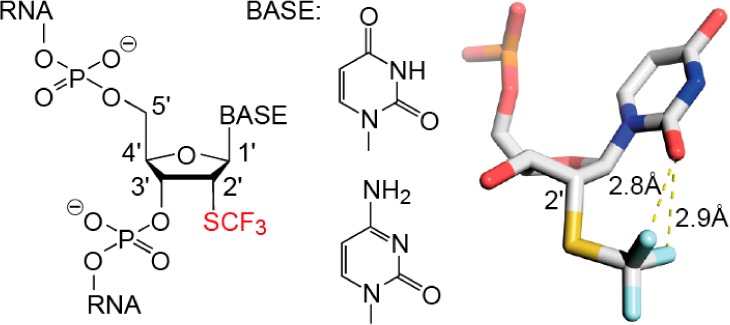
X-ray structure determinations showed that the 2′-SCF3 groups are well-defined in the electron density maps for both modified RNAs (Figure 6B,C). Superimpositions of both 2′-SCF3-modified RNA structures with the unmodified RNA revealed a root-mean-square deviation (rmsd) of 0.52 and 0.21 Å, thus showing that the 2′-SCF3 group does not significantly affect the overall RNA structure (Supporting Information, Figure S6A). Importantly, the 2′-SCF3 nucleosides were found in the same C3′-endo ribose conformations as observed in the structures of the unmodified RNA. Therefore, crystal packing must be made responsible to compensate for the energetic contributions that originate from the less favorable ribose pucker mode.
Detailed analysis of the RNA hydration pattern disclosed a displacement of several water molecules from the RNA minor groove in the vicinity of the 2′-SCF3 group (Supporting Information, Figure S6B). The hydrogen-bond acceptor capability of the 2′-SCF3 group, however, manifests in the participation to the well-defined hydration patterns (Figure 6 and Supporting Information, Figure S2B).
Encouraged by the X-ray structure solutions of 2′-SCF3 nucleosides in an RNA mismatch environment, we were wondering if crystallization of a 2′-SCF3 nucleoside would also be possible in a Watson–Crick base-paired region, despite the pronounced destabilizing effect that a 2′-SCF3 group exerts in solution. We therefore chose U2650 as an attractive position, not least because of our previous experience in structure solutions of modified SRL RNA with 2′-OCH3, 2′-SeCH3, and 2′-N3 at U2650.19,38 Although UV melting experiments of the 2′-SCF3 U2650-modified SRL RNA indicated destabilization compared to the unmodified counterpart, the Tm value was still significantly higher than the temperature used for crystallization trials (Supporting Information, Figure S5B). We indeed obtained well diffracting crystals of the Watson–Crick base pair forming 2′-SCF3 U2650 containing SRL RNA and were able to solve the structure at 1.2 Å resolution (Figure 6D). Comparable to the cases discussed above, crystal packing very likely compels the preferable C2′-endo conformation of single stranded 2′-SCF3 modified uridine into the observed C3′-endo U2650 conformation within the crystallized RNA double helix.
In all three structures, fluorine atoms of the 2′-SCF3 group closely approached the oxygen atom of the corresponding pyrimidine (O2). We do not think that the short distances observed (2.8–3.1 Å) are indicative of a halogen bond since fluorine (as opposed to chlorine, bromine, or iodine) usually retains a strongly electronegative electrostatic potential in biomolecules.39 More likely, fluorine atoms serve as hydrogen-bond acceptors in F···H–O-type interactions. Organic fluorine, however, is known to be a poor hydrogen acceptor,40,41 and in our specific case, most likely does not induce tautomeric forms of the pyrimidine nucleobase (2′-SC-F···H–O–C(2)=N(3)), though not completely excludable. We mention that 19F NMR spectroscopic experiments indicated a solvent-induced isotope shift for the 19F resonance in 5′-GU(2′-SCF3-U)CG (Supporting Information, Figure S7). However, we did not observe fractionated 19F resonances that would have to be expected for a H–O–C(2)=N(3) nucleobase tautomer with an exchangeable proton involved in a F···H–O-type interaction. Such fractionated 19F resonances were detected, for instance for 5-fluorocytidine in DNA and provided direct evidence for a pronounced F···H–N(C4) hydrogen bond.12
In this context, it is noteworthy that in former crystal structures of SRL RNA with 2′-OCH3 or 2′-SeCH3 at U2650, these modifications adopted the same orientation as observed here for the 2′-SCF3 group,38 and hence, the close vicinity of fluorine to the pyrimidine O2 might be a coincidence. Also, a recent X-ray structure of an A-form DNA duplex by Egli and co-workers, shows that the closely related 2′-SCH3 group does not differ in its orientation in the minor groove.42
Reflection and Concluding Remarks
In this study, we have explored and rationalized the structural basis of the 2′-SCF3 modification based on various chemical and biophysical methods, including NMR and high-resolution X-ray structure analysis of RNAs that carry the modification at distinct positions and in distinct base pair situations. While the 2′-SCF3 modification has only a minor impact on the thermodynamic stability of an RNA fold when it resides in a single-stranded region, it exerts a surprisingly high degree of destabilization if located in a Watson–Crick base paired helix. We have provided some experimental evidence that one reason for this behavior arises from the pronounced intrinsic preference for C2′-endo conformation of the 2′-SCF3 modified nucleoside. This argument is strengthened by a recent computational study that revealed that the C2′-endo conformation of a single nucleoside within a native A-form RNA duplex is significantly less stable (by 6 kcal mol−1) compared to the C3′-endo conformer.26 The large value becomes allegeable because the adoption of the C2′-endo pucker mode within an A-form RNA disrupts the planar base pair structure, therefore weakening stacking and hydrogen-bonding interactions.26
Many known ribose 2′ modifications, such as 2′-OCH3,17 2′-OCH2CH2OCH3,17 2′-OCF3,18 or 2′-F,17 increase double helix stability or leave it more or less unaltered (e.g., 2′-N3),19,20 while few (2′-CH3,21 2′-NH2,22 2′-SeCH3)23 are known to reduce stability (although to much less extent than the 2′-SCF3 modification does). At the nucleoside level, the stabilizing 2′-OH mimics firm up the C3′-endo sugar pucker, partly due to the strong gauche effect imparted by these modifications.17 The increased stabilities at the oligoribonucleotide level have therefore been reported to be due to conformational preorganization of the ribose for formation of A-form duplexes.17 However, a recent revisit of the 2′-fluoro modification with respect to the origins of the enhanced pairing affinity suggests that preorganization is not the only reason, but also there are enthalpy benefits from enhanced base-pairing and stacking interactions arising from the electronegative fluorine.43,44
In this context we note that the 2′-deoxy-2′-fluoro-β-d-arabino nucleic acid (2′-F-ANA) modification is an epimer of 2′-F-RNA, structurally identical to 2′-F-RNA in all respects with the single exception of the fluorine atom substitution at the 2′ position, which corresponds to the furanose form of arabinose.45,46 As a result, 2′-F-ANA is a structural mimic of DNA, preferentially adopting a C2′-endo sugar pucker.47,48 Nevertheless, 2′-F-ANA enhances binding to RNA complements. Certainly, a 2′-SCF3 nucleoside in C2′-endo conformation would cause significantly more steric interference within an A-form duplex. Additionally, it is likely that the 2′-SCF3 modification attenuates pairing strength and stacking interactions arising from the less electronegative sulfur, as reflected by the less favorable enthalpy contributions (Table 1).
Unfortunately, only little data on the impact of the closely related 2′-SCH3 modification on thermodynamics are available for a direct comparison.49 A short note, however, confirms that the 2′-SCH3 modification slightly destabilizes DNA/RNA and 2′-OCH3-RNA/RNA duplexes, by about 1.4 to 1.9 °C per insert.50 The influence of 2′-SCH3 is therefore much less compared to 2′-SCF3 and may indeed reflect a pronounced difference in electronegativity that can be expected for the methylated versus trifluoromethylated 2′-sulfur atoms.
We have recently highlighted the merits of ribonucleic acids with 2′-SCF3 groups to persue RNA folding processes, RNA-small molecule binding, and RNA-protein interactions, using 19F-NMR spectroscopy.16 The strong influence on Watson–Crick base pairing stability makes it advisible to use the label preferentially in single-stranded regions of the RNA under investigation. The advantage of the 2′-SCF3 label primarily lies in the three magnetically equivalent fluorine atoms that allow 19F NMR experiments to be performed at very low RNA concentrations; less material is needed and potential aggregation problems are minimized. The 2′-SCF3 group represents an isolated spin system, therefore proton decoupling (as advisible for 2′-F labeled RNA) is not required and consequently makes the label metrologically straightforward (for a direct comparison see Supporting Information, Figure S8). Accounting for an additional advantage in measurements of large RNA molecules or RNA–protein systems, 2′-SCF3 groups allow the prolongation of coherence lifetime based on transverse relaxation optimized spectroscopy (TROSY).
As final thought, nucleosides with strong destabilizing effects on Watson–Crick pairing have been developed for valuable applications in oligonucleotide therapeutics. Most prominent, is the highly flexible unlocked nucleic acid (UNA) (or “seconucleoside”) modification.51 UNA, missing the covalent C2′-C3′ bond of a ribose sugar, is not conformationally restrained, and can be used to influence oligonucleotide flexibility. UNA inserts reduce duplex Tm values by 5 to 10 °C per insert,51 they facilitate antisense strand selection as the RISC guide, and UNA modifications to the seed region of a siRNA guide strand can significantly reduce off target effects.52 A potential role for the 2′-SCF3 modification in antisense, siRNA, or aptamer applications, remains to be explored.
Materials and Methods
For the synthesis and characterization of 2′-SCF3 cytosine phosphor amidite C7 and its incorporation into RNA see the Supporting Information. NMR spectroscopic and ITC experiments are also described in the Supporting Material.
X-ray Crystallography
The 27-nucleotide SRL hairpin was crystallized as described.38 This sequence was chosen as a test case since crystallization conditions easily produce crystals that diffract well. Crystals were grown for 3 days at 20 °C for the unmodified SRL sequence, but several weeks were required for 2′-SCF3-U2656, 2′-SCF3-U2667, and 2′-SCF3-U2650 modified SRL. Crystals were cryoprotected for about 5 min in a reservoir solution containing 15% of glycerol and 3.5 M of ammonium sulfate and flash-frozen in liquid ethane for data collection. Crystals of 2′-SCF3-U2650 modified SRL grew as multicrystal clusters instead of single monocrystals. Very good data could however be collected using the highly focused beam of the X06SA beamline at the SLS synchrotron. Data were processed with the XDS Package.53 Structures were refined with PHENIX.54
Thermal Denaturation Studies
Absorbance versus temperature profiles were recorded at 250, 260, and 270 nm on a Cary-1 spectrometer equipped with a Peltier temperature control device. Each sequence was measured at five or six different concentrations ranging from ∼1 to 60 μM. RNAs were measured in buffer solutions of 10 mM Na2HPO4, pH 7.0, containing 150 mM NaCl. Data were collected after a complete cooling and heating cycle at a rate of 0.7 °C min–1. Melting transitions were reversible and essentially the same with respect to the three different wavelengths. For sample preparation, oligonucleotides were lyophylized to dryness, dissolved in the corresponding buffer from stock solutions and subsequently degassed. A layer of silicon oil was placed on the surface of the solution. ΔHvH and ΔSvH values for biomolecular melting transitions were obtained from plots of Tm–1 versus (ln c) plots where ΔHvH and ΔSvH are extracted from the slope and intercept of linear fits to the data. For monomolecular transitions, ΔHvH and ΔSvH were obtained from a two-state van’t Hoff analysis by fitting the shape of the individual α versus temperature curve.27,28
Acknowledgments
M.K. is an ESR fellow of the EU FP7Marie Curie ITN RNPnet program (289007). Karl Grubmayr is thanked for valuable discussions. Funding by the Austrian Science Foundation FWF (P21641, I1040 to R.M., I844 to C.K.) and the ‘Agence Nationale pour la Recherche’ (Grant ANR-12-BS07-0007-03 “ClickEnARN”) to E.E., is acknowledged. We thank V. Olieric for his support at the SLS synchrotron.
Supporting Information Available
Synthetic procedures and analysis data for the synthesis of phosphoramidite C7; table of 2′-SCF3 RNAs synthesized; 1H and 19F-NMR spectra of 2′-SCF3 RNAs; additional views and overlays of X-ray structures of 2′-SCF3 SRL RNA and hydration patterns. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Author Contributions
§ M.K. and L.J. contributed equally to this work.
Supplementary Material
References
- Liu L.; Byeon I. J.; Bahar I.; Gronenborn A. M. J. Am. Chem. Soc. 2012, 134, 4229–4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwani P.; Strandberg E.; Heidenreich N.; Bürck J.; Fanghänel S.; Ulrich A. S. J. Am. Chem. Soc. 2012, 134, 6512–6515. [DOI] [PubMed] [Google Scholar]
- Temme S.; Bönner F.; Schrader J.; Flögel U. WIRE Nanomed. Nanobiotechnol. 2012, 4, 329–343. [DOI] [PubMed] [Google Scholar]
- Kiviniemi A.; Virta P. J. Am. Chem. Soc. 2010, 132, 8560–8562. [DOI] [PubMed] [Google Scholar]
- Li C.; Wang G.-F.; Wang Y.; Creager-Allen R.; Lutz E. A.; Scronce H.; Slade K. M.; Ruf R. A. S.; Mehl R. A.; Pielak G. J. J. Am. Chem. Soc. 2010, 132, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moumné R.; Pasco M.; Prost E.; Lecourt T.; Micouin L.; Tisné C. J. Am. Chem. Soc. 2010, 132, 13111–13113. [DOI] [PubMed] [Google Scholar]
- Cobb S.; Murphy C. J. Fluorine Chem. 2009, 130, 132–143. [Google Scholar]
- Hennig M.; Scott L. G.; Sperling E.; Bermel W.; Williamson J. R. J. Am. Chem. Soc. 2007, 129, 14911–14921. [DOI] [PubMed] [Google Scholar]
- Olejniczak M.; Gdaniec Z.; Fischer A.; Grabarkiewicz T.; Bielecki L.; Adamiak R. W. Nucleic Acids Res. 2002, 30, 4241–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammann C.; Norman D. G.; Lilley D. M. J. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 5503–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W. C.; Horowitz J. Nucleic Acids Res. 1989, 17, 7241–7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffer B.; Kreutz C.; Rieder U.; Ebert M. O.; Konrat R.; Micura R. Nucleic Acids Res. 2009, 37, 7728–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luy B.; Merino J. P. J. Biomol. NMR 2001, 20, 39–47. [DOI] [PubMed] [Google Scholar]
- Reif B.; Wittmann V.; Schwalbe H.; Griesinger C.; Worner K.; Jahn-Hoffmann K.; Engels J.; Bermel W. Helv. Chim. Acta 1997, 80, 1952–1971. [Google Scholar]
- Kreutz C.; Kählig H.; Konrat R.; Micura R. Angew. Chem., Int. Ed. 2006, 45, 3450–3453. [DOI] [PubMed] [Google Scholar]
- Fauster K.; Kreutz C.; Micura R. Angew. Chem., Int. Ed. 2012, 51, 13080–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleavey G. F.; Damha M. J. Chem. Biol. 2012, 19, 937–954(and references cited therein). [DOI] [PubMed] [Google Scholar]
- Nishizono N.; Sumita Y.; Ueno Y.; Matsuda A. Nucleic Acids Res. 1998, 26, 5067–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauster K.; Hartl M.; Santner T.; Aigner M.; Kreutz C.; Bister K.; Ennifar E.; Micura R. ACS Chem. Biol. 2012, 7, 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigner M.; Hartl M.; Fauster K.; Steger J.; Bister K.; Micura R. ChemBioChem. 2010, 12, 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit C.; Bévierre M.-O.; De Mesmaeker A.; Altmann K.-H. Bioorg. Med. Chem. Lett. 1994, 4, 1969–1974. [Google Scholar]
- Aurup H.; Tuschl T.; Benseler F.; Ludwig J.; Eckstein F. Nucleic Acids Res. 1994, 22, 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzin Y.; Carrasco N.; Huang Z. Org. Lett. 2004, 6, 1099–1102. [DOI] [PubMed] [Google Scholar]
- Pitsch S.; Weiss P. A.; Jenny J.; Stutz A.; Wu X. Helv. Chim. Acta 2001, 84, 3773–3795. [Google Scholar]
- Wachowius F.; Höbartner C. ChemBioChem. 2010, 11, 469–480. [DOI] [PubMed] [Google Scholar]
- Li L.; Szostak J. W. J. Am. Chem. Soc. 2014, 136, 2858–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marky L. A.; Breslauer K. J. Biopolymers 1987, 26, 1601–1620. [DOI] [PubMed] [Google Scholar]
- Petersheim M.; Turner D. H. Biochemistry 1983, 22, 256–263. [DOI] [PubMed] [Google Scholar]
- Haziri A. I.; Leumann C. J. J. Org. Chem. 2012, 77, 5861–5869. [DOI] [PubMed] [Google Scholar]
- Altona C.; Sundaralingam M. J. J. Am. Chem. Soc. 1973, 95, 2333–2344. [DOI] [PubMed] [Google Scholar]
- Fürtig B.; Wenter P.; Reymond L.; Richter C.; Pitsch S.; Schwalbe H. J. Am. Chem. Soc. 2007, 129, 16222–16229. [DOI] [PubMed] [Google Scholar]
- Micura R.; Höbartner C. ChemBioChem. 2003, 4, 984–990. [DOI] [PubMed] [Google Scholar]
- Saenger W.Principles of Nucleic Acid Structure; Springer-Verlag: New York, 1984. [Google Scholar]
- Majlessi M.; Becker M. M. Nucleic Acids Res. 2008, 36, 2981–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim N. N.; Feig A. L. Methods 2009, 47, 198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky P. J.; Feig A. L. Biochemistry 2006, 46, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulecky P. J.; Feig A. L. Biopolymers 2006, 82, 38–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olieric V.; Rieder U.; Lang K.; Serganov A.; Schulze-Briese C.; Micura R.; Dumas P.; Ennifar E. RNA 2009, 15, 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffinger P.; Hays F. A.; Westhof E.; Ho P. S. Proc. Nat. Acad. Sci. U.S.A. 2004, 101, 16789–16794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunitz J. D. ChemBioChem. 2004, 5, 614–621. [DOI] [PubMed] [Google Scholar]
- Dunitz J. D.; Taylor R. Chem.—Eur. J. 1997, 3, 89–98. [Google Scholar]
- Pallan P. S.; Prakash T. P.; Li F.; Eoff R. L.; Manoharan M.; Egli M. Chem. Commun. 2009, 15, 2017–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallan P. S.; Greene E. M.; Jicman P. A.; Pandey R. K.; Manoharan M.; Rozners E.; Egli M. Nucleic Acids Res. 2011, 39, 3482–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra A.; Paolillo M.; Charisse K.; Manoharan M.; Rozners E.; Egli M. Angew. Chem., Int. Ed. 2012, 51, 11863–11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilds C. J.; Damha M. J. Nucleic Acids Res. 2000, 28, 3625–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damha M.; Wilds C.; Noronha A.; Brukner I.; Borkow G.; Arion D.; Parniak M. J. Am. Chem. Soc. 1998, 120, 12976–12977. [Google Scholar]
- Pintado N. M.; Deleavey G. F.; Portella G.; Campos-Olivas R.; Orozco M.; Damha M. J.; González C. Angew. Chem., Int. Ed. 2013, 52, 12065–12068. [DOI] [PubMed] [Google Scholar]
- Ikeda H.; Fernandez R.; Wilk A.; Barchi J. J. Jr; Huang X.; Marquez V. E. Nucleic Acids Res. 1998, 26, 2237–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu D.; Lind K. E.; Mohan V.; Manoharan M.; Ferguson D. M. Nucleic Acids Res. 1999, 27, 2189–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffey R. H.; Lesnik E.; Freier S.; Sanghvi Y. S.; Teng K.; Kawasaki A.; Guinosso C.; Wheeler P.; Mohan V.; Cook P. D., New Twists on Nucleic Acids, Structural Properties of Modified Nucleosides Incorporated into Oligonucleotides. In Carbohydrate Modifications in Antisense Research; Sanghvi Y. S., Cook P. D., Eds.; ACS Symposium Series, Vol. 580; American Chemical Society: Washington, DC, 1994; Chapter 14, pp 212–224. [Google Scholar]
- Campbell M. A.; Wengel J. Chem. Soc. Rev. 2011, 40, 5680–5689. [DOI] [PubMed] [Google Scholar]
- Vaish N.; Chen F.; Seth S.; Fosnaugh K.; Liu Y.; Adami R.; Brown T.; Chen Y.; Harvie P.; Johns R.; et al. Nucleic Acids Res. 2011, 39, 1823–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. J. Appl. Crystallogr. 1993, 26, 795–800. [Google Scholar]
- Adams P. D.; Grosse-Kunstleve R. W.; Hung L. W.; Ioerger T. R.; McCoy A. J.; Moriarty N. W.; Read R. J.; Sacchettini J. C.; Sauter N. K.; Terwilliger T. C. Acta Crystallogr., Sect. D 2002, 58, 1948–1954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.