Abstract
Background
The Onychophora are a probable sister group to Arthropoda, one of the most intensively studied animal phyla from a developmental perspective. Pioneering work on the fruit fly Drosophila melanogaster and subsequent investigation of other arthropods has revealed important roles for Wnt genes during many developmental processes in these animals.
Results
We screened the embryonic transcriptome of the onychophoran Euperipatoides kanangrensis and found that at least 11 Wnt genes are expressed during embryogenesis. These genes represent 11 of the 13 known subfamilies of Wnt genes.
Conclusions
Many onychophoran Wnt genes are expressed in segment polarity gene-like patterns, suggesting a general role for these ligands during segment regionalization, as has been described in arthropods. During early stages of development, Wnt2, Wnt4, and Wnt5 are expressed in broad multiple segment-wide domains that are reminiscent of arthropod gap and Hox gene expression patterns, which suggests an early instructive role for Wnt genes during E. kanangrensis segmentation.
Keywords: Development, Evolution, Segmentation, Segment polarity, Wnt signalling
Background
The phylum Onychophora is represented by only around 200 described species [1]. Like their probable sister group, the arthropods, onychophorans are segmented, a fact that is most obvious from the arrangement of up to 43 pairs of walking limbs on these animals. However, onychophorans differ from arthropods because they lack intersegmental ectodermal grooves, tagmosis is absent and their limbs are unsegmented [2]. Despite the great interest in, and growing understanding of, all aspects of arthropod biology, including the genetic regulation of segmentation (reviewed in, for example, [3-7]), relatively little is known about the onychophorans.
In Drosophila melanogaster, segmentation is under control of a hierarchic segmentation gene cascade that initially transforms aperiodic patterns of genetic information along the anterior-posterior body axis into a periodic pattern [8,9]. Comparative studies have revealed that at least some components of this hierarchical network are conserved and that the function of segment polarity genes in particular has been maintained during the evolution of arthropods and onychophorans [10-18]. The segment polarity genes act later in the hierarchy downstream of maternal effect genes, gap genes and pair rule genes, and regulate segment polarity and maintain segmental boundaries. The network of segment polarity genes includes morphogens, such as Hedgehog (Hh) and Wingless (wg/Wnt1).
The Wnt gene family comprises 13 subfamilies, of which 12 are found in protostomes, with Wnt3 having been lost in the lineage leading to these animals [19,20]. Preliminary studies of the Wnt gene repertoire in arthropods suggest some lineages have lost one or more Wnt genes in the course of evolution (summarized in [20]): for example, Wnt2 and Wnt4 appear to have been lost in insects.
To further study the role that Wnt genes play in development and evolution, we surveyed the repertoire of these genes in the onychophoran Euperipatoides kanangrensis and investigated their expression during its embryogenesis. We found that at least 11 of the predicted 12 Wnt genes found in protostomes are expressed during onychophoran ontogenesis. Our data suggest that onychophoran Wnt genes are likely to be involved in segment border formation or maintenance, intrasegmental patterning and possibly even the determination of segment identity. The latter function would not only represent an onychophoran-specific feature of Wnt gene function, but also suggest a role for these genes in segmentation beyond that of segment regionalization.
Methods
Animal husbandry and embryo preparation
Female specimens of E. kanangrensis were collected in Kanangra-Boyd National Park, New South Wales, Australia. To obtain all developmental stages, we dissected developing embryos of various stages in the months from September to December. Each female carries up to 100 embryos, representing a series of developing stages (sometimes even ranging from the one-cell stage up to the fully developed embryo). The chorion and vitelline membrane were removed by hand with Dumont size 5 forceps and directly afterwards the embryos were fixed in 4% formaldehyde in 0.1 M phosphate-buffered saline with 0.1% Tween-20 (PBST) (pH 7.4) for four to six hours at room temperature. Embryos were then dehydrated stepwise into 100% methanol and stored at −20°C for at least three weeks before being used for in-situ hybridization experiments.
PCR and gene cloning
RNA isolation and cDNA synthesis were described in [14]. Gene fragments of all Wnt gene orthologues described here were isolated by means of PCR with gene specific primers based on the sequences found in a sequenced embryonic transcriptome. For further information on the transcriptome see [18].
All Wnt gene fragments were cloned into the pCRII vector (Invitrogen, Carlsbad, CA, USA), and sequences were determined by means of Big Dye chemistry on an ABI3730XL analyzer by a commercial sequencing service (Macrogen, Amsterdam, The Netherlands). Sequences of the newly discovered E. kanangrensis Wnt genes are available from the EMBL nucleotide database under accession numbers HG529208 (Wnt2), HG529209 (Wnt4), HG529210 (Wnt5), HG529211 (Wnt6), HG529212 (Wnt7), HG529213 (Wnt9), HG529214 (Wnt10), HG529215 (Wnt11), HG529216 (Wnt16), and HG529217 (WntA).
In-situ hybridization, cell nuclei staining and data documentation
In-situ hybridization experiments were performed as described previously [21]. Cell nuclei were stained with 1 μg/ml DAPI (4-6-diamidino-2-phenylindole) in PBST for 20 minutes followed by several washing steps in PBST. Embryos were analyzed under a Leica dissection microscope equipped with a Leica DC100 digital camera. Brightness, contrast and colour values were adjusted if necessary, using the image processing software Adobe Photoshop CS2 (Version 9.0.1 for Apple Macintosh).
Phylogenetic analysis
The amino acid sequences of E. kanangrensis Wnt genes were aligned with those of Wnt sequence dataset 1 from [20] using T-Coffee followed by manual editing in SeaView [22,23].
Bayesian phylogenetic analyses were performed with MrBayes [24] using a fixed WAG amino acid substitution model with gamma-distributed rate variation across sites (with four rate categories). An unconstrained exponential prior probability distribution on branch lengths and an exponential prior for the gamma shape parameter for among-site rate variation was applied. The final topology was estimated using 1,100,000 cycles for the MCMCMC (metropolis-coupled Markov chain Monte Carlo) analysis with four chains and the chain-heating temperature set to 0.2. The Markov chain was sampled every 200 cycles. The starting trees for the chains were randomly selected. Clade support was assessed with posterior probabilities computed with MrBayes.
Results
The Wnt gene repertoire of E. kanangrensis
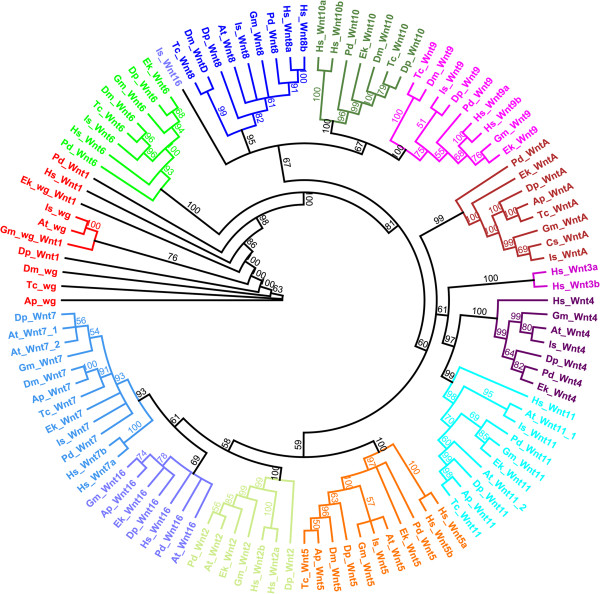
Phylogenetic analysis of E. kanangrensis Wnt amino acid sequences shows that this onychophoran has at least 11 Wnt genes representing the Wnt1, Wnt2, Wnt4, Wnt5, Wnt6, Wnt7, Wnt9, Wnt10, Wnt11, Wnt16 and WntA subfamilies (Figures 1 and 2). Therefore, E. kanangrensis has all Wnt genes reported in protostomes and arthropods except Wnt8 (Figure 1). To investigate the potential roles of these genes in comparison with other animals, we then examined their expression in E. kanangrensis embryos (see Figure 3 for an overview of early embryogenesis in this onychophoran).
Figure 1.
Phylogenetic analysis of Wnt genes. Maximum likelihood tree of Wnt amino acid sequences. The support value of each node is given as posterior probability (as a percentage) from the Bayesian analysis on the branches. Note that Is-Wnt16 is the only Wnt sequence that does not cluster with the respective orthologues. Included metazoan species are: Parasteatoda tepidariorum (At), Acyrthosiphon pisum (Ap), Cupiennius salei (Cs), Daphnia pulex (Dp), Drosophila melanogaster (Dm), Euperipatoides kanangrensis (Ek), Glomeris marginata (Gm), Homo sapiens (Hs), Ixodes scapularis (Is), Platynereis dumerilii (Pd) and Tribolium castaneum (Tc).
Figure 2.
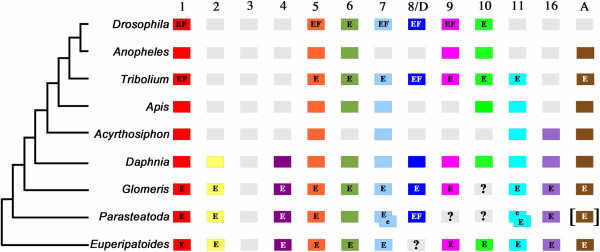
Repertoire of Wnt subfamilies in arthropods, an onychophoran and a lophotrochozoan. Grey boxes indicate putatively lost Wnt subfamilies. Question marks indicate Wnts that have not been found but cannot be defined as ‘missing’ because assigned genomes have not been sequenced or annotated. Duplicated Wnts are represented by overlapping boxes. Square brackets indicate that spider WntA was isolated from another spider, C. salei. E, embryonic expression patterns are available; e, embryonic expression has been studied but specific expression patterns could not be detected; F, functional data are available. Note that for T. castaneum all Wnts have been knocked down by means of RNAi, but phenotypes could only be detected for wg/Wnt1 and Wnt8/D. Wnt3 is present in deuterostomes and cnidarians, but has been lost in the lineage leading to the protostomes.
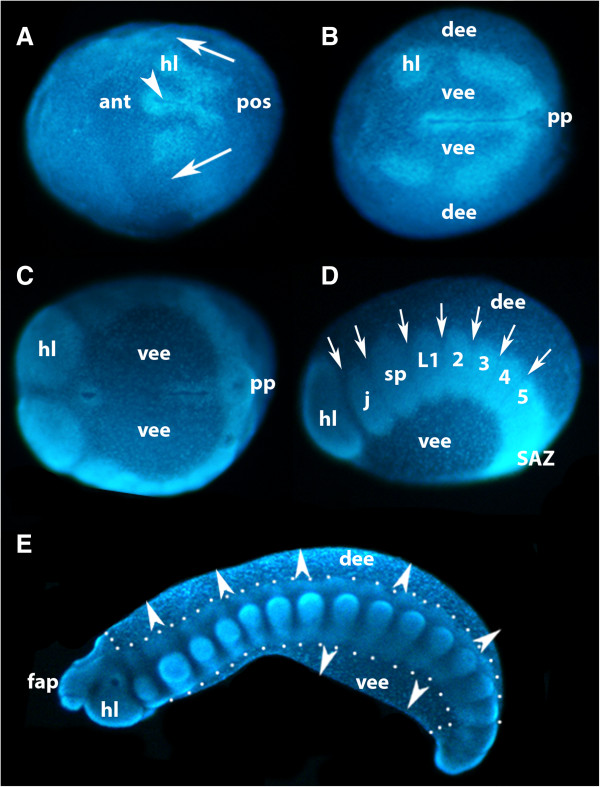
Figure 3.
Formation of the germ band in E. kanangrensis. A germ disc appears after the blastoderm has formed (not shown; [31]), in the centre of which forms a slit-like furrow (arrowhead in A) defining the AP axis. Anterior is to the left. (A) Stage 2 to 3 embryo, ventral view. Cells invaginating at the posterior pole move on either side of the indentation towards anterior (arrows), forming a split germ band. (B) Stage 5 embryo, ventral view. Split germ band subsequently moves towards the anterior and first coelomic pouches bud off from otherwise uniform halves of germ band. The distance between the furrow and the halves of the germ band has increased and ventral extraembryonic ectoderm covers the area between furrow and germ band on either side of the furrow; dorsal to the halves of the germ band is dorsal extraembryonic ectoderm. (C) Stage 8 embryo, ventral view. At later stages, the anterior extent of the split germ band (future head lobes) meets and fuses anterior to the anterior pole of the furrow. (D) Embryo of C, lateral view; dorsal is up. Coelomic pouches are recognizable by areas of higher cell density. As with short germ arthropods, anterior segments represent older (more developed) segments. In the posterior part of the split germ band (anterior to SAZ), coelomic pouches have not yet segregated from newly formed tissue. (E) Stage 15 embryo, lateral view; outline of embryo proper marked by dots. Dorsal and ventral embryonic tissue begins to grow out (arrowheads). 2 to 5, second to fifth walking-limb-bearing segment; ant, anterior; AP, anterior-posterior; dee, dorsal extraembryonic ectoderm; fap, frontal appendage; hl, head lobe; j, jaw; L1, first walking-limb-bearing segment; pos, posterior; pp, posterior pit region; SAZ, segment addition zone; sp, slime papilla; vee, ventral extraembryonic ectoderm.
Expression of E. kanangrensis wg/Wnt1
The expression of the E. kanangrensis wingless (wg/Wnt1) orthologue has been described previously [17]. It is expressed like a typical segment polarity gene in transverse stripes in the middle of each segment and anterior to the expression of engrailed (en) and hedgehog (hh) [18], as well as in the tips of all developing appendages [17].
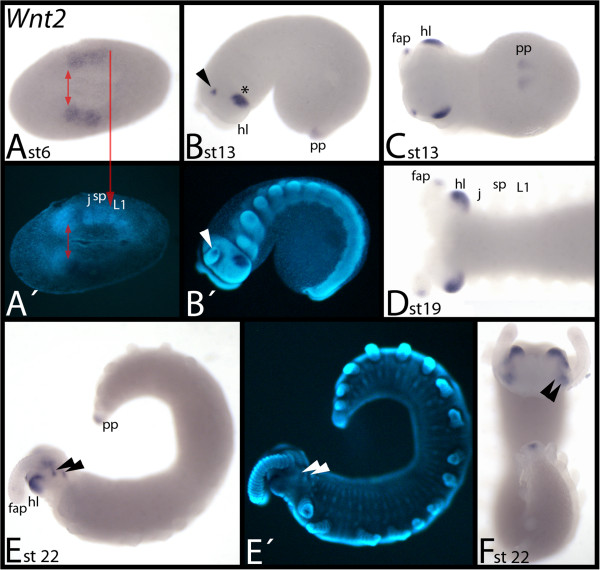
Expression of E. kanangrensis Wnt2
During early embryogenesis in E. kanangrensis, Wnt2 is expressed in broad domains covering the posterior of the future head lobes, and the primordia of the jaws and slime papillae-bearing segments (Figure 4A). At later stages, it is concentrated in the tips of the frontal appendages, the head lobes and in the posterior pit (Figure 4B-F). The expression in the head lobes is lateral and below the eyes. Expression in the tips of the frontal appendages is strong at first but weakens in older stages until it disappears in the oldest investigated stages (Figure 4B-F). Late during ontogenesis, additional expression appears on the dorsal side of the head lobes near the bases of the frontal appendages (Figure 4E, F).
Figure 4.
Expression of E. kanangrensis Wnt2. In all panels except (F) (anterior up), anterior is to the left. (A) Ventral view. Red double-arrow marks the anterior border of expression. Red arrow marks posterior border of expression. (B) Lateral view. Arrow points to expression in the tip of the frontal appendage. (C) Ventral view of embryo of (B). (D) Ventral view of the anterior of an embryo. (E) Lateral view. Arrowheads indicate dot-like expression posterior to the base of the frontal appendages. (F) Dorsal view. Embryo of (E). Arrowheads as in E. (A′), (B′) and (E′) represent DAPI staining of the embryos of (A), (B) and (E). fap, frontal appendage; hl, head lobe; j, jaw; L1, first walking-limb-bearing segment; pp, posterior pit region; sp, slime papilla; st, stage.
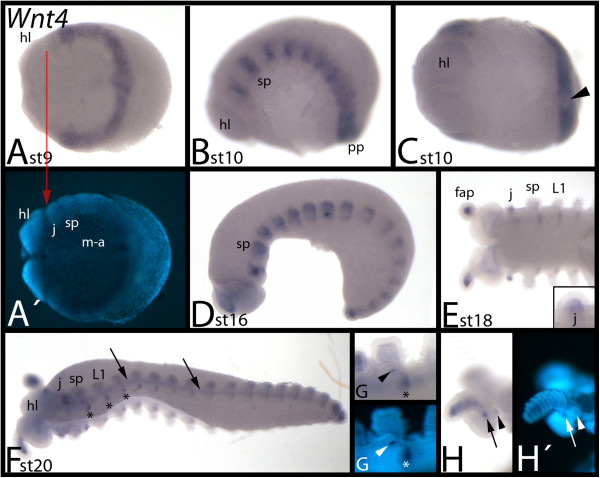
Expression of E. kanangrensis Wnt4
Wnt4 is first expressed ubiquitously in the posterior segments, including the posterior segment addition zone (SAZ) (Figure 5A); but the posterior pit region is free of transcripts (shown for a slightly older embryo in Figure 5C). There is also a sharp anterior expression boundary between the head lobes and the jaw-bearing segment (Figure 5A), and early expression in the jaw-bearing segment is weaker (Figure 5A, B). During subsequent stages, this band of expression transforms into a segmental pattern (Figure 5B). When the limbs form, Wnt4 is clearly expressed in the mesoderm of the frontal appendages (Figure 5D, E) and jaws (Figure 5E, F). However, the slime papillae and walking limbs do not express Wnt4. At later developmental stages, Wnt4 expression appears stepwise in a segmental pattern from anterior to posterior in a position ventral and posterior to the bases of the jaw, the slime papillae and the walking-limb-bearing segments (Figure 5E). This expression persists during later developmental stages (Figure 5F). At this point, a faint anterior-to-posterior stripe of expression appears ventral to the limbs (Figure 5F). The tissue in the developing openings to the salivary glands possibly expresses Wnt4 at late stages (Figure 5G); this, however, may represent nonspecific staining that sometimes occurs in this structure during late developmental stages. At stage 20, two spots of expression appear in each hemisphere of the head lobes posterior to the bases of the frontal appendages (Figure 5H).
Figure 5.
Expression of E. kanangrensis Wnt4. Anterior is to the left. (A) Ventral view. Red arrow marks the anterior border of expression. (B) Lateral view. (C) Embryo of (B), ventral view. Arrowhead indicates lack of expression in the posterior pit region. (D) Lateral view. (E) Anterior portion of embryo and close-up of jaw. Note that expression in the jaw is only in the mesoderm. (F) Ventrolateral view. Arrows indicate expression between the bases of the limbs. Asterisks mark expression in the ventral nervous system. (G) Embryo of (F). Close-up of slime-papillae-bearing segment. Arrowhead indicates staining in salivary gland. Asterisk marks expression in the ventral nervous system. (H) Embryo of (F). Close-up of head. Arrow and arrowhead indicate dot-like expression at base of frontal appendage. Note that expression in the frontal appendage is mesodermal. (A′), (G′) and (H′) represent DAPI staining of the embryos of (A), (G) and (H). fap, frontal appendage; hl, head lobe; j, jaw; L1, first walking-limb-bearing segment; m-a, mouth-anus furrow; pp, posterior pit region; sp, slime papilla; st, stage.
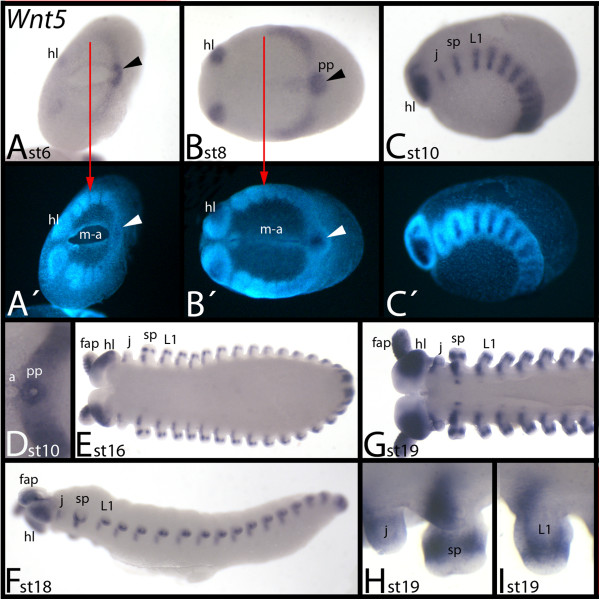
Expression of E. kanangrensis Wnt5
At early developmental stages, Wnt5 is expressed in the head lobes and posterior of the embryo with a sharp border between the slime-papillae-bearing segment and the first walking-limb-bearing segment (Figure 6A, B). At subsequent stages, the levels of expression increase in all Wnt5-expressing tissues, except the posterior pit. Wnt5 expression encircles the posterior pit and also extends towards the anterior into the posterior portion of the mouth-anus furrow but more weakly (Figure 6B). At around stage 10, Wnt5 is expressed in transverse stripes in the middle of each segment (Figure 6C). At this point, Wnt5 forms a ring of expression around the centre of the posterior pit (Figure 6D). At later stages, expression is in the anterior portion of the frontal appendages, jaws, slime papillae and walking limbs (Figure 6E-I). Expression, however, is restricted to the proximal region of the slime papillae and walking limbs, and for all segments this domain reaches into the tissue ventral to the bases of the limbs where the ventral nervous system will form. At around stage 19, spots of segmental expression appear ventral to the previously described expression; these spots appear in an anterior-to-posterior order, with the most anterior domains being located in the slime-papillae-bearing segments (Figure 6G). In addition to expression in the limbs described, Wnt5 is also expressed in a median ring in the slime papillae and the walking limbs (Figure 6E, G-I). Throughout development, a broad ventral and lateral domain of expression is also observed in each hemisphere of the head lobes.
Figure 6.
Expression of E. kanangrensis Wnt5. Anterior is to the left. (A) Ventral view. Red arrow marks the anterior border of expression (of the broad domain; note additional expression in the head lobes). Arrowhead indicates strong expression in the posterior pit region. (B) Ventral view. Arrow and arrowhead as in (A). (C) Lateral view. (D) Ventral view. Close-up of the posterior of the embryo of (C). Note the strong expression in a ring around the centre of the posterior pit region. (E) Ventral view. (G) Ventral view. Anterior part of an embryo. (F) Lateral view. (H) Close-up of a jaw and a slime papilla. (I) Close-up of a walking limb. (A′), (B′) and (C′) represent DAPI staining of the embryos of (A), (B) and (C). a, anus; fap, frontal appendage; hl, head lobe; j, jaw; L1, first walking-limb-bearing segment; m-a, mouth-anus furrow; pp, posterior pit region; sp, slime papilla; st, stage.
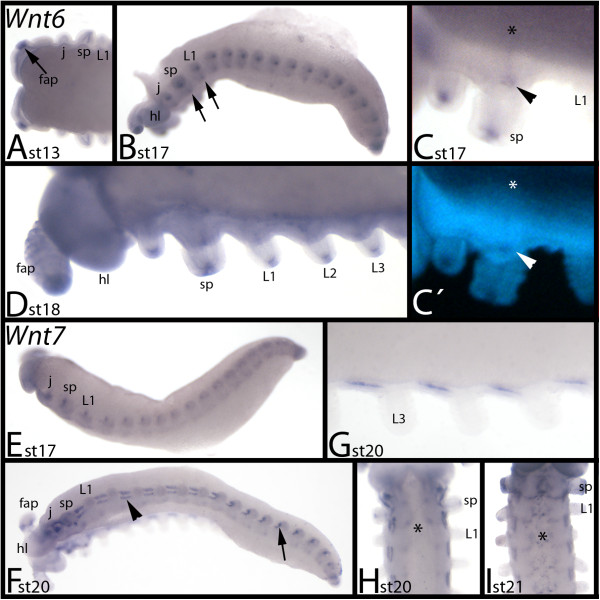
Expression of E. kanangrensis Wnt6
Wnt6 is not expressed before limb bud growth is observed (not shown). The first detectable expression is evident at stage 13, in the tips of the frontal appendages (Figure 7A). At later stages, expression appears in the tips of all other limbs (Figure 7B-D), and as very faint transverse segmental stripes in the centre of each segment (Figure 7B, C).
Figure 7.
Expression of E. kanangrensis Wnt6 and Wnt7. (A-D) Expression of Wnt6; (E-I) Expression of Wnt7. In (A-G), anterior is to the left; in (H) and (I), anterior is up. (A) Dorsal view. Arrow indicates expression in the tip of the developing frontal appendage. (B) Lateral view. Arrows indicate expression in the ventral nervous system. (C) Ventral view. Close-up of the jaw- and slime papillae-bearing segments. Arrow indicates staining in the salivary gland. Asterisk marks transverse ventral stripe. Note that it is level with the salivary gland. (D) Ventral view. Close-up of the anterior part of an embryo. (E) Lateral view. (F) Ventrolateral view. Arrowhead indicates expression between the limbs. Arrow indicates the same expression in a younger (more posterior) segment. Note the transformation of this expression domain from younger towards older segments. (G) Close-up of segmentally reiterated mesodermal expression in the trunk. (H) Ventral view. Asterisk marks expression in the ventral nervous system. (I) Ventral view. Asterisk marks expression in the ventral nervous system. (C′) represents DAPI staining of the embryo of (C). fap, frontal appendage; hl, head lobe; j, jaw; L1 to L3, first to third walking-limb-bearing segments; sp, slime papilla; st, stage.
Expression of E. kanangrensis Wnt7
Expression of Wnt7 is also absent from early developmental stages (not shown). The earliest expression is observed at the bases of all the appendages of the trunk segments (Figure 7E). At subsequent stages, this expression becomes stronger and eventually transforms into a series of short longitudinal stripes in the mesoderm between the bases of the limbs along the anterior-posterior axis of the embryo (Figure 7F, G). During late stages, segmental expression appears in the developing ventral nervous system (Figure 7H, I).
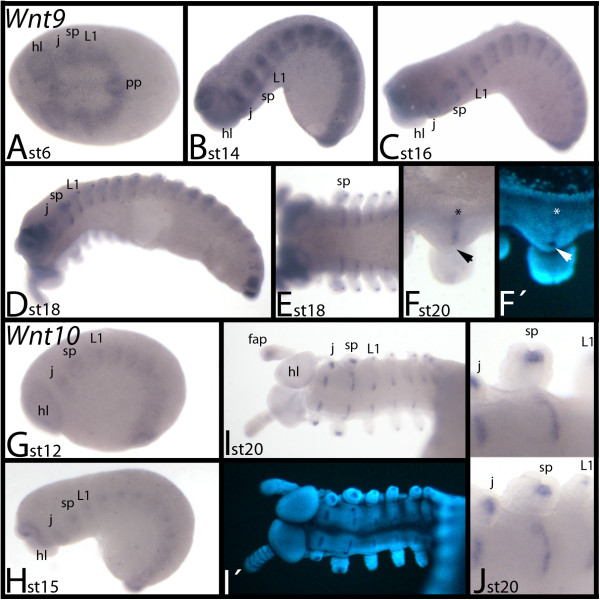
Expression of E. kanangrensis Wnt9
During early stages, Wnt9 is ubiquitously expressed (Figure 8A). Later, segmental expression is observed, which transforms into transverse stripes in the centre of the segments (Figure 8B, C). Subsequently, Wnt9 is also expressed in the tips of the limbs (except for the frontal appendages, in which Wnt9 is expressed throughout the mesoderm) (Figure 8D, E). In the slime-papillae-bearing segment Wnt9 expression marks the position of the salivary gland openings (Figure 8F).
Figure 8.
Expression of E. kanangrensis Wnt9 and Wnt10. (A-F) Expression of Wnt9. (G-J) Expression of Wnt10. Anterior is to the left. (A) Ventral view. Ubiquitous expression. (B) Lateral view. (C) Lateral view. (D) Ventrolateral view. (E) Ventral view. Anterior part of an embryo. (F) Ventral view. Close-up of the slime-papillae-bearing segment. Note the transverse stripe of expression (asterisk) that is level with the opening of the salivary gland (arrowhead) (cf. F′). (G) Lateral view. (H) Lateral view. (I) Ventral view, focused on the anterior of the embryo. (J) View of two slightly different angles of the same area of the embryo showing the slime-papillae-bearing segment. Note that the transverse stripe of expression is level with the posterior region of the salivary gland opening. (F′) and (I′) represent DAPI staining of the embryos of (F) and (I). fap, frontal appendage; hl, head lobe; j, jaw; L1, first walking-limb-bearing segment; pp, posterior pit region; sp, slime papilla; st, stage.
Expression of E. kanangrensis Wnt10
During early stages, Wnt10 is weakly expressed throughout E. kanangrensis embryos (not shown). Later, differential expression appears in the tips of the limb buds and as transverse stripes in the centre of the trunk segments (Figure 8G-J). However, expression in the tips of the frontal appendages is weaker than in the other appendages (Figure 8I). Segmental stripes of expression are located posteriorly adjacent to the openings of the salivary glands, which also express Wnt10 (Figure 8J).
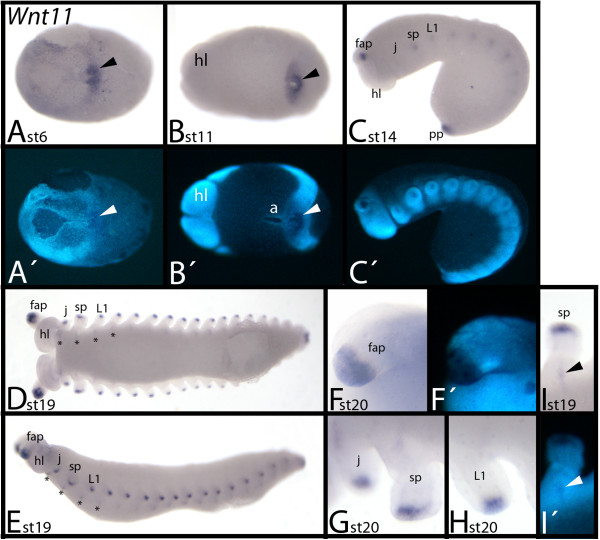
Expression of E. kanangrensis Wnt11
During early developmental stages, Wnt11 expression is detectable in the posterior pit region (Figure 9A). However, the indentation of the posterior pit does not express Wnt11, and expression does not extend into the future anus (Figure 9A, B). Later, expression appears in the tips of all appendages (Figure 9C). This expression persists throughout embryogenesis (Figure 9D-H). At stage 19, faint expression in segmental stripes appears in an anterior-to-posterior order in all trunk segments (Figure 9D, E, I).
Figure 9.
Expression of E. kanangrensis Wnt11. Anterior is to the left. (A) Ventral view. Arrow indicates strong expression in the posterior pit region. (B) Ventral view. Arrow indicates strong expression in the posterior pit region. (C) Lateral view. (D) Ventral view. Asterisks mark upcoming expression in segmental transverse stripes. (E) Embryo of (D), lateral view. Asterisks mark upcoming expression in segmental transverse stripes. (F) Close-up of frontal appendage. (G) Close-up of a jaw and a slime papilla. (H) Close-up of a walking limb. (I) Close-up of a slime-papillae-bearing segment, focused on the transverse stripe of expression. Note that this stripe is level with the opening of the salivary gland (arrowhead; cf. I′). (A′), (B′), (C′), (F′) and (I′) represent DAPI staining of the embryos of (A), (B), (C), (F) and (I). a, anus; fap, frontal appendage; hl, head lobe; j, jaw; L1, first walking-limb-bearing segment; pp, posterior pit region; sp, slime papilla; st, stage.
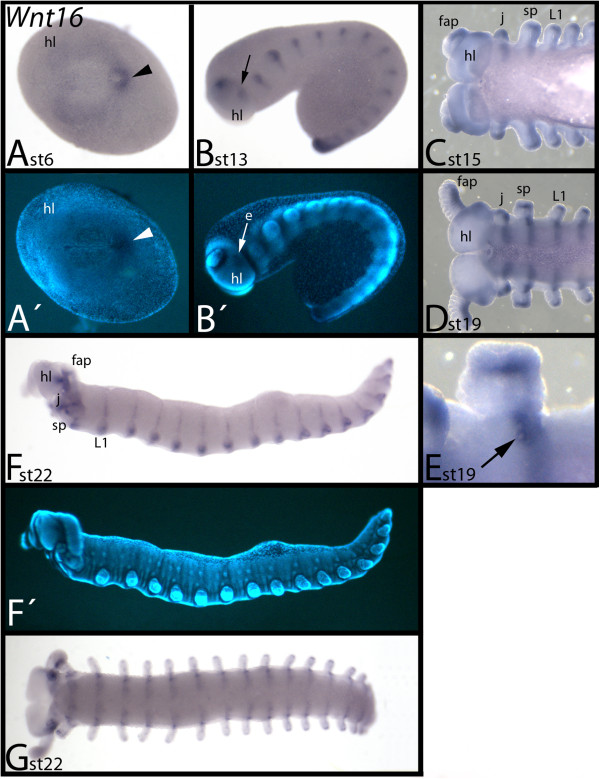
Expression of E. kanangrensis Wnt16
In early embryos, Wnt16 is expressed around the posterior pit region, and weakly but ubiquitously in all other tissue except the mouth-anus furrow (Figure 10A). During later stages, expression appears in the base of the frontal appendages, a more posterior region adjacent to the eye grooves and, in a segmental pattern, the trunk (Figure 10B). The segmental pattern then transforms into transverse stripes in dorsal (dorsal to the position of the limbs) and ventral (ventral to the position of the limbs) tissue (Figure 10C-G). This is different from the expression of other E. kanangrensis Wnt genes, which are expressed as stripes exclusively in ventral, but not dorsal, tissue. The ventral transverse stripes are broader than the comparable stripes of other Wnt genes and they cover the complete posterior compartment of the segments, including the openings of the salivary glands in the slime-papillae-bearing segments (Figure 10E). Wnt16 is also expressed in posterior tissue of the limbs and in the tips of the limbs (Figure 10C-E).
Figure 10.
Expression of E. kanangrensis Wnt16. Anterior is to the left. (A) Ventral view. Arrowhead indicates expression in the posterior pit region. (B) Lateral view. Arrow indicates expression posterior to the eye groove. (C) Ventral view. Anterior part of an embryo. (D) Ventral view. Anterior part of an embryo. (E) Close-up of a slime papillae-bearing segment. Note that the anterior border of the segmental stripe is level with the opening of the salivary gland (arrow). (F) Lateral view. Note the segmental stripes extending into tissue dorsal to the limbs. (G) Embryo of (F), ventral view. (A′), (B′) and (F′) represent DAPI staining of the embryos of (A), (B) and (F). e, eye groove; fap, frontal appendage; hl, head lobe; j, jaw; L1, first walking-limb-bearing segment; sp, slime papilla; st, stage.
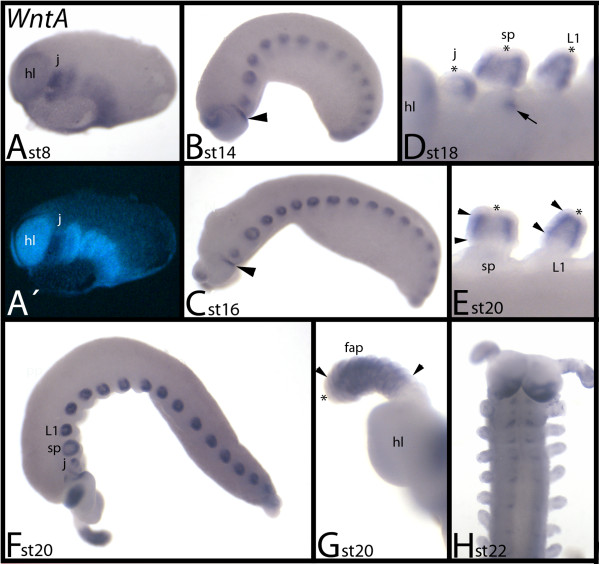
Expression of E. kanangrensis WntA
During early developmental stages, WntA is ubiquitously expressed, with the strongest expression observed in the jaw-bearing segment (Figure 11A). At later stages, it is expressed in a broad leg-gap-gene-like domain in all limbs (Figure 11B-G) (compare with expression of onychophoran limb gap genes in [20]). Only the tips of the limbs and the most proximal region do not express WntA. In the jaws, WntA is expressed in a posterior and central region (Figure 11D). The posterior rim of the head lobes expresses WntA (Figure 11B-D, F, H). During later stages, the openings of the salivary glands also exhibit expression (but this may represent an unspecific signal) (Figure 11D). WntA is only weakly expressed in the SAZ. At around stage 22, segmental expression appears in the ventral nervous system, and the domain of expression in the head lobes is enlarged (Figure 11H).
Figure 11.
Expression of E. kanangrensis WntA. In all panels (except for panel H, where anterior is up), anterior is to the left. (A) Ventrolateral view. (B) Lateral view. Arrowhead indicates expression at the posterior rim of the head lobe. (C) Lateral view. Arrowhead indicates expression at the posterior rim of the head lobe. (D) Close-up of the anterior appendages. Asterisks mark tips of jaw, slime papilla and walking limb that are free from expression. Arrow indicates staining in the opening of the salivary gland. (E) Close-up of slime papilla and walking limb of an older embryo (compare with D). Arrowheads mark anterior and posterior border of expression in the limbs. Asterisks mark tips of jaw, slime papilla and walking limb that are free from expression. (F) Lateral view. (G) Close-up of frontal appendage and head lobe. Arrowheads mark anterior and posterior border of expression in the limbs. Asterisks mark tips of jaw, slime papilla and walking limb that are free from expression. (H) Ventral view. Note developing expression in the ventral nervous system. (A′) represents DAPI staining of the embryo of (A). fap, frontal appendage; hl, head lobe; j, jaw; L1, first walking-limb-bearing segment; sp, slime papilla; st, stage.
Discussion
Wnt genes and segment identity
We observed that Wnt2, Wnt4, Wnt5 and Wnt11 are expressed in broad, several-segment-wide, domains with distinct anterior boundaries, and in the case of Wnt2 also a clear posterior boundary (Figure 12). These expression patterns are reminiscent of that of onychophoran and (to some extent) arthropod gap and Hox genes (Additional file 1: Figure S1) [25-28]. In particular, the early expression of Wnt2 is similar to that of anterior Hox genes and gap genes in arthropods, which provide each segment with its specific identity (summarized in [29,30]). The early expression patterns of E. kanangrensis Wnt genes thus suggest a possible role in providing segmental identity. Such a possible function, however, must be different from Hox and gap genes, which act as direct selector genes on the segments’ identity. The Wnt genes may act downstream of initial selector genes, such as the Hox genes and other hitherto undetected genes that act anterior to the Hox genes during development of the anterior body of onychophorans. Moreover, while the expression of onychophoran Hox genes is restricted to the slime-papillae-bearing segment and backwards, the anterior expression borders of two Wnt genes, Wnt2 and Wnt4, is located more anteriorly (Figure 12 and Additional file 1: Figure S1). Wnt genes may thus contribute to defining or maintaining segment identity of the walking-limb-bearing segments (expressing Wnt4 and Wnt5), the slime-papillae-bearing segment (expressing Wnt2 and Wnt4), and the jaw-bearing segment (expressing Wnt2 and Wnt4 (weakly)). The posterior region of the head lobes only expresses Wnt2 and the anterior portion of the head lobes is free of any Wnt gene expression.
Figure 12.
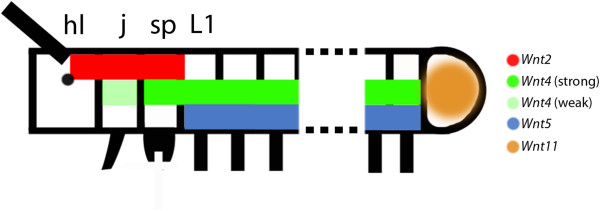
Gap-gene-like expression patterns. Depicted are the anterior and posterior parts of an embryo. Dashed lines indicate missing segments. Broad early multi-segment-wide domains of expression of Wnt genes are represented by coloured bars. hl, head lobe; j, jaw; L1, first walking-limb-bearing segment; sp, slime papilla.
Since the composition of the anterior head in onychophorans is still uncertain, the anterior border of Wnt2 expression within the head lobes indirectly raises the question of whether the head lobes represent two segments (or at least two independently patterned regions). So far, only one coelomic pouch has been identified in the developing head lobes (for example, [31]), and only one transverse stripe of hedgehog (hh) expression lies at the posterior rim of the head lobe [18].
Wnt genes in segmentation and segment regionalization
The arthropod wingless/Wnt1 (wg/Wnt1) gene is a classical segment polarity gene that is involved in maintaining segmental boundaries and intrasegmental patterning (reviewed in [32-34]). In D. melanogaster, other arthropods, and onychophorans, wg is expressed anterior adjacent to the expression of engrailed (en) (for example, [35,36,14,17]).
We observed that several onychophoran Wnt genes are expressed in typical segment polarity gene patterns (that is, in transverse segmental stripes), but that the intrasegmental position of these genes differs considerably (Figure 13): Three Wnt genes pattern the anterior (Wnt5), the median (Wnt16) and the posterior (Wnt4) region of each trunk segment, respectively (as indicated by the position of the limbs and the juxtaposed expression of en/hh and wg). Four onychophoran Wnt genes (wg/Wnt1, Wnt6, Wnt9 and Wnt11) are expressed within the Wnt16 domain in the centre of the onychophoran segments, and Wnt10 appears to be expressed posteriorly, adjacent to the wg/Wnt1 domain (Figure 13).
Figure 13.
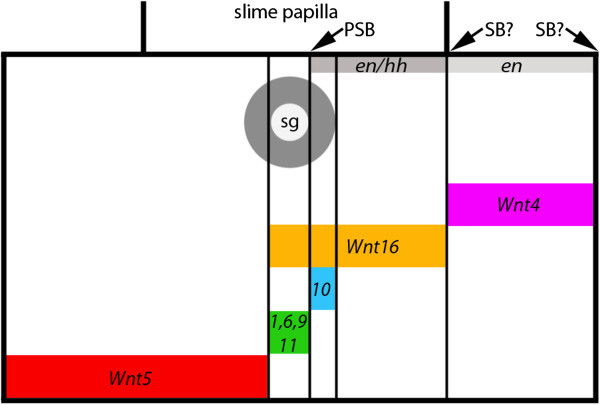
Segment polarity gene-like intrasegmental expression. The slime-papillae-bearing segment is represented (broad black lines) in embryos older than stage 10. The positions of the slime papillae are indicated by transverse broad black lines. Thin black lines mark intrasegmental borders of Wnt gene expression. The grey circle marks the position of the salivary gland. Arrows point to the position of the parasegment boundary and the two possible positions of the segmental boundary. Expression of Wnt genes is indicated by coloured bars. Positions of expression of the segment polarity genes engrailed (en) and hedgehog (hh) are indicated by dark grey (co-expression of en + hh) and light grey (expression of en) bars. Drawing is out of scale. PSB, parasegment boundary; SB?, possible position of segmental boundary; sg, salivary gland.
The onychophoran segment polarity gene orthologues are first expressed in their typical pattern in adjacent transverse stripes at around stage 10 [17,18]; note that en is expressed considerably earlier. However, most of the Wnt genes are expressed in segment polarity gene-like patterns only at later stages. The temporal delay of segmental patterning in comparison with segment polarity genes, such as en, wg/Wnt1 and hh[17,18], suggests that onychophoran Wnt genes are not generally involved in the determination of (morphologically invisible) intersegmental boundaries, but that their function is rather restricted to intrasegmental patterning.
All onychophoran segments are added from the posterior pit region, the SAZ. Since some of the Wnt genes are expressed in the SAZ, these genes may be involved in segment addition. The involvement of Wnt genes during segmentation has been demonstrated by the knocking down of components of the canonical Wnt gene network in insects [37-41]. In the basally branching insect Periplaneta americana, and in the spider Parasteatoda tepidariorum, the function of wg/Wnt1 and Wnt8, respectively, are indeed crucial for posterior segment addition [42-44]. In E. kanangrensis, we find that all Wnt genes except Wnt6 and Wnt7 are expressed in the SAZ. Of these, wg/Wnt1[17], Wnt2, Wnt4, Wnt5, Wnt11 and Wnt16 are all expressed prominently in the SAZ. Others, such as Wnt9, Wnt10 and WntA are only weakly expressed in the SAZ, or are expressed ubiquitously at some stages and thus also in the SAZ. Overall, these expression patterns suggest a function of onychophoran Wnt genes during segment addition, similar to that reported for insects and a spider.
Conserved and diverged aspects of Wnt expression in arthropods and onychophorans
Wnt1/wg is the best studied arthropod Wnt gene (reviewed in [45,46]). Its expression in transverse segmental stripes anterior adjacent to engrailed (en) is principally conserved in all arthropods [12,11,14], and even in E. kanangrensis[17,18]. The segment polarity gene-like function, however, has only been directly demonstrated for some insects [38,39,47].
Expression of Wnt2 has been studied in the millipede Glomeris marginata, the centipede Strigamia maritima and the spider P. tepidariorum[20,48,49]. Expression in the ocular region is conserved between E. kanangrensis and these arthropods. However, comparable expression in an early gap-gene-like domain, at the posterior end of the embryo, and in the frontal appendages of E. kanangrensis is not found in these arthropods.
Expression of Wnt4 has been studied in G. marginata, S. maritima and P. tepidariorum[20,48,49]. The expression profiles of Wnt4 in these arthropods are completely different. Expression in myriapods is observed throughout the developing embryo, whereas in P. tepidariorum, expression is restricted to the SAZ. None of the expression patterns of Wnt4 in myriapods and E. kanangrensis is the same. Reconstruction of the ancestral arthropod expression pattern of Wnt4 is therefore impossible based on the available data.
Expression of Wnt5 has been investigated in a number of arthropods, including D. melanogaster[50,51] and T. castaneum[52], the spiders C. salei[12,53] and P. tepidariorum[20], and the myriapods G. marginata[48] and S. maritima[49]. Wnt5 expression in the two spiders is virtually identical, and expression in the other arthropods is in many aspects comparable to that observed in these spiders: Wnt5 is expressed in the ventral nervous system, including the brain, transverse segmental stripes, and the labrum and the limb primordia (and later in the limbs). Expression in the brain, the limb primordia, and in the form of segmental stripes is conserved in E. kanangrensis. This strongly suggests an ancestral and conserved function for Wnt5 in the development of these tissues. This assumption is supported by the fact that Wnt5 is the only Wnt gene (besides wg/Wnt1) that is present in all hitherto studied arthropods (summarized in [20]; Figure 2). However, apparently lineage-specific expression of Wnt5 includes the expression observed in the heart of spiders and the early gap-gene-like expression in the onychophoran.
Wnt6 expression has been studied in D. melanogaster[54], T. castaneum[52], G. marginata[20] and S. maritima[49]. This work has shown that expression in the limbs and brain, and the segmental expression (probably associated with the central nervous system), is conserved among these arthropods. In E. kanangrensis, expression in the ventral nervous system and the limbs appears to be conserved, and may thus represent part of the ancestral expression pattern of Wnt6.
The embryonic expression of Wnt7 has been studied in D. melanogaster[50,55], T. castaneum[52], G. marginata[20], P. tepidariorum[20] and S. maritima[56,49]. In both insects, Wnt7 is expressed in a segmentally reiterated pattern; a comparable pattern is seen in S. maritima but not G. marginata. One of the two Wnt7 paralogues of P. tepidariorum is expressed in the SAZ, and this pattern is also seen in G. marginata, but not in the onychophoran, or in T. castaneum. In both myriapods, Wnt7 is expressed in the heart, and in the brain, but expression in the labrum is only seen in G. marginata, and expression in the antennae is only present in S. maritima. In summary, while some aspects of the expression of arthropod Wnt7 genes are conserved, none of these expression domains is observed in E. kanangrensis.
Expression of Wnt8 has been studied in D. melanogaster[57,58], where it is called WntD, and in T. castaneum[52], as well as in G. marginata[20] and P. tepidariorum[42]. In D. melanogaster, it is first expressed at both poles of the early blastoderm stage embryo. Later during ontogenesis, it is expressed in the mesectoderm and the ventral neurectoderm. In T. castaneum, Wnt8 is only expressed at the posterior pole in blastoderm stage embryos and in the ventral mesoderm in the SAZ. The early expression at the posterior pole and in the SAZ is also conserved in the spider, and functional studies have shown that Wnt8 is involved in posterior segment addition in both T. castaneum and P. tepidariorum[39,42]. The fact that Wnt8 is only expressed in the primordia of the ocular region and the mandibular segment of G. marginata was therefore unexpected [20], and contradicted the idea that Wnt8 could possibly play an ancestral and conserved role in arthropod segmentation [43].
We did not recover an E. kanangrensis Wnt8 in our surveys. It may be that Wnt8 was missed because it is expressed at a low level or because it is not expressed at all during ontogenesis. The possible lack, or nonexpression during ontogenesis, of onychophoran Wnt8, however, supports the possibility that Wnt8 is not an ancestral component of the segmentation machinery of arthropods and onychophorans.
Expression of Wnt9 has been studied in D. melanogaster[59], T. castaneum[52,49] and G. marginata[48]. In T. castaneum and S. maritima, Wnt9 is only expressed in a few cells in the gut. In G. marginata, this gene is transiently expressed in segment polarity gene-like segmental stripes, in the appendages including the labrum, and in the SAZ. Later, it is expressed in a dorsal segmental pattern and in the form of stripes in the dorsal extraembryonic tissue. D. melanogaster Wnt9 is also expressed in a segment polarity-like pattern and in the labrum. In E. kanangrensis, at least, expression in the tips of the appendages and the segment polarity gene-like expression are conserved, and this may indeed represent the ancestral expression profile of Wnt9.
Wnt10 expression has so far only been studied in D. melanogaster[54], S. maritima[49] and T. castaneum[52]. In D. melanogaster, it is expressed in the mesoderm, the developing gut and the central nervous system. In T. castaneum, it is expressed in the cephalic lobes, the appendages and in transverse segmental stripes anteriorly adjacent to the expression of engrailed (en). This expression is also conserved in S. maritima. Segmental expression and expression in the limbs (discussed later) appears to be conserved between T. castaneum, S. maritima and the onychophoran, and may thus represent part of the ancestral expression pattern of Wnt10. This scenario would mean that the expression profile of D. melanogaster Wnt10, however, is derived.
Wnt11 orthologues have been isolated and their expression investigated in T. castaneum[52], P. tepidariorum[20], G. marginata[20], and S. maritima[56,49]. In P. tepidariorum, there are two paralogues of Wnt11, but only one, Wnt11-2, is expressed in embryos. The expression profile of Wnt11 is similar in the onychophoran, the spider and the myriapods. In all species, Wnt11 is first expressed in the SAZ at the posterior region of the embryo. Later, expression appears in the tips of all appendages (except for S. maritima). However, segment polarity gene-like stripes of expression are only seen in E. kanangrensis. Only in T. castaneum is Wnt11 expressed in the heart.
Wnt11 is likely to play a conserved role during limb development as represented by the strong expression in the tips of the limbs, and segment addition, as represented by the strong expression in the posterior end of the embryos.
Expression of Wnt16 has been described in P. tepidariorum[20], S. maritima[49] and G. marginata[14,20] (in 2004, erroneously described as Wnt7). In both the spider and the myriapods, Wnt16 is expressed in transverse segmental stripes anterior and directly adjacent to the expression of en (somewhat unclear for S. maritima) suggesting that it is involved in segmental boundary formation. Wnt16 is also expressed in the developing brain and in the tips and ventral tissue of the limbs. The arthropod Wnt16-expression profile is conserved in E. kanangrensis. One important difference is, however, that the segmental expression of Wnt16 in E. kanangrensis reaches posteriorly into the domain of en expression.
Expression of WntA has been analyzed in T. castaneum[52], S. maritima[49] and G. marginata[14,20] (in 2004, erroneously described as Wnt5), and C. salei[20]. Expression in the mandibulate arthropods is comparable. It is strongly expressed in the developing brain, heart, limbs and central nervous system. In E. kanangrensis, WntA is similarly expressed in the brain (head lobe), the limbs and, at later stages, in the ventral nervous system; expression in the heart, however, is not observed in the onychophoran. Expression of WntA in the spider C. salei differs significantly from that in the other arthropods and the onychophoran. The only possibly conserved pattern of WntA is in the SAZ (apart from that, spider WntA is only expressed in small domains in the spinnerets and the chelicerae).
The onychophoran ‘segment’
Onychophorans represent segmented animals, although some of the key characteristics of the arthropods, such as full adult body segmentation with pronounced segmental indentations are not present. The latter is best interpreted as being primitive [60]. Since segmental indentations are lacking in onychophorans, it is difficult to determine the position of the segmental boundaries in the ectoderm. The best approximation may be given by the expression of segment polarity genes that determine segmental and parasegmental boundaries in arthropods [12,61].
The segment polarity gene network is conserved in arthropods (for example, [11,12,14,62]) and onychophorans [17,18]. The parasegmental boundary in arthropods lies at the interface between en and wg expressing cells, and the segmental boundary lies posterior to the expression of en (for example, [12,63]). This means that the parasegmental boundaries of onychophorans are located in the posterior of the limbs, exactly as in arthropods. Determination of the segmental boundaries by means of gene expression patterns is not that clear because the domain of en expression is broadened in ventral tissue [17] and the posterior border of each stripe thus lies posterior to that of hh[18], which is the direct downstream target of en in arthropods [15,64]. Therefore, the segmental boundaries lie either posterior to the en/hh domain and thus directly posterior to the limbs, or the segmental boundaries lie posterior to en and thus somewhat shifted towards the posterior (compare with Figure 13).
Wnt genes and limbs
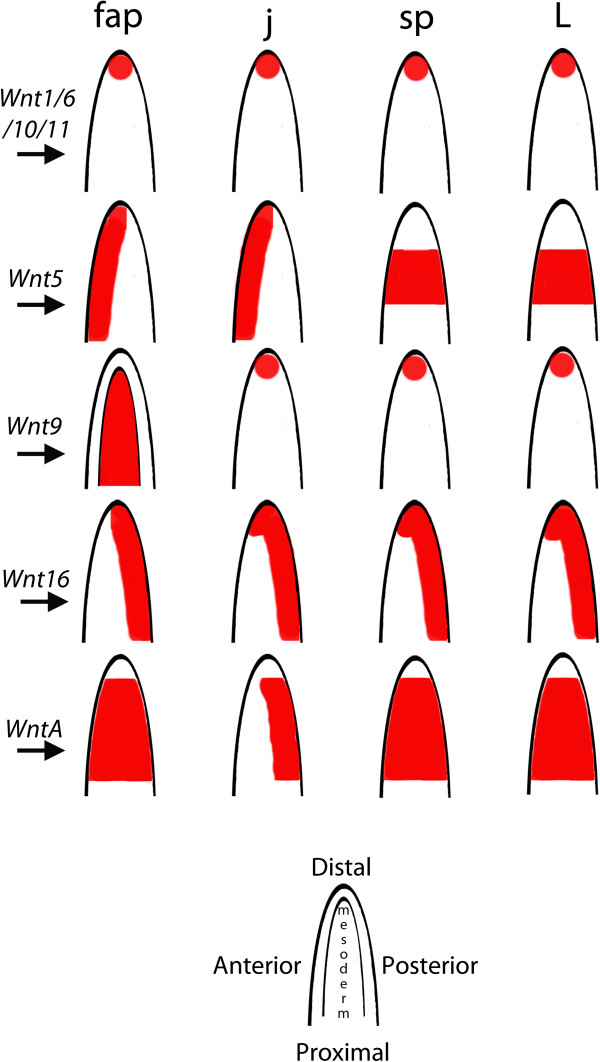
Comparative analyses of Wnt gene expression in a wide range of arthropods indicates that they play an important role in limb development. Typically, Wnt genes are expressed along the ventral side of the limbs [14,20,48,52,65,66]. It was, therefore, proposed that Wnt patterning could have a conserved and potentially combinatorial function in ventral limb patterning in arthropods, acting upstream of ventralizing limb genes, such as midline/H15 (mid/H15) [41,65,67-70] (but see [38] for a different opinion on the possibly conserved function of wg/Wnt1 in ventral limb development). The expression of wg/Wnt1 in the tips of the limbs in E. kanangrensis was thus somewhat unexpected [17]. This could have been explained by the potential presence of additional, but at the time unstudied, Wnt genes that could substitute for wg/Wnt1 in ventral tissue of the onychophoran limbs. Our comprehensive data on onychophoran Wnt gene expression now reveals that the expression of wg/Wnt1 in the tips of the limbs does not represent an exception. In arthropods, not only wg/Wnt1 but also a number of other Wnt genes are expressed along the ventral side of the limbs [20,48,52]. Recent studies suggest that Wnt genes and their receptors may act in combination or may have redundant functions, especially during segmentation and limb development, which would explain the relatively high number of Wnt genes with identical expression patterns in the limbs [41,52]. In the onychophoran, five Wnt genes are expressed in the tips of at least a subset of appendages (summarized in Figure 14). This implies a general change of expression from ‘all-ventral’ to ‘distal-only’. Whether this shift occurred in the lineage leading to the arthropods or whether it comprises an ancestral feature that has changed in the evolutionary line represented by onychophorans remains unclear and data from an outgroup, such as tardigrades, is required.
Figure 14.
Summary of the expression of E. kanangrensis Wnt genes in the appendages. Expression of Wnt genes in the limbs is shown in red. Expression is ectodermal, if not indicated differently (that is, Wnt9 in the frontal appendages is mesodermal). Note that mesodermal expression of Wnt4 in the frontal appendages and the jaws is not shown.
Interestingly, a recent study revealed two separate functions of wg/Wnt1 in limb development in T. castaneum[70]. One function concerns ventral limb patterning; the other concerns the proximodistal growth of the limbs. It is therefore likely that the expression seen in onychophorans is associated with limb growth, rather than with a function in dorsoventral patterning. This would mean that at least this component of dorsoventral patterning evolved in the lineage leading to the arthropods. The evolution of the dual function of Wnt genes during limb development may be reflected by the situation in the millipede G. marginata, where some Wnt genes are expressed strongly in the tips of the limbs, but only weakly along the ventral side of the limbs [48]. This implies that the expression in the tips as seen in the onychophoran may represent the ancestral state and that ventral expression evolved step by step within the arthropod lineage.
Conclusions
Wnt ligands play important roles during animal development. Our study shows that most of the protostome Wnt ligands are present in onychophorans, and that all of those present are expressed in distinct patterns throughout embryogenesis. It is surprising that eight of twelve Wnt genes are expressed in segmental stripes reminiscent of the expression of classical arthropod segment polarity genes, and that their combined expression covers the complete segments. This suggests that Wnt genes may play a more prominent role in segment regionalization than they do in arthropods, where the expression of Wnt genes is mostly restricted to anterior cells abutting the domain of engrailed (en) expression. Early expression of Wnt2, Wnt4 and Wnt5 in gap-gene-like and Hox-gene-like patterns suggests a contributing role of these genes in giving anterior segments their specific identities. Strong expression of most of the onychophoran Wnt genes in the posterior SAZ might be correlated with a role in germ band elongation or segmentation. Thus, it seems likely that Wnt genes are involved in segment formation, segment regionalization and the definition of segment identity in onychophorans. If these assumptions hold true, the role of Wnt genes in onychophoran segmentation would clearly extend their roles in arthropod segmentation.
Abbreviations
DAPI: 4-6-diamidino-2-phenylindole; MCMCMC: metropolis-coupled Markov chain Monte Carlo; PBST: phosphate-buffered saline with 0.1% Tween-20; PCR: polymerase chain reaction; SAZ: segment addition zone.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MH carried out most of the experiments, discussed the experimental outline and wrote part of the first draft of the manuscript. AS was involved in performing the phylogenetic analysis. GEB discussed the experimental outline, was involved in drafting the final version of the manuscript and initiated work on onychophoran segmentation. NP was involved in performing the phylogenetic analysis, assembled the E. kanangrensis transcriptome and was involved in drafting the final version of the manuscript. APM was involved in performing the phylogenetic analysis and was involved in drafting the final version of the manuscript. RJ was mainly responsible for the experimental outline, carried out part of the experiments, wrote part of the first draft of the manuscript and was involved in drafting the final version of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Schematic summary of early multiple-segment-wide expression domains of Wnt genes compared with the expression of Hox genes. This figure has been modified after [26]. Expression of Wnt genes is indicated by black, dark grey and light grey bars. Low level expression is indicated by thin bars and ‘w’. 1–15, first to fifteenth leg-bearing segments; abd-A, abdominal-A, Abd-B, Abdominal-B; Antp, Antennapedia; Dfd, Deformed; fap, frontal appendage; ftz, fushi-tarazu; hl, head lobe; j, jaw; lab, labial; pb, proboscipedia; SAZ, segment addition zone; Scr, Sex combs reduced; sp, slime papilla; Ubx, Ultrabithorax.
Contributor Information
Mattias Hogvall, Email: mattias.hogvall@geo.uu.se.
Anna Schönauer, Email: 12084765@brookes.ac.uk.
Graham E Budd, Email: graham.budd@pal.uu.se.
Alistair P McGregor, Email: amcgregor@brookes.ac.uk.
Nico Posnien, Email: nposnie@gwdg.de.
Ralf Janssen, Email: ralf.janssen@geo.uu.se.
Acknowledgements
This work has been supported by the Swedish Research Council (grant to GEB) and the Volkswagen Foundation (grant to NP).
We gratefully acknowledge the support of the New South Wales Government Department of Environment and Climate Change by provision of a permit SL100159 to collect onychophorans at Kanangra-Boyd National Park and to the Australian Government Department of the Environment, Water, Heritage and the Arts for export permits WT2009-4598 and WT2012-4704. The authors wish to thank Jean Joss, Rolf Ericsson, Robyn Stutchbury and, especially, Noel Tait, for their help during onychophoran collection.
References
- Oliveira Ide S, Read VM, Mayer G. A world checklist of Onychophora (velvet worms), with notes on nomenclature and status of names. Zookeys. 2012;211:1–70. doi: 10.3897/zookeys.211.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LI, Rota-Stabelli O, Edgecombe GD, Marchioro T, Longhorn SJ, Telford MJ, Philippe H, Rebecchi L, Peterson KJ, Pisani D. MicroRNAs and phylogenomics resolve the relationships of Tardigrada and suggest that velvet worms are the sister group of Arthropoda. Proc Natl Acad Sci U S A. 2011;108:15920–15924. doi: 10.1073/pnas.1105499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. Segmentation. Dev Cell. 2004;7:301–312. doi: 10.1016/j.devcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Damen WG. Evolutionary conservation and divergence of the segmentation process in arthropods. Dev Dyn. 2007;236:1379–1391. doi: 10.1002/dvdy.21157. [DOI] [PubMed] [Google Scholar]
- Peel AD. The evolution of developmental gene networks: lessons from comparative studies on holometabolous insects. Philos Trans R Soc Lond B Biol Sci. 2008;363:1539–1547. doi: 10.1098/rstb.2007.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman AD. Parallel evolution of segmentation by co-option of ancestral gene regulatory networks. Bioessays. 2010;32:60–70. doi: 10.1002/bies.200900130. [DOI] [PubMed] [Google Scholar]
- Andrioli LP. Toward new Drosophila paradigms. Genesis. 2012;50:585–598. doi: 10.1002/dvg.22019. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Kluding H, Jürgens G. Genes affecting the segmental subdivision of the Drosophila embryo. Cold Spring Harb Symp Quant Biol. 1985;50:145–154. doi: 10.1101/SQB.1985.050.01.020. [DOI] [PubMed] [Google Scholar]
- Ahzhanov A, Kaufman TC. Evolution of distinct expression patterns for engrailed paralogues in higher crustaceans (Malacostraca) Dev Genes Evol. 2000;210:493–506. doi: 10.1007/s004270000090. [DOI] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC. Exploring myriapod segmentation: the expression patterns of even-skipped, engrailed, and wingless in a centipede. Dev Biol. 2002;247:47–61. doi: 10.1006/dbio.2002.0683. [DOI] [PubMed] [Google Scholar]
- Damen WG. Parasegmental organization of the spider embryo implies that the parasegment is an evolutionary conserved entity in arthropod embryogenesis. Development. 2002;129:1239–1250. doi: 10.1242/dev.129.5.1239. [DOI] [PubMed] [Google Scholar]
- Simonnet F, Deutsch J, Queinnec E. hedgehog is a segment polarity gene in a crustacean and a chelicerate. Dev Genes Evol. 2004;214:345–537. doi: 10.1007/s00427-004-0435-z. [DOI] [PubMed] [Google Scholar]
- Janssen R, Prpic NM, Damen WG. Gene expression suggests decoupled dorsal and ventral segmentation in the millipede Glomeris marginata (Myriapoda: Diplopoda) Dev Biol. 2004;268:89–104. doi: 10.1016/j.ydbio.2003.12.021. [DOI] [PubMed] [Google Scholar]
- Farzana L, Brown SJ. Hedgehog signaling pathway function conserved in Tribolium segmentation. Dev Genes Evol. 2008;218:181–192. doi: 10.1007/s00427-008-0207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R. Segment polarity gene expression in a myriapod reveals conserved and diverged aspects of early head patterning in arthropods. Dev Genes Evol. 2012;222:299–309. doi: 10.1007/s00427-012-0413-9. [DOI] [PubMed] [Google Scholar]
- Eriksson BJ, Tait NN, Budd GE, Akam M. The involvement of engrailed and wingless during segmentation in the onychophoran Euperipatoides kanangrensis (Peripatopsidae: Onychophora) (Reid 1996) Dev Genes Evol. 2009;219:249–264. doi: 10.1007/s00427-009-0287-7. [DOI] [PubMed] [Google Scholar]
- Janssen R, Budd GE. Deciphering the onychophoran ‘segmentation gene cascade’: gene expression reveals limited involvement of pair rule gene orthologs in segmentation, but a highly conserved segment polarity gene network. Dev Biol. 2013;382:224–234. doi: 10.1016/j.ydbio.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Cho SJ, Valles Y, Giani VC Jr, Seaver EC, Weisblat DA. Evolutionary dynamics of the Wnt gene family: a lophotrochozoan perspective. Mol Biol Evol. 2010;27:1645–1658. doi: 10.1093/molbev/msq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R, Eriksson BJ, Budd GE, Akam M, Prpic NM. Gene expression patterns in an onychophoran reveal that regionalization predates limb segmentation in pan-arthropods. Evol Dev. 2010;12:363–372. doi: 10.1111/j.1525-142X.2010.00423.x. [DOI] [PubMed] [Google Scholar]
- Janssen R, Le Gouar M, Pechmann M, Poulin F, Bolognesi R, Schwager EE, Hopfen C, Colbourne JK, Budd GE, Brown SJ, Prpic NM, Kosiol C, Vervoort M, Damen WG, Balavoine G, McGregor AP. Conservation, loss, and redeployment of Wnt ligands in protostomes: implications for understanding the evolution of segment formation. BMC Evol Biol. 2010;10:374. doi: 10.1186/1471-2148-10-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Eriksson BJ, Tait NN, Budd GE, Janssen R, Akam M. Head patterning and Hox gene expression in an onychophoran and its implications for the arthropod head problem. Dev Genes Evol. 2010;220:117–122. doi: 10.1007/s00427-010-0329-1. [DOI] [PubMed] [Google Scholar]
- Janssen R, Eriksson BJ, Tait NN, Budd GE. Onychophoran Hox genes and the evolution of arthropod Hox gene expression. Front Zool. 2014;11:22. doi: 10.1186/1742-9994-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R, Damen WG. The ten Hox genes of the millipede Glomeris marginata. Dev Genes Evol. 2006;216:451–465. doi: 10.1007/s00427-006-0092-5. [DOI] [PubMed] [Google Scholar]
- Sharma PP, Schwager EE, Extavour CG, Giribet G. Hox gene expression in the harvestman Phalangium opilio reveals divergent patterning of the chelicerate opisthosoma. Evol Dev. 2012;14:450–463. doi: 10.1111/j.1525-142X.2012.00565.x. [DOI] [PubMed] [Google Scholar]
- Hughes CL, Kaufman TC. Hox genes and the evolution of the arthropod body plan. Evol Dev. 2002;4:459–499. doi: 10.1046/j.1525-142X.2002.02034.x. [DOI] [PubMed] [Google Scholar]
- Jaeger J. The gap gene network. Cell Mol Life Sci. 2011;68:243–274. doi: 10.1007/s00018-010-0536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson BJ, Tait NN. Early development in the velvet worm Euperipatoides kanangrensis Reid 1996 (Onychophora: Peripatopsidae) Arthropod Struct Dev. 2012;41:483–493. doi: 10.1016/j.asd.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingensmith J, Nusse R. Signaling by wingless in Drosophila. Dev Biol. 1994;166:396–414. doi: 10.1006/dbio.1994.1325. [DOI] [PubMed] [Google Scholar]
- Sanson B. Generating patterns from fields of cells. EMBO Rep. 2001;2:1083–1088. doi: 10.1093/embo-reports/kve255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves FC, DasGupta R. Function of the wingless signaling pathway in Drosophila. Methods Mol Biol. 2008;469:115–125. doi: 10.1007/978-1-60327-469-2_10. [DOI] [PubMed] [Google Scholar]
- Baker NB. Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: the spatial distribution of a transcript in embryos. EMBO J. 1987;6:1765–1773. doi: 10.1002/j.1460-2075.1987.tb02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P, Baker N, Martinez Arias A. Positive and negative regulation of segment polarity genes in the Drosophila blastoderm by the pair rule genes fushi tarazu and even skipped. Nature. 1988;331:73–75. doi: 10.1038/331073a0. [DOI] [PubMed] [Google Scholar]
- Miyawaki K, Mito T, Sarashina I, Zhang H, Shinmyo Y, Ohuchi H, Noji S. Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mech Dev. 2004;121:119–130. doi: 10.1016/j.mod.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Angelini DR, Kaufman TC. Functional analyses in the milkweed bug Oncopeltus fasciatus (Hemiptera) support a role for Wnt signaling in body segmentation but not appendage development. Dev Biol. 2005;283:409–423. doi: 10.1016/j.ydbio.2005.04.034. [DOI] [PubMed] [Google Scholar]
- Bolognesi R, Farzana L, Fischer TD, Brown SJ. Multiple Wnt genes are required for segmentation in the short-germ embryo of Tribolium castaneum. Curr Biol. 2008;18:1624–1629. doi: 10.1016/j.cub.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi R, Fischer TD, Brown SJ. Loss of Tc-arrow and canonical Wnt signaling alters posterior morphology and pair-rule gene expression in the short-germ insect, Tribolium castaneum. Dev Genes Evol. 2009;219:369–375. doi: 10.1007/s00427-009-0299-3. [DOI] [PubMed] [Google Scholar]
- Beermann A, Prühs R, Lutz R, Schröder R. A context-dependent combination of Wnt receptors controls axis elongation and leg development in a short germ insect. Development. 2011;38:2793–2805. doi: 10.1242/dev.063644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor AP, Pechmann M, Schwager EE, Feitosa NM, Kruck S, Aranda M, Damen WG. Wnt8 is required for growth-zone establishment and development of opisthosomal segments in a spider. Curr Biol. 2008;18:1619–1623. doi: 10.1016/j.cub.2008.08.045. [DOI] [PubMed] [Google Scholar]
- McGregor AP, Pechmann M, Schwager EE, Damen WG. An ancestral regulatory network for posterior development in arthropods. Commun Integr Biol. 2009;2:174–176. doi: 10.4161/cib.7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro JE, Pueyo JI, Couso JP. Interplay between a Wnt-dependent organizer and the Notch segmentation clock regulates posterior development in Periplaneta americana. Biol Open. 2013;2:227–237. doi: 10.1242/bio.20123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat S, Hopfen C, McGregor AP. The function and evolution of Wnt genes in arthropods. Arthropod Struct Dev. 2010;39:446–452. doi: 10.1016/j.asd.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Bejsovec A. Wingless/Wnt signaling in Drosophila: the pattern and the pathway. Mol Reprod Dev. 2013;80:882–894. doi: 10.1002/mrd.22228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober KA, Jockusch EL. The roles of wingless and decapentaplegic in axis and appendage development in the red flour beetle, Tribolium castaneum. Dev Biol. 2006;294:391–405. doi: 10.1016/j.ydbio.2006.02.053. [DOI] [PubMed] [Google Scholar]
- Janssen R, Posnien N. Identification and embryonic expression of Wnt2, Wnt4, Wnt5 and Wnt9 orthologs in the millipede Glomeris marginata (Myriapoda: Diplopoda) Gene Expr Patterns. 2014;14:55–61. doi: 10.1016/j.gep.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Hayden L, Arthur W. The centipede Strigamia maritima possesses a large complement of Wnt genes with diverse expression patterns. Evol Dev. [DOI] [PubMed]
- Russell J, Gennissen A, Nusse R. Isolation and expression of two novel Wnt/ wingless gene homologues in Drosophila. Development. 1992;115:475–485. doi: 10.1242/dev.115.2.475. [DOI] [PubMed] [Google Scholar]
- Fradkin LG, Noordermeer JN, Nusse R. The Drosophila Wnt protein DWnt-3 is a secreted glycoprotein localized on the axon tracts of the embryonic CNS. Dev Biol. 1995;168:202–213. doi: 10.1006/dbio.1995.1072. [DOI] [PubMed] [Google Scholar]
- Bolognesi R, Beermann A, Farzana L, Wittkopp N, Lutz R, Balavoine G, Brown SJ, Schroeder R. Tribolium Wnts: evidence for a larger repertoire in insects with overlapping expression patterns that suggest multiple redundant functions in embryogenesis. Dev Genes Evol. 2008;218:193–202. doi: 10.1007/s00427-007-0170-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen R, Damen WG. Diverged and conserved aspects of heart formation in a spider. Evol Dev. 2008;10:155–165. doi: 10.1111/j.1525-142X.2008.00223.x. [DOI] [PubMed] [Google Scholar]
- Janson K, Cohen ED, Wilder EL. Expression of DWnt6, DWnt10, and DFz4 during Drosophila development. Mech Dev. 2001;103:117–120. doi: 10.1016/S0925-4773(01)00323-9. [DOI] [PubMed] [Google Scholar]
- Llimargas M, Lawrence PA. Seven Wnt homologues in Drosophila: a case study of the developing tracheae. Proc Natl Acad Sci U S A. 2001;98:14487–14492. doi: 10.1073/pnas.251304398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden L, Arthur W. Expression patterns of Wnt genes in the venom claws of centipedes. Evol Dev. 2013;15:365–372. doi: 10.1111/ede.12044. [DOI] [PubMed] [Google Scholar]
- Ganguly A, Jiang J, Ip YT. Drosophila WntD is a target and an inhibitor of the Dorsal/Twist/Snail network in the gastrulating embryo. Development. 2005;132:3419–3429. doi: 10.1242/dev.01903. [DOI] [PubMed] [Google Scholar]
- Gordon MD, Dionne MS, Schneider DS, Nusse R. WntD is a feedback inhibitor of Dorsal/NF-κB in Drosophila development and immunity. Nature. 2005;437:746–749. doi: 10.1038/nature04073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graba Y, Gieseler K, Aragnol D, Laurenti P, Mariol MC, Berenger H, Sagnier T, Pradel J. DWnt-4, a novel Drosophila Wnt gene acts downstream of homeotic complex genes in the visceral mesoderm. Development. 1995;121:209–218. doi: 10.1242/dev.121.1.209. [DOI] [PubMed] [Google Scholar]
- Budd GE. Tardigrades as ‘stem-group arthropods’: the evidence from the Cambrian fauna. Zool Anz. 2001;240:265–279. doi: 10.1078/0044-5231-00034. [DOI] [Google Scholar]
- van den Heuvel M, Klingensmith J, Perrimon N, Nusse R. Cell patterning in the Drosophila segment: engrailed and wingless antigen distributions in segment polarity mutant embryos. Dev Suppl. 1993. pp. 105–114. [PubMed]
- Janssen R, Budd GE, Damen WG, Prpic NM. Evidence for Wg-independent tergite boundary formation in the millipede Glomeris marginata. Dev Genes Evol. 2008;218:361–370. doi: 10.1007/s00427-008-0231-2. [DOI] [PubMed] [Google Scholar]
- Larsen CW, Hirst E, Alexandre C, Vincent JP. Segment boundary formation in Drosophila embryos. Development. 2003;130:5625–5635. doi: 10.1242/dev.00867. [DOI] [PubMed] [Google Scholar]
- Tabata T, Eaton S, Kornberg TB. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 1992;6:2635–2645. doi: 10.1101/gad.6.12b.2635. [DOI] [PubMed] [Google Scholar]
- Prpic NM, Tautz D. The expression of the proximodistal axis patterning genes Distal-less and dachshund in the appendages of Glomeris marginata (Myriapoda: Diplopoda) suggests a special role of these genes in patterning the head appendages. Dev Biol. 2003;260:97–112. doi: 10.1016/S0012-1606(03)00217-3. [DOI] [PubMed] [Google Scholar]
- Prpic NM. Homologs of wingless and decapentaplegic display a complex and dynamic expression profile during appendage development in the millipede Glomeris marginata (Myriapoda: Diplopoda) Front Zool. 2004;1:6. doi: 10.1186/1742-9994-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prpic NM, Janssen R, Damen WG, Tautz D. Evolution of dorsal-ventral axis formation in arthropod appendages: H15 and optomotor-blind/bifid-type T-box genes in the millipede Glomeris marginata (Myriapoda: Diplopoda) Evol Dev. 2005;7:51–57. doi: 10.1111/j.1525-142X.2005.05006.x. [DOI] [PubMed] [Google Scholar]
- Janssen R, Feitosa NM, Damen WG, Prpic NM. The T-box genes H15 and optomotor-blind in the spiders Cupiennius salei, Tegenaria atrica and Achaearanea tepidariorum and the dorsoventral axis of arthropod appendages. Evol Dev. 2008;10:143–154. doi: 10.1111/j.1525-142X.2008.00222.x. [DOI] [PubMed] [Google Scholar]
- Svendsen PC, Formaz-Preston A, Leal SM, Brook WJ. The Tbx20 homologs midline and H15 specify ventral fate in the Drosophila melanogaster leg. Development. 2009;136:2689–2693. doi: 10.1242/dev.037911. [DOI] [PubMed] [Google Scholar]
- Grossmann D, Scholten J, Prpic NM. Separable functions of wingless in distal and ventral patterning of the Tribolium leg. Dev Genes Evol. 2009;219:469–479. doi: 10.1007/s00427-009-0310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic summary of early multiple-segment-wide expression domains of Wnt genes compared with the expression of Hox genes. This figure has been modified after [26]. Expression of Wnt genes is indicated by black, dark grey and light grey bars. Low level expression is indicated by thin bars and ‘w’. 1–15, first to fifteenth leg-bearing segments; abd-A, abdominal-A, Abd-B, Abdominal-B; Antp, Antennapedia; Dfd, Deformed; fap, frontal appendage; ftz, fushi-tarazu; hl, head lobe; j, jaw; lab, labial; pb, proboscipedia; SAZ, segment addition zone; Scr, Sex combs reduced; sp, slime papilla; Ubx, Ultrabithorax.