Abstract
Background
Schistosoma japonicum is one of the major causative agents of schistosomiasis. The pairing of males and females leads to female sexual maturation and maintains this mature state. However, the mechanisms by which pairing facilitates sexual maturation are yet to be investigated.
Methods
Parasites isolated from single- and double-sex cercariae-infected mice were analyzed by Solexa to uncover pair-regulated miRNA profiles. To reveal the biological functions of differentially expressed miRNAs among the samples, we predicted the target genes of these differentially expressed miRNAs and compared the gene expression between 23-d-old female schistosomula from double-sex infections (23DSI) and 23-d-old female schistosomula from single-sex infections (23SSI) by analyzing digital gene expression profiling (DGE). KEGG pathway analysis was used to investigate the relevant biological processes of these target genes to understand the significance of differentially expressed miRNAs after pairing.
Results
The differentially expressed miRNA profiles of female 18- and 23-d post-single- and double-sex infections were analysed by Solexa. Similar miRNA profiles were observed in 18SSI and 18DSI, with the presence of identically expressed high-abundance miRNA, such as miRNA-1, miRNA-71b-5p and let-7. By contrast, in 23DSI and 23SSI, most of these high-abundance miRNAs were down-regulated. Furthermore, among all samples, bantam was distinctly up-regulated in 23 DSI, and miR-1, miR-71, miR-7-5p, and miR-7 were distinctly up-regulated in 23SSI. The transcriptomes of 23DSI and 23SSI revealed that the predicted target genes of miRNA-1, miRNA-71, miRNA-7, and miR-7-5p were associated with the ribonucleoprotein complex assembly and microtubule-based process. Conversely, the predicted target genes of bantam were related to the embryo development, development of primary sexual characteristics and regulation of transcription. KEGG pathway analysis revealed that in unpaired females, the highly-expressed miRNA-1, miRNA-71, miRNA-7, and miR-7-5p only inhibited the limited pathways, such as proteasome and ribosome assembly. Meanwhile, in paired mature females, highly-expressed bantam inhibited more biological pathways, such as the citrate cycle, glycolysis, fatty acid biosynthesis and RNA degradation.
Conclusions
The differentially expressed miRNAs between 23SSI and 23DSI and their different functions indicated that more genes or metabolic pathways in paired mature females were inhibited than those in unpaired ones. The results suggested that after pairing, specific miRNAs regulated gene expression to lead to female sexual maturation.
Keywords: Schistosoma japonicum, Pairing, Solexa, miRNA, bantam, miRNA-1, miRNA-71, miRNA-7, miR-7-5p
Background
Schistosomiasis is a chronic and debilitating parasitic disease caused by blood flukes of the genus Schistosoma. Over 200 million people are infected, and close to 800 million are at risk of this parasitic disease [1]. No successful vaccine is available for this disease. Praziquantel is the only drug used to treat schistosomiasis but is ineffective against young worms. Moreover, the drug resistance of the blood flukes to praziquantel has been reported [2]. Thus, identifying novel drug targets and developing alternative therapeutic strategies are urgent and necessary. In the hosts, males and females pair and eventually develop into adult worms, accompanied by remarkable morphological and molecular changes throughout their life cycle [3-7]. Previous studies have proposed that male contact is necessary for ovary and vitelline gland development. Moreover, this interaction is ultimately linked to the sexual maturation and maintenance of the mature state of females [8-10]. In evolutionary terms, schistosomes are likely to have evolved from hermaphroditic flukes to dioecious trematodes with an increasingly complete functional separation of the sexes. During the process, females still need the constant pairing contact with males to reach sexual maturation. To date, the exact mechanisms by which males influence female maturation are still unknown. Different factors have been suggested to be involved in schistosome sexual maturation, including physical or tactile contact [8,11], nutrition [12,13], and chemical stimuli [14,15]. However, the mechanisms by which pairing facilitates female development are yet to be investigated.
MicroRNAs (miRNAs) are a class of small non-coding RNAs (18–25 nucleotides long) generated from endogenous transcripts that form hairpins [16], functioning in transcriptional and post-transcriptional regulation of gene expression [17,18]. The binding of miRNA to a target mRNA typically results in gene silencing through translational repression and target mRNA degradation [19]. Recently, miRNAs have been reported to be involved in translational activation [20] and heterochromatin formation [21]. Through these actions, miRNAs regulate gene expression during development, differentiation, proliferation, death, and metabolism in many organisms [22]. Recently, some schistosome miRNAs have been identified [23-29], which provided some insight into the role of miRNAs in schistosome development.
During the development of S. japonicum, males and females begin to pair about 18 d post-infection, and the female begins to lay eggs about 24 d post-infection [30]. The role miRNAs play after pairing remains unknown. In this study, the expression profile of miRNAs of S. japonicum before and after pairing were investigated. Moreover, based on the analysis of their predicted targets, the different and specific functional requirements before and after pairing will be determined based on the novel profiles of miRNAs.
Methods
Ethics statement
This study was carried out in strict accordance with the recommendations of the Regulations for the Administration of Affairs Concerning Experimental Animals of the State Science and Technology Commission. The protocol was approved by the Internal Review Board of Tongji University School of Medicine.
Unisexual and paired infections
Oncomelania hupensis snails were obtained from the Jiangsu Institute of Schistosome Diseases, Jiangsu Province, China. To obtain single-sex female worms, the snails were exposed to a single miracidium, which was generated from eggs acquired from the liver of infected rabbits or mice. Approximately 100 to 150 freshly shed cercariae were used to percutaneously infect each mouse. Schistosomula were recovered by perfusion within 18 d and 23 d post-infections. The worms were washed in cold saline solution and checked by microscopy for possible undesirable mixed-sex infections. We separated single-sex female worms and froze them at −80°C until further processing of the samples.
To obtain double-sex female worms, about 100 to 150 multiple cercariae freshly shed by snails were used to percutaneously infect each mouse. The mice were sacrificed 18 d and 23 d post-infection, respectively. Females were recovered by washing with cold saline solution. The 23-d-old females were carefully separated from the paired worms under a microscope. All samples were frozen until further processing.
RNA extraction and small RNA library construction and sequencing
Total RNA was extracted using Trizol reagent (Invitrogen Life Technologies) according to the manufacturer’s instructions. RNA concentration and purity were evaluated spectrophotometrically at 260 nm and 280 nm, respectively, using a NanoDrop ND1000 spectrophotometer and an Agilent 2100 Bioanalyzer (Agient Technologics, Palo Alto, CA). RNA samples were stored at −80°C.
The construction of small RNA libraries was carried out as described in the Additional file 1. Briefly, RNAs with sizes ranging from 18–30 nucleotides (nt) were excised and purified and ligated to 3′ and 5′ adaptors, and further converted into 62–75 nt single-stranded cDNAs. The cDNAs were amplified using Illumina’s 3′ adaptor reverse primer and 5′ adaptor forward primers. The purified PCR products were sequenced by an Illumina Genome Analyzer at the BGI (Beijing Genomics Institute, Shenzhen, China).
Mapping sequence reads to the reference genome
All raw datasets produced by deep sequencing from the libraries (23DSI, 23SSI, 18DSI, and 18SSI) were analysed as follows. Clean reads were obtained after removal of the low quality reads, insert null reads, adaptor null reads, reads with ployA tail, and reads shorter than 18 nt. The small RNAs with sizes ranging from 18 nt to 30 nt were used for further analyses. All identical sequences were counted and merged as unique sequences, herein referred to as sequence tags. The unique reads along with their associated read counts were mapped to the S. japonicum genome sequences (http://www.chgc.sh.cn/japonicum/Resources.html) using the programme SOAP [31]. The small RNAs which matched with known rRNAs, tRNAs, small nuclear RNAs (snRNAs), and small nucleolar RNAs (snoRNAs) deposited in the Rfam database (ftp://selab.janelia.org/pub/Rfam/) and NCBI GenBank (http://www.ncbi.nlm.nih.gov/GenBank) were excluded. All unique sequences were utilised for the BLASTN search against the miRNA database (miRBase 18.0) (http://www.mirbase.org/) to identify conserved miRNAs (Additional file 1).
Bioinformatic analysis of S. japonicum small RNAs
Small RNA tags were aligned to the miRNA precursor/mature miRNA of corresponding species in miRBase18. Detailed information of the alignment, including the structure of known miRNA precursor, and length and count of tags from the sample, among others, were collected. To make every unique small RNA mapped to only one annotation, we followed the following priority rule: rRNA etc. (in which Genbank > Rfam) > known miRNA > repeat > exon > intron3. The total rRNA proportion should be less than 40%. The expression of the miRNAs in four samples were visualised by plotting the Log2-ratio figure and the scatter plot. The procedures adopted were as follows: (1) the expression of miRNA in two samples (control and treatment) were normalised to get the expression in transcript per million (TPM) with the normalisation formula (normalised expression = actual miRNA count/total count of clean reads × 1000000); and (2) the fold-change and P-value were calculated from the normalised expression, and log2ratio and scatter plots were generated using the fold-change formula (fold change = log2 (treatment/control)).
To understand the molecular function of differentially expressed miRNAs, we used the algorithms PicTar [32] and TargetScan [33] to predict their target mRNAs. All predicted target genes were evaluated by the scoring system and the criteria described by Chi et al. [34]. Sequences with total scores less than 3.0 points were considered as miRNA potential targets. Based on the digital gene expression profiling (DGE) analysis and negative correlation analysis between miRNA profiles and genes expression profiles, the false positive miRNA targets were further excluded.
Identification of differentially expressed genes and pathways in 23DSI and 23SSI
DGE was used to analyse the whole genome gene expression in 23DSI and 23SSI. Raw reads were filtered to obtain high quality data in the Tag-seq libraries of 23DSI and 23SSI. All of the clean tags were mapped to the S. japonicum genome (http://www.chgc.sh.cn/japonicum/Resources.html) (predicted coding genes). The clean tags mapped to the reference sequences from multiple genes were filtered, and the remaining clean tags were designed and annotated as unambiguous clean tags. The initial counts of the clean tags of each gene were normalised (transcripts per million) to obtain the normalised gene expression [35,36]. All differentially expressed genes were mapped to terms in the GO database, to identify significantly enriched GO terms in DEGs compared with the genome background. Pathway enrichment analysis was used to identify significantly enriched metabolic pathways or signal transduction pathways in DEGs compared with the whole genome background. Detailed pathway information was determined with the KEGG database. More details are shown in Additional file 2. The regulatory effect of differentially expressed miRNA in worms after pairing were predicted by analysing the functions of their predicted target genes and by comparing the proportion of their predicted target genes in differentially expressed genes.
MiRNA quantification by quantitative RT-PCR analysis
To ensure specificity of the PCR amplification of the cDNAs, we designed the primers based on the following criteria: predicted melting temperatures of 66 ± 2°C, limited self-complementarity, and primer length ranging from 20–22 nt. The first cDNA strand was synthesised from 0.15 μg total RNA using PrimeScript® RT reagent Kit (Perfect Real Time) (Takara Code: DRR037A). Stem-loop qRT-PCR was performed to quantify the sex-biased expressed miRNAs. Stem-loop RT primers were designed to reverse-transcribe the target miRNAs into cDNAs using total RNAs isolated from female worms (from the same samples used for Solexa sequencing). The 20 μl reaction RT system contained 2 μl of total RNA (0.15 μg), 2 μl (1 μM) of each individual stem-loop RT primer, 4 μl 5× PrimeScript® Buffer (for Real Time), 1 μl PrimeScript® RT Enzyme Mix I, and 11 μl RNase Free dH2O.
The following were designed as the RT primers of the miRNAs: Sja-mir-71 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTCACTAC3′; Sja-bantam 5′GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCAGCTT3′; and Sja-mir-1 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTCATACC3′. The primer 5′ TATGGAACGCTTCACGATTTTG3′ was designed for the RT of U6. The cDNAs were synthesised by incubation for 30 min at 16°C, 30 min at 42°C, and 15 min at 70°C. The products were amplified using SYBR® Premix Ex Taq™ (Tli RNaseH Plus) (Takara Code: RR420A) in an ABI Prism 7300 sequence detection system (Applied Biosystems) according to the manufacturer’s instruction. Our system contained 2 μl of cDNA from the RT reaction product (1:50 dilution), 5 μl of 2× SYBR Premix Ex Taq™ (TaKaRa), and 0.4 μl of 10 μM forward and reverse primers. The following were designed as the forward primers of the miRNAs: Sja-mir-71 5′ATGGTTCGTGGGTGAAAGACGATGGT3′; Sja-mir-1 5′ ATGGTTCGTGGGTGGAATGTAAAGAAGTATGG3′; and Sja-bantam 5′ ATGGTTCGTGGGTGAGATCGCGATTAAA3′. The primer 5′GCAGGGTCCGAGGTATTC3′ was used as the common reverse primer. The forward primer 5′CGGCGGTACATATACTAAAAT3′ and reverse primer 5′AACGCTTCACGATTTTGCGTA3′ were used to amplify the U6 gene as an endogenous control within each sample. Relative levels of gene expression were calculated using the 2-ΔΔCt method. Each sample was analysed for primer dimer, contamination, or mispriming by inspection of their dissociation curves.
Statistical analysis
Results were presented as mean ± standard deviation from at least three independent experiments. Statistical analyses were performed using one-way ANOVA and Student’s t-test. A value of P < 0.05 was considered statistically significant.
Results
Screening of miRNAs in S. japonicum from both single- and double-sex female worms at 18 d and 23 d post-infection
To understand the effect of pairing on the development of female S. japonicum, we constructed cDNA libraries derived from 18–30 nt long RNAs isolated from both single- and double-sex female worms at 18 and 23 d post-infection. We sequenced the RNAs using an Illumina (Solexa) DNA sequencer, yielding a total of 14 333 070, 14 425 899, 14 624 020, and 12 268 947 clean reads corresponding to 98.92%, 97.27%, 99.07%, and 97.82% of high-quality reads for 18-d-old female schistosomula from 18DSI, 23DSI, 18SSI, and 23SSI, respectively. The sequencing tags were merged, and the expression level of each unique tag was normalised to TPM as previously described [37-39]. Our results showed that the majority of miRNA were between 20 nt to 24 nt in long (Figure 1A).
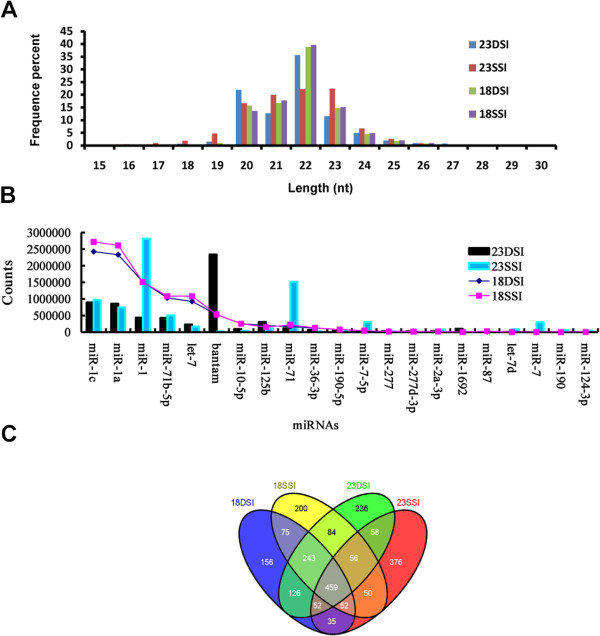
Figure 1.
Comparison of the miRNA profiles from single- and double-sex female worms at 18 d and 23 d post-infection. A. Length, distribution, and abundance of small RNA tags of 18DSI, 18SSI, 23DSI, and 23SSI. B. miRNA profiles of 23DSI and 23SSI, in comparison with those of 18DSI and 18SSI. Different characteristics are shown among four profiles. C. The quantity of exclusive and common miRNAs among different samples. The Venn diagram is created with an online Venn diagram maker. (http://bioinfogp.cnb.csic.es/tools/venny/index.html)
Differential profiles of miRNAs in female Schistosoma japonicum from both single- and double-sex female worms at 18 and 23 d post-infection
Similar miRNA profiles were observed between 18SSI and 18DSI (Figure 1B). They shared 829 commonly known miRNAs (Figure 1C). The miRNAs with high-abundance, such miRNA-1c, miRNA-1a, miRNA-1, miRNA-71b-5p, let-,7 and so on showed identical expression. By contrast, the levels of all of these high-abundance miRNAs were down-regulated in 23DSI and 23SSI compared with 18 DSI or 18SSI (Figure 1B). Only the amount of bantam in 23DSI was up-regulated far more than that in 18 DSI, 18SSI, or 23 SSI. Compared with 18SSI, 23DSI shared a little more known miRNA with 18DSI (Figure 1C). Similarly, 23SSI shared more known miRNA with 18SSI than with 18DSI. Similar characteristics were shared between the miRNA profiles of 23SSI and 23DSI. For example, their levels of miR-1c, miR-1a, miR-1, miRNA-71b-5p, and let-7 were far lower than those in 18 DSI or 18SSI. Moreover, they shared 625 commonly known miRNAs (Figure 1C). Almost half of the miRNAs of 23DSI and 23SSI were identical. However, the expression of a few miRNAs in the worms exhibited distinct differences. For example, the higher expression of bantam was observed only in 23DSI, whereas higher expression of miR-1, miR-71, miR-7-5p, and miR-7 manifested only in 23SSI (Figure 1B).
Confirmation of differentially expressed miRNAs by quantitative RT–PCR analysis
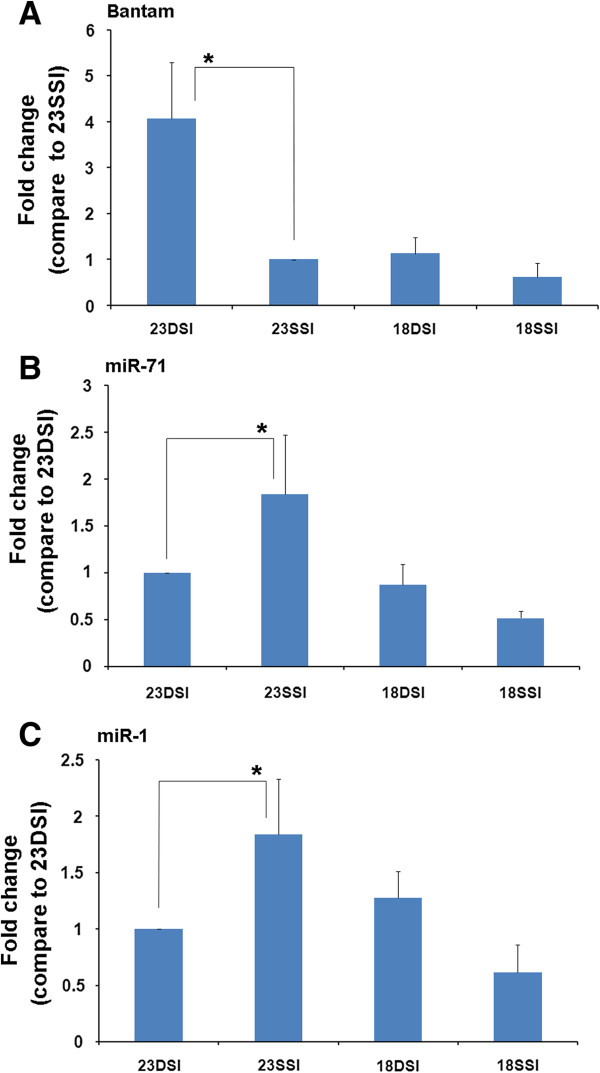
To confirm the differentially expressed miRNAs in 23DSI, 23SSI, 18DSI, and 18SSI, bantam, miRNA-1, and miR-71 were selected for quantitative RT–PCR analysis. The results of the RT-PCR showed that bantam was more abundant in 23DSI than in 23SSI, 18DSI, and 18SSI (Figure 2A). Similarly, higher amount of miR-71 (Figure 2B) and miR-1 (Figure 2C) were observed in 23SSI than in 23DSI. For the three miRNAs, quantitative RT–PCR analysis showed consistent expression with the Solexa analysis.
Figure 2.
Quantification of miRNAs dominantly expressed in 18DSI, 18SSi, 23DSI, and 23SSI. A. Bantam, with respect to 23SSI, was significantly up-regulated in 23DSI (P < 0.01). B. miR-71, with respect to 23DSI, was significantly up-regulated in 23SSI (P < 0.05). C. miR-1, with respect to 23DSI, was significantly up-regulated in 23SSI (P < 0.01).
Differential expression of the predicted target genes of bantam and miRNA-1-miRNA-71-miRNA-7-5p- miR-7 between samples from 23 DSI and 23SSI
To analyse the effect of the differential expression of miRNAs on female development after pairing, we sequenced the libraries of 23DSI and 23SSI, predicted the target genes of miRNA-1-miRNA-71-miRNA-7-miR-7-5p (Additional file 3: Table S1) and bantam (Additional file 4: Table S2), and analysed the differential expression of these genes in 23DSI compared with 23SSI. To reduce the false positives in the target gene prediction, we just analysed the predicted targets identified in the differentially expressed genes between 23DSI and 23SSI. Among the differentially expressed genes of 23DSI, we found that in the down-regulated genes of 23DSI, many predicted targets of bantam regulated protein metabolic process, mitotic spindle organisation, embryo development, glucose catabolic process, RNA metabolic process, apoptosis, hexose metabolic process, gene expression, mRNA transport, nematode larval development, multicellular organismal aging, development of primary sexual characteristics, translation, regulation of transcription, signalling, membrane lipid metabolic process, and the glucose metabolic process, among others (Table 1 and Additional file 5: Table S3). By contrast, in the up-regulated genes of 23DSI, the predicted target genes of miR-1-miR-71-miR-7-miR-7-5p appeared to regulate the ribonucleoprotein complex assembly, cellular protein complex assembly, microtubule-based process, response to oxidative stress, multicellular organismal aging, respiratory electron transport chain, pyrimidine ribonucleoside triphosphate biosynthetic process, positive regulation of epithelial cell differentiation, positive regulation of cell proliferation, apoptosis, energy coupled proton transport, electron transport chain, ATP synthesis-coupled proton transport, anatomical structure formation involved in morphogenesis, ribonucleoprotein complex biogenesis, mitotic cell cycle, larval development, microtubule polymerisation or depolymerisation, female gamete generation, regulation of transcription from RNA polymerase II promoter, and imaginal disc development, among others (Table 2 and Additional file 6: Table S4).
Table 1.
Selected predicted target genes of bantam in the down-regulated genes in 23DSI
| Gene | RawIntensity-23SSI | RawIntensity-23DSI | log2 Ratio(23DSI/23SSI) | P-Value | FDR | GO Component | GO Function | GO Process |
|---|---|---|---|---|---|---|---|---|
| Sjc_0013630|Sjp_0013630|SJC_S000059.548289 |
3987 |
208 |
−4.25 |
0 |
0 |
- |
- |
gi|257206034|emb|CAX82668.1|/1.09956e-27/Troponin T |
| Sjc_0007940|Sjp_0007940|SJC_S000025.694320 |
3316 |
400 |
−3.04 |
0 |
0 |
GO:0004175//endopeptidase activity |
GO:0019222//regulation of metabolic process;GO:0019538//protein metabolic process |
gi|30995341|gb|AAO59414.2|/0/cathepsin B endopeptidase |
| Sjc_0018120|Sjp_0018120|SJC_S000088.269899 |
3165 |
1412 |
−1.15 |
0 |
0 |
GO:0005488//binding;GO:0005198//structural molecule activity |
GO:0007052//mitotic spindle organization;GO:0010467//gene expression |
gi|226489394|emb|CAX75841.1|/1.27333e-22/Ribosomal protein L11 |
| Sjc_0083460|Sjp_0083460|SJC_S001708.6042 |
2501 |
328 |
−2.92 |
0 |
0 |
GO:0016740//transferase activity |
GO:0009987//cellular process;GO:0009790//embryo development |
gi|308507601|ref|XP_003115984.1|/0/hypothetical protein CRE_08793 |
| Sjc_0112990|Sjp_0112990|SJC_S007878.4178 |
1851 |
267 |
−2.78 |
0 |
0 |
GO:0004175//endopeptidase activity |
GO:0019538//protein metabolic process |
gi|56753605|gb|AAW25005.1|/3.77535e-176/SJCHGC02852 protein |
| Sjc_0031630|Sjp_0031630|SJC_S000203.86734 |
1450 |
109 |
−3.72 |
0 |
0 |
GO:0016832//aldehyde-lyase activity |
GO:0006007//glucose catabolic process |
gi|226475766|emb|CAX71973.1|/0/Aldolase |
| Sjc_0068090|Sjp_0068090|SJC_S000902.40949 |
1040 |
445 |
−1.21 |
0 |
0 |
GO:0016857//racemase and epimerase activity, acting on carbohydrates and derivatives;GO:0048037//cofactor binding |
GO:0016070//RNA metabolic process;GO:0006915//apoptosis;GO:0019318//hexose metabolic process;GO:0010467//gene expression;GO:0051028//mRNA transport |
gi|226487130|emb|CAX75430.1|/0/putative UDP-galactose-4-epimerase |
| Sjc_0038080|Sjp_0038080|SJC_S000276.119101 |
985 |
321 |
−1.6 |
0 |
0 |
GO:0016670//oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor |
GO:0008152//metabolic process |
gi|76156276|gb|AAX27495.2|/0/SJCHGC06250 protein |
| Sjc_0006800|Sjp_0006800|SJC_S000021.979701 |
935 |
434 |
−1.09 |
0 |
0 |
GO:0004175//endopeptidase activity |
GO:0019538//protein metabolic process |
gi|226476888|emb|CAX72314.1|/0/cathepsin D (lysosomal aspartyl protease) |
| Sjc_0103990|Sjp_0103990|SJC_S004191.40055 |
664 |
260 |
−1.34 |
0 |
0 |
- |
- |
gi|256090158|ref|XP_002581079.1|/0/eukaryotic translation initiation factor 2c |
| Sjc_0044670|Sjp_0044670|SJC_S000376.73872 |
583 |
146 |
−1.98 |
0 |
0 |
GO:0005515//protein binding;GO:0000166//nucleotide binding |
GO:0009790//embryo development;GO:0002119//nematode larval development;GO:0010259//multicellular organismal aging;GO:0045137//development of primary sexual characteristics |
gi|226469288|emb|CAX70123.1|/1.05716e-112/heat shock protein 90 kDa alpha |
| Sjc_0092310|Sjp_0092310|SJC_S002488.15565 |
574 |
11 |
−5.69 |
0 |
0 |
GO:0005488//binding |
GO:0006412//translation |
gi|238658344|emb|CAZ29402.1|/0/expressed protein |
| Sjc_0060730|Sjp_0060730|SJC_S000689.17829 |
490 |
26 |
−4.22 |
0 |
0 |
GO:0015175//neutral amino acid transmembrane transporter activity;GO:0015291//secondary active transmembrane transporter activity |
GO:0015804//neutral amino acid transport |
gi|226468260|emb|CAX69807.1|/0/solute carrier family 6 (neurotransmitter transporter, glycine) |
| Sjc_0087030|Sjp_0087030|SJC_S001927.503 |
447 |
75 |
−2.56 |
0 |
0 |
GO:0005488//binding;GO:0016301//kinase activity |
GO:0009987//cellular process |
gi|226487670|emb|CAX74705.1|/0/putative Elongation factor 2 kinase (eEF-2 kinase) |
| Sjc_0053950|Sjp_0053950|SJC_S000536.83029 |
331 |
19 |
−4.11 |
0 |
0 |
GO:0005488//binding;GO:0003712//transcription cofactor activity |
GO:0050896//response to stimulus;GO:0006355//regulation of transcription, DNA-dependent;GO:0023052//signaling |
gi|56753834|gb|AAW25114.1|/9.32058e-50/SJCHGC01061 protein |
| Sjc_0094720|Sjp_0094720|SJC_S002677.1428 |
304 |
12 |
−4.65 |
0 |
0 |
- |
GO:0006643//membrane lipid metabolic process |
gi|56758970|gb|AAW27625.1|/0/SJCHGC01869 protein |
| Sjc_0101270|Sjp_0101270|SJC_S003878.693 | 100 | 12 | −3.05 | 0 | 0 | GO:0016868//intramolecular transferase activity, phosphotransferases | GO:0006006//glucose metabolic process | gi|257206128|emb|CAX82715.1|/2.08691e-122/putative phosphoglucomutase 2 |
Table 2.
Selected predicted target genes of miR-1- miR-71- miR-7- miR-7-5p in the up-regulated genes in 23DSI
| Gene | RawIntensity-23SSI | RawIntensity-23DSI | log2 Ratio(23DSI/23SSI) | P-Value | FDR | GO Component | GO Function | GO Process |
|---|---|---|---|---|---|---|---|---|
| Sjc_0070710|Sjp_0070710|SJC_S000983.72365 |
6928 |
14676 |
1.1 |
0 |
0 |
GO:0005198//structural molecule activity |
GO:0006412//translation;GO:0022618//ribonucleoprotein complex assembly |
gi|226475894|emb|CAX72037.1|/1.92699e-63/40S ribosomal protein S17 |
| Sjc_0032370|Sjp_0032370|SJC_S000210.378287 |
1955 |
11833 |
2.61 |
0 |
0 |
GO:0016705//oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen;GO:0016209//antioxidant activity |
GO:0008152//metabolic process;GO:0019725//cellular homeostasis |
gi|226489432|emb|CAX75860.1|/2.80977e-109/Thioredoxin peroxidase |
| Sjc_0004280|Sjp_0004280|SJC_S000011.664038 |
4263 |
10833 |
1.36 |
0 |
0 |
GO:0017111//nucleoside-triphosphatase activity |
GO:0043623//cellular protein complex assembly;GO:0009207;GO:0007017//microtubule-based process |
gi|256087763|ref|XP_002580033.1|/0/alpha tubulin |
| Sjc_0066500|Sjp_0066500|SJC_S000837.212969 |
4057 |
10368 |
1.37 |
0 |
0 |
GO:0003676//nucleic acid binding;GO:0005198//structural molecule activity |
GO:0010467//gene expression |
gi|226475302|emb|CAX71939.1|/3.31945e-63/small subunit ribosomal protein S20e |
| Sjc_0054240|Sjp_0054240|SJC_S000544.55028 |
4128 |
8942 |
1.13 |
0 |
0 |
GO:0015078//hydrogen ion transmembrane transporter activity |
GO:0006979//response to oxidative stress;GO:0010259//multicellular organismal aging;GO:0022904//respiratory electron transport chain;GO:0017062//respiratory chain complex III assembly |
gi|29841122|gb|AAP06135.1|/2.29565e-72/SJCHGC00717 protein |
| Sjc_0009310|Sjp_0009310|SJC_S000031.859578 |
1125 |
6729 |
2.59 |
0 |
0 |
GO:0005198//structural molecule activity |
GO:0010467//gene expression |
gi|226478448|emb|CAX78482.1|/3.46982e-91/putative ribosomal protein L18a |
| Sjc_0058770|Sjp_0058770|SJC_S000642.39790 |
998 |
4195 |
2.08 |
0 |
0 |
GO:0032559;GO:0016776//phosphotransferase activity, phosphate group as acceptor |
GO:0009209//pyrimidine ribonucleoside triphosphate biosynthetic process;GO:0006796//phosphate metabolic process;GO:0030858//positive regulation of epithelial cell differentiation;GO:0008284//positive regulation of cell proliferation;GO:0006915//apoptosis |
gi|189502914|gb|ACE06838.1|/5.59719e-79/unknown |
| Sjc_0046710|Sjp_0046710|SJC_S000412.116878 |
1504 |
4011 |
1.43 |
0 |
0 |
- |
- |
gi|226472284|emb|CAX77178.1|/8.40244e-140/ribosomal protein L7a |
| Sjc_0075950|Sjp_0075950|SJC_S001243.77896 |
603 |
3843 |
2.68 |
0 |
0 |
GO:0003723//RNA binding;GO:0005198//structural molecule activity |
GO:0010467//gene expression |
gi|29841451|gb|AAP06483.1|/3.70409e-88/similar to NM_057813 ribosomal protein L9 in Ictalurus punctatus |
| Sjc_0091980|Sjp_0091980|SJC_S002467.11983 |
921 |
3113 |
1.77 |
0 |
0 |
GO:0017111//nucleoside-triphosphatase activity;GO:0032561 |
GO:0043623//cellular protein complex assembly;GO:0009207;GO:0007017//microtubule-based process |
gi|226487270|emb|CAX75500.1|/0/Tubulin alpha-1 chain |
| Sjc_0059730|Sjp_0059730|SJC_S000664.27965 |
704 |
2801 |
2.01 |
0 |
0 |
GO:0019829//cation-transporting ATPase activity;GO:0015078//hydrogen ion transmembrane transporter activity;GO:0046872//metal ion binding;GO:0032559;GO:0043498//cell surface binding;GO:0015662//ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism;GO:0004888//transmembrane receptor activity |
GO:0015988//energy coupled proton transport, against electrochemical gradient;GO:0006929//substrate-dependent cell migration;GO:0022900//electron transport chain;GO:0015986//ATP synthesis coupled proton transport;GO:0006897//endocytosis;GO:0030641//regulation of cellular pH;GO:0048646//anatomical structure formation involved in morphogenesis;GO:0006839//mitochondrial transport |
gi|226487050|emb|CAX75390.1|/0/ATP synthase, H + transporting, mitochondrial F1 complex, beta polypeptide |
| Sjc_0116280|Sjp_0116280|SJC_S008728.7876 |
857 |
2290 |
1.43 |
0 |
0 |
GO:0005198//structural molecule activity |
GO:0022613//ribonucleoprotein complex biogenesis;GO:0006412//translation |
gi|226469644|emb|CAX76652.1|/4.83022e-166/deoxyribonuclease |
| Sjc_0082530|Sjp_0082530|SJC_S001652.6464 |
659 |
1887 |
1.53 |
0 |
0 |
GO:0015662//ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism;GO:0005515//protein binding;GO:0032559 |
GO:0000278//mitotic cell cycle;GO:0044267//cellular protein metabolic process;GO:0006909//phagocytosis |
gi|56754732|gb|AAW25551.1|/0/SJCHGC06338 protein |
| Sjc_0071910|Sjp_0071910|SJC_S001024.53841 |
605 |
1823 |
1.6 |
0 |
0 |
GO:0017111//nucleoside-triphosphatase activity;GO:0032561 |
GO:0000278//mitotic cell cycle;GO:0035556//intracellular signal transduction;GO:0002164//larval development;GO:0031109//microtubule polymerization or depolymerization;GO:0007292//female gamete generation;GO:0051169//nuclear transport;GO:0015031//protein transport;GO:0009790//embryo development |
gi|256085737|ref|XP_002579070.1|/2.37251e-121/ran |
| Sjc_0051340|Sjp_0051340|SJC_S000485.44512 | 235 | 1710 | 2.88 | 0 | 0 | GO:0003677//DNA binding;GO:0005515//protein binding | GO:0016568//chromatin modification;GO:0002759;GO:0006357//regulation of transcription from RNA polymerase II promoter;GO:0007444//imaginal disc development | gi|29841393|gb|AAP06425.1|/3.25519e-81/SJCHGC00614 protein |
KEGG pathway analysis
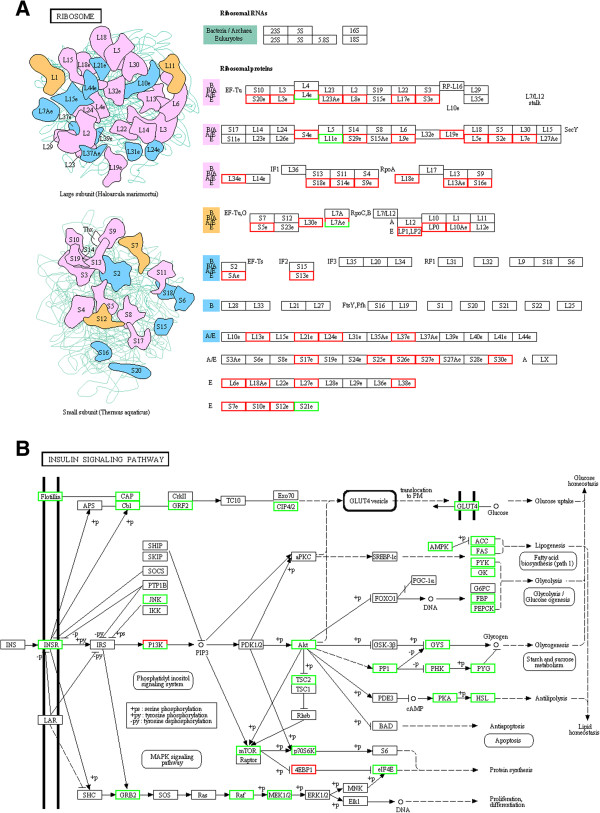
KEGG pathway analysis was used to investigate the characteristics of the biochemical and metabolic processes involved with the differentially expressed genes. We found that these predicted target genes were involved in various metabolic processes, such as proteasome, porphyrin metabolism, ribosome, DNA replication, oxidative phosphorylation, pyrimidine metabolism, phagosome, folate biosynthesis, purine metabolism protein processing in the endoplasmic reticulum, aminoacyl-tRNA biosynthesis, protein digestion and absorption, basal transcription factors, glycerophospholipid metabolism, oocyte meiosis, neurotrophin signalling pathway, ubiquitin mediated proteolysis, RNA transport, peroxisome, basal transcription factors, apoptosis, glycerophospholipid metabolism, glycolysis/gluconeogenesis, citrate cycle, pentose phosphate pathway, fatty acid biosynthesis, and the insulin signalling pathway (Table 3). The predicted target genes of bantam hardly participated in the proteasome, porphyrin metabolism, ribosome, whereas more predicted target genes of miR-1-miR-71-miR-7-miR-7-5p were involved in these process. For instance, in ribosome assembly, 15 of 49 detected genes in this metabolic process were predicted as the target genes of miR-1-miR-71-miR-7-miR-7-5p, whereas only 1 of 49 genes was the predicted target gene of bantam (Figure 3A). However, none of the predicted target genes of miR-1-miR-71-miR-7-miR-7-5p are involved the citrate cycle, gastric acid secretion, glycolysis/gluconeogenesis, protein digestion and absorption, aminoacyl-tRNA biosynthesis, fatty acid biosynthesis, and the pentose phosphate pathway. Moreover, few of the predicted target genes of miR-1-miR-71-miR-7-miR-7-5p participated in the peroxisome, RNA degradation, mRNA surveillance pathway, axon guidance, basal transcription factors, apoptosis, glycerophospholipid metabolism, insulin signalling pathway, lysosome, regulation of actin cytoskeleton, and endocytosis. By contrast, more predicted target genes of bantam were involved in these processes. For example, among the 50 genes of the insulin signalling pathway, most of which were down-regulated in 23DSI. Out of the 50 genes, 33 were the predicted target genes of bantam (Figure 3B), whereas only 2 were predicted target genes of miR-1-miR-71-miR-7-miR-7-5p. In addition, in some processes such as DNA replication and oxidative phosphorylation, both participated in these processes on account of their similar amount of predicted target genes.
Table 3.
Comparison of predicted target genes participating in regulating metabolic processes between miR-1- miR-71-miR-7-miR-7-5p and bantam
| Metabolic processes in KEGG | Detected genes participating in the process | Rate of predicted target genes of miR-1,miR-71,miR-7 and miR-7-5p | Rate of predicted target genes of bantam |
|---|---|---|---|
| Proteasome |
15 |
8(53%) |
2 (13%) |
| Porphyrin metabolism |
10 |
4 (40%) |
3 (30%) |
| Ribosome |
49 |
15 (31%) |
1 (2%) |
| DNA replication |
15 |
4 (27%) |
3 (20%) |
| Oxidative phosphorylation |
39 |
10 (26%) |
11 (28%) |
| Pyrimidine metabolism |
24 |
6 (25%) |
11 (46%) |
| Phagosome |
49 |
9 (18%) |
26 (53%) |
| Folate biosynthesis |
6 |
1 (17%) |
3 (50%) |
| Purine metabolism |
38 |
6 (16%) |
20 (53%) |
| Protein processing in endoplasmic reticulum |
68 |
9 (13%) |
32 (47%) |
| Oocyte meiosis |
32 |
4 (12%) |
17 (53%) |
| Neurotrophin signaling pathway |
25 |
3 (12%) |
14 (56%) |
| Ubiquitin mediated proteolysis |
54 |
6 (11%) |
27 (50%) |
| RNA transport |
62 |
6 (10%) |
33 (53%) |
| Peroxisome |
11 |
1 (9%) |
4 (36%) |
| RNA degradation |
35 |
3 (8%) |
22 (63%) |
| mRNA surveillance pathway |
35 |
3 (8%) |
19 (54%) |
| Axon guidance |
36 |
3 (8%) |
25 (69%) |
| Basal transcription factors |
15 |
1 (7%) |
11 (73%) |
| Apoptosis |
15 |
1 (7%) |
9 (60%) |
| Glycerophospholipid metabolism |
22 |
1 (5%) |
12 (55%) |
| Insulin signaling pathway |
50 |
2 (4%) |
33 (66%) |
| Lysosome |
61 |
2 (3%) |
37 (61%) |
| Regulation of actin cytoskeleton |
98 |
3 (3%) |
57 (58%) |
| Endocytosis |
68 |
2 (3%) |
39 (57%) |
| Citrate cycle (TCA cycle) |
13 |
0 (0%) |
8 (62%) |
| Gastric acid secretion |
19 |
0 (0%) |
11 (58%) |
| Glycolysis/Gluconeogenesis |
23 |
0 (0%) |
11 (48%) |
| Protein digestion and absorption |
21 |
0 (0%) |
13 (62%) |
| Aminoacyl-tRNA biosynthesis |
17 |
0 (0%) |
10 (59%) |
| Fatty acid biosynthesis |
3 |
0 (0%) |
2 (67%) |
| pentose phosphate pathway | 9 | 0 (0%) | 6 (67%) |
Figure 3.
KEGG pathway analysis. A. Ribosome assembly. B. Insulin signalling pathway. Compared with the same genes in 23SSI, the up- and down-regulated genes are indicated with red and green colour in 23DSI, respectively.
Discussion
Pairing of Schistosoma japonicum initiates female development, leading to female sexual maturation, and the maintenance of this mature state. miRNAs are post-transcriptional regulators of growth and development in both plants and animals [18]. A comparison of the miRNA profiles of worms before and after pairing can provide insight into the mechanism by which pairing promotes female sexual maturation. During the development of S. japonicum, males and females just begin to pair about 18 d post-infection. At this stage, only some begin to pair. In this article, 18 d post-infection was also considered as the stage when pairing begins. Paired-females from double-sex infections begin to lay eggs about 24 d post-infection [30]. To rule out the influences of the egg, the 23-d-old female was considered herein as the earliest stage of sexual maturation after pairing. We compared the miRNA profiles among 18DSI, 18SSI, 23DSI, and 23SSI. We distinguished the effects of the pairing instead of the development from 18 d to 23 d post-infection. We found the level of high-abundance miRNAs such as miR-1, miR-1a, miR-1c, miR-71b-5p, and let-7 to be higher in 18DSI and 18SSI than in 23DSI and 23SSI. A similar miRNA profile between 18DSI and 18SSI was reasonable because both of them stayed nearly at the same stage at 18 d post-infection. Although some paired, they likely paired for a short while, indicating that males did not play a distinct role in paired-females.
At 5 d post-pairing, 23DSI was significantly longer and thicker than 23SSI. Moreover, 23 DSI had a dark brown colour because of the accumulation of larger amounts of schistosome hemozoin in their guts. By contrast, 23SSI was smaller and hardly accumulated schistosoma hemozoin in their guts [5]. Interestingly, these distinct morphological changes appeared to bring about limited changes in the miRNA profiles. However, these limited differentially depressed miRNAs were possibly associated with some significant developmental events regarding sexual maturation. We found that the expression profile of miRNA in 23DSI is a little similar to that of 23SSI. In particular, nearly all high-abundance miRNAs, such as miR-1c, miR-1a, miR-10-5p, miR-71b-5p, and let-7, were down-regulated in both, compared with 18DSI or 18SSI. Only several high-abundance miRNAs differentially expressed between 23DSI and 23 SSI, such as bantam, miR-1, miR-71, miR-7, and miR-7-5p. These results suggested that high-abundance miRNAs such as miR-1c, miR-1a, miR-10-5p, miR-71b-5p, and let-7 were closely related to the development of 18 d-old females before pairing, whereas during the development from 18 d to 23 d, all of these high-abundance miRNAs were down-regulated not only in 23 DSI, but also in 23SSI. This trend indicated that the up- or down-regulation of all of these high-abundance miRNAs were not related to the pairing but only to female development. However, the up-regulation of bantam in 23DSI was associated not only with pairing but also likely played an essential role in female sexual maturation.
To investigate the role of these differentially expressed miRNAs in 23 DSI and 23SSI, we analysed their predicted target genes and relevant functions. Moreover, KEGG pathway analysis was used to investigate the various metabolic processes involved in these predicted target genes. We found that the target genes of bantam were involved in widely different metabolic processes, such as in peroxisome, RNA degradation, mRNA surveillance pathway, axon guidance, basal transcription factors, apoptosis, glycerophospholipid metabolism, insulin signalling pathway, lysosome, regulation of actin cytoskeleton, citrate cycle, gastric acid secretion, glycolysis/gluconeogenesis, protein digestion and absorption, aminoacyl-tRNA biosynthesis, and fatty acid biosynthesis. In 23DSI, the up-regulation of bantam likely inhibited a large number of genes or pathways involved within a wide range of biological functions, including glycometabolism, lipid metabolism, nucleic acid metabolism, protein digestion and utilisation, and other biological processes. This result was consistent with findings on gene expression analysis [5].
In unpaired females (23SSI), bantam was notably not up-regulated, whereas miR-1, miR-71, miR-7, and miR-7-5p were significantly up-regulated. By contrast, in paired females (23DSI), the above mentioned miRNAs were not up-regulated, suggesting that the functions of the target genes of miR-1-miR-71-miR-7-miR-7-5p were required in paired females. We found that the target genes of miR-1-miR-71-miR-7-miR-7-5p, such as ribosomal protein genes (CAX72037.1, CAX71939.1, CAX78482.1, CAX77178.1, AAP06483.1, CAX77387.1, CAX72859.1, CAX70956.1, CAX71543.1, CAX83047.1, CAX70121.1) (Additional file 6: Table S4), thioredoxin peroxidase (CAX75860.1), tubulin (XP_002580033.1, CAX75788.1, CAX75500.1, CAX71989.1, CAX76110.1), ATP synthase- H + transporting (CAX75390.1, CAX76063.1), and cytochrome c oxidase (CAX74747.1, CAX76589.1), among others, were significantly up-regulated. In particular, various ribosomal protein genes were regulated by miR-1-miR-71-miR-7-miR-7-5p. These results suggested that miR-1, miR-71, miR-7, and miR-7-5p played an essential role in regulating ribosomal assembly. Hence, enhancing the ribosomal assembly leads to the enhancement of protein expression, suggesting that, although more genes and biological processes were inhibited in paired females, enhanced ribosomal assembly likely maintains the supply of a large amount of special proteins for sexual maturation and egg production. In 23 DSI, the high level of bantam and low levels of miR-1, miR-71, miR-7, and miR-7-5p possibly regulated and organised a specific gene expression profile for sexual maturation and egg production by inhibiting and strengthening specific gene expression and metabolic processes. Our previous research has shown that genes coding for proteins such as phosphoglycerate mutase, superoxide dismutase, egg antigen, ribosomal protein, ferritin-1 heavy chain, and eukaryotic translation initiation factor 2 were significantly up-regulated in 23DSI. These genes function in glycolysis, antioxidant defense, protein biosynthesis, egg formation, iron transport and utilisation, and translational regulation (data not shown). The above mentioned proteins appeared to be more necessary for egg production. Other previous studies have compared differential gene expression between females and males, and similar results have also been reported. For example, egg antigen [5,40,41], ferritin-1 [5,40,42,43], ribosomal proteins [5,40,44], ATPase [44,45], cathepsin [45], extracellular superoxide dismutase [5,43,44], cytochrome C oxidase [5], tyrosinase [43], mucin-like protein [46], fs800 [47], and adenylosuccinate lyase [5], among others, are often detected in females. These up-regulated genes in paired females are considered to play roles in female sexual maturation and egg production. Only some of them belong to the predicted target genes of differentially expressed miRNAs, suggesting that miRNAs of 23DSI do not regulate all genes relate to sexual maturation and egg production.
Previous studies have reported sexual differences in the gene expression of schistosomes [5,6,40,43,48-50]. However, how pairing induces changes in the gene expression of females is unclear. Although miRNAs do not regulate all genes in organisms, evidence provided by miRNA analyses in the present study indicated that pairing likely limited the expression of non-essential genes through increasing the expression of bantam and specific genes by maintaining miR-1, miR-71, miR-7, and miR-7-5p at relatively low levels. Interestingly, miR-71 was located on the female W chromosome, suggesting mir-71 be involved in female sex-specific functions [51]. Thus, the change of the abundance of miR-71 in worms maybe plays a key role in female development. In addition, mass MALDI-TOF-TOF MS analyses have also shown that fatty acid-binding protein and phosphoglycerate mutase can be detected in paired 19-d-old females, and ribosomal protein LP1 [52] and ribosomal protein L30 [53] can be detected in paired 42-d-old females. These results further revealed that these up-regulated genes in 23 DSI detected by DGE and predicted as target genes of differentially expressed miRNAs can be translated to proteins, such as ribosomal proteins. Thus, the regulation of ribosome assembly by miRNA likely played a significant role in sexual maturation and egg production in paired females.
After pairing, the up-regulation of bantam was potentially capable of inhibiting a wide range of genes or biological processes. Furthermore, the low abundance of miR-1, miR-71, miR-7, and miR-7-5p in 23DSI compared with 23SSI was likely capable of promoting specific gene expression. The function of their target genes and relevant biological processes were consistent with evidence observed from gene expression experiments. These results suggested that pairing facilitated female sexual maturation and egg production by regulating miRNA profiles, and thus, gene expression.
Conclusions
The differentially expressed miRNAs between 23SSI and 23DSI and their different functions indicated that more genes or metabolic pathways in paired mature females were inhibited than those in unpaired immature females. The results suggested that specific miRNAs regulated gene expression to lead to female sexual maturation after pairing.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Collected material, bioinformatic analysis and quantitative RT-PCR analysis: JS, SW, CL, YR, JW. Analyzed the data and performed statistical evaluation: JS, SW. Wrote the paper: JS. All authors read and approved the final version of the manuscript.
Supplementary Material
Analysis of differentially expressed miRNAs.
Analysis of differentially expressed genes.
Predicted target genes of miR-1-miR-71-miR-7-miR-7-5p in Schistosoma japonicum.
Predicted target genes of bantam in Schistosoma japonicum.
Predicted target genes of bantam in down-regulated genes in 23DSI.
Predicted target genes of miR-1-miR-71-miR-7-miR-7-5p in up-regulated genes in 23DSI.
Contributor Information
Jun Sun, Email: swksj@tongji.edu.cn.
Suwen Wang, Email: wswtjzlx2012@hotmail.com.
Chen Li, Email: 133449096@qq.com.
Yijiu Ren, Email: 804745919@qq.com.
Jinqiang Wang, Email: 867031496@qq.com.
Acknowledgements
We would like to thank Pan W.Q. for his valuable advice. We are grateful for discussions and comments from Xiao S.H., Zhang Q.F. and Xu X.D. This research was supported by the National Natural Science Foundation of China (No.81071383).
References
- Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis. 2006;6:411–425. doi: 10.1016/S1473-3099(06)70521-7. [DOI] [PubMed] [Google Scholar]
- Cioli D. Praziquantel: is there real resistance and are there alternatives? Curr Opin Infect Dis. 2000;13:659–663. doi: 10.1097/00001432-200012000-00014. [DOI] [PubMed] [Google Scholar]
- Sun J, Hu W, Li C. Beyond heme detoxification: a role for hemozoin in iron transport in S. japonicum. Parasitol Res. 2013;112:2983–2990. doi: 10.1007/s00436-013-3470-8. [DOI] [PubMed] [Google Scholar]
- Williams DL, Sayed AA, Bernier J, Birkeland SR, Cipriano MJ, Papa AR, McArthur AG, Taft A, Vermeire JJ, Yoshino TP. Profiling Schistosoma mansoni development using serial analysis of gene expression (SAGE) Exp Parasitol. 2007;117:246–258. doi: 10.1016/j.exppara.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick JM, Hoffmann KF. Dioecious Schistosoma mansoni express divergent gene repertoires regulated by pairing. Int J Parasitol. 2006;36:1081–1089. doi: 10.1016/j.ijpara.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Vermeire JJ, Taft AS, Hoffmann KF, Fitzpatrick JM, Yoshino TP. Schistosoma mansoni: DNA microarray gene expression profiling during the miracidium-to-mother sporocyst transformation. Mol Biochem Parasitol. 2006;147:39–47. doi: 10.1016/j.molbiopara.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Walker AJ. Insights into the functional biology of schistosomes. Parasit Vectors. 2011;4:203. doi: 10.1186/1756-3305-4-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popiel I, Basch PF. Reproductive development of female Schistosoma mansoni (Digenea: Schistosomatidae) following bisexual pairing of worms and worm segments. J Exp Zool. 1984;232:141–150. doi: 10.1002/jez.1402320117. [DOI] [PubMed] [Google Scholar]
- Clough ER. Morphology and reproductive organs and oogenesis in bisexual and unisexual transplants of mature Schistosoma mansoni females. J Parasitol. 1981;67:535–539. doi: 10.2307/3280487. [DOI] [PubMed] [Google Scholar]
- Shaw JR. Schistosoma mansoni: pairing in vitro and development of females from single sex infections. Exp Parasitol. 1977;41:54–65. doi: 10.1016/0014-4894(77)90129-1. [DOI] [PubMed] [Google Scholar]
- Armstrong JC. Mating Behavior and Development of Schistosomes in the Mouse. J Parasitol. 1965;51:605–616. doi: 10.2307/3276242. [DOI] [PubMed] [Google Scholar]
- Basch PF. Why do schistosomes have separate sexes? Parasitol Today. 1990;6:160–163. doi: 10.1016/0169-4758(90)90339-6. [DOI] [PubMed] [Google Scholar]
- Gupta BC, Basch PF. The role of Schistosoma mansoni males in feeding and development of female worms. J Parasitol. 1987;73:481–486. [PubMed] [Google Scholar]
- Shaw JR, Marshall I, Erasmus DA. Schistosoma mansoni: in vitro stimulation of vitelline cell development by extracts of male worms. Exp Parasitol. 1977;42:14–20. doi: 10.1016/0014-4894(77)90056-X. [DOI] [PubMed] [Google Scholar]
- Haseeb MA. Schistosoma mansoni: females enhance [14C]-tyrosine incorporation in males maintained in vitro. J Helminthol. 1998;72:123–126. doi: 10.1017/S0022149X00016291. [DOI] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Muñoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Xue X, Sun J, Zhang Q, Wang Z, Huang Y, Pan W. Identification and characterization of novel microRNAs from Schistosoma japonicum. PLoS One. 2008;3:e4034. doi: 10.1371/journal.pone.0004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Hao P, Chen H, Hu W, Yan Q, Liu F, Han ZG. Genome-wide identification of Schistosoma japonicum microRNAs using a deep-sequencing approach. PLoS One. 2009;4:e8206. doi: 10.1371/journal.pone.0008206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xue X, Sun J, Luo R, Xu X, Jiang Y, Zhang Q, Pan W. An “in-depth” description of the small non-coding RNA population of Schistosoma japonicum schistosomulum. PLoS Negl Trop Dis. 2010;4:e596. doi: 10.1371/journal.pntd.0000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Cai P, Jiang N, Wang H, Chen Q. Identification and characterization of microRNAs and endogenous siRNAs in Schistosoma japonicum. BMC Genomics. 2010;11:55. doi: 10.1186/1471-2164-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Jin Y. MicroRNAs: potentially important regulators for schistosome development and therapeutic targets against schistosomiasis. Parasitology. 2012;139:669–679. doi: 10.1017/S0031182011001855. [DOI] [PubMed] [Google Scholar]
- Simões MC, Lee J, Djikeng A, Cerqueira GC, Zerlotini A, Da Silva-Pereira RA, Dalby AR, LoVerde P, El-Sayed NM, Oliveira G. Identification of Schistosoma mansoni microRNAs. BMC Genomics. 2011;12:47. doi: 10.1186/1471-2164-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco A, Kozomara A, Hui JH, Emery AM, Rollinson D, Griffiths-Jones S, Ronshaugen M. Sex-biased expression of microRNAs in Schistosoma mansoni. PLoS Negl Trop Dis. 2013;7:e2402. doi: 10.1371/journal.pntd.0002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YX, Yang HZ. Physiological studies on the post-cercarial development of Schistosoma japonicum. ACTA Zoologica Sinica. 1980;26:32–41. [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Kennell JA, MacDougald OA. Wnt signaling inhibits adipogenesis through beta-catenin-dependent and -independent mechanisms. J Biol Chem. 2005;280:24004–24010. doi: 10.1074/jbc.M501080200. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Chi X, Yang Q, Chen X, Wang J, Pan L, Chen M, Yang Z, He Y, Liang X, Yu S. Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing. PLoS One. 2011;6:e27530. doi: 10.1371/journal.pone.0027530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoen PA T, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, De Menezes RX, Boer JM, Van Ommen GJ, Den Dunnen JT. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy AS, Morin RD, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M, Marra MA. Next-generation tag sequencing for cancer gene expression profiling. Genome Res. 2009;19:1825–1835. doi: 10.1101/gr.094482.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Tej SS, Vu TH, Haudenschild CD, Agrawal V, Edberg SB, Ghazal H, Decola S. The use of MPSS for whole-genome transcriptional analysis in Arabidopsis. Genome Res. 2004;14:1641–1653. doi: 10.1101/gr.2275604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, Roth R, George D, Eletr S, Albrecht G, Vermaas E, Williams SR, Moon K, Burcham T, Pallas M, DuBridge RB, Kirchner J, Fearon K, Mao J, Corcoran K. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Huang J, Hao P, Zhang YL, Deng FX, Deng Q, Hong Y, Wang XW, Wang Y, Li TT, Zhang XG, Li YX, Yang PY, Wang HY, Han ZG. Discovering multiple transcripts of human hepatocytes using massively parallel signature sequencing (MPSS) BMC Genomics. 2007;8:207. doi: 10.1186/1471-2164-8-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann KF, Johnston DA, Dunne DW. Identification of Schistosoma mansoni gender-associated gene transcripts by cDNA microarray profiling. Genome Biol. 2002;3:RESEARCH0041. doi: 10.1186/gb-2002-3-8-research0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Rekosh DM, LoVerde PT. Schistosoma mansoni p48 eggshell protein gene: characterization, developmentally regulated expression and comparison to the p14 eggshell protein gene. Mol Biochem Parasitol. 1992;52:39–52. doi: 10.1016/0166-6851(92)90034-H. [DOI] [PubMed] [Google Scholar]
- Grevelding CG, Sommer G, Kunz W. Female-specific gene expression in Schistosoma mansoni is regulated by pairing. Parasitology. 1997;115:635–640. doi: 10.1017/S0031182097001728. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JM, Johansen MV, Johnston DA, Dunne DW, Hoffmann KF. Gender-associated gene expression in two related strains of Schistosoma japonicum. Mol Biochem Parasitol. 2004;136:191–209. doi: 10.1016/j.molbiopara.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Waisberg M, Lobo FP, Cerqueira GC, Passos LK, Carvalho OS, Franco GR, El-Sayed NM. Microarray analysis of gene expression induced by sexual contact in Schistosoma mansoni. BMC Genomics. 2007;8:181. doi: 10.1186/1471-2164-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moertel L, Gobert GN, McManus DP. Comparative real-time PCR and enzyme analysis of selected gender-associated molecules in Schistosoma japonicum. Parasitology. 2008;135:575–583. doi: 10.1017/S0031182008004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menrath M, Michel A, Kunz W. A female-specific cDNA sequence of Schistosoma mansoni encoding a mucin-like protein that is expressed in the epithelial cells of the reproductive duct. Parasitology. 1995;111:477–483. doi: 10.1017/S0031182000065987. [DOI] [PubMed] [Google Scholar]
- Reis MG, Kuhns J, Blanton R, Davis AH. Localization and pattern of expression of a female specific mRNA in Schistosoma mansoni. Mol Biochem Parasitol. 1989;32:113–119. doi: 10.1016/0166-6851(89)90062-5. [DOI] [PubMed] [Google Scholar]
- Dillon GP, Feltwell T, Skelton JP, Ashton PD, Coulson PS, Quail MA, Nikolaidou-Katsaridou N, Wilson RA, Ivens AC. Microarray analysis identifies genes preferentially expressed in the lung schistosomulum of Schistosoma mansoni. Int J Parasitol. 2006;36:1–8. doi: 10.1016/j.ijpara.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Gobert GN, McInnes R, Moertel L, Nelson C, Jones MK, Hu W, McManus DP. Transcriptomics tool for the human Schistosoma blood flukes using microarray gene expression profiling. Exp Parasitol. 2006;114:160–172. doi: 10.1016/j.exppara.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Moertel L, McManus DP, Piva TJ, Young L, McInnes RL, Gobert GN. Oligonucleotide microarray analysis of strain- and gender-associated gene expression in the human blood fluke, Schistosoma japonicum. Mol Cell Probes. 2006;20:280–289. doi: 10.1016/j.mcp.2006.02.002. [DOI] [PubMed] [Google Scholar]
- De Souza GM, Muniyappa MK, Carvalho SG, Guerra-Sá R, Spillane C. Genome-wide identification of novel microRNAs and their target genes in the human parasite Schistosoma mansoni. Genomics. 2011;98:96–111. doi: 10.1016/j.ygeno.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Hong Y, Sun A, Zhang M, Gao F, Han Y, Fu Z, Shi Y, Lin J. Proteomics analysis of differentially expressed proteins in schistosomula and adult worms of Schistosoma japonicum. Acta Trop. 2012;126:1–10. doi: 10.1016/j.actatropica.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Cheng GF, Lin JJ, Feng XG, Fu ZQ, Jin YM, Yuan CX, Zhou YC, Cai YM. Proteomic analysis of differentially expressed proteins between the male and female worm of Schistosoma japonicum after pairing. Proteomics. 2005;5:511–521. doi: 10.1002/pmic.200400953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of differentially expressed miRNAs.
Analysis of differentially expressed genes.
Predicted target genes of miR-1-miR-71-miR-7-miR-7-5p in Schistosoma japonicum.
Predicted target genes of bantam in Schistosoma japonicum.
Predicted target genes of bantam in down-regulated genes in 23DSI.
Predicted target genes of miR-1-miR-71-miR-7-miR-7-5p in up-regulated genes in 23DSI.