Abstract
In recent history, the nematode Caenorhabditis elegans has provided a compelling platform for the discovery of novel antimicrobial drugs. In this protocol, we present an automated, high-throughput C. elegans pathogenesis assay, which can be used to screen for anti-infective compounds that prevent nematodes from dying due to Pseudomonas aeruginosa. New antibiotics identified from such screens would be promising candidates for treatment of human infections, and also can be used as probe compounds to identify novel targets in microbial pathogenesis or host immunity.
Keywords: Caenorhabditis elegans, Pseudomonas aeruginosa, high-throughput screening, pathogenesis model, antibiotic, antimicrobial, drug discovery, liquid killing
INTRODUCTION
It is imperative to identify new classes of antibacterials for the treatment of pathogens with intrinsically low antibiotic susceptibility such as Pseudomonas aeruginosa, and for drug-resistant pathogens such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE). However, in recent history, the use of traditional, in vitro screening methods to identify novel antibiotics has been largely unsuccessful and is unlikely to meet the expected need for the foreseeable future (Lewis, 2013). We describe here the use of a Caenorhabditis elegans whole-animal pathogenesis model, which allows screening for compounds that block the ability of a pathogen to kill the worms. This model offers distinct advantages for antimicrobial discovery as it provides the possibility to identify compounds with no antimicrobial activity in vitro, but which may target functions only important for in vivo survival or virulence. These compounds would include prodrugs, activators of host innate immunity, or repressors of pathogen virulence factors. Additionally, by screening for compounds that promote survival of infected worms, this method eliminates the many compounds that are toxic or ineffective in vivo due to poor pharmacokinetics. Finally, whole-organism screening allows assaying of complex phenotypes such as immunity, where interaction between multiple organs and tissues is required, without many of the ethical and financial concerns that come with the use of vertebrate animals.
In recent years, the bacteriovorous nematode C. elegans has become a prominent model organism in the study of microbial pathogenesis, innate immunity, and drug discovery, particularly in the search for novel antimicrobial drugs. C. elegans can be infected with a growing list of Gram-positive and Gram-negative bacteria and fungal pathogens, many of which are clinically relevant (Sifri et al., 2005). The first microbe shown to be able to infect and kill C. elegans was P. aeruginosa, an opportunistic, nosocomial pathogen of immunocompromised individuals (Tan et al., 1999a). P. aeruginosa strain PA14 was isolated from a burn patient and can kill C. elegans in several medium-dependent ways. PA14 can kill under low-osmolarity conditions via an active infection by colonizing and dividing in the intestine, a mechanism called “Slow Killing” (Tan et al., 1999a). Under high-osmolarity conditions, PA14 kills nematodes via “Fast Killing”, a toxin-mediated mechanism that depends on low molecular weight diffusible toxins of the pyocyanin/phenazine class (Mahajan-Miklos et al., 1999). The methods for setting up Slow Killing and Fast Killing assays have been previously described (Powell and Ausubel, 2008).
Here, we present the protocol for a “Liquid Killing” assay, which is designed specifically for high-throughput screening of chemical libraries for anti-infective compounds. A workflow of the assay is presented in Figure 1. A small compound screen using the described Liquid Killing assay and subsequent work revealed that pyoverdin, a bacterial siderophore synthesized by PA14, disrupts host iron homeostasis and is critical for killing in this liquid-based C. elegans pathogenesis assay (Kirienko et al., 2013).
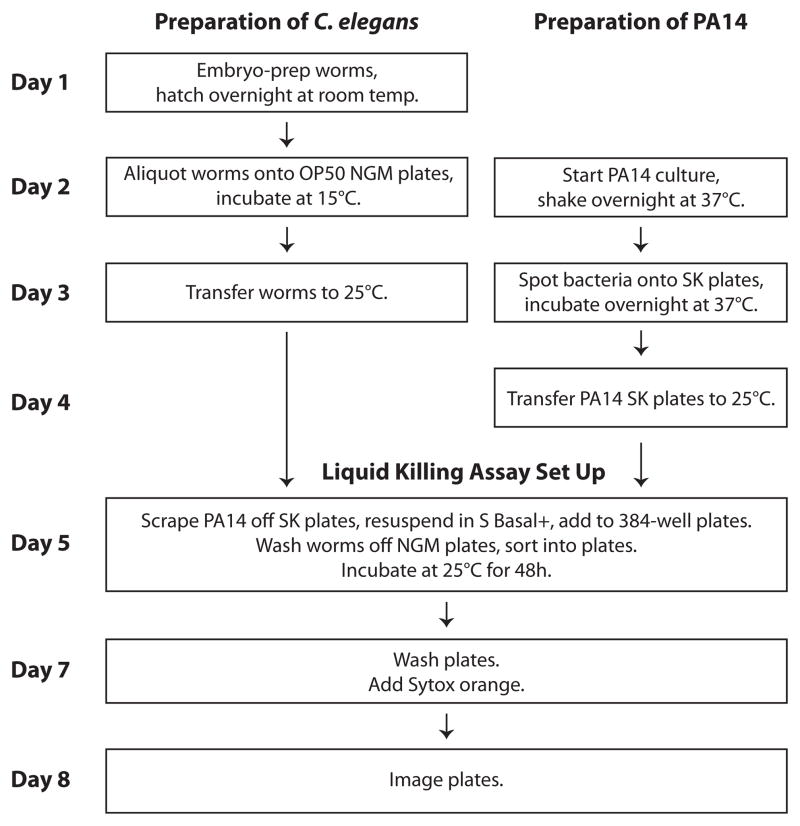
Fig. 1.
Workflow of high-throughput, liquid-based Pseudomonas aeruginosa infection assay. The methodology used for the LK assay is diagrammed, with steps involving worm preparation on the left and bacterial preparation on the right. Steps involving both are centered.
Liquid Killing is a modification of the Slow Killing assay in which PA14 is grown on agar and then transferred to a liquid-based medium in 384-well microtiter plates. The development of a liquid killing assay allows automation of liquid and worm handling, image acquisition, and analysis, thereby permitting high-throughput small molecule screening. The individual steps and automation concepts presented in this protocol are broadly applicable to other types of small molecule screening with C. elegans.
BASIC PROTOCOL 1: P. AERUGINOSA LIQUID KILLING ASSAY IN C. ELEGANS
Materials
-
Gravid C. elegans strain SS104 glp-4(bn2ts) (Beanan and Strome, 1992)
Can be ordered from the Caenorhabditis Genetics Center: (http://www.cbs.umn.edu/research/resources/cgc) P. aeruginosa strain PA14 (Rahme et al., 1995)
10 cm LB agar plates
LB broth
10 cm SK agar plates
10 cm OP50 NGM agar plates
M9 buffer
Worm bleaching solution
S Basal buffer
Liquid Kill media
Sytox® Orange (Life Technologies, Catalog #S-34861) solution
15 mL conical tubes
Sterile bacteria spreader
Sterile cell scraper (e.g., Small Cell Scraper #3010, Corning)
Gas permeable membrane (e.g., Breathe-Easy, Diversified Biotech)
384-well microplate (e.g., Corning #3712 Sterile, black with flat/clear-bottom, polystyrene, TC-treated with lid)
Clinical centrifuge (e.g., Thermo Scientific CL2 centrifuge)
Dissecting microscope (e.g., Nikon SMZ660)
Rotator (e.g., Thermo Scientific Labquake tube rotator)
Spectrophotometer (e.g., Beckman Coulter DU730 UV/Vis spectrophotometer)
Multichannel pipette or reagent dispenser (e.g., Thermo Scientific Multidrop Combi)
Union Biometrica COPAS BioSort large particle sorter
Microplate mixer (e.g., Eppendorf MixMate)
Microplate washer (e.g., BioTek ELx405)
Automated microscope (e.g., Molecular Devices ImageXpress Micro)
Image analysis software (e.g., CellProfiler, www.cellprofiler.org)
Preparation of P. aeruginosa
-
1
Before the start of the assay, streak P. aeruginosa PA14 bacteria from a frozen stock onto an LB agar plate. Grow at 37°C for 12–18 hours and store the streaked plate at 4°C for no longer than a week.
PA14 must always be streaked fresh from a frozen stock prior to each experiment as the strain loses virulence with passage on plates. -
2
Three days prior to setting up assay plates, inoculate 3–5 mL of LB broth with a single colony of PA14. Grow with agitation at 37°C for 12–16 hours.
PA14 has a tendency to form phenotypic variants that are less virulent in C. eleganskilling assays when grown in a static culture. Be sure to grow liquid cultures with sufficient aeration. Also, do not grow bacteria in liquid culture for longer than 16 hours at 37°C as the bacteria will begin to lyse. -
3
Pipet 350 μL of saturated culture onto 10 cm SK agar plates and spread the culture using a sterile spreader to cover the plate completely. Allow the plates to dry at room temperature for 10–15 minutes. Incubate spread plates at 37°C for 24 hours.
-
4
Transfer the plates to 25°C or room temperature and incubate for an additional 24 hours.
Preparation of C. elegans
-
5
Four days prior to setting up assay plates, prepare a synchronized worm population: Wash off gravid glp-4(bn2ts) hermaphrodites with many fertilized embryos in the gonad from up to four 10 cm E. coli OP50 NGM plates with M9 buffer into a 15 mL conical tube.
glp-4(bn2ts) mutant worms animals are maintained at 15°C and are sterile at the restrictive temperature of 25°C. glp-4(bn2ts) are used for the Liquid Killing Assay to prevent matricidal hatching of progeny inside the parent worm that can occur in liquid growth conditions. Also, progeny that hatch externally hinders scoring of the initial population of worms. -
6
Spin the worms in a clinical centrifuge at 1500 x g for 30 seconds to pellet; aspirate the supernatant until approximately 1.5 mL worm solution remain.
-
7
Add 2.5 mL of worm bleaching solution. Shake vigorously.
-
8
Monitor the lysis of the worms with a dissecting microscope by laying the 15 mL conical tube flat on the microscope stage and observing worms directly in the tube every ~15 seconds. Once 50% of the worms have broken in half releasing their embryos, quickly add 11 mL of M9 buffer. The duration of the bleach treatment will be 3–5 minutes on average, although the timing is highly dependent on the amount of bacteria and number of worms in the tube, and the freshness of the bleaching solution. Do not allow the bleach treatment to proceed longer than 7 minutes or the embryos will begin to die.
-
9
Pellet the embryos in a clinical centrifuge at 1500 x g for 30 seconds and aspirate the supernatant.
-
10
Quickly wash the embryos 3 times by adding 14 mL of M9 buffer, shaking to resuspend the embryos, and centrifuging to pellet the embryos. After three washes, resuspend the embryos in 5 mL of M9 buffer in a 15 mL conical tube.
In order to confirm that the embryo preparation has been successful, a small aliquot (1–2 μls) of the embryo suspension can be spotted onto an agar plate or an agar plate lid for observation with the dissecting scope. Most of the worm sample should be embryos (oval, ~50 μm in length and ~25 μm in diameter), with occasional parts of adult worm carcasses. -
11
Rotate 15 mL tubes on a rotator at room temperature overnight. The embryos will hatch and larval development will arrest at the L1 larval stage in the absence of food.
-
12
To confirm that most of the embryos have hatched after the overnight incubation, the 15 mL conical tube can be viewed with a dissecting microscope. The majority of worm sample should be L1 larvae (long, thin worms ~250 μm in length) with some unhatched, dead embryos.
-
13
Determine the density of L1s in the L1 suspension by removing a small sample (1–2 μls) and spotting on an agar plate or agar plate lid. View the spot with a dissecting microscope and count the number of L1s. The density of L1s will depend on the number of worms that were processed into the original embryo preparation. A density of ~30–50 L1s/μl is a good concentration at which to move on to the next step.
-
14
Aliquot ~5,000–6,000 L1s onto each 10 cm OP50 NGM plate and make sure that the plates are dry before putting them in the incubator. Incubate ~16h at 15°C followed by ~48h at 25°C. After the incubation at 15°C and then at 25°C, worms should be young adults and ready to add to the assay plates as described in Step 5 of the following section. There should be little attrition and 5,000 L1s should yield an equivalent number of young adult animals.
Young adults are the preferred stage for Liquid Killing Assays. L4 larval animals die more quickly than young adults and also settle more slowly during wash steps, causing a greater number of worms to be lost.
Set up of Liquid Killing Assay
-
14
Using a cell scraper, scrape P. aeruginosa bacteria from SK plates, and resuspend in M9 buffer. Measure OD600 to determine bacterial density.
-
15
Prepare 4X bacterial solution by diluting the bacterial suspension with S Basal buffer to OD600=0.12. The final concentration of OD600 in each well should be 0.03.
-
16
Combine one part diluted bacterial solution with three parts Liquid Kill media.
-
17
Use a multichannel pipette or an automated reagent dispenser (e.g., Thermo Scientific Multidrop Combi) to add 50 μL/well of diluted bacteria in Liquid Kill media to 384-well plates.
-
18
Wash worms off OP50 NGM plates (from Step 8 of the previous section, Preparation of C. elegans) with M9 buffer into a 50 mL conical tube. Allow them to gravitationally settle (~5 minutes). Aspirate liquid down to ~5 mL. Wash three times with S Basal buffer.
For the wash steps, allow the worms to settle to the bottom of the conical tube rather than use centrifugation. Centrifugation will also pellet debris, which could then clog the COPAS BioSort large particle sorter. -
19
Use the Union Biometrica COPAS BioSort worm sorter to dispense 15–18 worms per well of the 384-well assay plate prepared in Step 17 of “Set-up of Liquid Killing Assay”.
Alternatively, worms can be added manually using a multichannel pipette. However, manually dispensing the worms will cause the number of worms in each well to be more variable (+/− 10 worms), which may increase the variability in the level of killing and may also cause difficulties with image analysis if the density of worms is too high. In order to reduce the variability in the number of worms dispensed, the worm suspension should always be mixing and not stationary so that worms do not settle while they are being pipetted. -
20
After addition of worms, seal plates with a gas permeable membrane (Breathe-Easy, Diversified Biotech) and incubate at 25°C with 80–85% humidity without agitation for ~48 hours.
-
21
To stop killing, wash plates five times with S Basal buffer using a microplate washer (e.g., BioTek ELx405). Prior to the first wash cycle, let worms settle to the bottom of the plates (~2–5 minutes), aspirate all but 20 μL per well, and shake the plates using a microplate vortexer (e.g., Eppendorf MixMate). Each following wash cycle should consist of a dispense step that fills wells to the top, a waiting step for the worms to settle, and an aspiration step to aspirate all but 20 μL of volume per well.
-
22
Add 50 μL of Sytox® orange solution in each well for a final dye concentration of 0.7μM and stain for 16–24h.
If there is a high level of background staining, the plates should be washed twice with S Basal after staining. Under these conditions, Sytox orange stains only dead worms.
Image acquisition and analysis
Because a great volume of data will be generated with image acquisition and analysis, it is important to consider organization and storage of screening data prior to starting a large-scale screen. Depending on the size of the screen, it may be prudent to create a database to store the raw image data, in addition to any outputs from the image analysis. A web-based interface for retrieving and viewing data may also be useful.
-
23
Using an automated microscope (e.g., Molecular Devices ImageXpress Micro), capture transmitted light and TRITC (535 nm excitation, 610 nm emission) fluorescent images with a 2X objective.
The use of a 2x objective should allow capture the area of an entire well with a single image and also allow image capture from an entire plate in the same z plane without the use of autofocus functions. -
24
For small-scale experiments, images can be scored manually by counting the number of stained (dead) worms per well in the fluorescence image and total worms per well in the transmitted light image. For example, if you have 5 stained worms out of 15 total worms, the fraction dead is 0.33, or 33%.
-
25
For large-scale experiments, unbiased, automated scoring methods, such as the use of the open-source, free image analysis software CellProfiler (www.cellprofiler.org, (Carpenter et al., 2006)) can be utilized to calculate the percentage of dead worms in each well (see Fig. 1).
-
26The first critical step in assay validation is Z′ factor determination (Zhang et al., 1999). For this purpose, an experiment should be carried out with a 384-well plate divided evenly between negative (DMSO) and positive (i.e., 200 μg/mL gentamycin) controls. The Z′ factor is calculated as follows:
Where,
M1 is the mean of the negative control.
M2 is the mean of the positive control.
SD1 is the standard deviation of the negative control.
SD2 is the standard deviation of the positive control.
The assay is appropriately optimized when the Z′ factor > 0.5.
M1 and M2 are calculated with different data values depending on the method used to score the assays. If the assays were scored by eye, M1 and M2 would be calculated using the fraction dead/total worms. If the assays were scored using CellProfiler as described above, M1 and M2 would be calculated using the fraction Sytox area/transmitted light area.
REAGENTS AND SOLUTIONS
Use ultrapure water (e.g., MilliQ ddH2O) for the preparation of all media and buffers. Unless otherwise noted, sterilize buffers and media by autoclaving at 121°C for 20 minutes for volumes less than 1 L; for larger batches, adjust autoclave time accordingly to ensure sterility. Store at room temperature unless otherwise noted.
10 cm LB agar plates
Mix 10 g Bacto-tryptone, 5 g Bacto-yeast extract, 10 g NaCl, and 15 g agar in 1 L ddH2O. Autoclave. Cool agar to 55°C and pour into 10 cm sterile Petri dishes (typically ~20 mL per dish). Dry plates at room temperature overnight, then store at 4°C in a covered plastic box for up to 3 months.
LB broth
Dissolve 10 g Bacto-tryptone, 5 g Bacto-yeast extract, and 10 g NaCl in 1 L ddH2O. Autoclave.
10 cm Slow Killing (SK) agar plates
Mix 3.5 g Bacto-Peptone, 3 g NaCl, and 18 g Bacto-Agar in 1 L ddH2O. Autoclave, then cool to 55°C. Slowly add the following sterile solutions: 1 mL 1 M MgSO4, 25 mL 1 M KH2PO4, pH 6, 1 mL 1M CaCl2, and 1 mL 5 mg/mL cholesterol in ethanol. Pour agar into sterile 10 cm sterile Petri dishes (typically ~20 mL). Dry plates at room temperature overnight, then store at 4°C in a covered plastic box for up to 1 month.
10 cm OP50 Nematode Growth Medium (NGM) agar plates
Mix 2.5 g Bacto-Peptone, 3 g NaCl, and 18 g Bacto-Agar in 1 L ddH2O. Autoclave, then cool to 55°C. Slowly add the following sterile solutions: 1 mL 1 M MgSO4, 25 mL 1 M KH2PO4, pH 6, 1 mL 1M CaCl2, and 1 mL 5 mg/mL cholesterol in ethanol. Pour agar into sterile 10 cm sterile Petri dishes (typically ~20 mL) and store at 4°C in a covered plastic box for several months. Before use, add 1 mL of an OP50 E. coli overnight culture concentrated 20X in S Basal and dry plates in a biosafety hood.
M9 buffer
Dissolve 3 g KH2PO4, 6 g Na2HPO4, and 5 g NaCl in 1 L of ddH2O. Autoclave, cool to 55°C, and add 1 mL of sterile 1 M MgSO4.
Worm bleaching solution
Mix 20 mL NaOCl solution (available chlorine = 10–15%), 16 mL 3M NaOH, 64 mL H2O. Store at 4°C in the dark for up to 1 month.
S Basal buffer
0.1M NaCl, 0.05M potassium phosphate (pH 6.0). Autoclave.
Liquid Kill media
Prepare S Basal+ buffer by supplementing 1 L of sterile S Basal buffer with 1 mL 5 mg/mL cholesterol in ethanol immediately before use; S Basal+ buffer should not be stored overnight. Prepare SK media as described above, omitting the agar and add the MgSO4, KH2PO4, CaCl2, and cholesterol immediately before use. Mix 2 parts SK broth with 1 part S Basal+ buffer.
Sytox® Orange solution
Immediately before use, dilute Sytox® Orange (5mM in DMSO) to 0.98μM in S Basal buffer.
COMMENTARY
Background Information
For over forty years, the nematode C. elegans has been a powerful model organism used for the study of a wide range of biological processes. With the discovery that many of the genetic pathways that govern its development are highly conserved with humans, C. elegans has been instrumental in addressing fundamental questions in several fields of research, including neurobiology, development, and aging. This small worm has a quick generation time, is easy to propagate, and has an array of forward and reverse genetics tools available (Riddle et al., 1997; Wood, 1988). There are many publications that describe the biology of C. elegans and its general husbandry. The online publication WormBook (http://www.wormbook.org/) is a comprehensive review of C. elegans biology with a section devoted to methods used in nematode experimentation (Girard et al., 2007). The website WormBase (http://www.wormbase.org/) is a compendium of useful information about the genome and biology of C. elegans (Stein et al., 2001). Essentially, WormBase is a database of curated information on every predicted gene in the genome, including RNAi and mutant phenotypes and expression data. These Internet resources are useful references for both novice and experienced C. elegans researchers.
Because worms can be cultured in liquid and can fit comfortably in 96- or 384-well microtiter plates, it is possible to use them to carry out high-throughput screens. Several publications describe methods specifically for high-throughput chemical screening in C. elegans. Of note, Moy et al., present a high-throughput C. elegans infection screen with E. faecalis and Burns et al., describe compound screening for a variety of worm phenotypes and demonstrates a method for target identification (Burns et al., 2006; Moy et al., 2009). Wählby et al., present the CellProfiler WormToolbox, a set of tools for automated image analysis of high-throughput, high-content C. elegans assays (Wahlby et al., 2012). In the image analysis methods presented here, we quantify survival by measuring total worm and Sytox stained areas. With the use of the WormToolbox, it may be possible to determine worm death on an individual worm basis.
In this unit, we describe how C. elegans can be used as a model host to screen for antimicrobials effective against the pathogen P. aeruginosa. Because this infection system is well established, there is a wealth of information regarding both the pathogen and host, which are advantages when probing the mechanism of action of any promising antimicrobial compounds. For example, the novel anti-infective could target virulence factors of PA14. For P. aeruginosa in the liquid-killing assay described here, these virulence factors include KinB (a two-component histidine kinase that regulates motility, virulence factor production, and biofilm formation) and pyoverdin, a siderophore or iron chelator (Kirienko et al., 2013). Additionally, the antimicrobial compounds can be used as a tool to discover novel virulence factors of Pseudomonas. Importantly, many pathogen mutations that diminish killing of C. elegans also diminish pathogenesis in mammalian hosts (Tan et al., 1999a; Tan et al., 1999b), validating C. elegans as a surrogate host in which pathogen virulence factors required for mammalian pathogenicity can be identified.
In addition to reducing the virulence of PA14 in vivo, anti-infective compounds identified using the P. aeruginosa–C. elegans liquid killing assay may promote survival of C. elegans by stimulating host immunity. As with the information available regarding Pseudomonas factors important for pathogenicity, there is an entire field of research dedicated to the study of C. elegans innate immunity. We and others have shown that C. elegans respond to bacterial pathogens by the activation of a variety of genes, homologues of which are known to be involved in antimicrobial responses in mammals. Although C. elegans does not have cell-mediated adaptive immunity, key features of mammalian innate immunity are also found in C. elegans (Pukkila-Worley and Ausubel, 2012). This includes a highly-conserved p38 MAPK signaling cassette (Kim et al., 2002) and an upstream TIR-domain containing protein homologous to the human SARM protein, which are both part of the basal resistance to pathogen attack (Couillault et al., 2004; Liberati et al., 2004). In addition to basal resistance, worms also mount a specific immune response for each pathogen to respond to individual infection mechanisms. The immune response against P. aeruginosa in liquid medium involves a hypoxic response (Kirienko et al., 2013). In addition to utility of anti-infectives as drugs, those that target host immunity can be used as probe compounds to identify and dissect conserved components of the innate immune system.
With this protocol, we have explored killing of C. elegans by P. aeruginosa. A remarkably large number of other human microbial pathogens kill worms, including Serratia marcescens (Mallo et al., 2002), Salmonella enterica (Aballay et al., 2000; Labrousse et al., 2000), Yersinia pestis (Darby et al., 2002), Enterococcus faecalis (Garsin et al., 2001), Staphylococcus aureus (Garsin et al., 2001), Streptococcus pneumoniae (Garsin et al., 2001; Mylonakis et al., 2002), Cryptococcus neoformans (Mylonakis et al., 2002) and Candida albicans (Breger et al., 2007). Each of these organisms has been studied in the C. elegans model by simply replacing the normal food source, a lawn of E. coli strain OP50, with the pathogen in question and monitoring the survival of the nematodes. For most of these infection models, the virulence factors required for pathogenesis and the genes involved in the specific immune response mounted for each pathogen are known. The methods presented here can be readily adapted to develop infection assays in liquid with many pathogens other than P. aeruginosa (See Critical Parameters) and the individual automation steps can be applied to a broad range of high-throughput C. elegans screening assays.
Critical Parameters
It is critical that a fresh streak of PA14 is used to start a culture and that the proper growing conditions are used to grow PA14 in liquid and on agar.
The glp-4(bn2ts) worms should appear phenotypically wild type and healthy before use in experiments. If worms are passaged for many months, especially if there is an opportunity for population bottlenecks, it is possible that the strain could accumulate unwanted mutations that may affect the outcome of the assay. When not actively being used, worms can be maintained stably on a plate without food in a “starved” or stasis condition for many months.
A crucial parameter when setting up the LK plate is the PA14 bacterial concentration in each well. The final OD600 in each well should be 0.03, which is approximately the lowest titer that kills most of the worms in 24 – 48 hours. If the bacterial concentration is too low, there will not be sufficient killing of worms by PA14. Conversely, if the bacterial concentration is too high, compounds with relatively weak anti-infective activity may not be identified.
Many of the critical parameters in setting up a high-throughput infection assay depend on the choice of pathogen. When considering a different pathogen, there are a number of variables to consider. Some pathogens, such as E. faecalis, cause persistent infections in C. elegans, meaning they will permanently colonize and expand in the worm intestine (Garsin et al., 2001). Others, such as P. aeruginosa, are not persistent in C. elegans and the infection can be cleared (i.e. the intestinal pathogen can be excreted) if infected worms are removed from the pathogen before a time where the damage caused by the assault is so extensive that the worms cannot recover (Tan et al., 1999a). If the pathogen of choice is persistent, worms can be infected prior to adding them to assay plates, but with those that are not persistent, the pathogen must be added with the worms in the assay plates for the duration of the infection. Also, each pathogen kills with different kinetics. If the killing takes over a week, as with the adherent invasive Escherichia coli LF82 (Simonsen et al., 2011), parameters such as temperature or the genetic background of the worm may need to be adjusted to speed up the killing process and make the timing amenable to high-throughput screening. Generally, increasing the temperature and using a worm strain that is more sensitive to killing by the pathogen of choice can cause the killing to proceed faster. For persistent infections, worms can be infected for a longer period of time before adding to the assay plate, which will also cause the worms to die more quickly in the assay plates.
Anticipated Results
Ideally, 50 to 75% of worms in the DMSO negative control wells will be dead, while less than 10% of worms in a positive control well with 200 μg/mL gentamycin will be dead at the 48 hour time point. A typical Z′ factor for one 384-well plate is 0.65.
Time Considerations
From the day that worm propagation is started, it will take ~8 days to complete the assay.
Also, it is important to consider the rate-limiting steps for the experiment, as it will affect the overall throughput of the screen (i.e., the number of plates that can be processed with each experiment and the number of experiments that can be set up per week). The rate-limited steps for this protocol are the worm sorting and plate imaging, with each step requiring ~10–15 minutes/plate. We have found that it is feasible to process 20–25 plates per day, and reasonable to set up an experiment each week so that 2 experiments are running concurrently during the same week.
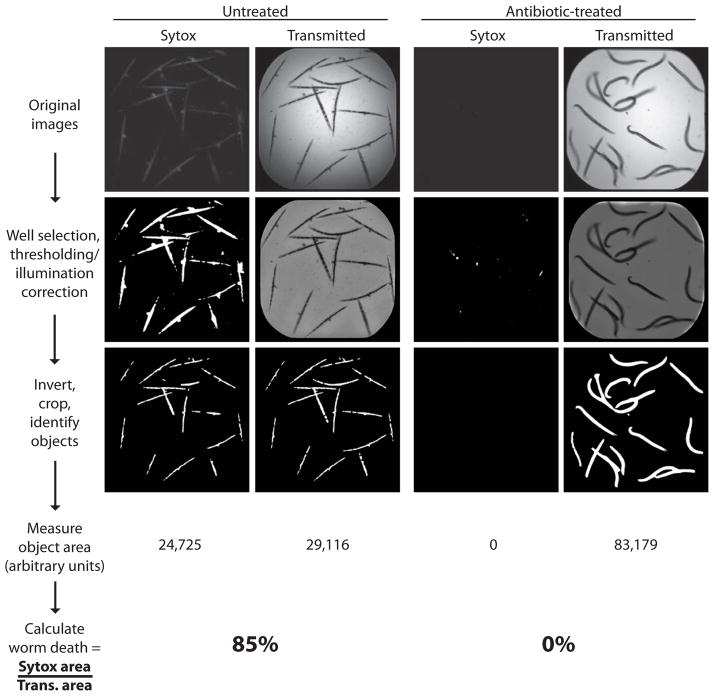
Fig. 2.
Small-molecule inhibition of pathogen-induced worm death: quantification using CellProfiler. Worms in 384-well plates are incubated with Sytox® orange, which specifically stains dead worms. Top row shows raw fluorescent Sytox® orange and bright field images of an untreated and an antibiotic-treated well. The images were analyzed using CellProfiler with a pipeline of image processing and analysis modules. The results of several processing steps are shown. The images in the middle row show the results of well selection, correction for uneven illumination of the bright field images and thresholding for the Sytox® images to remove background fluorescence. Images in the bottom row show the result of inverting the bright field image, identifying worms and cropping the worm area. Finally, the total area of fluorescent and bright field images are measured and are used to calculate worm death in each well.
Vokes and Carpenter, in Unit 14.17 of Current Protocols in Molecular Biology, describe in detail the procedure for setting up and using CellProfiler (Vokes and Carpenter, 2008).
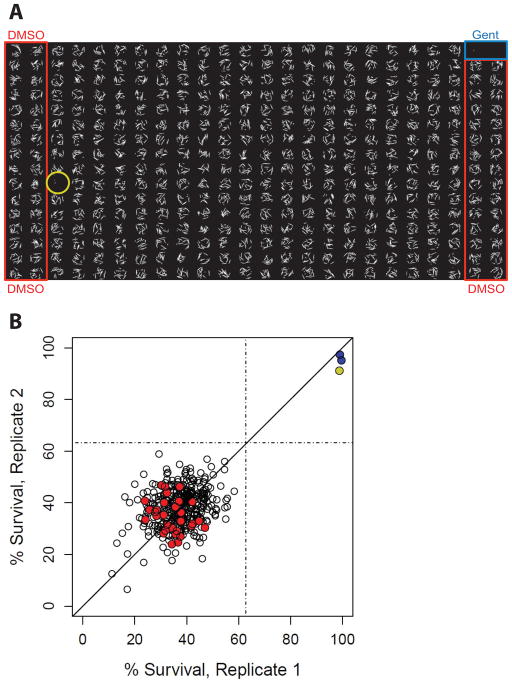
Fig. 3.
Example of screening plate. A) Fluorescence micrographs of every well of a 384-well plate tiled in a single image. Columns outlined in blue and red are gentamicin (positive) and DMSO (negative) controls, respectively. Each sample well (not outlined) contains a different small molecule that is being screened for anti-infective properties with the PA14 Liquid Killing assay. Circled in yellow is a compound that showed significant rescue, as demonstrated by the absence of stained worms. B) A scatter plot shows the percent survival in each of the two replicates for the example plate. Red circles are wells containing DMSO, blue circles are gentamicin wells, and test compounds are shown as open circles. The compound showing significant rescue is in yellow at the upper right.
Footnotes
Literature Cited
- Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Current biology: CB. 2000;10:1539–1542. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- Beanan MJ, Strome S. Characterization of a germ-line proliferation mutation in C. elegans. Development. 1992;116:755–766. doi: 10.1242/dev.116.3.755. [DOI] [PubMed] [Google Scholar]
- Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, Mylonakis E. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS pathogens. 2007;3:e18. doi: 10.1371/journal.ppat.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns AR, Kwok TC, Howard A, Houston E, Johanson K, Chan A, Cutler SR, McCourt P, Roy PJ. High-throughput screening of small molecules for bioactivity and target identification in Caenorhabditis elegans. Nature protocols. 2006;1:1906–1914. doi: 10.1038/nprot.2006.283. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nature immunology. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Darby C, Hsu JW, Ghori N, Falkow S. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature. 2002;417:243–244. doi: 10.1038/417243a. [DOI] [PubMed] [Google Scholar]
- Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, Calderwood SB, Ausubel FM. A simple model host for identifying Gram-positive virulence factors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10892–10897. doi: 10.1073/pnas.191378698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard LR, Fiedler TJ, Harris TW, Carvalho F, Antoshechkin I, Han M, Sternberg PW, Stein LD, Chalfie M. WormBook: the online review of Caenorhabditis elegans biology. Nucleic acids research. 2007;35:D472–475. doi: 10.1093/nar/gkl894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan MW, Ausubel FM. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- Kirienko NV, Kirienko DR, Larkins-Ford J, Wahlby C, Ruvkun G, Ausubel FM. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell host & microbe. 2013;13:406–416. doi: 10.1016/j.chom.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse A, Chauvet S, Couillault C, Kurz CL, Ewbank JJ. Caenorhabditis elegans is a model host for Salmonella typhimurium. Current biology: CB. 2000;10:1543–1545. doi: 10.1016/s0960-9822(00)00833-2. [DOI] [PubMed] [Google Scholar]
- Lewis K. Platforms for antibiotic discovery. Nature reviews Drug discovery. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, Ausubel FM. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6593–6598. doi: 10.1073/pnas.0308625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, Ewbank JJ. Inducible antibacterial defense system in C. elegans. Current biology: CB. 2002;12:1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- Moy TI, Conery AL, Larkins-Ford J, Wu G, Mazitschek R, Casadei G, Lewis K, Carpenter AE, Ausubel FM. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS chemical biology. 2009;4:527–533. doi: 10.1021/cb900084v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15675–15680. doi: 10.1073/pnas.232568599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Ausubel FM. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods Mol Biol. 2008;415:403–427. doi: 10.1007/978-1-59745-570-1_24. [DOI] [PubMed] [Google Scholar]
- Pukkila-Worley R, Ausubel FM. Immune defense mechanisms in the Caenorhabditis elegans intestinal epithelium. Current opinion in immunology. 2012;24:3–9. doi: 10.1016/j.coi.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- Riddle DL, Blumenthal T, Meyer BJ, Priess JR. Introduction to C. elegans. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. 2. Cold Spring Harbor; NY: 1997. [Google Scholar]
- Sifri CD, Begun J, Ausubel FM. The worm has turned--microbial virulence modeled in Caenorhabditis elegans. Trends in microbiology. 2005;13:119–127. doi: 10.1016/j.tim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Simonsen KT, Nielsen G, Bjerrum JV, Kruse T, Kallipolitis BH, Moller-Jensen J. A role for the RNA chaperone Hfq in controlling adherent-invasive Escherichia coli colonization and virulence. PloS one. 2011;6:e16387. doi: 10.1371/journal.pone.0016387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L, Sternberg P, Durbin R, Thierry-Mieg J, Spieth J. WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic acids research. 2001;29:82–86. doi: 10.1093/nar/29.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 1999a;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proceedings of the National Academy of Sciences of the United States of America. 1999b;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes MS, Carpenter AE. Using CellProfiler for automatic identification and measurement of biological objects in images. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. Unit 14. Chapter 14. 2008. p. 17. [DOI] [PubMed] [Google Scholar]
- Wahlby C, Kamentsky L, Liu ZH, Riklin-Raviv T, Conery AL, O’Rourke EJ, Sokolnicki KL, Visvikis O, Ljosa V, Irazoqui JE, Golland P, Ruvkun G, Ausubel FM, Carpenter AE. An image analysis toolbox for high-throughput C. elegans assays. Nature methods. 2012;9:714–716. doi: 10.1038/nmeth.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 1988. [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of biomolecular screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Key Reference
- Kirienko NV, Kirienko DR, Larkins-Ford J, Wählby C, Ruvkun G, Ausubel FM. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe. 2013;13(4):406–16. doi: 10.1016/j.chom.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]